Abstract
The transcriptional feedback circuit, which is at the core of the suprachiasmatic nucleus (SCN) circadian (i.e., 24 h) clock, is tightly coupled to both external entrainment cues, such as light, as well as rhythmic cues that arise on a system-wide level within the SCN. One potential signaling pathway by which these cues are conveyed to the molecular clock is the CREB/CRE transcriptional cascade. In this study, we employed a tetracycline-inducible CREB repressor mouse strain, in which approximately 60% of the SCN neurons express the transgene, to test CREB functionality in the clock and its effects on overt rhythmicity. We show that attenuated CREB signaling in the SCN led to a significant reduction in light-evoked clock entrainment. An examination of circadian timing revealed that CREB repressor mice exhibited normal free-running rhythms in the absence of external lighting cues. However, under conditions of constant light, which typically leads to a lengthening of the circadian period, CREB repressor mice exhibited a dramatic arrhythmic phenotype, which could be reversed with doxycycline. At a cellular level, the repression of CREB led to a significant reduction in both the expression of the circadian clock proteins PERIOD1 and PERIOD2 and the clock output hormones AVP and VIP. Together, these data support the idea that the CRE transcriptional pathway orchestrates transcriptional events that are essential for both the maintenance of SCN timing and light entrainment of the circadian clock.
Keywords: CREB, suprachiasmatic nucleus, light, circadian
The master mammalian circadian (i.e., 24 h) clock located in the suprachiasmatic nucleus (SCN) regulates a diverse set of biochemical and physiological processes (Lowrey and Takahashi, 2000; Welsh et al., 2010). Central to the clock timing process is a cell autonomous transcription/translation/posttranslational feedback loop centered on a limited set of “clock” genes (Gekakis et al., 1998; Bunger et al., 2000; Shearman et al., 2000; Eide et al., 2005; Shirogane et al., 2005). Importantly, both the period and the amplitude of this core molecular clock oscillator, as well as the efficacy of clock cell communication, which is a key event in the generation of coherent SCN rhythms, are influenced by changes in the cellular environment (Hastings et al., 2008). Thus, one central issue for the field is the identification of cellular signaling mechanisms by which external stimuli affect these clock timing processes. In this regard, one pathway that is of particular interest is the CREB (cAMP response element-binding protein)/CRE transcriptional pathway.
CREB is a member of the basic helix-loop-helix family of transcription factors, which include ATF1-4 and CREM. CREB transactivation is a complex multistep process that appears to be initiated by the phosphorylation of Ser 133 via a number of Ca2+ as well as cAMP-activated kinase pathways (Impey and Goodman, 2001). This event allows the KID domain of CREB to bind to the scaffolding protein CBP (CREB-binding protein). This physical association between CREB and CBP stabilizes the basal transcriptional complex and thus facilitates gene transcription (Mayr and Montminy, 2001). With respect to SCN physiology, the potential importance of CREB was revealed in a series of reports showing that the pathway is activated by photic input (Ginty et al., 1993; Ding et al., 1997; von Gall et al., 1998; Obrietan et al., 1999; Gau et al., 2002) and plays a significant role in light-induced clock entrainment (Gau et al., 2002; Tischkau et al., 2003). Interestingly, CREB activation has been shown to be under control of a circadian oscillator (Belvin et al., 1999; Obrietan et al., 1999; Gau et al., 2002). This, in addition to findings showing that a number of core clock genes as well as clock-controlled genes are targets of the CREB/CRE pathway (Travnickova-Bendova et al., 2002; Deutsch et al., 1988; Montminy and Bilezikjian, 1987), raised the distinct possibility that CREB plays a key role in SCN physiology.
To begin to assess the role of CREB in the clock, we generated a transgenic mouse strain in which A-CREB, a dominant-negative form of CREB, is expressed in a tetracycline-inducible manner in the SCN. This mosaic mouse strain, in which a subset of SCN neurons expresses the transgene, exhibits deficits in clock entrainment and is arrhythmic under constant light/free-running conditions. These data, coupled with our cellular analysis of CREB function in the SCN, indicate a significant role for the CREB/CRE pathway in SCN clock physiology.
MATERIALS AND METHODS
Animals
Details regarding the generation of the A-CREB-eGFP (here referred to as “A-CREB::tTA”) transgenic mice are found in Lee et al. (2007b). αCaMKII-tTA (designated “tTA”) mice were generated by Mayford et al. (1996) and acquired from Jackson Laboratory (Bar Harbor, ME). The A-CREB strain was generated in a C57/Bl6 background, and αCaMKII-tTA has been bred into the C57/Bl6 background for at least 12 generations. Genotyping information can be found in the Supplementary Methods section. Transgene expression was not repressed via doxycycline at any point prior to the experimental manipulations described here. Of note, histological examination with Fluoro-Jade B, a marker of dead and dying cells, and cresyl violet staining did not detect neuronal loss in any brain region of A-CREB::tTA transgenic mice (data not shown), thus indicating that tonic transgene expression does not affect neuronal maturation or the overall health of the animals. All experimental procedures were approved by the Ohio State University Animal Care and Use Committee.
Behavior Analysis and Doxycycline Treatment
For circadian locomotor activity analysis, age- (10–14 weeks) and gender-matched A-CREB::tTA and control tTA littermates were transferred to cages equipped with running wheels (15-cm diameter), and binned (5 min) wheel rotation data were automatically recorded to PC-running Vital View (MiniMitter Corp., Bend, OR) data acquisition software. To assess the effect of the transgene on free-running rhythms, mice were entrained to a 12-h light:12-h dark cycle (LD cycle) for 10 days and then switched to either continuous darkness (DD cycle) or constant light (LL). During the day, luminescence was provided with a fluorescent white light (~200 lux at midcage level). Animals received a once-weekly food and water replenishment and a change of bedding. Cage maintenance under DD conditions was performed at various times during the subjective night using a dim red light (<3 lux) source. To block A-CREB expression, doxycycline (200 ug/mL) was added into the drinking water, which was replenished every 3 days. χ2 periodogram analysis (Refinetti, 1993) was performed using VBScript software (Circadian Rhythm Laboratory, University of South Carolina: http://www.circadian.org/periodogram.html) to determine the presence of significant circadian locomotor rhythms. For data sets deemed statistically significant, regression analysis was performed using the Actiview software program (MiniMitter Corp.) to determine the free-running period. Data were collected on the days denoted in the figures. As part of our analysis of A-CREB::tTA transgenic mice, we also examined total daily activity under LD, LL, and DD conditions.
To assess the entraining effects of light, mice were transferred to DD for 10 days and then received a light pulse (50 or 100 lux, 15 min) at circadian time 15 (CT15). The least squares linear regression approach described by Daan and Pittendrigh (1976) was used to assess the entraining effects of light. Thus, activity onset was calculated for the 6 days preceding light treatment and 3 to 10 days after light treatment. The difference in the projected versus the actual activity onset between the 2 lines was determined to be the phase shift. Group data were expressed as mean phase shift ± standard error of the mean (SEM). Significance was assessed via 1-way ANOVA. p < 0.05 was accepted as statistically significant.
Immunohistochemistry
Mice were dark-adapted for 2 days and killed under dim red light via cervical dislocation at multiple time points over a 24-h period. Brains were then isolated and immersed in oxygenated physiological saline and thick-sectioned (600 μm) with a vibratome. Tissue was then fixed in 4% formaldehyde (PFA), incubated in 30% sucrose overnight, and then thin-cut (40 μm) using a freezing microtome. For DAB staining, sections were initially washed (3x) in phosphate-buffered saline supplemented with 0.1% Triton X-100 (PBST: 10 min), then incubated in 0.3% H2O2 in PBST (15 min). Next, tissue was blocked (1 h) in 5% goat serum/PBS and incubated (overnight, 4 °C) with antibodies against the following: mPER1 (rabbit polyclonal IgG, 1:10,000 dilution; provided by Dr. Steven Reppert, University of Massachusetts), mPER2 (1:500 dilution; Alpha Diagnostic Intl. Inc., San Antonio, TX), VIP (rabbit polyclonal IgG, 1:1000 dilution; Santa Cruz Biotech, Santa Cruz, CA), AVP (porcine polyclonal IgG, 1:1000 dilution; Peninsula Labs, San Carlos, CA), or GFP (rabbit polyclonal IgG, 1:5000 dilution; acquired from Dr. Luc G. Berthiaume, University of Alberta, Canada). Next, sections were incubated (2 h) at room temperature in biotinylated secondary antibodies (1:300 dilution; Vector Laboratories, Burlingame, CA) and then placed in an avidin/biotin HRP complex for 1 h (prepared according to the manufacturer’s instructions). The signal was visualized by the addition of DAB-nickel– intensified substrate (Vector Laboratories) and mounted on gelatin-coated slides and cover-slipped with Permount media (Fisher Scientific, Houston, TX). Images of diaminobenzidine (DAB)–labeled sections were captured using a 16-bit digital camera (Micromax YHS 1300; Princeton Instruments, Trenton, NJ) connected to a Leica DM IRB microscope (Nussloch, Germany).
Immunohistochemistry Quantitation
All data were quantified using MetaMorph software (Universal Imaging Corporation, West Chester, PA). For the analysis of mPer1, mPER2, VIP, and AVP expression from DAB-labeled tissues, images containing the central SCN were captured using a 10× objective. A drawing tool was used to demarcate the outline of the SCN, and the mean intensity values were obtained. A digital oval (150 × 200 pixels) was then placed in the adjacent ipsilateral hypothalamic region and the average value determined. The SCN to lateral hypothalamic ratio was generated and averaged across 3 central SCN sections per animal. Mean group data were then determined, and significance was assessed using the 2-tailed Student t test. All data are expressed as the mean ± SEM. p < 0.05 was accepted as statistically significant.
Fluoro-Jade B Histochemistry
Cell death was assayed using Fluoro-Jade B (FJB) labeling. Initially, A-CREB::tTA mice were maintained under LL conditions (7 days: 400 lux) and then transcardially perfused (~CT12), and brain tissue was thin-cut as described above. Next, 40-μm-thick coronal sections through the SCN were washed in water and incubated with potassium permaganate (0.06% for 10 min). Sections were then incubated (20 min) with FJB (0.001%; Histo-Chem Inc., Jefferson, AR) in 0.1% acetic acid. Finally, tissue was immersed in xylene and cover-slipped with DPX.
As a control for the FJB labeling approach, tissue was also processed from an animal that was challenged with an excitotoxic dose of pilocarpine. To this end, the adult male C57BL/6 mouse initially received an intraperitoneal (i.p.) injection of atropine methyl nitrate (1 mg/kg); then 30 min later, it received a second i.p. injection of pilocarpine hydrochloride (325 mg/kg). The mouse achieved status epilepticus (SE) (defined as a continuous motor seizure over a 3-h period). Two days later, the animal was sacrificed and fixed, and brain tissue was thin-cut and labeled for FJB as described above.
RESULTS
To address the functional role of CREB-mediated transcription in SCN clock timing, we generated a transgenic mouse strain that drives the expression of the CREB repressor A-CREB in a tetracycline-inducible manner (Fig. 1A and Suppl. Fig. S1A). By dimerizing with the basic region of CREB, and thereby blocking the ability of CREB to bind to the CRE, A-CREB functions as a potent inhibitor of CREB-dependent transcription (Ahn et al., 1998). The A-CREB construct was designed to express enhanced green fluorescent protein (GFP) from an internal ribosomal entry site (IRES), thereby allowing clear detection of cells that express the transgene construct (Fig. 1A–C). A-CREB transgenic founders were crossed to mice expressing tTA via the αCaMKII promoter, which has been used predominately to drive transgene expression in forebrain neurons (Mayford et al., 1996). Thus far, we have used these bitransgenic mice to examine the role of CREB in a number of pathophysiological experimental paradigms, including seizure-induced cell death, and Huntington-mediated striatal pathology (Lee et al., 2007b, 2009; Choi et al., 2009). In these studies, we have shown that the A-CREB transgene leads to marked attenuation of CRE-regulated gene expression (Lee et al., 2007b, 2009; Choi et al., 2009).
Figure 1.
A-CREB transgene expression in the SCN. (A) Schematic outline of the bitransgenic system used to drive the expression of A-CREB and the transgene marker GFP. (B) Representative immunohistochemical labeling for GFP in a coronal brain section from an A-CREB/GFP::tTA bitransgenic mouse (denoted “A-CREB::tTA”) and a monotransgenic tTA mouse (tTA) mouse. Arrows denote the locations of the SCN. Scale bar: 1 mm. (C) GFP immunofluorescent labeling of A-CREB::tTA bitransgenic tissue reveals marked transgene expression in the SCN. Scale bar: 125 μm. 3V = third ventricle. (D) The A-CREB transgene does not alter SCN cell viability. Representative Fluoro-Jade B (FJB) labeling of A-CREB::tTA bitransgenic tissue from a mouse that was maintained under constant light for 7 days. Of note, FJB-labeled cells were not detected. As a control, we provide representative FJB labeling from the piriform cortex collected 2 days following the pilocarpine-induced repetitive seizure activity. Note the large number of FJB-labeled cells. Scale bar: 300 μm.
Of interest, we noted that a subset of founder lines (2/6) exhibited robust transgene expression within the SCN (Fig. 1B). This contrasts with other regions of the diencephalon, where little transgene expression was detected. Within the SCN, the A-CREB transgene was broadly expressed (Fig. 1C); hence, it does not appear to be localized to a distinct neuronal population (e.g., VIP-positive cells, calbindin-positive cells). Quantitative analysis revealed mosaicism of transgene expression in the SCN; thus, GFP was detected in approximately 60% of central SCN neurons (Suppl. Fig. S1B). Hence, these animals should exhibit a potent attenuation of CREB function in this subset of the neurons, whereas CRE-mediated gene expression should be intact in the nontransgenic cell population (~40% of neurons). Given our prior work showing that A-CREB increases vulnerability to neuronal cell stress, we also tested whether cell death could be detected in the SCN of A-CREB::tTA mice. To this end, SCN tissue from animals maintained on constant light (400 lux) for 7 days (see below) was labeled with Fluoro-Jade B, a highly sensitive histochemical marker of cell death and degeneration. Under this condition, cell death was not detected in the SCN or in any other regions of the CNS. As a validation of the sensitivity of the labeling procedure, FJB-labeled tissue from the piriform cortex of a mouse exhibiting pilocarpine-induced repetitive seizure activity is presented in Figure 1D.
Given that a number of studies have implicated CREB as a key effector of light-induced clock resetting, we initially examined the photic entrainment phenotype of A-CREB/GFP::tTA bitransgenic (denoted “A-CREB::tTA”) mice. To this end, A-CREB::tTA mice and tTA monotransgenic littermates (denoted “tTA”) were placed in a standard 12-h light:12-h dark cycle (L/D: 200 lux) and then were allowed to free-run under constant darkness (DD). Wheel-running activity was used as an overt measure of clock timing. Of note, A-CREB::tTA mice showed normal entrainment to the LD cycle (Fig. 2A) and exhibited an equivalent amount of locomotor activity to tTA littermates (Fig. 2C). Further, under DD, both the amount of running (Fig. 2A–C) and the free-running period of A-CREB::tTA mice (23.50 ± 0.24 h, N = 11) were indistinguishable from tTA littermates (23.47 ± 0.28 h, N = 7).
Figure 2.
The A-CREB transgene represses light-induced phase delaying of the circadian clock. (A) Representative double-plotted actographs of wheel-running activity in tTA and A-CREB::tTA mice. Mice were dark-adapted (DD) and then exposed to light (50 lux, 15 min [asterisks]). Regression lines approximate the phase-shifting effects of light. (B) Statistical analysis of the light-induced phase delay. Numbers in bars denote the number of animals examined for each condition. (C) Average locomomotor activity (i.e., wheel rotations) of A-CREB::tTA and tTA mice under both LD and DD conditions.
Next, the entraining effects of early night light were examined. To this end, we found that a 50-lux light pulse administered at CT15 led to a 150.00 ± 9.54-min phase delay in tTA mice, whereas A-CREB::tTA mice exhibited an attenuated 73.50 ± 33.0-min delay (Fig. 2B). Interestingly, in A-CREB::tTA mice, doubling the light intensity to 100 lux was still not as effective as a 50-lux pulse in tTA littermates (Fig. 2B). Together, these data indicate that CREB plays a key role in coupling photic input to circadian clock entrainment. To provide a molecular correlate to the behavioral data, we examined light-evoked immediate early gene expression in A-CREB::tTA transgenic mice. Supplementary Figure S2 reveals that an early night light pulse triggered JunB and cFos expression in A-CREB::tTA transgenic SCN tissue; however, the level of induction was blunted relative to monotransgenic animals. These findings indicate that A-CREB attenuates the light-actuated transcriptional response in the SCN.
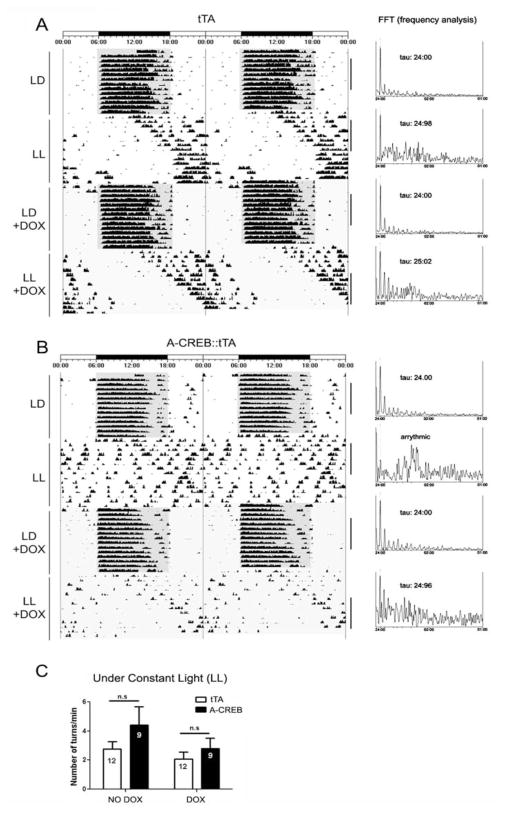
Several lines of evidence raise the possibility that CREB-mediated signaling affects SCN clock timing. For example, CRE-mediated gene expression is rhythmically regulated in the SCN (Obrietan et al., 1998). Likewise, the robustness of SCN rhythms is altered by cAMP, which is a key regulator of CREB phosphorylation (O’Neill et al., 2008). Further, CREB has been shown to affect the transcription of a number of neuropeptides, which have been implicated in circadian physiology (Aton et al., 2005; Maywood et al., 2006; Li et al., 2009). Given that A-CREB did not affect free-running rhythms under DD, we examined whether the free-running rhythm under constant light would be affected. The constant light (LL) paradigm has been used extensively to modulate the SCN timing mechanism and thus reveals important underlying functional features of the clock. For example, constant light results in a behavioral splitting phenotype, depression of free-running activity, or under high-intensity conditions, arrhythmia (Aschoff, 1960; Daan and Pittendrigh, 1976). Interestingly, these effects of constant light appear to occur at a system level. Along these lines, cellular communication within the SCN is affected, whereas the inherent timekeeping capacity of SCN neurons is not altered (Ohta et al., 2005). Here we examined whether SCN timing under LL was altered by the attenuation of CREB-dependent signaling. For these experiments, A-CREB::tTA mice and tTA littermates were entrained to a 12-h LD cycle as described above and then transferred to constant light conditions (200 lux). In tTA mice, constant light led to the expected lengthening of the circadian period and a sharp drop in total activity (Fig. 3A). In contrast, a large number of A-CREB::tTA bitransgenic mice exhibited a rapid fragmentation of circadian activity under LL (Fig. 3B). χ2 periodogram analysis revealed a complete loss of circadian activity in 6 of 9 A-CREB::tTA mice, whereas 12 of 12 monotransgenic littermates maintained rhythmicity (Suppl. Table S1). Of the 3 A-CREB::tTA mice that maintained statistically significant rhythmicity under LL, 2 mice showed marked fragmentation of activity rhythms; only one mouse exhibited consolidated long period locomotor activity, similar to the tTA mice (data not shown). Together, these data indicate a role for CREB in the maintenance of circadian clock timing. Of note, the total amount of wheel-running activity (both before and during doxycycline administration) was not significantly different between the tTA monotransgenic and A-CREB::tTA mice (Fig. 3C).
Figure 3.
Constant light triggers arrhythmia in A-CREB::tTA transgenic mice. Represenative actograph trace from a tTA (A) and A-CREB::tTA transgenic (B) mouse under LD and constant light (LL) conditions. Note the lengthening of tau (relative to the DD condition in Fig. 2) in the tTA mouse. In contrast, the A-CREB::tTA mouse exhibited a pronounced disruption on circadian locomotor output under LL. At the end of the first LL period, animals were transferred to LD, and the drinking water was supplemented with doxycycline to suppress transgene expression. Subsequent transfer to LL revealed a long tau phenotype in the A-CREB::tTA mouse, thus indicating the reversible nature of A-CREB– mediated arrhythmia. FFT analysis and the associated tau for the indicated blocks of time (black vertical bars) are shown to the right of the panels. (C) The A-CREB transgene does not affect overall locomotor activity under constant light (LL). Mean ± SEM daily wheel rotation per minute is represented on the Y-axis for A-CREB::tTA and the tTA lines under both control conditions (no doxycycline: NO DOX) and with doxycycline (DOX) added to the drinking water (p > 0.05). n.s. = not significant.
Next, we tested whether the effects of A-CREB could be reversed by application of doxycycline to the drinking water. These experiments address 2 distinct questions. First, is there a development effect of the transgene that compromises the functionality of the SCN? Second, can reactivation of CREB signaling reinstate rhythms in an arrhythmic animal?
For these assays, the free-running rhythm under LL without doxycycline treatment was profiled (as noted above). Mice were returned to an LD cycle, and the drinking water (of both genotypes) was supplemented with 200 μg/mL of doxycycline to suppress transgene expression. Recent work reported that the same dose of doxycycline leads to a rapid repression of tTA-mediated gene expression in the SCN (Hong et al., 2007) (Suppl. Fig. S3C). Following 2 weeks in LD, mice were transitioned to LL. As expected, tTA mice exhibited a marked lengthening of the free-running rhythm. Strikingly, A-CREB::tTA mice also exhibited a long tau and maintained a clear rhythm of locomotor activity throughout the experimental condition (Fig. 3), thus revealing that the arrhythmic A-CREB phenotype was reversible. We also tested whether doxycycline administration under LL can reverse arrhythmia in A-CREB::tTA mice. Hence, 10 days into LL, drinking water was supplemented with doxycycline. Under this condition, 2 of 5 mice re-established a rhythmic pattern of locomotor activity (Suppl. Fig. S3A and B), whereas the rest of the animals remained arrhythmic. Together, these data strongly support a role for CREB in the maintenance of circadian clock timing. Further, the ability to reverse the effects of A-CREB indicates that repression of CREB-dependent transcription does not lead to a permanent dysregulation of SCN clock physiology.
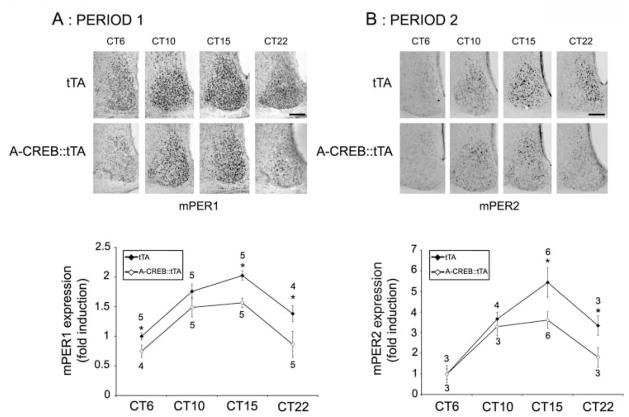
To gain insight into a possible mechanism by which CREB influences the clock, we examined the expression of the core clock proteins PERIOD1 and PERIOD2 as well as the expression of the clock-regulated peptides arginine vasopressin (AVP) and vasoactive intestinal peptide (VIP). An examination of Period gene expression was based on promoter analysis studies that identified CREB-binding sites in the 5′ regulatory regions of both genes (Travnickova-Bendova et al., 2002); an examination of AVP and VIP was based on work showing that both peptides influence the clock and are under the control of the CREB/CRE transcriptional pathway (Deutsch et al., 1988; Pardy et al., 1992; Aton et al., 2005; Maywood et al., 2006; Li et al., 2009). For the examination of PERIOD proteins, mice were dark-adapted for 2 days, and then animals were sacrificed during the subjective day (CT6 and CT10) and the subjective night (CT15 and CT22). We chose to examine PERIOD protein expression under DD conditions rather than under LL free-running conditions because, unlike LL, A-CREB::tTA mice maintain circadian rhythmicity under DD, and thus, relative transgene expression can be plotted over a 24-h cycle. Immunolabeling of central SCN sections revealed significantly lower expression of PERIOD1 and PERIOD2 in A-CREB::tTA versus the tTA mice (Fig. 4A and B) at multiple points across the circadian cycle.
Figure 4.
A-CREB alters clock gene expression. tTA and A-CREB::tTA mice were dark-adapted and sacrificed at the noted circadian times, and SCN tissue was immunolabeled for PERIOD1 (A) or PERIOD2 (B). Representative PERIOD labeling is shown (top), and quantitative protein expression over the circadian cycle is shown (bottom). Values next to each data point indicate the number of animals examined; asterisks note statistically significant differences (p < 0.05). All data were normalized to CT6 tTA values, which were set equal to 1. Scale bars: 125 μm.
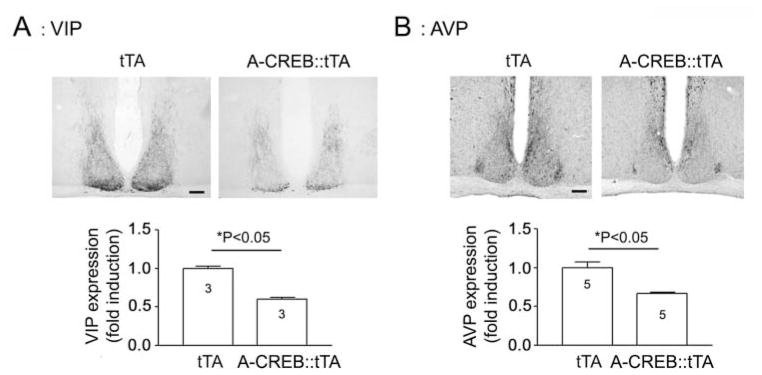
To examine AVP and VIP expression, A-CREB::tTA mice and tTA littermates were sacrificed at CT15, and central SCN-containing sections were immunolabeled for the 2 peptides. Representative data reveal a significant attenuation in both AVP and VIP in the CREB::tTA versus the tTA monotransgenic mice (Fig. 5A and B). Of note, the topographical distribution of both peptides did not appear to be affected by CREB repression. Thus, in A-CREB::tTA mice, VIP was enriched in the ventral SCN, and AVP expression was predominantly located within the dorsal and lateral SCN. Interestingly, double labeling for GFP and AVP revealed that strongly transgenic cells expressed relatively low levels of AVP (Suppl. Fig. S4). These data provide cellular-level support for immunohistochemical examination of the effects of A-CREB on AVP expression. Together, these results indicate that CREB-dependent transcription influences both the core clock as well as clock-control gene expression, thus providing potential clues regarding how CREB influences overt rhythmicity.
Figure 5.
Representative immunolabeling for VIP (A) and AVP (B) at CT15 (top) and quantitative analysis (bottom). Data were normalized to tTA values, which were set equal to 1. Please see the Methods section for an explanation of the quantitative analysis. Scale bar: 125 μm.
DISCUSSION
The data reported here support previous work showing that light couples to the clock via the CREB/CRE pathway and reveal a hitherto unknown role of CREB in circadian clock physiology. With respect to the light entrainment data, the A-CREB mice exhibited a significant decrease in the early night phase delay. These data are consistent with a prior study that employed a CRE decoy oligodeoxynucleotide to sequester CREB and in turn suppress light entrainment of the clock in vivo (Tischkau et al., 2003), and with work showing that phosphorylation of CREB at Ser 142 influences light-evoked clock entrainment (Gau et al., 2002). Interestingly, our data revealed an approximately 50% attenuation of the light-evoked phase delay. This attenuated light entrainment phenotype could be interpreted in several ways. First, attenuation (rather than complete inhibition) might result from the mosaicism of the A-CREB transgene. Hence, given that the transgene was expressed in approximately 60% of the cells, it is likely that robust light-evoked CRE-mediated gene expression was preserved in the subset of retinoreceptive SCN neurons that did not express the transgene. A second possibility is that CRE-mediated gene expression is one of several independent cellular signaling events that function in an additive manner to regulate clock entrainment. Along these lines, several studies have indicated roles for a number of ostensibly CREB-independent transcriptional, translational, and posttranslational events in clock entrainment (Coogan and Piggins, 2003; Oster et al., 2003; Jakubcakova et al., 2007; Lee et al., 2007a; Masubuchi et al., 2010).
Although the precise mechanism by which CREB contributes to overt rhythm generation is not known, there are 2 leading possibilities. The first is that CREB-dependent transcription facilitates robust cycling of the core clock transcriptional loop. Thus, in A-CREB–expressing SCN cells, a damped molecular rhythm could lead to an overall weakening of the SCN circadian clock; under the desynchronizing influence of the LL paradigm, this could result in a rapid loss of a coherent clock-output signal. This interpretation is supported by work showing that CREB-dependent transcription exhibits a circadian oscillation (Obrietan et al., 1999; O’Neill et al., 2008) and that disruption of upstream signaling events (i.e., adenylyl cyclase activity), which are tightly coupled to CREB transactivation, dampens SCN molecular clock amplitude, as assessed via period2-luciferase rhythms (O’Neill et al., 2008). Interestingly, in Drosophila, disruption of CRE-mediated transcription results in a shortened circadian period and a reduction in the amplitude of the period circadian rhythm (Belvin et al., 1999). Consistent with these observations, we detected a reduction in the amplitude of the PERIOD1 and PERIOD2 rhythm in A-CREB transgenic mice. At a molecular/mechanistic level, the most parsimonious explanation for these results is that CREB directly facilitates the rhythmic, E-box–dependent transcription of core clock genes. Of note, a number of clock genes, including period1 and period2, contain CREs within their regulatory region, and with respect to period1, reporter studies have shown tight regulation via the CREB/CRE transcriptional pathway (Travnickova-Bendova et al., 2002).
The second potential mechanism involves a decrease in the expression of clock-output genes, whose protein products serve as coupling cues within the SCN. For example, a number of rhythmically regulated neurohormones, including somatostatin, AVP, and VIP (Kalsbeek et al., 1995; Ban et al., 1997; Yang et al., 1994; Dardente et al., 2004), contain one or more CREs within their promoter regions (Deutsch et al., 1988; Montminy and Bilezikjian, 1987; Pardy et al., 1992), and thus, abrogation of CREB-dependent transcription could reduce their expression. Of note, prior work has shown that AVP expression is positively regulated by CLOCK/BMAL1 transactivation (Jin et al., 1999). Thus, the observation reported here, showing that CREB repression leads to a reduction in PER expression, might lead one to predict an upregulation of AVP (as a result of the derepression of CLOCK/BMAL1). However, our data reveal an opposite effect (i.e., a reduction in AVP expression). This finding, coupled with work showing that AVP transcription is regulated by CREB, suggests that the CRE plays a more prominent role than the E-box in the regulation of AVP in the SCN.
A number of studies have revealed a key role for peptidergic signaling in the coupling of autonomous SCN clock cells. Along these lines, VIP has been shown to maintain SCN rhythmicity by synchronizing pacemaker neurons (Aton et al., 2005; Maywood et al., 2006). Likewise, work by Li et al. (2009) suggests that, in addition to functioning as a clock output, vasopressin may also function within the SCN to coordinate individual or groups of cellular oscillators. Here we found that the expression of both AVP and VIP is decreased in A-CREB transgenic mice, thus providing a potential mechanism for the weakened oscillator efficacy. Of note, both the expression and rhythmicity of AVP and VIP are markedly affected by constant light, with VIP losing rhythmicity within a week of LL and AVP exhibiting a residual rhythm at 3 weeks in LL (Isobe and Nishino, 1998). The approximately 35% reduction in either VIP or AVP reported here may not be sufficient to elicit arrhythmia under LL; however, the attenuated/altered expression of multiple peptides within the SCN could work in combination to affect SCN clock neuron synchronization.
The potential mechanisms outlined above may not be mutually exclusive but rather could work in combination to mediate the effects of CREB in the clock. Along these lines, the disruption of peptidergic content within the SCN would likely alter Gs-cAMP signaling, which in turn would influence the amplitude and phasing of the core clock oscillator (O’Neill et al., 2008). Likewise, disruption of the oscillator (via abrogation of CREB-mediated clock gene transactivation) would likely alter the expression of clock-controlled genes, including the aforementioned peptides. Thus, the dysregulation of both of these intertwined processes (Hastings et al., 2008) could be required to trigger clock desynchronization.
Interestingly, in a subset of the A-CREB mice, LL-induced arrhythmia could be reversed by doxycycline administration. At this point, we can only speculate on the mechanism underlying the reinstatement of rhythms. Experiments that examine single cell clock gene rhythms and/or peptidergic output may provide important clues as to how overt rhythmicity is restored following A-CREB repression.
The lack of a significant effect of A-CREB on DD free-running rhythms indicates that abrogation of CREB-dependent transcription in a subset of SCN cells is not sufficient to affect clock timing in the absence of an additional desynchronizing event, such as constant light. This lack of an effect may, in part, result from the mosaicism of neuronal transgene expression. In some respects, this effect may be similar to the mosaic Clock mutant mouse aggregation data described by Low-Zeddies and Takahashi (2001). In those studies, varying combinations of CLOCK mutant and wild-type SCN neurons in chimeric mice led to a range of phenotypes, depending on the proportion of mutant SCN neurons. These data revealed that coherent circadian rhythms arise from the integration of clock cell phenotypes and, with respect to our data, raise the possibility that a higher percentage of A-CREB transgenic neurons would have led to a more pronounced circadian phenotype. At this point, it is worth noting that a logical extension of this hypothesis is that the reported phenotype might only have been revealed with a mosaic transgene expression pattern, which allows for photic entrainment. Hence, if CREB transcription were completely blocked in the SCN (which might lead to uncoupling of light and the clock), then the LL phenotype would not be revealed.
Together, the data presented here support the idea that the CRE transcriptional pathway plays an important role in orchestrating events that are essential for both the maintenance of robust SCN timing and light-induced clock entrainment. Additional studies examining clock cell coupling and cell autonomous clock rhythms will be required to reveal the precise mechanism by which CREB affects SCN physiology.
Supplementary Material
Acknowledgments
This work was supported by an NSF grant (IBN-0090974) and grants from the NIH (MH62335 and NS067409). The authors thank Heather Dziema and James Stone for technical assistance.
Footnotes
Supplementary material for this article is available on the Journal of Biological Rhythms website: http://jbr.sagepub.com/supplemental.
References
- Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:867–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschoff J. Exogenous and endogenous components in circadian rhythms. Cold Spring Harb Symp Quant Biol. 1960;25:11–28. doi: 10.1101/sqb.1960.025.01.004. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Y, Shigeyoshi Y, Okamura H. Development of vasoactive intestinal peptide mRNA rhythm in the rat suprachiasmatic nucleus. J Neurosci. 1997;17:3920–3931. doi: 10.1523/JNEUROSCI.17-10-03920.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–787. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Lee B, Cho HY, Reyes IB, Pu XA, Saido TC, Hoyt KR, Obrietan K. CREB is a key regulator of striatal vulnerability in chemical and genetic models of Huntington’s disease. Neurobiol Dis. 2009;36:259. doi: 10.1016/j.nbd.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coogan AN, Piggins HD. Circadian and photic regulation of phosphorylation of ERK1/2 and Elk-1 in the suprachiasmatic nuclei of the Syrian hamster. J Neurosci. 2003;23:3085–3093. doi: 10.1523/JNEUROSCI.23-07-03085.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daan S, Pittendrigh CS. A functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol. 1976;106:267–290. [Google Scholar]
- Dardente H, Menet JS, Challet E, Tournier BB, Pévet P, Masson-Pévet M. Daily and circadian expression of neuropeptides in the suprachiasmatic nuclei of nocturnal and diurnal rodents. Brain Res Mol Brain Res. 2004;124:143–151. doi: 10.1016/j.molbrainres.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Deutsch P, Hoeffler JP, Jameson JL, Lin JC, Habener JF. Structural determinants for transcriptional activation by cAMP-responsive DNA elements. J Biol Chem. 1988;263:18466–18472. [PubMed] [Google Scholar]
- Ding JM, Faiman LE, Hurst WJ, Kuriashkina LR, Gillette MU. Resetting the biological clock: mediation of nocturnal CREB phosphorylation via light, glutamate, and nitric oxide. J Neurosci. 1997;17:667–675. doi: 10.1523/JNEUROSCI.17-02-00667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gau D, Lemberger T, von Gall C, Kretz O, Le Minh N, Gass P, Schmid W, Schibler U, Korf HW, Schütz G. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron. 2002;34:245–253. doi: 10.1016/s0896-6273(02)00656-6. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Ginty DD, Kornhauser JM, Thompson MA, Bading H, Mayo KE, Takahashi JS, Greenberg ME. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Hong HK, Chong JL, Song W, Song EJ, Jyawook AA, Schook AC, Ko CH, Takahashi JS. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS Genet. 2007;3:e33. doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Goodman RH. CREB signaling: timing is everything. Sci STKE. 2001;82:pe1. doi: 10.1126/stke.2001.82.pe1. [DOI] [PubMed] [Google Scholar]
- Isobe Y, Nishino H. AVP rhythm in the suprachiasmatic nucleus in relation to locomotor activity under constant light. Peptides. 1998;19:827–832. doi: 10.1016/s0196-9781(98)00021-7. [DOI] [PubMed] [Google Scholar]
- Jakubcakova V, Oster H, Tamanini F, Cadenas C, Leitges M, van der Horst GT, Eichele G. Light entrainment of the mammalian circadian clock by a PRKCA-dependent posttranslational mechanism. Neuron. 2007;54:831–843. doi: 10.1016/j.neuron.2007.04.031. [DOI] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- Kalsbeek A, Buijs R, Engelmann M, Wotjak C, Landgraf R. In vivo measurement of a diurnal variation in vasopressin release in the rat suprachiasmatic nucleus. Brain Res. 1995;682:75–82. doi: 10.1016/0006-8993(95)00324-j. [DOI] [PubMed] [Google Scholar]
- Lee B, Almad A, Butcher GQ, Obrietan K. Protein kinase C modulates the phase-delaying effects of light in the mammalian circadian clock. Eur J Neurosci. 2007a;26:451–462. doi: 10.1111/j.1460-9568.2007.05664.x. [DOI] [PubMed] [Google Scholar]
- Lee B, Dziema H, Lee KH, Choi YS, Obrietan K. CRE-mediated transcription and COX-2 expression in the pilocarpine model of status epilepticus. Neurobiol Dis. 2007b;25:80–91. doi: 10.1016/j.nbd.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Cao R, Choi YS, Cho HY, Rhee AD, Hah CK, Hoyt KR, Obrietan K. The CREB/CRE transcriptional pathway: protection against oxidative stress-mediated neuronal cell death. J Neurochem. 2009;108:1251–1265. doi: 10.1111/j.1471-4159.2008.05864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JD, Burton KJ, Zhang C, Hu SB, Zhou QY. Vasopressin receptor V1a regulates circadian rhythms of locomotor activity and expression of clock-controlled genes in the suprachiasmatic nuclei. Am J Physiol Regul Integr Comp Physiol. 2009;296:R824–R830. doi: 10.1152/ajpregu.90463.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of the mammalian circadian system: photic entrainment, circadian pacemaker mechanisms, and posttranslational regulation. Annu Rev Genet. 2000;34:533–562. doi: 10.1146/annurev.genet.34.1.533. [DOI] [PubMed] [Google Scholar]
- Masubuchi S, Gao T, O’Neill A, Eckel-Mahan K, Newton AC, Sassone-Corsi P. Protein phosphatase PHLPP1 controls the light-induced resetting of the circadian clock. Proc Natl Acad Sci U S A. 2010;107:1642–1647. doi: 10.1073/pnas.0910292107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate MAP kinase activation in the suprachiasmatic nuclei. Nat Neurosci. 1998;1:693–700. doi: 10.1038/3695. [DOI] [PubMed] [Google Scholar]
- Obrietan K, Impey S, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem. 1999;274:17748–17756. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- Ohta H, Yamazaki S, McMahon DG. Constant light desynchronizes mammalian clock neurons. Nat Neurosci. 2005;8:267–269. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H, Werner C, Magnone MC, Mayser H, Feil R, Seeliger MW, Hofmann F, Albrecht U. cGMP-dependent protein kinase II modulates mPer1 and mPer2 gene induction and influences phase shifts of the circadian clock. Curr Biol. 2003;13:725–733. doi: 10.1016/s0960-9822(03)00252-5. [DOI] [PubMed] [Google Scholar]
- Pardy K, Adan RA, Carter DA, Seah V, Burbach JP, Murphy D. The identification of a cis-acting element involved in cyclic 3′,5′-adenosine monophosphate regulation of bovine vasopressin gene expression. J Biol Chem. 1992;267:21746–21752. [PubMed] [Google Scholar]
- Refinetti R. Comparison of six methods for the determination of the period of circadian rhythms. Physiology and Behavior. 1993;54:869–875. doi: 10.1016/0031-9384(93)90294-p. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- Tischkau SA, Mitchell JW, Tyan SH, Buchanan GF, Gillette MU. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J Biol Chem. 2003;278:718–723. doi: 10.1074/jbc.M209241200. [DOI] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gall C, Duffield GE, Hastings MH, Kopp MD, Dehghani F, Korf HW, Stehle JH. CREB in the mouse SCN: a molecular interface coding the phase adjusting stimuli light, glutamate, PACAP, and melatonin for clockwork access. J Neurosci. 1998;18:10389–10397. doi: 10.1523/JNEUROSCI.18-24-10389.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tominaga K, Otori Y, Fukuhara C, Tokumasu A, Inouye S. Day-night variation of preprosomatostatin messenger RNA level in the suprachiasmatic nucleus. Mol Cell Neurosci. 1994;5:97–102. doi: 10.1006/mcne.1994.1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.