Abstract
Background
The popularity of a large number of microarray applications has in cancer research led to the development of predictive or prognostic gene expression profiles. However, the diversity of microarray platforms has made the full validation of such profiles and their related gene lists across studies difficult and, at the level of classification accuracies, rarely validated in multiple independent datasets. Frequently, while the individual genes between such lists may not match, genes with same function are included across such gene lists. Development of such lists does not take into account the fact that genes can be grouped together as metagenes (MGs) based on common characteristics such as pathways, regulation, or genomic location. Such MGs might be used as features in building a predictive model applicable for classifying independent data. It is, therefore, demanding to systematically compare independent validation of gene lists or classifiers based on metagene or individual gene (SG) features.
Methods
In this study we compared the performance of either metagene-or single gene-based feature sets and classifiers using random forest and two support vector machines for classifier building. The performance within the same dataset, feature set validation performance, and validation performance of entire classifiers in strictly independent datasets were assessed by 10 times repeated 10-fold cross validation, leave-one-out cross validation, and one-fold validation, respectively. To test the significance of the performance difference between MG- and SG-features/classifiers, we used a repeated down-sampled binomial test approach.
Results
MG- and SG-feature sets are transferable and perform well for training and testing prediction of metastasis outcome in strictly independent data sets, both between different and within similar microarray platforms, while classifiers had a poorer performance when validated in strictly independent datasets. The study showed that MG- and SG-feature sets perform equally well in classifying independent data. Furthermore, SG-classifiers significantly outperformed MG-classifier when validation is conducted between datasets using similar platforms, while no significant performance difference was found when validation was performed between different platforms.
Conclusion
Prediction of metastasis outcome in lymph node–negative patients by MG- and SG-classifiers showed that SG-classifiers performed significantly better than MG-classifiers when validated in independent data based on the same microarray platform as used for developing the classifier. However, the MG- and SG-classifiers had similar performance when conducting classifier validation in independent data based on a different microarray platform. The latter was also true when only validating sets of MG- and SG-features in independent datasets, both between and within similar and different platforms.
Keywords: microarray, classification, metagenes, breast cancer
Background
Microarray gene expression analysis has in several studies been applied to elucidate the relation between clinical outcome and gene expression patterns in breast cancer and has demonstrated improvement of recurrence prediction.1–14 In some studies, genes in such profiles might be fully or partially missing in the test data used for validation due to the choice of microarray platform or the presence of missing values associated with a given probe. Furthermore, an obtained gene list can have none or few genes in common with other gene lists addressing the same clinical outcome,15,16 due to usage of different microarray platforms, different methods for measuring mRNA expression levels, variation in patient sampling,15 lab variation/measurement noise,17 and differences in data processing such as different normalization methods.18 Furthermore, a wide array of feature selection methods is available for gene selection, which also affects the constitution of such final gene lists.15 These feature selection methods encompass filter approaches; selection of top features from a ranked list of genes; wrapper methods where model selection algorithms are wrapped in the search process of feature subsets (ie, the Gini index in random forest);19 and embedded methods where the feature selection is an integrated part of the classification method, such as iteratively eliminating redundant features with minimal information regarding classification performance.20 More recent approaches to gene selection include recursive feature elimination based on support vector machines (SVM-RFE). This approach uses the coefficient of the weight vector to compute a feature ranking score, from which features with the smallest ranking scores are built into the model, for example, a leave-out-N number of genes approach.21 The advanced version combines SVM-RFE with a minimum-redundancy and maximum-relevancy filter, where relevance of each feature is determined by the mutual information among genes and class labels, and the redundancy is given by the mutual information among the genes.22
In addition to the above mentioned factors, the choice of classification method also impacts the final model and gene lists. Furthermore, the validations of such gene lists in independent data are very heterogeneous, with the majority testing significant differences in survival, which barely reflect the actual classification accuracy, while few studies conducts validations in terms of classification accuracies.
To overcome the above obstacles, individual genes could be considered part of a larger network, that is, their expression being controlled by the expression level of other genes or that a group of genes belong to a specific pathway performing a well-defined task. These genes may be controlled by the same transcription factor or located in the same chromosomal region. Such grouping has been collected in public databases such as the Kyoto Encyclopedia of Genes and Genomes (KEGG)23 the Molecular Signature Database (MsigDB),24 and the Gene Ontology database (GO).25 In relation to breast cancer, for example, cell cycle upregulation or deregulation of other pathways are associated with metastasis2,3,26,27 Furthermore, it has been shown that metastasis progression28 and tumor grading29 in breast cancer are associated with accumulated mutations in several genes, leading to amplification or inactivation of genes, and even large genomic losses or gains in specific chromosomal regions affecting gene expression levels.
Our previous studies showed that the expression levels of such specific entities, called metagenes (MGs), are significantly associated with metastatic outcome in breast cancer across eight different datasets.30,31 Several studies have defined metagene/gene modules derived from microarray data using various methods such as penalized matrix decomposition which clusters similar genes but without similar expression profiles32 hierarchical clustering,33 correlation,34 or combining a priori protein-protein interactions with microarray gene expression data defining interaction networks as features.35,36 Few studies have attempted to use such predefined gene sets for prediction models. One such study used a compendia of microarray cancer genes for defining metagene/gene modules by performing hierarchical clustering of these genes expressions and seeding genes within the clusters into gene sets annotated in the public databases.33 A second study defined metagene/gene modules as sets of significantly correlated or anticorrelated genes combined with prior information about the genes.34 One of the strengths of using gene sets as features is that this circumvents the necessity of sharing all genes between studies. Furthermore, grouping the genes together also reduces the dimensionality of the datasets and thus functions as feature reduction. Therefore, profiles consisting of MGs might be used for developing predictive classifiers that can be validated in independent data.
This study systematically assesses and compares the performance of MG- and SG-(single gene) feature sets and MG- and SG-classifiers extracted from the same samples in predicting metastasis outcome among lymph node–negative breast cancer patients who have not been treated with adjuvant therapy.
These comparisons were first made by model building and classification within the same dataset using 10-fold cross validation. Furthermore, the comparisons were also done across datasets in two ways: (1) application of the entire classifier on the test data and (2) only the features from the classifier are transferred to the test data for model building and testing and evaluated by leave-one-out cross validation (LOOCV). In each case, we also examined two possible scenarios. In the first, the validations were conducted between studies using the same microarray platform (Affymetrix classifier/feature set validated on an independent Affymetrix dataset), while the second, encompassed validations across studies with different platforms (Agilent-developed classifier/feature set validated on an independent Affymetrix dataset).
Results
Features and models
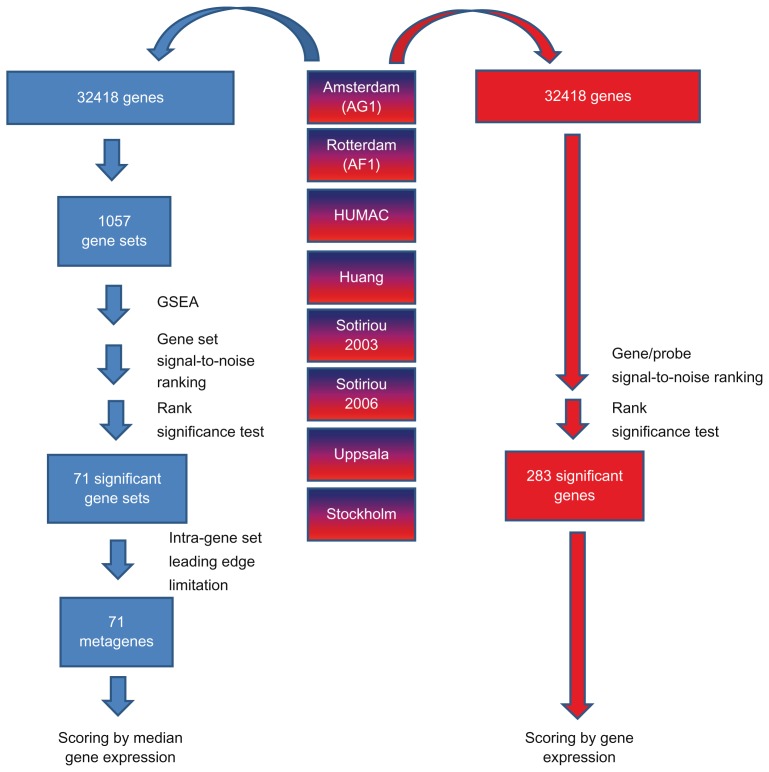
The 71 metagenes used in this study wer determined as gene sets covering similar biological pathways having common transcription factor binding sites or genes located in the same chromosomal region (Supplementary Table 1), which in previous studies have proven to be associated with breast cancer metastasis across eight different datasets using a rank-based method (Fig. 1).30,31 An overview of the eight datasets is shown in Table 1. The final MGs consist, on average, of 21 genes, with the smallest MGs consisting of only 2 genes and the largest, of 65 genes. This rank-based method was also applied to each gene across the same eight datasets, which led to identification of 283 rank-significant SGs (Supplementary Table 2) shared by the four datasets to be used later. Amongst the 283 SGs, 119 genes were also present on the gene lists underlying the MGs (data not shown). These 71 MGs and 283 SGs were used for selecting the optimal MG- and SG-feature sets and development of their corresponding MG- and SG-classifiers.
Figure 1.
Metagene and single gene selection procedure.
Notes: MGs (blue) and SGs (red) were both derived from the same eight breast cancer gene expression datasets. These covered 32418 genes. 1057 gene lists was defined from these 32418 genes/probes. These were subjected to gene set enrichment analysis (GSEA), ranked within each dataset according to their signal-to-noise ratio, and their across dataset mean rank calculated. This mean rank was significance tested as described in the Materials and Methods section, resulting in 71 metagenes that were scored by the median gene expression of the GSEA leading edge genes. The single genes were selected by directly ranking each gene/probe across the datasets and subsequently following the same procedure as for the metagenes, resulting in 283 significant single genes. The measure for each single is the gene expression level associated with each gene.
Table 1.
Overview of datasets.
| Dataset | Chip | Probes (K) | Patients | Outcome | Treatment | Define MG | Define SG | Train | Test | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Amsterdam | Agilent/Rosetta | 25 | 295, N+, N− | DM | None, et, ct | √ | √ | [14] | ||
| Amsterdam (AG1) (subset of the above) | Agilent/Rosetta | 25 | 151, N− | DM | None | √ | √ | √ | [14] | |
| Rotterdam (AF1) | Affymetrix HG-133A | 22 | 286, N− | DM | None | √ | √ | √ | [3] | |
| HUMAC | Spotted oligonucleotides | 29 | 60, N− | ME | None | √ | √ | [7] | ||
| Huang | Affymetrix 95av2 | 12 | 52, N+ | RE | Ct | √ | √ | [13] | ||
| Sotiriou 2003 | Spotted cDNA | 7.6 | 99, N+/N− | RE | Et, ct | √ | √ | [1] | ||
| Sotiriou 2006 | Affymetrix HG-133A | 22 | 179, N+/N− | DM | Et | √ | √ | [12] | ||
| Uppsala | Affymetrix HG-133A+B | 44 | 236, N+/N− | DF | None, ct, et | √ | √ | [52] | ||
| Stockholm | Affymetrix HG-133A+B | 44 | 159, N+/N− | RE | None, ct, et | √ | √ | [11] | ||
| TRANSBIG (AF2) | Affymetrix HG-133A | 22 | 147, N− | DM | None | √a | √ | [35] | ||
| Mainz (AF3) | Affymetrix HG-133A | 22 | 200, N− | DM | None | √b | √ | [53] |
Notes: The table shows name of dataset, microarray chip, number of probes, patients, outcome, patient treatment, datasets used to define features and for training and testing and the references.
designate used as training only when validating AF3;
designate used for training only when validating AF2.
Abbreviations: K, thousands; N+ and N−, node-positive and -negative patients; DM, distant metastasis; ME, metastasis; RE, relapse; DF, death from breast cancer; et, endocrine therapy; ct, chemo therapy; none, no adjuvant therapy.
Two (AG1 and AF1) of the eight datasets used to define the features were, therefore, solely used for training purposes (Table 1). The AF1 corresponds to the Affymetrix-based Rotterdam dataset and AG1 is a subset of the Agilent-based Amsterdam dataset. Both dataset containing only node-negative samples. The AF2 and AF3 datasets are both based on the Affymetrix platform and function as test sets for classifiers and feature sets developed and selected by the AG1 and AF1 datasets (Table 1). Furthermore, AF2 was used for training to validate on AF3 and vice versa (Table 1). The following three classification methods were used for model building: random forest (RF) and support vector machines with a radial-based kernel (R-SVM) or a sigmoid-based kernel (S-SVM). These were optimized to achieve the best mean of sensitivity and specificity, referred to as balanced accuracy (bAcc).
In order to build the models, the SGs or MGs within each training dataset were ranked according to their random forest importance values. For a given feature, this value reports the standardized drop in prediction accuracy when the class labels are permuted.37 This rank was then used for model building by subsequently adding one feature at a time in a top-down forward wrapper approach starting with the top two features. To avoid creating bias during gene selection and training of the final classifier and on classification performance, 10 times repeated 10-fold cross validation bAcc was used as optimization measure, as this optimization metric has previously been shown to give an excellent bias-variance balance.38 The above described procedure led to a total of 24 models, with 12 composed of MG and SG features, respectively (Table 2). In the metagene models, each metagene contributes one input value calculated as the median expression level of the genes underlying that particular metagene. We, therefore, considered a metagene model composed of four metagenes to have four inputs or four features. These MG- and SG-models varied in complexity, consisting of 4 to 122 features (Table 2). Comparison of the number of features in each model displays a slightly higher complexity of SG-models with MG-models ranging from 4 to 67 features having, on average, 26 features per model (Table 2), while SG-models varied from 14 to 122 features with an average of 34 features per model (Table 2). This suggests that each SG-feature is less informative compared to the MG-features and thus a larger number of SG-features is a requirement for reaching optimal performance.
Table 2.
The number of metagene and single genes features in the 24 models.
| Dataset | AG1 | AF1 | AF2 | AF3 | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||
| Features method | #MG | #SG | #MG | #SG | #MG | #SG | #MG | #SG |
| RF | 4 | 21 | 15 | 21 | 14 | 14 | 21 | 26 |
| R-SVM | 18 | 20 | 57 | 25 | 5 | 22 | 10 | 71 |
| S-SVM | 29 | 17 | 67 | 35 | 9 | 19 | 64 | 122 |
Internal performance of MGand SG-models
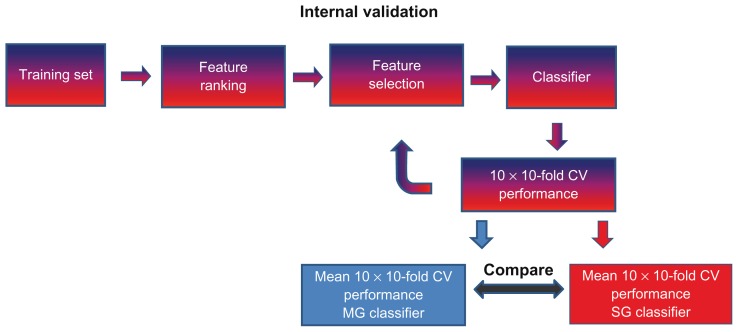
To reduce variability and complexity and keeping validation parameters as constant as possible, the performance of MG- and SG-models were evaluated within the same dataset from which they were initially developed. This internal model performance was evaluated using 10 times repeated 10-fold cross validation (Fig. 2). This validation scheme partitions the training data into 10 nearly equally sized folds. Subsequently, 10 iterations of training and validation are performed. During each of these iterations, a different fold of the training data is left out for validation, and the remaining folds are used for learning. The mean accuracy of all 10 folds validated is thus the 10-fold cross validated accuracy of the model. By repeating this process 10 times, a more robust and unbiased estimation of the generalization performance is obtained.39 It should, therefore, also be noted that the individual classification performances are artificially elevated due to information leakage caused by using the entire dataset for ranking. However, as metagenes are a simple linear combination of single genes, we assume that the comparisons between the MG- and SG-model performances are similarly affected by this leakage.
Figure 2.
The internal dataset validation procedure.
Notes: For both types of features, the entire training set was used to rank each feature by the random forest importance value. This rank was used for feature selection adding one feature at a time starting from top 2 to top 71 (for MGs) or top 283 (for SGs), thus testing a classifier with a fixed number of features in each round. The performance of the classifier was evaluated using 10 times repeated 10-fold cross validation. Using the same combination of training data and classification method, the mean 10 times repeated 10-fold cross validation of the MG-classifier and SG-classifier were compared with each other.
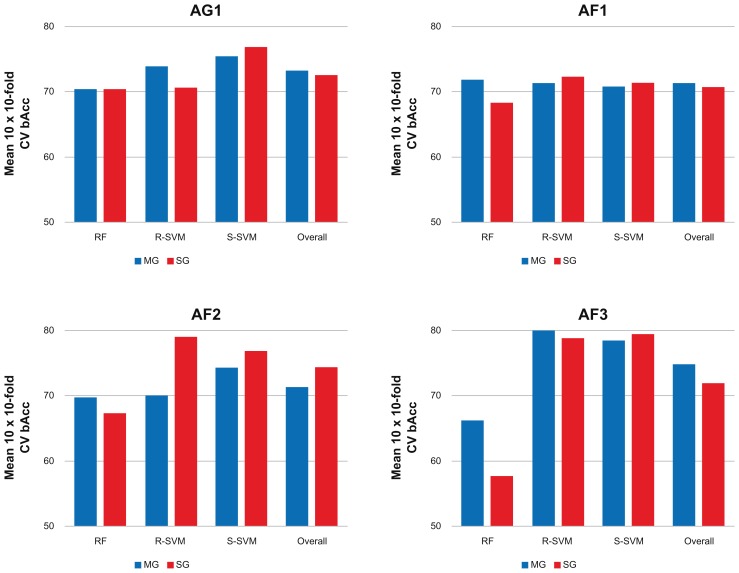
As indicated by the internal classification performances, the models predicted outcome with high accuracy (Fig. 3). Comparison across the three classification methods within the majority of each training dataset showed that MG-models perform slightly better than SG-models (Fig. 3). However, these differences were only minor and nonsignificant, suggesting that these results tend to converge to a common optimization level independent of feature type.
Figure 3.
Internal classification performance.
Notes: The 10 times repeated 10-fold cross validation balanced accuracies (bAcc) within the four datasets, AG1, AF1, AF2, and AF3, using random forest (RF), support vector machines with a radial (R-SVM) or sigmoid-kernel (S-SVM), or across the three classification methods (Overall) are shown in blue and red respectively.
Feature set transferability
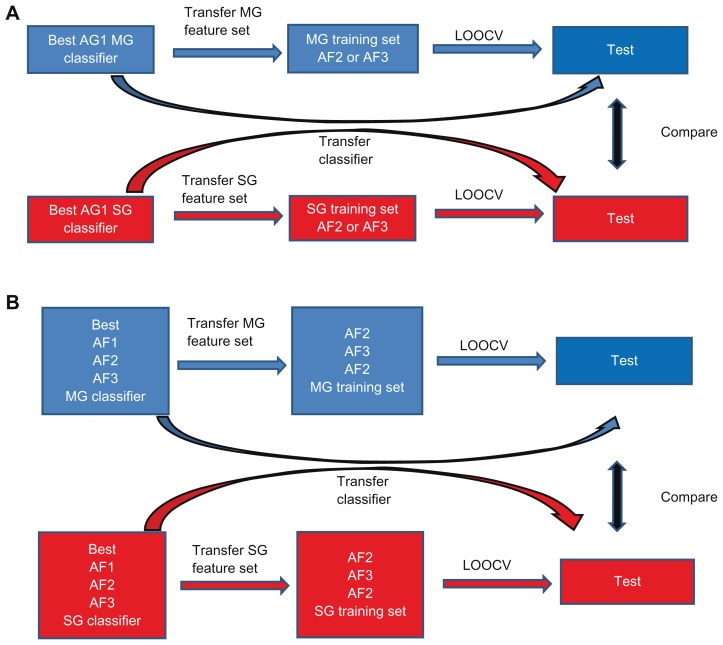
The feature sets within the MG- and SG-models were then transferred to independent datasets to examine if these features could be used to build a model for predicting outcome within the test data as illustrated in Figure 4A and B. The transferability was assessed by leave-one-out cross validation (LOOCV). Briefly, in LOOCV, one sample is left out at a time and used for testing, while the remaining samples are used for training a classifier, which is used to classify the left out sample. This process is repeated until all samples have been left out for testing.
Figure 4.
Between study classifier or feature set validation. (A) Between different platform validations. The best classifiers developed from the training set (AG1) are either directly (transfer classifier) applied and validated in the independent test data (AF2 or AF3) or the features only from the best classifier are used within the test data for model building and testing by leave-one-out cross validation (LOOCV). In each case comparison of MG- (blue) and SG-classifier (red) or feature set performance is conducted using the same training data, classification method and test data. (B) Between similar platform validations. The best classifiers developed from the training set (AF1, AF2, or AF3) are either directly (Transfer classifier) applied and validated in the independent test data (AF2 or AF3) or the features only from the best classifier are used within the test data for model building and testing by leave-one-out cross validation (LOOCV). In each case, comparison of MG- and SG-classifier or feature set performance is conducted using the same training data, classification method, and test data.
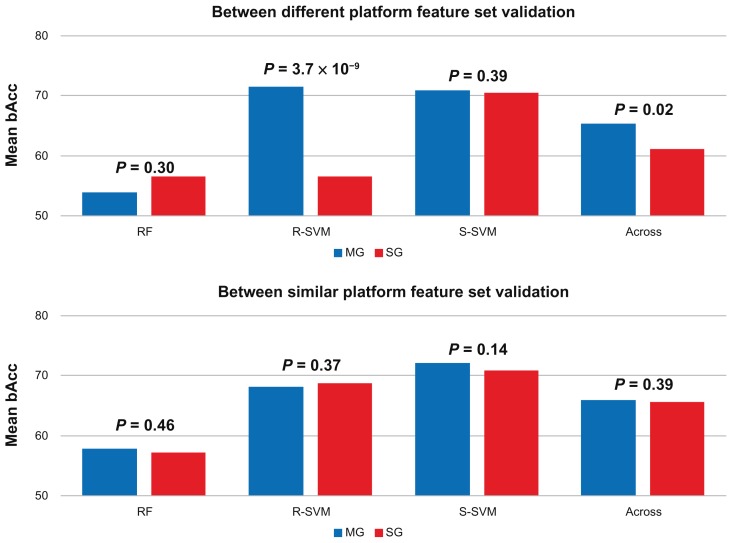
The results show that transfer of the feature sets are able to classify samples with a mean LOOCV bAcc ranging from 54.0% to 72.0%, and where all the MG- and SG-gene lists have a mean LOOCV bAcc of 65.7% and 63.4%, respectively (Fig. 5). This suggests that the majority of these gene lists perform significantly better than random and that the selected MG- and SG-feature sets, being optimal in one dataset, display transferability across studies and can train and build a predictive model in independent samples.
Figure 5.
Exported feature set classification performance.
Notes: The mean balanced accuracies for the between platform validation (AG1 vs. AF2 or AF3) and the within similar platform validations (AF1 vs. AF2 or AF3 or AF2 vs. AF3, and vice versa) using random forest (RF), support vector machines with a radial (R-SVM) or sigmoid-kernel (S-SVM), or across the three classification methods (Across) are shown in blue and red respectively. The P values show the significance in classification between down-sampled testing using exported MG- and SG-feature sets for model building and testing in independent data.
Comparison of classifier performance based on MG- and SG-feature sets in independent datasets
Comparison of the classification performance between the transferred MG- and SG-feature sets between different platforms (AG1-feature sets validated in AF2 or AF3) (Fig. 4A) showed MG features significantly outperformed SG-features using R-SVM (P = 3.7 × 10−9). This is also true when comparing across the three classification methods (P = 0.02) (Fig. 5). However no significant difference in performance was found when using RF (P = 0.30) or S-SVM (P = 0.39). This could suggest that the overall effect is solely due to the significance associated with the R-SVM method (Fig. 5). Comparison of the similar-platform performances (AF1-feature sets validated on AF2 or AF3, AF2-feature sets validated on AF3, and AF3-feature sets validated on AF2) (Fig. 4B), revealed that MG- and SG-features performed equally well for all three classification methods used (Fig. 5).
Classifier transferability and performance
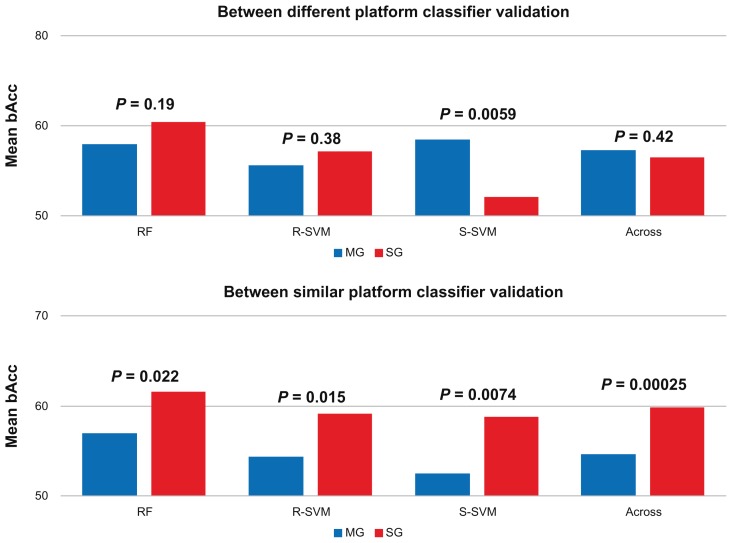
We next used the entire classifier developed in the training sets, based on the features and rules associated with the classifier, to classify the independent samples in the entire test data and, therefore, to determine if the classifier can be exported and used in the test data (Fig. 4A and B). These results showed that the classifiers are less transferable than the feature sets, which is reflected by the weak mean bAcc ranging from 52.5% to 61.5%, with MG- and SG-classifiers having an overall mean bAcc of 56.0% and 58.2%, respectively (Fig. 6).
Figure 6.
Exported classifier classification performance.
Notes: The mean balanced accuracies for the between platform classifier validation (AG1 vs. AF2 or AF3) and the within similar platform classifier validations (AF1 vs. AF2 or AF3 or AF2 vs. AF3, and vice versa) using random forest (RF), support vector machines with a radial (R-SVM) or sigmoid-kernel (S-SVM), or across the three classification methods (Across) are shown in blue and red respectively. The P values show the significance in classification between MG- and SG-classifiers in terms of down-sampled testing.
Comparison of classifier performance
Comparison of MG- and SG-classifiers external validation performance, when the classifier was developed on the Agilent dataset (AG1) and validated on the Affymetrix datasets (AF2 and AF3), showed that MG- and SG-classifiers performed equally well, with the exception of using S-SVM, which favored the MG-classifiers (Fig. 6) (P = 0.006). Comparison of classifier performance across the three methods revealed that MG-classifiers performed slightly better than SG-profiles (Fig. 6). However, this difference was not statistically significant (P = 0.42).
Comparison of MG- and SG-classifiers developed from an Affymetrix dataset and validated on another independent Affymetrix dataset showed that SG-classifiers performed significantly better than MG-classifiers for each of the three classification methods (RF, P = 0.02; R-SVM, P = 0.015; S-SVM, P = 0.007) and across the three classification methods (P < 0.001).
Discussion
A strong prediction model should be robust, reproducible, and ideally exportable to allow validation in independent datasets. In this study, predictive models based on metagenes or single genes being significantly associated with breast cancer metastasis outcome were developed. The metagene features are composed of gene lists sharing a biological consensus, while the single genes are features representing the expression of one gene. The study examined the transferability of MG- and SG-feature sets and classifiers and compared their classification performance on independent datasets. The genes present in the MG-feature sets were always present in the validation, as we had assured that the single genes and metagenes were shared by all four datasets. However, such sharing of gene lists between classifiers derived from different studies is rarely the case. The reason that different studies derive gene expression based classifiers having few or no genes in common is probably due to the selection bias caused by the microarray platform used for measuring gene expression, patient sampling, and the way the data were analyzed. In this respect, a study by Ein-Dor and coworkers showed that the same dataset could derive classifiers having comparable performances but differing substantially with respect to which genes are contained in these classifiers, thus also leading to a selection bias.15 In our study, the problem with missing common genes was circumvented by using features shared by all four datasets used in the study.
We found that both MG- and SG-feature sets could be transferred to independent datasets and were able to build a predictive model achieving good results for classifying node-negative breast cancer metastasis outcome.
The finding that the SG-feature sets are exportable agrees well with few predictive gene lists being exportable to independent data. One such gene list is the 70-gene Mammaprint prognosis gene list embedded in the 70-gene correlation classifier originally developed to predict metastasis within five years (defined as poor prognosis), or no metastasis within five years (good prognosis).2 Transfer of this 70-gene Mammaprint list for model training and testing in independent datasets have proved successful using the original correlation classification method7,40 or by SVM,7 while another study only showed random performance when validating this gene list in independent data.35 Due to missing genes, other studies have only validated a subset of the 70 genes successfully by correlation,8 VFI-classification,8 and centroid classification.41 A second frequently validated gene list is the 76-gene list underlying the 76-gene classifier based on a regression determined risk score trained to predict distant metastasis within five years among lymph node–negative breast cancer patients.3 Transfer of the 76-gene list for model building and testing in fully independent datasets have been proven with limited success in one study by using SVM achieving balanced accuracies of 37% to 64%,35 while a subset of 46 of the 76 genes performed well in predicting metastasis from low-malignant breast cancer, achieving balanced accuracy of 75%.27 A third list is a “wound signature” containing 512 genes, which is able to predict increased risk of metastasis in three types of cancers, including breast cancer,42 The entire wound signature gene list has been transferred to independent data for model building and testing, obtaining results in the range of our transferred SG-gene lists using decision trees,43 but also using a subset of 252 genes has provided similar performance when predicting metastasis outcome among low-malignant breast cancer patients using SVM.27 A fourth study derived a 70-gene Cox-ranked gene list, which was developed to achieve optimal significant hazard ratios with respect to survival analysis in a cohort of patients with early breast cancer.5 Subsets of this list were transferred to two independent datasets for validation by model training and testing using nearest centroid classification obtaining balanced accuracies of 72% and 59%.5
Validation of the classifiers with some of the gene lists mentioned above using the original classification method and cutoffs for class/outcome prediction has been conducted for the 70-gene profile achieving balanced accuracies of 63%,44 while validation of the 76-gene profile has achieved balanced accuracies of 53% to 65%,45 59%,44 and 62%.46 These reported balanced classification accuracies are similar or fairly lower than those obtained by the MG- and SG-classifiers in our study. Although, these MG- and SG-classifier performances are far from optimal to be used in the clinic, they do perform better for long-term outcome prediction than clinicopathological risk criteria illustrated by the findings from three studies. The study by Daemen and coworkers showed that the St. Gallen, NIH, and the Nottingham prognostic index assessors predicted metastasis outcome in a group of 147 breast cancer patients with balanced accuracies of 51%, 52%, and 57%.47 Another study by Sun and coworkers demonstrated that the St. Gallen assessor predicted 5-year relapse free survival in 97 node-negative breast cancer patients with only 51% balanced accuracy.48 Furthermore, the study by Schmidt and coworkers showed that the St. Gallen assessor predicted 5-year and 10-year distant metastasis free survival with a balanced accuracy of 57% and 54%, respectively, and that the Adjuvant! Online algorithm predicted the same endpoints with 56% bAcc in 410 node-negative patients.49
Interestingly, although the 76-gene profile was developed to predict outcome among lymph node–negative patients, as in our study, the majority of MG- and SG-classifiers obtained in our study actually performed slightly better than those reported by the above studies. However, the reported low performance is likely caused by the classifiers being specific to the dataset from which they were developed, and the results, therefore, cannot be generalized. Despite the fact that the primary tumors from both the training and test sets all are lymph node–negative, they might still not be very representative of each other, due to, for example, biological variation, follow-up time of the studies (affecting outcome-coding), and cross-study/lab variation, thus impairing classifier external classification performance. The lack of classifier transferability has been shown in other studies, for example, in the classification of normal versus tumor tissue50 and in the prediction of pathologically complete response to breast cancer chemotherapy treatment.51 This stresses the importance of homogeneity between the samples used for building classifiers and those used for validation.
We evaluated and compared the classification performance between MG- and SG-models within the same datasets. We found that MG- and SG-models had equal performance during internal model building and performance testing, with a trend of MG-models performing slightly better than SG-models. However, the model sizes could imply that SG-models need to contain more features than the MG-models to obtain a similar performance, suggesting that the individual MG-features are more informative than each SG-feature.
The study found no significant difference in performance only between exported MG- and SG-feature sets when used for training and testing on independent datasets. In this setup, the models based on transferred features are trained and tested in the same independent dataset, rendering the measurements underlying the intratraining and testing iterations comparable. One explanation for this similar performance could be that there is a great deal of overlap between the genes constituting the metagenes and those underlying the list of 283 single genes, as the 119 genes are shared. A second explanation could concern the way the metagenes were defined and scored. In our study, the MGs cover lists of genes having a consensus. The first advantage of being defined as such is that the metagenes are conserved and robust across microarray platforms. A second advantage is that the metagenes are narrowed down to the gene set enrichment analysis (GSEA) leading edge genes, thereby picking the best genes within the predefined lists. However, the limitations of the metagenes are that they only consider interactions with members within the defined metagene, but do not take interactions with genes beyond the members of the defined pathways and gene sets. Furthermore, although the human genome has been sequenced, there are still many genes with unknown functions, and, thus, these have still not been annotated to a specified functional gene set. Such undiscovered networks might be discovered by single gene classifiers composed of mixed biological predictors, which might be more easily discovered when using the same microarray technology, reducing the variation caused by the shift in microarray platforms.
In this study, the scoring of the metagenes was based on the median expression values among the GSEA leading edge within each predefined metagene. The fact that only a weak difference in performance between MG- and SG-models was detected suggests that the definition of metagenes and/or their expression calculation might not be able to detect such a significant difference in performance, at least not for metastasis outcome among lymph node–negative breast cancer patients. Other studies have scored gene module/metagene activity in a different way compared with our median expression score by either using the arithmetic mean,52 the sum of discrete values within each module/set,33 the expression values of a median gene in a module/gene set (defined as the gene within the gene set having the smallest sum of distances to other genes within the given gene set/module),53 average rank score (calculated as the average rank of the relative expression levels in a pathway normalized by the total number of genes),54 or by probabilistic inference using log-likelihood ratios.55
Another reason could be that exported MG- and SG-feature sets used for training and testing, and that we only used random forest and support vector machines as classification methods. The above confinement implied, as some of our unpublished results suggest, that classifiers based on random forest and support vector machines have a better classification performance when validated in completely independent datasets compared with other classification methods such as logistic regression or neural networks. In this respect, an option is to validate if the MG- and SG-feature set performances also are similar when using other classification methods.
Interestingly, other studies have found a similarly slight performance difference between models composed of gene modules or individual genes. These studies have also addressed breast cancer metastasis outcome, but using a different module definition and scoring. The gene modules in one study were defined as a subset of genes from gene compendia with correlated expression across arrays and showed that classifiers consisting of gene set modules had a slightly better classification performance, compared with classifiers of individual genes.33 A second similar study, comparing the performance of classifiers consisting of gene sets defined by MsigDB, with classifiers consisting of individual genes, found that the two groups of classifiers had similar performance, but that gene set classifiers were more stable.56 A third study by Blazadonakis and coworkers57 used the 70-gene Mammaprint gene list2 and a previously determined 59-gene gene list58 to extract the presence of significant gene ontology biological processes (GOBPs). The underlying genes in each of these GOBPS, beyond those present in the gene lists, were used to construct of pool of genes for constructing new gene-lists. Interestingly, comparison of classification performance between the original gene-list and the GOBP-derived gene lists in model training and testing within the validation datasets showed that using the GOBP-derived genelist performed slightly better than the gene lists from which they were derived.57 However, compared with our study, the validations in these three studies were confined to using a single classification method (Bayes classifier),33 or centroid classification.56,57 Also, the predicted outcome differed compared with our study, being either defined as a “good” or “bad” outcome relative to 5-year time to metastasis33,56 or 5-year breast cancer survival.57
The results from applying the classifiers developed in the training sets directly upon the test sets revealed that SG-classifiers trained on an Affymetrix dataset and validated on an independent Affymetrix dataset performed significantly better than MG-classifiers. This suggests that the single gene expression values are better for defining classification functions to classify independent data. Therefore, although the genes underlying each MG have been limited to the leading edge, signals from highly predictive genes might be diluted within the metagenes by the median expression scoring across the metagenes, and therefore lose predictive power compared with sets of single genes, which has been picked by feature selection, making each of them highly predictive. This performance difference could also be due to classifier functions/rules based on single gene expression measurements being more transferable than leading edge median expression measurements.
In contrast, no significant difference in performance was found between the MG- and SG-classifiers when the classifier was trained on an Agilent dataset and tested on an Affymetrix dataset. This suggests that when switching platforms, measurements underlying the classifier functions and rules differ significantly between the training and test set, that is, the Agilent and Affymetrix measurements being defined as log ratios and log intensities, respectively. The finding that the MG- and SG-classifiers are equally impaired suggests that Agilent-defined rules are not applicable to an Affymetrix dataset either when using median expression measures or individual gene expression measures, which agrees well with a previous finding showing poor correlations between corresponding measurements conducted on the Agilent and Affymetrix platforms.59 Interestingly, a previous study has shown that Agilent log ratios show a bigger variability compared with Affymetrix log intensities, even after correcting the Agilent variance using log ratios.60 The higher Agilent variability suggests that it would be less feasible to conduct Agilent to Agilent validations compared with Affymetrix to Affymetrix validations.
Conclusions
In this study we compared the performance of metagene-or single gene-based feature sets and classifiers. As the function of the genes within breast cancer predictive profiles are frequently conserved, but not the individual genes, as we expected, gene sets having a biological consensus would both have predictive power and potentially also better validation performance than classical single gene lists when validated in new samples. Surprisingly, the metagene- and single gene-based features had equal performance. When comparing classifier performance in independent datasets, we found only a significant difference between MG- and SG-classifier performances when validation was conducted on datasets measured upon the same microarray platform from which the classifiers were developed. In this situation, SG-classifiers significantly outperformed MG-classifiers.
Methods
Datasets used in this study
This study used a total of ten different datasets. Eight datasets were used for defining the metagene and single gene features. These samples samples from the studies, 1,11–14 and samples from the Gene Expression Omnibus (GEO-) series GSE2034,3 GSE4796,7 GSE3494.61 In the further study, the GSE2034 (abbreviated AF1) and a subset of 151 node-negative samples from the Amsterdam dataset by van de Vijver14 (abbreviated AG1) were used for internal validation within the same dataset by 10 times repeated 10-fold cross validation. Furthermore, these two datasets were also used for defining gene sets used to build and train a classifier within the independent test datasets and also for building classifiers to be validated in our independent datasets. The following samples from two datasets were used as independent test datasets: 147 samples from GSE739040 (abbreviated AF2) and all samples from GSE1112162 (abbreviated AF3).
Dataset processing
The normalizations performed in the studies were retained because the authors found these methods optimal for the eight datasets and because initial ranking of metagenes and single genes was performed separately in each data set. Furthermore, the normalizations of the four datasets, AG1, AF1, AF2, and AF3, were retained for the reasons mentioned above. However, all four datasets were standardized, having a mean of zero and a standard deviation of one. Calculations and classification were also conducted using the R-environment. For random forest and support vector machines, we used the randomForest and e1071 packages, respectively.
Single gene and metagene features
To determine which single genes should be used to build single gene-based gene expression profiles, we focussed on the eight publicly available datasets used in our two previous studies.30,31 The determination of single genes was done by applying the microarray meta-analysis described in our previous study30 upon the same individual gene expression values of each individual probe/gene used to derive the metagenes in the eight datasets. This method ranks each individual gene in each dataset according to its signal-to-noise ratio (SNR). The SNR finds the features that will discriminate between two classes by calculating a score that gives the highest score to those features whose expression levels differ most on average in the two groups while favoring those with small deviations in scores in the respective classes. The SNR for a feature j is calculated as:
| (1) |
In this formula, XA, j, XB, j are the mean gene expression of class A (metastasis group) and class B (nonmetastasis group) and sA,j, sB,j are their associated standard deviations for feature j. In this setting, features that obtain the most positive values are correlated with the metastasis class, while the most negative values are most correlated with the nonmetastasis class.
Following gene ranking within each dataset, the meta-analysis calculates the genes’ mean rank across datasets and determines if this mean rank is significantly high or low, according to a significance cutoff at FDR ≤ 0.05. Using gene symbols, 283 genes (Supplementary Table 2) were found significant and shared by the AG1, AF1, AF2, and AF3 datasets and used in the further analysis. The MGs are 71 selected gene sets covering a specific biological pathway, chromosomal region, or a gene set sharing a DNA transcription factor binding motif. In a previous meta-analysis across eight breast cancer microarray datasets using individual gene enrichment score ranks calculated by GSEA. The 71 metagenes were shown to be associated with breast cancer metastasis30,31 These are listed in Supplementary Table 1. The gene sets covering biological pathways are defined by KEGG (http://www.genome.ad.jp/KEGG), GenMapp (http://www.genmapp.org), and Biocarta (http://www.biocarta.com), while the transcription factor motifs are collected from TransFac (http://www.gene-regulation.com) and sets of genes regulated by the same microRNA from the mirBase (http://microrna.sanger.ac.uk). The gene sets belonging to these were defined using bioinformatic prediction as described in our previous study.30 However, as only a fraction of the genes within each gene set display differential expression between the classes, we limited the final gene sets to those that constituted the leading edge in a GSEA analysis. These leading edge genes are considered to be the core of genes driving the enrichment signal.63 These leading edge genes thus form each of the final metagenes, and the score of each metagene is defined as the median expression of these leading edge genes.
Classifier building
SGs or MGs within each training dataset were ranked according to their random forest importance value. For each feature, this value reports the standardized drop in prediction accuracy when the class labels are imputed.37 For each feature, this rank was used for model building by subsequently adding one feature at a time in a top-down forward-selection wrapper-approach starting with the top two features. To avoid creating bias during gene selection and training of the final classifier or on classification performance, we used 10 times repeated 10-fold cross validation accuracies as a performance measure as this metric has previously been shown to give an excellent bias-variance balance.38 In this study, the models were developed to achieve the best mean sensitivity and specificity thus forcing the overall accuracy to give a balanced sensitivity and specificity. Three different classification methods were used for model building which included Random Forest (RF)37 and SVM with a radial-based kernal (R-SVM) and a sigmoid-based kernel (S-SVM).64 As all the classification methods have hyperparameters, we determined the optimal combination of these parameters using a grid search built into the 10-fold cross validation procedure. For this purpose the tune command from the e1071 R-package was applied. In random forest, we optimized the number of trees in the forest (ntree) from settings of 2000, 3000, 4000, and 5000 trees and the number of subselected predictors for node-splitting (mtry) with settings of 1, 0.5 times the number of features), 1 times the square root of features, 2 times the number of features, and the total number of features. In all support vector machines, the slack variable penalizing cost parameter (C) was optimized using settings of 0.01, 0.1, 1, and 10. The γ-parameter controlling the spreading of samples in feature space was optimized with the settings 0.001, 0.01, 0.1, times the the square root of features
Classification performance assessment
We compared the bAcc of SG- and MG-profiles developed from the four datasets (AG1, AF1, AF2, and AF3) at three levels. At each level, we report the classification accuracy as the mean of sensitivity and specificity, which is referred to as the balanced accuracy (bAcc). Within the same dataset (AG1 or AF1) this was done by 10 times repeated 10-fold cross validation classification accuracies. For each combination of either MG or SG and classification method, this led to generation of 100 different models. Therefore, the internal performance for either MG-or SG-models was defined as the mean performance of these 100 models. When transferring features (defined by AG1, AF1, AF2, or AF3) for classifier building and testing in independent data, leave-one-out cross validation bAcc was used as a performance metric. Although the 10 times repeated 10-fold cross validation is known to be a robust performance metric, we wanted to classify each patient in the validation datasets one by one to mimic the clinical situation and, therefore, the LOOCV bAcc was chosen as a performance metric.
When transferring the best trained classifier (defined by AG1, AF1, AF2, or AF3) from the training sets to classify the independent samples, the classification corresponding to one-fold validation upon all test samples is reported. During transfer of features and classifiers, we examined between different platform and between similar platform classification performances. The between platform (Agilent validated on Affymetrix) covers AG1 validations on AF2 or AF3. The similar platform covers validation of using the same microarray platform as the external validation set (Affymetrix validated on Affymetrix), that is, AF1 on AF2 or AF3, but also AF2 validated on AF3 and vice versa.
Endpoint/outcome
Several studies that have developed classifiers predicting a dichotomous endpoint, have all optimized classifier performance using a particular longitudinal cutoff,2,3,8,22 thus excluding patients not experiencing an event within the longitudinal cutoff and excluding patients experiencing events after this particular time point. These two circumstances could enhance the performance of these classifiers with respect to classifying early metastasis events, but they are prone to perform poorly when classifying late metastasis events.40,46 However, a study by Thomassen and coworkers used full follow-up time for classifier development, addressing if a patient would ever develop a metastasis for the length of the entire study7 and thus have the strength of giving equal weight to both early and late metastasis events. Furthermore, time-to-event analysis sometimes can be misleading when considering classification, and transformation of time-to-event into an binary outcome can blur prediction of the classes.65 In addition, genes being significantly correlated with survival time are not always optimal for classification.66
The outcome differs in the eight datasets used for defining the single genes and metagenes, that is, local and regional recurrences are included in some studies. However, non-metastatic relapses constitute a minority in clinical cohorts. Therefore, the outcome is defined as metastasis or no metastasis after time of diagnosis.
Comparison of external validation performance
To test the significance of the performance difference between MG- and SG-features/classifiers, we used a repeated down-sampling approach consisting of the following five steps: (1) The MG- and SG-features/classifiers classification results upon the entire test data were initially converted into a balanced test-result by down-sampling. Down-sampling obtains a class-balanced dataset from an imbalanced dataset by removing a subset of randomly selected samples from the majority class, where the number of samples removed equals the sample size of the minor class. In this study the majority class is the non-metastasis class. (2) The number of samples correctly classified by MGs but incorrectly by SGs and vice versa is counted; (3) the significance of the difference in these counts in determined using a χ2 test;67 (4) The P value of this test is stored and the steps 2 to 4 are repeated 1000 times; and (5) the median P value from the 1000 tests is reported as the significance between the MGs and SGs.
Supplementary Tables
Table S1.
List of the 71 metagenes.
| Metagene | Type | # genes |
|---|---|---|
| 12q13 | Chromosome region | 28 |
| 14q24 | Chromosome region | 18 |
| 16q22 | Chromosome region | 23 |
| 16q24 | Chromosome region | 14 |
| 17q23 | Chromosome region | 13 |
| 17q25 | Chromosome region | 16 |
| 1p31 | Chromosome region | 14 |
| 1q42 | Chromosome region | 24 |
| 20q11 | Chromosome region | 10 |
| 20q13 | Chromosome region | 29 |
| 5q14 | Chromosome region | 6 |
| 5q33 | Chromosome region | 7 |
| 8p21 | Chromosome region | 14 |
| 8q22 | Chromosome region | 12 |
| 8q24 | Chromosome region | 21 |
| ACTINYPATHWAY | Biological pathway | 14 |
| AMINOACYL_TRNA_BIOSYNTHESIS | Biological pathway | 8 |
| ARAPPATHWAY | Biological pathway | 5 |
| ATRBRCAPATHWAY | Biological pathway | 10 |
| BETA_ALANINE_METABOLISM | Biological pathway | 11 |
| CELL_CYCLE_KEGG | Biological pathway | 39 |
| DNA_REPLICATION_REACTOME | Biological pathway | 19 |
| EGFPATHWAY | Biological pathway | 8 |
| ELECTRON_TRANSPORT_CHAIN | Biological pathway | 39 |
| ERBB2_GRB7 | Biological pathway | 2 |
| FATTY_ACID_METABOLISM | Biological pathway | 20 |
| FRUCTOSE_AND_MANNOSE_METABOLISM | Biological pathway | 10 |
| G2PATHWAY | Biological pathway | 11 |
| GCCATNTTG_V$YY1_Q6 | Transcription factor binding motif | 65 |
| GLEEVECPATHWAY | Biological pathway | 7 |
| GLYCEROLIPID_METABOLISM | Biological pathway | 14 |
| GLYCOLYSIS_AND_GLUCONEOGENESIS | Biological pathway | 12 |
| GPCRPATHWAY | Biological pathway | 8 |
| HISTIDINE_METABOLISM | Biological pathway | 11 |
| Il-12 | Biological pathway | 8 |
| MRNA_PROCESSING_REACTOME | Biological pathway | 24 |
| MRPPATHWAY | Biological pathway | 3 |
| NUCLEAR_RECEPTORS | Biological pathway | 12 |
| OXIDATIVE_PHOSPHORYLATION | Biological pathway | 26 |
| PDGFPATHWAY | Biological pathway | 7 |
| PENTOSE_PHOSPHATE_PATHWAY | Biological pathway | 11 |
| PPARAPATHWAY | Biological pathway | 10 |
| PROTEASOME_DEGRADATION | Biological pathway | 18 |
| PURINE_METABOLISM | Biological pathway | 28 |
| PYRIMIDINE_METABOLISM | Biological pathway | 23 |
| RNA_TRANSCRIPTION_REACTOME | Biological pathway | 9 |
| S1P_SIGNALING | Biological pathway | 6 |
| S1P54_01 | Biological pathway | 53 |
| TGASTMAGC_V$NFE2_01 | Transcription factor binding motif | 35 |
| TNFR2 | Biological pathway | 9 |
| TOLLPATHWAY | Biological pathway | 10 |
| UBIQUITIN_MEDIATED_PROTEOLYSIS | Biological pathway | 2 |
| V$AP1_01 | Transcription factor binding motif | 39 |
| V$AP2_Q3 | Transcription factor binding motif | 33 |
| V$ARNT_02 | Transcription factor binding motif | 34 |
| V$BACH1_01 | Transcription factor binding motif | 50 |
| V$CETS1P54_01 | Transcription factor binding motif | 53 |
| V$COUP_DR1_Q6 | Transcription factor binding motif | 29 |
| V$E2F_Q6_01 | Transcription factor binding motif | 52 |
| V$ELK1_02 | Transcription factor binding motif | 38 |
| V$ER_Q6_02 | Transcription factor binding motif | 25 |
| V$GABP_B | Transcription factor binding motif | 20 |
| V$HIF1_Q5 | Transcription factor binding motif | 27 |
| V$MYCMAX_B | Transcription factor binding motif | 54 |
| V$NFY_Q6 | Transcription factor binding motif | 22 |
| V$NRF1_Q6 | Transcription factor binding motif | 35 |
| V$NRF2_01 | Transcription factor binding motif | 35 |
| V$SP1_Q6_01 | Transcription factor binding motif | 26 |
| V$USF2_Q6 | Transcription factor binding motif | 34 |
| VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION | Biological pathway | 15 |
| VEGFPATHWAY | Biological pathway | 9 |
Notes: The first column shows the name of the metagenes. The second column shows whether the metagene covers a biological pathway, chromosomal region or genes sharing a specific transcription factor binding motif. # genes lists the number of genes underlying the final metagene.
Table S2.
List of the 283 single genes.
| Gene symbol | Description |
|---|---|
| ABCA5 | ATP-binding cassette, sub-family A (ABC1), member 5 |
| ABCA8 | ATP-binding cassette, sub-family A (ABC1), member 8 |
| ABCC10 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| ABCC5 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| ABTB2 | Ankyrin repeat and BTB (POZ) domain containing 2 |
| ACD | Adrenocortical dysplasia homolog (mouse) |
| ADFP | Adipose differentiation-related protein |
| ADH1B | Alcohol dehydrogenase IB (class I), beta polypeptide |
| ADRA2A | Adrenergic, alpha-2A-, receptor |
| ADRM1 | Adhesion regulating molecule 1 |
| ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 |
| ALDH2 | Aldehyde dehydrogenase 2 family (mitochondrial) |
| ALDH6A1 | Aldehyde dehydrogenase 6 family, member A1 |
| APOD | Apolipoprotein D |
| ARHGEF6 | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| ATP1B3 | ATPase, Na+/K+ transporting, beta 3 polypeptide |
| ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| ATP9A | ATPase, Class II, type 9A |
| AURKB | Aurora kinase B |
| BARD1 | BRCA1 associated RING domain 1 |
| BCL2 | B-cell CLL/lymphoma 2 |
| BCL2L1 | BCL2-like 1 |
| BRCA1 | Breast cancer 1, early onset |
| BUB1 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) |
| BUB1B | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) |
| C6 | Complement component 6 |
| C7ORF24 | Chromosome 7 open reading frame 24 |
| CACNA1D | Calcium channel, voltage-dependent, L type, alpha 1D subunit |
| CARS | Cysteinyl-tRNA synthetase |
| CAT | Catalase |
| CCNA2 | Cyclin A2 |
| CCNB1 | Cyclin B1 |
| CCNB2 | Cyclin B2 |
| CCNE2 | Cyclin E2 |
| CCNF | Cyclin F |
| CCT5 | Chaperonin containing TCP1, subunit 5 (epsilon) |
| CCT6A | Chaperonin containing TCP1, subunit 6A (zeta 1) |
| CD44 | CD44 molecule (Indian blood group) |
| CDC2 | Cell division cycle 2, G1 to S and G2 to M |
| CDC20 | CDC20 cell division cycle 20 homolog (S. cerevisiae) |
| CDC25B | Cell division cycle 25B |
| CDC25C | Cell division cycle 25C |
| CDC34 | Cell division cycle 34 |
| CDC45L | CDC45 cell division cycle 45-like (S. cerevisiae) |
| CDK8 | Cyclin-dependent kinase 8 |
| CDKN3 | Cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) |
| CDO1 | Cysteine dioxygenase, type I |
| CENPE | Centromere protein E, 312 kDa |
| CENPF | Centromere protein F, 350/400 ka (mitosin) |
| CH25H | Cholesterol 25-hydroxylase |
| CHAF1B | Chromatin assembly factor 1, subunit B (p60) |
| CIRBP | Cold inducible RNA binding protein |
| CKAP5 | Cytoskeleton associated protein 5 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 |
| CNN3 | Calponin 3, acidic |
| CNTN1 | Contactin 1 |
| CP | Ceruloplasmin (ferroxidase) |
| CREBL2 | CAMP responsive element binding protein-like 2 |
| CRIM1 | Cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| CSE1L | CSE1 chromosome segregation 1-like (yeast) |
| CSTF1 | Cleavage stimulation factor, 3′ pre-RNA, subunit 1, 50 kDa |
| CTPS | CTP synthase |
| CTSD | Cathepsin D (lysosomal aspartyl peptidase) |
| CTSL | Cathepsin L |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 |
| CYP4B1 | Cytochrome P450, family 4, subfamily B, polypeptide 1 |
| CYP4F12 | Cytochrome P450, family 4, subfamily F, polypeptide 12 |
| DDIT4 | DNA-damage-inducible transcript 4 |
| DDX39 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| DLG7 | Discs, large homolog 7 (Drosophila) |
| DLX2 | Distal-less homeobox 2 |
| DOCK1 | Dedicator of cytokinesis 1 |
| DPT | Dermatopontin |
| DUSP1 | Dual specificity phosphatase 1 |
| DUSP4 | Dual specificity phosphatase 4 |
| DYRK2 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| EBP | Emopamil binding protein (sterol isomerase) |
| EDG1 | Endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 |
| EGR2 | Early growth response 2 (Krox-20 homolog, Drosophila) |
| ELOVL5 | ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3- like, yeast) |
| ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) |
| EPHX2 | Epoxide hydrolase 2, cytoplasmic |
| ESPL1 | Extra spindle poles like 1 (S. cerevisiae) |
| EVPL | Envoplakin |
| EXO1 | Exonuclease 1 |
| EZH2 | Enhancer of zeste homolog 2 (Drosophila) |
| F3 | Coagulation factor III (thromboplastin, tissue factor) |
| FADD | Fas (TNFRSF6)-associated via death domain |
| FANCG | Fanconi anemia, complementation group G |
| FAS | Fas (TNF receptor superfamily, member 6) |
| FBLN1 | Fibulin 1 |
| FBLN5 | Fibulin 5 |
| FCER1A | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| FEN1 | Flap structure-specific endonuclease 1 |
| FGL2 | Fibrinogen-like 2 |
| FLJ22531 | – |
| FMO2 | Flavin containing monooxygenase 2 (non-functional) |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog |
| FOXM1 | Forkhead box M1 |
| FRZB | Frizzled-related protein |
| FUCA1 | Fucosidase, alpha-L-1, tissue |
| GABARAP | GABA(A) receptor-associated protein |
| GAD1 | Glutamate decarboxylase 1 (brain, 67 kDa) |
| GALK1 | Galactokinase 1 |
| GEM | GTP binding protein overexpressed in skeletal muscle |
| GGCX | Gamma-glutamyl carboxylase |
| GLA | Galactosidase, alpha |
| GLI1 | Glioma-associated oncogene homolog 1 (zinc finger protein) |
| GMPS | Guanine monphosphate synthetase |
| GNG11 | Guanine nucleotide binding protein (G protein), gamma 11 |
| GNG12 | Guanine nucleotide binding protein (G protein), gamma 12 |
| GPSM2 | G-protein signalling modulator 2 (AGS3-like, C. elegans) |
| GRIK1 | Glutamate receptor, ionotropic, kainate 1 |
| GSTM3 | Glutathione S-transferase M3 (brain) |
| GUK1 | Guanylate kinase 1 |
| GYS2 | Glycogen synthase 2 (liver) |
| H2AFZ | H2A histone family, member Z |
| HIST1H3D | Histone cluster 1, H3d |
| HMGB2 | High-mobility group box 2 |
| HMMR | Hyaluronan-mediated motility receptor (RHAMM) |
| HNMT | Histamine N-methyltransferase |
| HNRPAB | Heterogeneous nuclear ribonucleoprotein A/B |
| HNRPH2 | Heterogeneous nuclear ribonucleoprotein H2 (H′) |
| HPN | Hepsin (transmembrane protease, serine 1) |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) |
| IFNGR2 | Interferon gamma receptor 2 (interferon gamma transducer 1) |
| IGFBP4 | Insulin-like growth factor binding protein 4 |
| IQGAP2 | IQ motif containing GTPase activating protein 2 |
| ITM2A | Integral membrane protein 2A |
| ITPR1 | Inositol 1,4,5-triphosphate receptor, type 1 |
| JAK2 | Janus kinase 2 (a protein tyrosine kinase) |
| KCTD12 | Potassium channel tetramerisation domain containing 12 |
| KIF11 | Kinesin family member 11 |
| KIF13B | Kinesin family member 13B |
| KIF14 | Kinesin family member 14 |
| KIF2C | Kinesin family member 2C |
| KIFC1 | Kinesin family member C1 |
| KIAA0101 | KIAA0101 |
| KIAA0247 | KIAA0247 |
| KIAA0286 | – |
| KIAA0319 | KIAA0319 |
| LAMA2 | Laminin, alpha 2 (merosin, congenital muscular dystrophy) |
| LARP1 | La ribonucleoprotein domain family, member 1 |
| LEP | Leptin (obesity homolog, mouse) |
| LIG1 | Ligase I, DNA, ATP-dependent |
| LMNB1 | Lamin B1 |
| LMO2 | LIM domain only 2 (rhombotin-like 1) |
| LPHN2 | Latrophilin 2 |
| LPL | Lipoprotein lipase |
| LRIG1 | Leucine-rich repeats and immunoglobulin-like domains 1 |
| LRP2 | Low density lipoprotein-related protein 2 |
| MAD2L1 | MAD2 mitotic arrest deficient-like 1 (yeast) |
| MAPRE1 | Microtubule-associated protein, RP/EB family, member 1 |
| MARS | Methionine-tRNA synthetase |
| MCM3 | MCM3 minichromosome maintenance deficient 3 (S. cerevisiae) |
| MCM6 | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) |
| MCM7 | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) |
| MEIS1 | Meis1, myeloid ecotropic viral integration site 1 homolog (mouse) |
| MELK | Maternal embryonic leucine zipper kinase |
| MGP | Matrix Gla protein |
| MKI67 | Antigen identified by monoclonal antibody Ki-67 |
| MN1 | Meningioma (disrupted in balanced translocation) 1 |
| MRPL12 | Mitochondrial ribosomal protein L12 |
| MT2A | Metallothionein 2A |
| MTHFD2 | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| MVD | Mevalonate (diphospho) decarboxylase |
| MYBL2 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 |
| NCOA1 | Nuclear receptor coactivator 1 |
| NDUFA9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39 kDa |
| NEDD9 | Neural precursor cell expressed, developmentally down-regulated 9 |
| NEK2 | NIMA (never in mitosis gene a)-related kinase 2 |
| NME5 | Non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase) |
| NNAT | Neuronatin |
| NP | Nucleoside phosphorylase |
| NR3C2 | Nuclear receptor subfamily 3, group C, member 2 |
| NTRK2 | Neurotrophic tyrosine kinase, receptor, type 2 |
| NUDT1 | Nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| NUP155 | Nucleoporin 155 kDa |
| NUP62 | Nucleoporin 62 kDa |
| NVL | Nuclear VCP-like |
| OMD | Osteomodulin |
| P4HA2 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide II |
| PDCD4 | Programmed cell death 4 (neoplastic transformation inhibitor) |
| PDE4A | Phosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) |
| PDZRN3 | PDZ domain containing RING finger 3 |
| PFKP | Phosphofructokinase, platelet |
| PHLDA2 | Pleckstrin homology-like domain, family A, member 2 |
| PIN1 | Protein (peptidylprolyl cis/trans isomerase) NIMA-interacting 1 |
| PIP | Prolactin-induced protein |
| PIR | Pirin (iron-binding nuclear protein) |
| PKMYT1 | Protein kinase, membrane associated tyrosine/threonine 1 |
| PLK4 | Polo-like kinase 4 (Drosophila) |
| PLP2 | Proteolipid protein 2 (colonic epithelium-enriched) |
| PNMA2 | Paraneoplastic antigen MA2 |
| PNRC1 | Proline-rich nuclear receptor coactivator 1 |
| POLD1 | Polymerase (DNA directed), delta 1, catalytic subunit 125 kDa |
| POLR2H | Polymerase (RNA) II (DNA directed) polypeptide H |
| POLS | Polymerase (DNA directed) sigma |
| PRAME | Preferentially expressed antigen in melanoma |
| PSD3 | Pleckstrin and Sec7 domain containing 3 |
| PSMB3 | Proteasome (prosome, macropain) subunit, beta type, 3 |
| PSMB7 | Proteasome (prosome, macropain) subunit, beta type, 7 |
| PSMD1 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| PSMD11 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| PTDSR | Phosphatidylserine receptor |
| PTGER3 | Prostaglandin E receptor 3 (subtype EP3) |
| PTGER4 | Prostaglandin E receptor 4 (subtype EP4) |
| PTPRT | Protein tyrosine phosphatase, receptor type, T |
| PTTG1 | Pituitary tumor-transforming 1 |
| QDPR | Quinoid dihydropteridine reductase |
| RABGGTA | Rab geranylgeranyltransferase, alpha subunit |
| RABIF | RAB interacting factor |
| RAD51 | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) |
| RAD51AP1 | RAD51 associated protein 1 |
| RAE1 | RAE1 RNA export 1 homolog (S. pombe) |
| RALA | v-ral simian leukemia viral oncogene homolog A (ras related) |
| RBMS3 | RNA binding motif, single stranded interacting protein |
| RDBP | RD RNA binding protein |
| RECQL4 | RecQ protein-like 4 |
| RFC3 | Replication factor C (activator 1) 3, 38 kDa |
| RFC4 | Replication factor C (activator 1) 4, 37 kDa |
| RGS5 | Regulator of G-protein signalling 5 |
| RICS | – |
| RNASEH2A | Ribonuclease H2, subunit A |
| RRM1 | Ribonucleotide reductase M1 polypeptide |
| RRM2 | Ribonucleotide reductase M2 polypeptide |
| RTN1 | Reticulon 1 |
| SAC3D1 | SAC3 domain containing 1 |
| SC5DL | Sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, fungal)-like |
| SDS | Serine dehydratase |
| SEC14L2 | SEC14-like 2 (S. cerevisiae) |
| SEC61G | Sec61 gamma subunit |
| SELE | Selectin E (endothelial adhesion molecule 1) |
| SEMA3E | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| SERPINA1 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| SF3B4 | Splicing factor 3b, subunit 4, 49 kDa |
| SFRP4 | Secreted frizzled-related protein 4 |
| SFRS5 | Splicing factor, arginine/serine-rich 5 |
| SH3BGRL | SH3 domain binding glutamic acid-rich protein like |
| SIAHBP1 | – |
| SIX1 | Sine oculis homeobox homolog 1 (Drosophila) |
| SLBP | Stem-loop (histone) binding protein |
| SLC14A1 | Solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| SLC16A3 | Solute carrier family 16, member 3 (monocarboxylic acid transporter 4) |
| SLC25A1 | Solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| SLC4A7 | Solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| SLIT2 | Slit homolog 2 (Drosophila) |
| SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| SORBS2 | Sorbin and SH3 domain containing 2 |
| SORL1 | Sortilin-related receptor, L(DLR class) A repeats-containing |
| SPAG5 | Sperm associated antigen 5 |
| SPRY2 | Sprouty homolog 2 (Drosophila) |
| SSPN | Sarcospan (Kras oncogene-associated gene) |
| SSRP1 | Structure specific recognition protein 1 |
| STC2 | Stanniocalcin 2 |
| STMN1 | Stathmin 1/oncoprotein 18 |
| SURF2 | Surfeit 2 |
| TACSTD1 | Tumor-associated calcium signal transducer 1 |
| TAT | Tyrosine aminotransferase |
| TBCD | Tubulin-specific chaperone d |
| TGFB3 | Transforming growth factor, beta 3 |
| TIMELESS | Timeless homolog (Drosophila) |
| TIMM17B | Translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| TLR3 | Toll-like receptor 3 |
| TOP2A | Topoisomerase (DNA) II alpha 170 kDa |
| TPX2 | TPX2, microtubule-associated, homolog (Xenopus laevis) |
| TRIP13 | Thyroid hormone receptor interactor 13 |
| TROAP | Trophinin associated protein (tastin) |
| TUBA1 | Tubulin, alpha 1 |
| TXN | Thioredoxin |
| TXNIP | Thioredoxin interacting protein |
| TXNRD1 | Thioredoxin reductase 1 |
| TYRP1 | Tyrosinase-related protein 1 |
| UBE2C | Ubiquitin-conjugating enzyme E2C |
| UBE2V2 | Ubiquitin-conjugating enzyme E2 variant 2 |
| WDHD1 | WD repeat and HMG-box DNA binding protein 1 |
| WFDC2 | WAP four-disulfide core domain 2 |
| WWP2 | WW domain containing E3 ubiquitin protein ligase 2 |
| XPOT | Exportin, tRNA (nuclear export receptor for tRNAs) |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| ZNF238 | Zinc finger protein 238 |
| ZWINT | ZW10 interactor |
| AASS | Aminoadipate-semialdehyde synthase |
Notes: Gene symbol shows the gene symbol of the 283 single genes. Description shows their name.
Table S3.
Internal result.
| Internal results | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 73 | 69 | 71 | 80 | 65 | 73 | 71 | 76 | 74 | 75 | 70 | 73 |
| SG | 85 | 53 | 69 | 85 | 59 | 72 | 69 | 79 | 74 | 80 | 64 | 72 |
Table S4.
Exported feature set performance (different platform).
| Feature set different platforms | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 14 | 95 | 55 | 70 | 74 | 72 | 70 | 74 | 72 | 51 | 81 | 66 |
| SG | 35 | 79 | 57 | 48 | 66 | 57 | 72 | 69 | 71 | 52 | 71 | 62 |
Notes: The table shows the mean sensitivity, specificity and balanced accuracies for feature sets defined by AG1 and validated in AF2 and AF3, using either metagenes (MG) or single genes (SG) as features and using random forest (RF), support vector machine with a radial-based kernel (R-SVM) or a sigmoid kernel (S-SVM). Across shows the mean of the results across the three classification methods.
Table S5.
Exported feature set performance (similar platform).
| Feature set similar platforms | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 71 | 46 | 59 | 68 | 69 | 69 | 73 | 72 | 73 | 71 | 62 | 67 |
| SG | 76 | 39 | 58 | 67 | 71 | 69 | 71 | 71 | 71 | 71 | 60 | 66 |
Notes: The table shows the mean sensitivity, specificity and balanced accuracies for external validation of feature sets covering the following validation: Feature sets defined by AF1 and validated in AF2 and AF3. Feature sets defined by AF2 and validated in AF3 and vice versa, using either metagenes (MG) or single genes (SG) as features and using random forest (RF), support vector machine with a radial-based kernel (R-SVM) or a sigmoid kernel (S-SVM). Across shows the mean of the results across the three classification methods.
Table S6.
Exported classifier performance (different platform).
| Feature set different platforms | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 42 | 75 | 59 | 26 | 85 | 56 | 33 | 85 | 59 | 34 | 82 | 58 |
| SG | 56 | 67 | 62 | 49 | 66 | 58 | 33 | 71 | 52 | 46 | 68 | 58 |
Notes: The table shows the mean sensitivity, specificity and balanced accuracies for classifiers defined by AG1 and validated in AF2 and AF3, using either metagenes (MG) or single genes (SG) as features and using random forest (RF), support vector machine with a radial-based kernel (R-SVM) or a sigmoid kernel (S-SVM).
Table S7.
Exported classifier performance (similar platform).
| Feature set Similar platforms | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 53 | 61 | 57 | 67 | 42 | 55 | 62 | 43 | 53 | 61 | 49 | 55 |
| SG | 65 | 58 | 62 | 65 | 53 | 59 | 58 | 60 | 59 | 63 | 57 | 60 |
Notes: The table shows the mean sensitivity, specificity and balanced accuracies for external validation of classifiers covering the following validation: Feature sets defined by AF1 and validated in AF2 and AF3. Feature sets defined by AF2 and validated in AF3 and vice versa, using either metagenes (MG) or single genes (SG) as features and using random forest (RF), support vector machine with a radial-based kernel (R-SVM) or a sigmoid kernel (S-SVM).
Acknowledgements
This work was partially funded by the Ministry of Public Health, the Danish Cancer Society, the Clinical Institute at University of Southern Denmark, and the Human Microarray Center associated with the department of Clinical Genetics at Odense University Hospital. Furthermore, we acknowledge authors and people who made the microarray datasets used in this study publicly available.
List of abbreviations used
- AF1
Affymetrix dataset 1 (Rotterdam dataset)
- AF2
Affymetrix dataset 2 (Transbig dataset)
- AF3
Affymetrix dataset 3 (Mainz dataset)
- AG1
Agilent dataset 1 (node-negative Amsterdam samples)
- bAcc
balanced accuracy
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GOBP
Gene Ontology Biological Process
- GSEA
gene set enrichment analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LOOCV
leave-one-out cross validation
- MG
metagenes
- MsigDB
Molecular Signature Database
- mRNA
messenger RNA
- RF
random forest
- R-SVM
support vector machine with a radialbasis kernel
- SG
single gene
- S-SVM
support vector machine with a sigmoid kernel.
Footnotes
Author Contributions
Designed the study: MB, MT, QT, TK. Performed scripting, calculations, and data analysis: MB. Developed methods for statistical analysis: MB, QT. Provided the metagene datasets: MT. Wrote the manuscript: MB. Proofread the manuscript: MT, QT, TK.
Funding
Author(s) disclose no funding sources.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission.
References
- 1.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100:10393–8. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van ‘t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 4.Finak G, Bertos N, Pepin F, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 5.Naderi A, Teschendorff AE, Barbosa-Morais NL, et al. A gene-expression signature to predict survival in breast cancer across independent data sets. Oncogene. 2007;26:1507–16. doi: 10.1038/sj.onc.1209920. [DOI] [PubMed] [Google Scholar]
- 6.Tan Q, Thomassen M, Kruse TA. Feature selection for predicting tumor metastases in microarray experiments using paired design. Cancer Inform. 2007;3:213–8. [PMC free article] [PubMed] [Google Scholar]
- 7.Thomassen M, Tan Q, Eiriksdottir F, Bak M, Cold S, Kruse TA. Prediction of metastasis from low-malignant breast cancer by gene expression profiling. Int J Cancer. 2007;120:1070–5. doi: 10.1002/ijc.22449. [DOI] [PubMed] [Google Scholar]
- 8.Karlsson E, Delle U, Danielsson A, et al. Gene expression variation to predict 10-year survival in lymph-node-negative breast cancer. BMC Cancer. 2008;8:254. doi: 10.1186/1471-2407-8-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starmans MH, Krishnapuram B, Steck H, et al. Robust prognostic value of a knowledge-based proliferation signature across large patient microarray studies spanning different cancer types. Br J Cancer. 2008;99:1884–90. doi: 10.1038/sj.bjc.6604746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damasco C, Lembo A, Somma MP, Gatti M, Di CF, Provero P. A signature inferred from Drosophila mitotic genes predicts survival of breast cancer patients. PLoS One. 2011;6:e14737. doi: 10.1371/journal.pone.0014737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calza S, Hall P, Auer G, et al. Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res. 2006;8:R34. doi: 10.1186/bcr1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 13.Huang E, Cheng SH, Dressman H, et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361:1590–6. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 14.van de Vijver MJ, He YD, Van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 15.Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–8. doi: 10.1093/bioinformatics/bth469. [DOI] [PubMed] [Google Scholar]
- 16.Yu JX, Sieuwerts AM, Zhang Y, et al. Pathway analysis of gene signatures predicting metastasis of node-negative primary breast cancer. BMC Cancer. 2007;7:182. doi: 10.1186/1471-2407-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu Y, Stolovitzky G, Klein U. Quantitative noise analysis for gene expression microarray experiments. Proc Natl Acad Sci U S A. 2002;99:14031–6. doi: 10.1073/pnas.222164199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sontrop HM, Moerland PD, van den Ham R, Reinders MJ, Verhaegh WF. A comprehensive sensitivity analysis of microarray breast cancer classification under feature variability. BMC Bioinformatics. 2009;10:389. doi: 10.1186/1471-2105-10-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz-Uriarte R, Alvarez de AS. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–17. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 21.Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Machine Learning. 2002;46:389–422. [Google Scholar]
- 22.Mundra PA, Rajapakse JC. SVM-RFE with MRMR filter for gene selection. IEEE Trans Nanobioscience. 2010;9:31–7. doi: 10.1109/TNB.2009.2035284. [DOI] [PubMed] [Google Scholar]
- 23.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pawitan Y, Bjohle J, Amler L, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7:R953–64. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomassen M, Tan Q, Eiriksdottir F, Bak M, Cold S, Kruse TA. Comparison of gene sets for expression profiling: prediction of metastasis from low-malignant breast cancer. Clin Cancer Res. 2007;13:5355–60. doi: 10.1158/1078-0432.CCR-07-0249. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Martens JW, Yu JX, et al. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res. 2009;69:3795–801. doi: 10.1158/0008-5472.CAN-08-4596. [DOI] [PubMed] [Google Scholar]
- 29.Buness A, Kuner R, Ruschhaupt M, Poustka A, Sultmann H, Tresch A. Identification of aberrant chromosomal regions from gene expression microarray studies applied to human breast cancer. Bioinformatics. 2007;23:2273–80. doi: 10.1093/bioinformatics/btm340. [DOI] [PubMed] [Google Scholar]
- 30.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies metastatic pathways and transcription factors in breast cancer. BMC Cancer. 2008;8:394. doi: 10.1186/1471-2407-8-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomassen M, Tan Q, Kruse TA. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res Treat. 2009;113:239–49. doi: 10.1007/s10549-008-9927-2. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Zheng CH, Liu JX, Wang HQ. Discovering the transcriptional modules using microarray data by penalized matrix decomposition. Comput Biol Med. 2011;41:1041–50. doi: 10.1016/j.compbiomed.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 33.van Vliet MH, Klijn CN, Wessels LF, Reinders MJ. Module-based outcome prediction using breast cancer compendia. PLoS One. 2007;2:e1047. doi: 10.1371/journal.pone.0001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia M, Millat-Carus R, Bertucci F, Finetti P, Birnbaum D, Bidaut G. Interactome-Transcriptome integration for predicting distant metastasis in breast cancer. Bioinformatics. 2012;28(5):672–8. doi: 10.1093/bioinformatics/bts025. [DOI] [PubMed] [Google Scholar]
- 36.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 38.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining Inference and Prediction. 2nd ed. New York, NY: Springer; 2009. [Google Scholar]
- 39.Kohavi R. A study of cross-validation and boot-strap for accuracy estimation and model selection. Proceedings of The Fourteenth International Joint Conference on Artificial Intelligence. 1995;2:1137–43. [Google Scholar]
- 40.Buyse M, Loi S, Van’t Veer L, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–92. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 41.Blazadonakis ME, Zervakis ME, Kafetzopoulos D. Integration of gene signatures using biological knowledge. Artif Intell Med. 2011;53:57–71. doi: 10.1016/j.artmed.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 42.Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A. 2005;102:3738–43. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haibe-Kains B, Desmedt C, Piette F, et al. Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics. 2008;9:394. doi: 10.1186/1471-2164-9-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haibe-Kains B, Desmedt C, Sotiriou C, Bontempi G. A comparative study of survival models for breast cancer prognostication based on microarray data: does a single gene beat them all? Bioinformatics. 2008;24:2200–8. doi: 10.1093/bioinformatics/btn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–14. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 47.Daemen A, Gevaert O, De MB. Integration of clinical and microarray data with kernel methods. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:5411–5. doi: 10.1109/IEMBS.2007.4353566. [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Goodison S, Li J, Liu L, Farmerie W. Improved breast cancer prognosis through the combination of clinical and genetic markers. Bioinformatics. 2007;23:30–7. doi: 10.1093/bioinformatics/btl543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt M, Victor A, Bratzel D, et al. Long-term outcome prediction by clinicopathological risk classification algorithms in node-negative breast cancer—comparison between Adjuvant!, St. Gallen, and a novel risk algorithm used in the prospective randomized Node-Negative-Breast Cancer-3 (NNBC-3) trial. Ann Oncol. 2009;20:258–64. doi: 10.1093/annonc/mdn590. [DOI] [PubMed] [Google Scholar]
- 50.Warnat P, Eils R, Brors B. Cross-platform analysis of cancer microarray data improves gene expression based classification of phenotypes. BMC Bioinformatics. 2005;6:265. doi: 10.1186/1471-2105-6-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stec J, Wang J, Coombes K, et al. Comparison of the predictive accuracy of DNA array-based multigene classifiers across cDNA arrays and Affymetrix GeneChips. J Mol Diagn. 2005;7:357–67. doi: 10.1016/s1525-1578(10)60565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Z, Zhang T, Li X, et al. Towards precise classification of cancers based on robust gene functional expression profiles. BMC Bioinformatics. 2005;6:58. doi: 10.1186/1471-2105-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mi Z, Shen K, Song N, et al. Module-based prediction approach for robust inter-study predictions in microarray data. Bioinformatics. 2010;26:2586–93. doi: 10.1093/bioinformatics/btq472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang H, Cheng C, Zhang W. Average rank-based score to measure deregulation of molecular pathway gene sets. PLoS One. 2011;6:e27579. doi: 10.1371/journal.pone.0027579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su J, Yoon BJ, Dougherty ER. Accurate and reliable cancer classification based on probabilistic inference of pathway activity. PLoS One. 2009;4:e8161. doi: 10.1371/journal.pone.0008161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abraham G, Kowalczyk A, Loi S, Haviv I, Zobel J. Prediction of breast cancer prognosis using gene set statistics provides signature stability and biological context. BMC Bioinformatics. 2010;11:277. doi: 10.1186/1471-2105-11-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blazadonakis ME, Zervakis ME, Kafetzopoulos D. Integration of gene signatures using biological knowledge. Artif Intell Med. 2011;53:57–71. doi: 10.1016/j.artmed.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 58.Blazadonakis ME, Zervakis M. The linear neuron as marker selector and clinical predictor in cancer gene analysis. Comput Methods Programs Biomed. 2008;91:22–35. doi: 10.1016/j.cmpb.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 59.Kuo WP, Jenssen TK, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–12. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- 60.Pedotti P, ‘t Hoen PA, Vreugdenhil E, et al. Can subtle changes in gene expression be consistently detected with different microarray platforms? BMC Genomics. 2008;9:124. doi: 10.1186/1471-2164-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt M, Bohm D, von TC, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–13. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 63.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics. 2007;23:3251–3. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 64.Vapnik V. The Nature of Statistical Learning Theory. New York, NY: Springer; 1995. [Google Scholar]
- 65.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99:147–57. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 66.Chiu SH, Chen CC, Lin TH. Using support vector regression to model the correlation between the clinical metastases time and gene expression profile for breast cancer. Artif Intell Med. 2008;44:221–31. doi: 10.1016/j.artmed.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 67.Demsar J. Statistical comparisons of classifiers over multiple data sets. Journal of Machine Learning Research. 2006;7:1–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
List of the 71 metagenes.
| Metagene | Type | # genes |
|---|---|---|
| 12q13 | Chromosome region | 28 |
| 14q24 | Chromosome region | 18 |
| 16q22 | Chromosome region | 23 |
| 16q24 | Chromosome region | 14 |
| 17q23 | Chromosome region | 13 |
| 17q25 | Chromosome region | 16 |
| 1p31 | Chromosome region | 14 |
| 1q42 | Chromosome region | 24 |
| 20q11 | Chromosome region | 10 |
| 20q13 | Chromosome region | 29 |
| 5q14 | Chromosome region | 6 |
| 5q33 | Chromosome region | 7 |
| 8p21 | Chromosome region | 14 |
| 8q22 | Chromosome region | 12 |
| 8q24 | Chromosome region | 21 |
| ACTINYPATHWAY | Biological pathway | 14 |
| AMINOACYL_TRNA_BIOSYNTHESIS | Biological pathway | 8 |
| ARAPPATHWAY | Biological pathway | 5 |
| ATRBRCAPATHWAY | Biological pathway | 10 |
| BETA_ALANINE_METABOLISM | Biological pathway | 11 |
| CELL_CYCLE_KEGG | Biological pathway | 39 |
| DNA_REPLICATION_REACTOME | Biological pathway | 19 |
| EGFPATHWAY | Biological pathway | 8 |
| ELECTRON_TRANSPORT_CHAIN | Biological pathway | 39 |
| ERBB2_GRB7 | Biological pathway | 2 |
| FATTY_ACID_METABOLISM | Biological pathway | 20 |
| FRUCTOSE_AND_MANNOSE_METABOLISM | Biological pathway | 10 |
| G2PATHWAY | Biological pathway | 11 |
| GCCATNTTG_V$YY1_Q6 | Transcription factor binding motif | 65 |
| GLEEVECPATHWAY | Biological pathway | 7 |
| GLYCEROLIPID_METABOLISM | Biological pathway | 14 |
| GLYCOLYSIS_AND_GLUCONEOGENESIS | Biological pathway | 12 |
| GPCRPATHWAY | Biological pathway | 8 |
| HISTIDINE_METABOLISM | Biological pathway | 11 |
| Il-12 | Biological pathway | 8 |
| MRNA_PROCESSING_REACTOME | Biological pathway | 24 |
| MRPPATHWAY | Biological pathway | 3 |
| NUCLEAR_RECEPTORS | Biological pathway | 12 |
| OXIDATIVE_PHOSPHORYLATION | Biological pathway | 26 |
| PDGFPATHWAY | Biological pathway | 7 |
| PENTOSE_PHOSPHATE_PATHWAY | Biological pathway | 11 |
| PPARAPATHWAY | Biological pathway | 10 |
| PROTEASOME_DEGRADATION | Biological pathway | 18 |
| PURINE_METABOLISM | Biological pathway | 28 |
| PYRIMIDINE_METABOLISM | Biological pathway | 23 |
| RNA_TRANSCRIPTION_REACTOME | Biological pathway | 9 |
| S1P_SIGNALING | Biological pathway | 6 |
| S1P54_01 | Biological pathway | 53 |
| TGASTMAGC_V$NFE2_01 | Transcription factor binding motif | 35 |
| TNFR2 | Biological pathway | 9 |
| TOLLPATHWAY | Biological pathway | 10 |
| UBIQUITIN_MEDIATED_PROTEOLYSIS | Biological pathway | 2 |
| V$AP1_01 | Transcription factor binding motif | 39 |
| V$AP2_Q3 | Transcription factor binding motif | 33 |
| V$ARNT_02 | Transcription factor binding motif | 34 |
| V$BACH1_01 | Transcription factor binding motif | 50 |
| V$CETS1P54_01 | Transcription factor binding motif | 53 |
| V$COUP_DR1_Q6 | Transcription factor binding motif | 29 |
| V$E2F_Q6_01 | Transcription factor binding motif | 52 |
| V$ELK1_02 | Transcription factor binding motif | 38 |
| V$ER_Q6_02 | Transcription factor binding motif | 25 |
| V$GABP_B | Transcription factor binding motif | 20 |
| V$HIF1_Q5 | Transcription factor binding motif | 27 |
| V$MYCMAX_B | Transcription factor binding motif | 54 |
| V$NFY_Q6 | Transcription factor binding motif | 22 |
| V$NRF1_Q6 | Transcription factor binding motif | 35 |
| V$NRF2_01 | Transcription factor binding motif | 35 |
| V$SP1_Q6_01 | Transcription factor binding motif | 26 |
| V$USF2_Q6 | Transcription factor binding motif | 34 |
| VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION | Biological pathway | 15 |
| VEGFPATHWAY | Biological pathway | 9 |
Notes: The first column shows the name of the metagenes. The second column shows whether the metagene covers a biological pathway, chromosomal region or genes sharing a specific transcription factor binding motif. # genes lists the number of genes underlying the final metagene.
Table S2.
List of the 283 single genes.
| Gene symbol | Description |
|---|---|
| ABCA5 | ATP-binding cassette, sub-family A (ABC1), member 5 |
| ABCA8 | ATP-binding cassette, sub-family A (ABC1), member 8 |
| ABCC10 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 |
| ABCC5 | ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| ABTB2 | Ankyrin repeat and BTB (POZ) domain containing 2 |
| ACD | Adrenocortical dysplasia homolog (mouse) |
| ADFP | Adipose differentiation-related protein |
| ADH1B | Alcohol dehydrogenase IB (class I), beta polypeptide |
| ADRA2A | Adrenergic, alpha-2A-, receptor |
| ADRM1 | Adhesion regulating molecule 1 |
| ALDH1A1 | Aldehyde dehydrogenase 1 family, member A1 |
| ALDH2 | Aldehyde dehydrogenase 2 family (mitochondrial) |
| ALDH6A1 | Aldehyde dehydrogenase 6 family, member A1 |
| APOD | Apolipoprotein D |
| ARHGEF6 | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| ATP1B3 | ATPase, Na+/K+ transporting, beta 3 polypeptide |
| ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| ATP9A | ATPase, Class II, type 9A |
| AURKB | Aurora kinase B |
| BARD1 | BRCA1 associated RING domain 1 |
| BCL2 | B-cell CLL/lymphoma 2 |
| BCL2L1 | BCL2-like 1 |
| BRCA1 | Breast cancer 1, early onset |
| BUB1 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) |
| BUB1B | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) |
| C6 | Complement component 6 |
| C7ORF24 | Chromosome 7 open reading frame 24 |
| CACNA1D | Calcium channel, voltage-dependent, L type, alpha 1D subunit |
| CARS | Cysteinyl-tRNA synthetase |
| CAT | Catalase |
| CCNA2 | Cyclin A2 |
| CCNB1 | Cyclin B1 |
| CCNB2 | Cyclin B2 |
| CCNE2 | Cyclin E2 |
| CCNF | Cyclin F |
| CCT5 | Chaperonin containing TCP1, subunit 5 (epsilon) |
| CCT6A | Chaperonin containing TCP1, subunit 6A (zeta 1) |
| CD44 | CD44 molecule (Indian blood group) |
| CDC2 | Cell division cycle 2, G1 to S and G2 to M |
| CDC20 | CDC20 cell division cycle 20 homolog (S. cerevisiae) |
| CDC25B | Cell division cycle 25B |
| CDC25C | Cell division cycle 25C |
| CDC34 | Cell division cycle 34 |
| CDC45L | CDC45 cell division cycle 45-like (S. cerevisiae) |
| CDK8 | Cyclin-dependent kinase 8 |
| CDKN3 | Cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) |
| CDO1 | Cysteine dioxygenase, type I |
| CENPE | Centromere protein E, 312 kDa |
| CENPF | Centromere protein F, 350/400 ka (mitosin) |
| CH25H | Cholesterol 25-hydroxylase |
| CHAF1B | Chromatin assembly factor 1, subunit B (p60) |
| CIRBP | Cold inducible RNA binding protein |
| CKAP5 | Cytoskeleton associated protein 5 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 |
| CNN3 | Calponin 3, acidic |
| CNTN1 | Contactin 1 |
| CP | Ceruloplasmin (ferroxidase) |
| CREBL2 | CAMP responsive element binding protein-like 2 |
| CRIM1 | Cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| CSE1L | CSE1 chromosome segregation 1-like (yeast) |
| CSTF1 | Cleavage stimulation factor, 3′ pre-RNA, subunit 1, 50 kDa |
| CTPS | CTP synthase |
| CTSD | Cathepsin D (lysosomal aspartyl peptidase) |
| CTSL | Cathepsin L |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1 |
| CYP4B1 | Cytochrome P450, family 4, subfamily B, polypeptide 1 |
| CYP4F12 | Cytochrome P450, family 4, subfamily F, polypeptide 12 |
| DDIT4 | DNA-damage-inducible transcript 4 |
| DDX39 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| DLG7 | Discs, large homolog 7 (Drosophila) |
| DLX2 | Distal-less homeobox 2 |
| DOCK1 | Dedicator of cytokinesis 1 |
| DPT | Dermatopontin |
| DUSP1 | Dual specificity phosphatase 1 |
| DUSP4 | Dual specificity phosphatase 4 |
| DYRK2 | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| EBP | Emopamil binding protein (sterol isomerase) |
| EDG1 | Endothelial differentiation, sphingolipid G-protein-coupled receptor, 1 |
| EGR2 | Early growth response 2 (Krox-20 homolog, Drosophila) |
| ELOVL5 | ELOVL family member 5, elongation of long chain fatty acids (FEN1/Elo2, SUR4/Elo3- like, yeast) |
| ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) |
| EPHX2 | Epoxide hydrolase 2, cytoplasmic |
| ESPL1 | Extra spindle poles like 1 (S. cerevisiae) |
| EVPL | Envoplakin |
| EXO1 | Exonuclease 1 |
| EZH2 | Enhancer of zeste homolog 2 (Drosophila) |
| F3 | Coagulation factor III (thromboplastin, tissue factor) |
| FADD | Fas (TNFRSF6)-associated via death domain |
| FANCG | Fanconi anemia, complementation group G |
| FAS | Fas (TNF receptor superfamily, member 6) |
| FBLN1 | Fibulin 1 |
| FBLN5 | Fibulin 5 |
| FCER1A | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| FEN1 | Flap structure-specific endonuclease 1 |
| FGL2 | Fibrinogen-like 2 |
| FLJ22531 | – |
| FMO2 | Flavin containing monooxygenase 2 (non-functional) |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog |
| FOXM1 | Forkhead box M1 |
| FRZB | Frizzled-related protein |
| FUCA1 | Fucosidase, alpha-L-1, tissue |
| GABARAP | GABA(A) receptor-associated protein |
| GAD1 | Glutamate decarboxylase 1 (brain, 67 kDa) |
| GALK1 | Galactokinase 1 |
| GEM | GTP binding protein overexpressed in skeletal muscle |
| GGCX | Gamma-glutamyl carboxylase |
| GLA | Galactosidase, alpha |
| GLI1 | Glioma-associated oncogene homolog 1 (zinc finger protein) |
| GMPS | Guanine monphosphate synthetase |
| GNG11 | Guanine nucleotide binding protein (G protein), gamma 11 |
| GNG12 | Guanine nucleotide binding protein (G protein), gamma 12 |
| GPSM2 | G-protein signalling modulator 2 (AGS3-like, C. elegans) |
| GRIK1 | Glutamate receptor, ionotropic, kainate 1 |
| GSTM3 | Glutathione S-transferase M3 (brain) |
| GUK1 | Guanylate kinase 1 |
| GYS2 | Glycogen synthase 2 (liver) |
| H2AFZ | H2A histone family, member Z |
| HIST1H3D | Histone cluster 1, H3d |
| HMGB2 | High-mobility group box 2 |
| HMMR | Hyaluronan-mediated motility receptor (RHAMM) |
| HNMT | Histamine N-methyltransferase |
| HNRPAB | Heterogeneous nuclear ribonucleoprotein A/B |
| HNRPH2 | Heterogeneous nuclear ribonucleoprotein H2 (H′) |
| HPN | Hepsin (transmembrane protease, serine 1) |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 (Lesch-Nyhan syndrome) |
| IFNGR2 | Interferon gamma receptor 2 (interferon gamma transducer 1) |
| IGFBP4 | Insulin-like growth factor binding protein 4 |
| IQGAP2 | IQ motif containing GTPase activating protein 2 |
| ITM2A | Integral membrane protein 2A |
| ITPR1 | Inositol 1,4,5-triphosphate receptor, type 1 |
| JAK2 | Janus kinase 2 (a protein tyrosine kinase) |
| KCTD12 | Potassium channel tetramerisation domain containing 12 |
| KIF11 | Kinesin family member 11 |
| KIF13B | Kinesin family member 13B |
| KIF14 | Kinesin family member 14 |
| KIF2C | Kinesin family member 2C |
| KIFC1 | Kinesin family member C1 |
| KIAA0101 | KIAA0101 |
| KIAA0247 | KIAA0247 |
| KIAA0286 | – |
| KIAA0319 | KIAA0319 |
| LAMA2 | Laminin, alpha 2 (merosin, congenital muscular dystrophy) |
| LARP1 | La ribonucleoprotein domain family, member 1 |
| LEP | Leptin (obesity homolog, mouse) |
| LIG1 | Ligase I, DNA, ATP-dependent |
| LMNB1 | Lamin B1 |
| LMO2 | LIM domain only 2 (rhombotin-like 1) |
| LPHN2 | Latrophilin 2 |
| LPL | Lipoprotein lipase |
| LRIG1 | Leucine-rich repeats and immunoglobulin-like domains 1 |
| LRP2 | Low density lipoprotein-related protein 2 |
| MAD2L1 | MAD2 mitotic arrest deficient-like 1 (yeast) |
| MAPRE1 | Microtubule-associated protein, RP/EB family, member 1 |
| MARS | Methionine-tRNA synthetase |
| MCM3 | MCM3 minichromosome maintenance deficient 3 (S. cerevisiae) |
| MCM6 | MCM6 minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) |
| MCM7 | MCM7 minichromosome maintenance deficient 7 (S. cerevisiae) |
| MEIS1 | Meis1, myeloid ecotropic viral integration site 1 homolog (mouse) |
| MELK | Maternal embryonic leucine zipper kinase |
| MGP | Matrix Gla protein |
| MKI67 | Antigen identified by monoclonal antibody Ki-67 |
| MN1 | Meningioma (disrupted in balanced translocation) 1 |
| MRPL12 | Mitochondrial ribosomal protein L12 |
| MT2A | Metallothionein 2A |
| MTHFD2 | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| MVD | Mevalonate (diphospho) decarboxylase |
| MYBL2 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 |
| NCOA1 | Nuclear receptor coactivator 1 |
| NDUFA9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39 kDa |
| NEDD9 | Neural precursor cell expressed, developmentally down-regulated 9 |
| NEK2 | NIMA (never in mitosis gene a)-related kinase 2 |
| NME5 | Non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase) |
| NNAT | Neuronatin |
| NP | Nucleoside phosphorylase |
| NR3C2 | Nuclear receptor subfamily 3, group C, member 2 |
| NTRK2 | Neurotrophic tyrosine kinase, receptor, type 2 |
| NUDT1 | Nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| NUP155 | Nucleoporin 155 kDa |
| NUP62 | Nucleoporin 62 kDa |
| NVL | Nuclear VCP-like |
| OMD | Osteomodulin |
| P4HA2 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide II |
| PDCD4 | Programmed cell death 4 (neoplastic transformation inhibitor) |
| PDE4A | Phosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) |
| PDZRN3 | PDZ domain containing RING finger 3 |
| PFKP | Phosphofructokinase, platelet |
| PHLDA2 | Pleckstrin homology-like domain, family A, member 2 |
| PIN1 | Protein (peptidylprolyl cis/trans isomerase) NIMA-interacting 1 |
| PIP | Prolactin-induced protein |
| PIR | Pirin (iron-binding nuclear protein) |
| PKMYT1 | Protein kinase, membrane associated tyrosine/threonine 1 |
| PLK4 | Polo-like kinase 4 (Drosophila) |
| PLP2 | Proteolipid protein 2 (colonic epithelium-enriched) |
| PNMA2 | Paraneoplastic antigen MA2 |
| PNRC1 | Proline-rich nuclear receptor coactivator 1 |
| POLD1 | Polymerase (DNA directed), delta 1, catalytic subunit 125 kDa |
| POLR2H | Polymerase (RNA) II (DNA directed) polypeptide H |
| POLS | Polymerase (DNA directed) sigma |
| PRAME | Preferentially expressed antigen in melanoma |
| PSD3 | Pleckstrin and Sec7 domain containing 3 |
| PSMB3 | Proteasome (prosome, macropain) subunit, beta type, 3 |
| PSMB7 | Proteasome (prosome, macropain) subunit, beta type, 7 |
| PSMD1 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| PSMD11 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| PTDSR | Phosphatidylserine receptor |
| PTGER3 | Prostaglandin E receptor 3 (subtype EP3) |
| PTGER4 | Prostaglandin E receptor 4 (subtype EP4) |
| PTPRT | Protein tyrosine phosphatase, receptor type, T |
| PTTG1 | Pituitary tumor-transforming 1 |
| QDPR | Quinoid dihydropteridine reductase |
| RABGGTA | Rab geranylgeranyltransferase, alpha subunit |
| RABIF | RAB interacting factor |
| RAD51 | RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) |
| RAD51AP1 | RAD51 associated protein 1 |
| RAE1 | RAE1 RNA export 1 homolog (S. pombe) |
| RALA | v-ral simian leukemia viral oncogene homolog A (ras related) |
| RBMS3 | RNA binding motif, single stranded interacting protein |
| RDBP | RD RNA binding protein |
| RECQL4 | RecQ protein-like 4 |
| RFC3 | Replication factor C (activator 1) 3, 38 kDa |
| RFC4 | Replication factor C (activator 1) 4, 37 kDa |
| RGS5 | Regulator of G-protein signalling 5 |
| RICS | – |
| RNASEH2A | Ribonuclease H2, subunit A |
| RRM1 | Ribonucleotide reductase M1 polypeptide |
| RRM2 | Ribonucleotide reductase M2 polypeptide |
| RTN1 | Reticulon 1 |
| SAC3D1 | SAC3 domain containing 1 |
| SC5DL | Sterol-C5-desaturase (ERG3 delta-5-desaturase homolog, fungal)-like |
| SDS | Serine dehydratase |
| SEC14L2 | SEC14-like 2 (S. cerevisiae) |
| SEC61G | Sec61 gamma subunit |
| SELE | Selectin E (endothelial adhesion molecule 1) |
| SEMA3E | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| SERPINA1 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| SF3B4 | Splicing factor 3b, subunit 4, 49 kDa |
| SFRP4 | Secreted frizzled-related protein 4 |
| SFRS5 | Splicing factor, arginine/serine-rich 5 |
| SH3BGRL | SH3 domain binding glutamic acid-rich protein like |
| SIAHBP1 | – |
| SIX1 | Sine oculis homeobox homolog 1 (Drosophila) |
| SLBP | Stem-loop (histone) binding protein |
| SLC14A1 | Solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| SLC16A3 | Solute carrier family 16, member 3 (monocarboxylic acid transporter 4) |
| SLC25A1 | Solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| SLC4A7 | Solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| SLIT2 | Slit homolog 2 (Drosophila) |
| SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| SORBS2 | Sorbin and SH3 domain containing 2 |
| SORL1 | Sortilin-related receptor, L(DLR class) A repeats-containing |
| SPAG5 | Sperm associated antigen 5 |
| SPRY2 | Sprouty homolog 2 (Drosophila) |
| SSPN | Sarcospan (Kras oncogene-associated gene) |
| SSRP1 | Structure specific recognition protein 1 |
| STC2 | Stanniocalcin 2 |
| STMN1 | Stathmin 1/oncoprotein 18 |
| SURF2 | Surfeit 2 |
| TACSTD1 | Tumor-associated calcium signal transducer 1 |
| TAT | Tyrosine aminotransferase |
| TBCD | Tubulin-specific chaperone d |
| TGFB3 | Transforming growth factor, beta 3 |
| TIMELESS | Timeless homolog (Drosophila) |
| TIMM17B | Translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| TLR3 | Toll-like receptor 3 |
| TOP2A | Topoisomerase (DNA) II alpha 170 kDa |
| TPX2 | TPX2, microtubule-associated, homolog (Xenopus laevis) |
| TRIP13 | Thyroid hormone receptor interactor 13 |
| TROAP | Trophinin associated protein (tastin) |
| TUBA1 | Tubulin, alpha 1 |
| TXN | Thioredoxin |
| TXNIP | Thioredoxin interacting protein |
| TXNRD1 | Thioredoxin reductase 1 |
| TYRP1 | Tyrosinase-related protein 1 |
| UBE2C | Ubiquitin-conjugating enzyme E2C |
| UBE2V2 | Ubiquitin-conjugating enzyme E2 variant 2 |
| WDHD1 | WD repeat and HMG-box DNA binding protein 1 |
| WFDC2 | WAP four-disulfide core domain 2 |
| WWP2 | WW domain containing E3 ubiquitin protein ligase 2 |
| XPOT | Exportin, tRNA (nuclear export receptor for tRNAs) |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| ZNF238 | Zinc finger protein 238 |
| ZWINT | ZW10 interactor |
| AASS | Aminoadipate-semialdehyde synthase |
Notes: Gene symbol shows the gene symbol of the 283 single genes. Description shows their name.
Table S3.
Internal result.
| Internal results | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 73 | 69 | 71 | 80 | 65 | 73 | 71 | 76 | 74 | 75 | 70 | 73 |
| SG | 85 | 53 | 69 | 85 | 59 | 72 | 69 | 79 | 74 | 80 | 64 | 72 |
Table S4.
Exported feature set performance (different platform).
| Feature set different platforms | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 14 | 95 | 55 | 70 | 74 | 72 | 70 | 74 | 72 | 51 | 81 | 66 |
| SG | 35 | 79 | 57 | 48 | 66 | 57 | 72 | 69 | 71 | 52 | 71 | 62 |
Notes: The table shows the mean sensitivity, specificity and balanced accuracies for feature sets defined by AG1 and validated in AF2 and AF3, using either metagenes (MG) or single genes (SG) as features and using random forest (RF), support vector machine with a radial-based kernel (R-SVM) or a sigmoid kernel (S-SVM). Across shows the mean of the results across the three classification methods.
Table S5.
Exported feature set performance (similar platform).
| Feature set similar platforms | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 71 | 46 | 59 | 68 | 69 | 69 | 73 | 72 | 73 | 71 | 62 | 67 |
| SG | 76 | 39 | 58 | 67 | 71 | 69 | 71 | 71 | 71 | 71 | 60 | 66 |
Notes: The table shows the mean sensitivity, specificity and balanced accuracies for external validation of feature sets covering the following validation: Feature sets defined by AF1 and validated in AF2 and AF3. Feature sets defined by AF2 and validated in AF3 and vice versa, using either metagenes (MG) or single genes (SG) as features and using random forest (RF), support vector machine with a radial-based kernel (R-SVM) or a sigmoid kernel (S-SVM). Across shows the mean of the results across the three classification methods.
Table S6.
Exported classifier performance (different platform).
| Feature set different platforms | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 42 | 75 | 59 | 26 | 85 | 56 | 33 | 85 | 59 | 34 | 82 | 58 |
| SG | 56 | 67 | 62 | 49 | 66 | 58 | 33 | 71 | 52 | 46 | 68 | 58 |
Notes: The table shows the mean sensitivity, specificity and balanced accuracies for classifiers defined by AG1 and validated in AF2 and AF3, using either metagenes (MG) or single genes (SG) as features and using random forest (RF), support vector machine with a radial-based kernel (R-SVM) or a sigmoid kernel (S-SVM).
Table S7.
Exported classifier performance (similar platform).
| Feature set Similar platforms | RF | R-SVM | S-SVM | Across | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| Method | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc | Sen | Spe | bAcc |
| MG | 53 | 61 | 57 | 67 | 42 | 55 | 62 | 43 | 53 | 61 | 49 | 55 |
| SG | 65 | 58 | 62 | 65 | 53 | 59 | 58 | 60 | 59 | 63 | 57 | 60 |
Notes: The table shows the mean sensitivity, specificity and balanced accuracies for external validation of classifiers covering the following validation: Feature sets defined by AF1 and validated in AF2 and AF3. Feature sets defined by AF2 and validated in AF3 and vice versa, using either metagenes (MG) or single genes (SG) as features and using random forest (RF), support vector machine with a radial-based kernel (R-SVM) or a sigmoid kernel (S-SVM).