Abstract
Variation in responsiveness to bitter-tasting compounds has been associated with differences in alcohol consumption. One strong genetic determinant of variation in bitter taste sensitivity is alleles of the TAS2R gene family, which encode chemosensory receptors sensitive to a diverse array of natural and synthetic compounds. Members of the TAS2R family, when expressed in the gustatory system, function as bitter taste receptors. To better understand the relationship between TAS2R function and alcohol consumption, we asked if TAS2R variants are associated with measures of alcohol consumption in a head and neck cancer patient cohort. Factors associated with increased alcohol intake are of strong interest to those concerned with decreasing the incidence of cancers of oral and pharyngeal structures. We found a single nucleotide polymorphism (SNP) located within the TAS2R13 gene (rs1015443 [C1040T, Ser259Asn]), which showed a significant association with measures of alcohol consumption assessed via the Alcohol Use Disorders Identification Test (AUDIT). Analyses with other SNPs in close proximity to rs1015443 suggest that this locus is principally responsible for the association. Thus, our results provide additional support to the emerging hypothesis that genetic variation in bitter taste receptors can impact upon alcohol consumption.
Key words: alcohol consumption; alcohol use disorders identification test (AUDIT), bitter taste; behavioral genetics; taste receptor; TAS2R; alcohol consumption; alcohol consumption; alcohol consumption; alcohol consumption
Introduction
Cancers of the oral and pharyngeal structures have a high incidence worldwide and a grim prognosis (Zygogianni et al. 2011). Lifestyle risk factors play a major role in the development of these carcinomas. Alcohol intake, particularly in combination with smoking, is a significant lifestyle risk factor associated with the incidence of these cancers (Hayes RB et al. 1999; Anantharaman et al. 2011; Groome et al. 2011; Lambert et al. 2011; Zygogianni et al. 2011). Thus, factors associated with increased alcohol intake are of strong interest to those concerned with decreasing the incidence of these malignancies.
One of the many factors thought to influence alcohol intake is its perceived taste. Researchers have attempted to perceptually categorize the taste evoked by ethanol. Results from such experiments suggest that ethanol evokes both sweet and bitter percepts, depending on concentration (Di Lorenzo et al. 1986; Lawrence and Kiefer 1987; Kiefer and Lawrence 1988; Kiefer and Mahadevan 1993; Lanier et al. 2005; Blizard 2007). Sensitivity to the sweet component may contribute substantially to preference for alcohol, while sensitivity to the bitter component may negatively influence alcohol consumption. Indeed, research suggests that individuals who possess enhanced perception of bitter taste, as defined by sensitivity to the bitter tasting chemical 6-n-propylthiouracil (PROP), consume alcoholic beverages less frequently than individuals who are less sensitive to the compound (Intranuovo and Powers 1998; Duffy et al. 2004; Basson et al. 2005; Lanier et al. 2005).
In addition to influencing alcohol intake, a large body of research indicates that PROP-defined bitter taste sensitivity also affects the intake of certain bitter-tasting foods, including specific fruits and vegetables (Fischer et al. 1961; Glanville and Kaplan 1965; Jerzsa-Latta et al. 1990; Drewnowski et al. 1997, 1998, 1999, 2000; Dinehart et al. 2006; Tepper et al. 2009; Duffy et al. 2010; Feeney et al. 2011). Members of the T2R family of GPCRs have been shown to function as taste receptors for chemicals that humans and mammals perceive as bitter tasting (Chandrashekar et al. 2000; Mueller et al. 2005; Meyerhof et al. 2010). T2R receptors are encoded by members of the TAS2R gene family. The principal genetic determinants of phenotypic variation in PROP taste sensitivity are alleles of the gene TAS2R38 (Kim et al. 2003; Bufe et al. 2005). Indeed, variation in TAS2R38 is also associated with vegetable intake (Sacerdote et al. 2007; Duffy et al. 2010). Moreover, polymorphisms in or near TAS2R38, as well as the gene TAS2R16, have been associated with measures of alcohol consumption (Duffy et al. 2004; Hinrichs et al. 2006; Wang et al. 2007; Hayes JE et al. 2011). There are 25 functional TAS2R genes in the human genome (Kim et al. 2005; Meyerhof 2005). However, the influence of genetic variation in bitter taste receptor genes on alcohol consumption has been limited to the assessment of just a few genes (i.e., TAS2R38 and TAS2R16). Since many TAS2R receptors are broadly tuned (Meyerhof et al. 2010), it is entirely possible that other TAS2Rs may influence alcohol intake.
Because of the observed role that bitter taste plays in alcohol intake, we hypothesized that allelic variation in individual TAS2Rs could significantly impact upon alcohol consumption. Thus, we initiated a candidate gene study to identify specific sequence variants in TAS2R genes that are associated with measures of alcohol intake, using a cohort of head and neck cancer patients. These patients have relatively high premorbid levels of alcohol intake and may possess genetic factors that predispose them to increased levels of alcohol consumption, making head and neck patients an ideal group. This is one of the first studies attempting to link a known risk factor (alcohol intake) to allelic variations in bitter taste receptors in a head and neck cancer clinical population.
Materials and methods
Head and neck cancer patients
This study was conducted according to the principles of the 1975 Declaration of Helsinki and Good Clinical Practice guidelines and was approved by the University of Florida Institutional Review Board. A convenience sample of 173 (126 men and 47 women) head and neck cancer patients was recruited from clinics at the University of Florida. Sixty-five percent were newly diagnosed head and neck cancer patients. The characteristics of this group have been detailed elsewhere (Logan et al. 2010, also see Table 1).
Table 1 .
Characteristics of cancer patients
| N = 173 | |
| Sex | |
| Men | 126 |
| Women | 47 |
| Race | |
| White | 94% (N = 162) |
| Other | 6% (N = 11) |
| Education | |
| <high school | 7% |
| High school diploma or GED | 31% |
| Some college | 26% |
| Bachelor’s degree or higher | 36% |
| Age | 60.7±13.4 |
DNA extraction and genetic analysis
DNA was extracted from leukocytes from whole blood following manufacturer’s instructions (PureGene system, Qiagen), with occasional modification for hemolyzed samples. Purified DNA samples were stored at 4°C in 1xTE (Tris 10mM and EDTA 1mM, pH 8.0) until analyzed. When necessary, DNA was genome-amplified using the Illustra GenomiPhi V2 D amplification kit from GE Lifesciences. Genotypes were determined using vendor-supplied TaqMan SNP assays from Applied Biosystems with the plates read on an ABI Prism 7900 HT (Applied Biosystems/Life Technologies).
We identified candidate SNPs in coding and regulatory regions from the Entrez SNP database and from the literature (e.g. Kim et al. 2005). In total, 23 TAS2R-associated SNPs were genotyped (Table 2). SNP frequencies in our sample were similar to values seen in reference data sets. Three additional haplotype-tagging SNPs (r 2 ≥ 0.8), identified from the HapMap (International Hapmap Consortium 2005), were genotyped for the purpose of defining linkage disequilibrium (LD; bottom of Table 2).
Table 2.
SNP genotyping statistics and results of association analyses#
| Chromosome,position (kb) | SNP ID | Associated/nearest gene | Call rate(%) | HWE P Value | Major/minor allele | MAF | SNP type | Six ormore | Per day | Drink containing alcohol |
|---|---|---|---|---|---|---|---|---|---|---|
| 7, 122625 | rs1308724 | TAS2R16 | 93.3 | 0.055 | G/C | 0.38 | Noncoding | 0.49 | 0.87 | 0.08 |
| 7, 122630 | rs846672 | TAS2R16 | 93.8 | 0.561 | C/A | 0.35 | Noncoding | 0.61 | 0.52 | 0.91 |
| 7, 141464 | rs765007 | TAS2R3 | 97.3 | 0.613 | C/T | 0.47 | 5’ UTR | 0.11 | 0.15 | 0.42 |
| 7, 141479 | rs2234001 | TAS2R4 | 95.1 | 0.554 | C/G | 0.48 | V96L | 0.35 | 0.69 | 0.28 |
| 7, 141490 | rs2234012 | TAS2R5 | 96.4 | 0.032 | G/A | 0.13 | 5’ UTR | 0.46 | 0.24 | 0.37 |
| 7, 141673 | rs10246939 | TAS2R38 | 96.4 | 0.067 | T/C | 0.46 | I296V | 0.72 | 0.76 | 0.87 |
| 7, 141673 | rs1726866 | TAS2R38 | 98.2 | 0.266 | T/C | 0.46 | V262A | 0.10 | 0.34 | 0.57 |
| 7, 141673 | rs713598 | TAS2R38 | 99.6 | 0.095 | C/G | 0.43 | P49A | 0.03 | 0.68 | 0.0008 |
| 7, 142881 | rs4726600 | TAS2R39 | 97.3 | 0.518 | G/A | 0.23 | Noncoding | 0.32 | 0.93 | 0.12 |
| 7, 142920 | rs10260248 | TAS2R40 | 95.1 | 0.012 | C/A | 0.09 | S187Y | 0.86 | 0.79 | 0.95 |
| 7, 142921 | rs534126 | TAS2R40 | 97.8 | 0.934 | C/T | 0.35 | Noncoding | 0.51 | 0.43 | 0.19 |
| 7, 143168 | rs12666496 | TAS2R41 | 97.3 | 0.019 | A/T | 0.26 | Noncoding | 0.90 | 0.77 | 0.37 |
| 7, 143175 | rs1404635 | TAS2R41 | 96.0 | 0.255 | G/A | 0.29 | T63T | 0.21 | 1.00 | 0.08 |
| 12, 10954 | rs619381 | TAS2R7 | 98.7 | 0.910 | C/T | 0.12 | M304I | 0.69 | 0.13 | 0.34 |
| 12, 10959 | rs1548803 | TAS2R8 | 97.8 | 0.138 | T/C | 0.42 | L183L | 0.01 | 0.04 | 0.55 |
| 12, 10978 | rs10845219 | TAS2R10 | 97.8 | 0.244 | C/T | 0.42 | Noncoding | 0.02 | 0.05 | 0.72 |
| 12, 10982 | rs4763216 | TAS2R10 | 98.7 | 0.236 | G/C | 0.42 | Noncoding | 0.00 | 0.02 | 0.38 |
| 12, 11061 | rs1015443 | TAS2R13 | 98.2 | 0.027 | C/T | 0.42 | S259N | 0.0002 | 0.001 | 0.06 |
| 12, 11092 | rs7138535 | TAS2R14 | 95.5 | 0.021 | T/A | 0.24 | G38G | 0.26 | 0.42 | 0.41 |
| 12, 11139 | rs1376251 | TAS2R50 | 95.5 | 0.055 | C/T | 0.31 | C203Y | 0.56 | 0.49 | 0.65 |
| 12, 11150 | rs10845281 | TAS2R20 | 97.3 | 0.235 | T/C | 0.33 | I236V | 0.49 | 0.95 | 0.23 |
| 12, 11174 | rs10772420 | TAS2R19 | 94.6 | 0.230 | A/G | 0.47 | C299R | 0.06 | 0.08 | 0.18 |
| 12, 11182 | rs11612527 | TAS2R31 | 95.5 | 0.078 | T/A | 0.22 | Noncoding | 0.81 | 0.18 | 0.78 |
| 12, 11000 | rs1047699* | PRR4 | 91.1 | 0.481 | C/T | 0.21 | G43R | 0.62 | 0.45 | 0.86 |
| 12, 11000 | rs1063193* | PRR4 | 90.0 | 0.434 | T/C | 0.47 | Q109R | 0.98 | 0.65 | 0.72 |
| 12, 11036 | rs10492098* | PRH1 | 91.3 | 0.009 | A/G | 0.43 | intronic | 0.003 | 0.005 | 0.06 |
kb, kilobases; MAF, minor allele frequency.
*SNPs genotyped after the first 23 SNPs; used for LD analysis.
#Raw, unadjusted p values are presented in the table.
Measures
Measures were drawn from the 10-item Alcohol Use Disorders Identification Test (AUDIT; Saunders et al. 1993). The AUDIT was developed by the World Health Organization as a simple method of screening for excessive drinking. We evaluated the impact of genetic variation in taste receptor SNPs on responses to three questions from the AUDIT: “How often do you have a drink containing alcohol?” (DRINKSCONTAININGALCOHOL); “How many drinks containing alcohol do you have on a typical day when you are drinking?” (PERDAY); and “How often do you have six or more drinks on one occasion? (SIXORMORE).”
Analysis
The genetic association analyses were performed using SNP &Variation Suite 7.4 (Golden Helix, Bozeman). All SNPs were analyzed for extreme deviation from Hardy–Weinberg equilibrium (p < 0.001) using chi-square test. SNPs with less than 90% complete genotyping information were excluded from analyses. Single marker associations were tested using linear regression including age, sex, and smoking status (i.e., 1 = never smoked, 2 = past smoker, 3 = current smoker) as covariates in an additive model (i.e., heterozygotes expressing intermediate phenotypes: xx = 0, Xx = 1, XX = 2).
Genetic association can be confounded by population stratification (Hamer and Sirota 2000). Therefore, our data was corrected by principal component analysis to control for the possibility of stratification (Price et al. 2006). For our initial association analyses, in addition to the observed p values, we present Bonferroni adjusted values for the interested reader. There is, however, an active debate as to whether multiple comparison adjustments are appropriate for exploratory studies (Perneger 1998; Bender and Lange 2001). Analysis of associations related to TAS2R16 (i.e., rs1308724 and rs846672) and TAS2R38 (i.e., rs10246939, rs1726866 and rs713598) were based on a priori hypotheses (Duffy et al. 2004; Hinrichs et al. 2006; Wang et al. 2007; Hayes JE et al. 2011). Thus, no multiple comparisons adjustments were made. Pairwise LD between the SNPs and haplotype block analysis was computed using Haploview 4.2 (Barrett et al. 2005). Haplotype blocks were defined by 95% confidence bounds on D' (Gabriel et al. 2002).
Results
We first asked whether any variants in and around select TAS2Rs, previously associated with measures of alcohol consumption, were correlated with reported alcohol intake in our cancer patients. We observed that a nonsynonymous coding SNP located in the gene TAS2R38 showed significant association with the measure DRINKSCONTAININGALCOHOL (p = 0.0008) but not with PERDAY or SIXORMORE. The C allele of rs713598, the major allele in the cohort, is strongly associated with increased taste sensitivity to PROP (Kim et al. 2003; Bufe et al. 2005) and decreased alcohol consumption in our head and neck cancer cohort.
Next we asked whether any other TAS2R SNPs were associated with measures of alcohol consumption from the AUDIT. We found one SNP, located on chromosome 12, that showed significant associations with the measures PERDAY and SIXORMORE. Variation in the nonsynonymous coding SNP rs1015443, located in the gene TAS2R13, was significantly associated with the measure PERDAY (p = 0.0011; Bonferroni value = 0.03 after correction for multiple comparisons; Table 3) and with SIXORMORE (p = 0.000231; Bonferroni value = 0.005; Table 3). By and large, subjects homozygous for the major allele (CC carriers) consumed alcoholic beverages less frequently relative to heterozygotes and those homozygous for the minor allele (Table 4).
Table 3 .
Perday and sixormore p values for rs1015443* and rs10492098#
| SNP/Genotype | Perday | Sixormore |
|---|---|---|
| rs1015443 | 0.001 | 0.0002 |
| rs10492098 | 0.005 | 0.003 |
*Number of subjects per rs1015443 genotype (TT = 42; CT = 61; CC = 66).
#Number of subjects per rs10492098 genotype (GG = 41; GA = 57; AA = 60).
Table 4 .
Phenotype averages* by rs1015443 genotype#
| Phenotype | TT | CT | CC |
|---|---|---|---|
| PERDAY | 1.50±0.93 | 1.57±1.01 | 1.41±0.85 |
| SIXORMORE | 1.75±1.32 | 1.66±1.09 | 1.57±1.06 |
*Mean ± standard deviation. These values are unadjusted for the variance associated with the covariates used in the regression models (age, sex, and smoking status).
#Number of subjects per genotype (TT = 42; CT = 61; CC = 66).
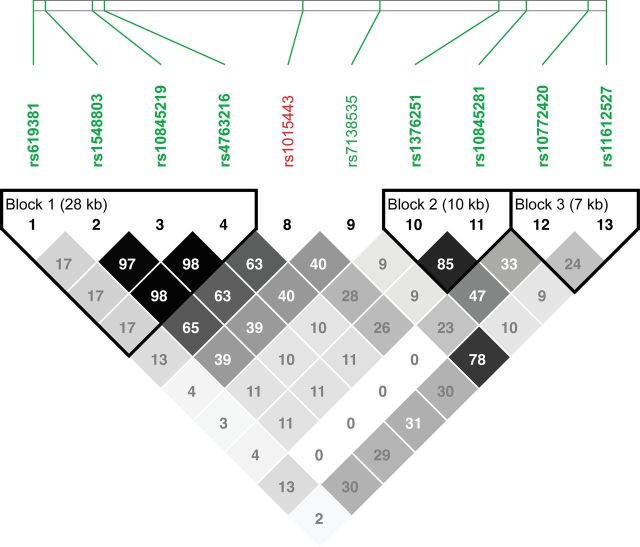
To define the extent of LD around rs1015443, we used the other taste receptor-associated SNPs that we genotyped both upstream and downstream of TAS2R13 on chromosome 12 (Figure 1). We identified three LD blocks. The first (LD Block 1) contains four SNPs (rs619381, rs1548803, rs10845219, and rs4763216) and is found ~79kb upstream of rs1015443, extending for ~28kb (Figure 1). SNP rs1015443 (TAS2R13) displays moderate LD with the SNPs in this block (r 2 = 0.13–0.65) but significantly less than the threshold for inclusion in the LD block. Indeed, none of these SNPs were associated with any of the measures of alcohol intake. The five other SNPs in this region show low levels of LD with rs1015443, although four of them form two small LD blocks at the distal end of this cluster (LD Blocks 2 and 3; Figure 1).
Figure 1 .
Pairwise LD (r 2) among nine SNPs located both upstream and downstream of rs1015443 on chromosome 12. r 2 values × 100 are indicated within squares, with darker shades indicating higher r 2 values.
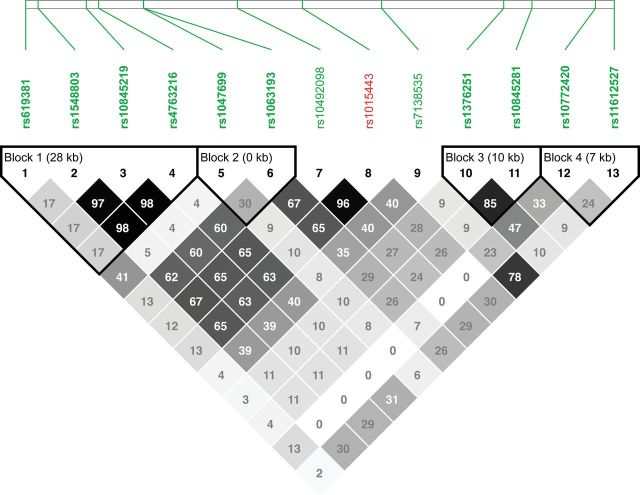
Because of the large intervals between rs1015443 and the SNPs located in LD block 1, we sought to further refine the extent of LD in this region by examining several additional SNPs in the genes located directly upstream of TAS2R13. The coding sequences of two genes, proline-rich protein HaeIII subfamily 1 (PRH1) and proline rich 4 (PRR4), are located in this region. Thus, we identified haplotype-tagging SNPs within each of these genes; two SNPs were identified for PRR4 (rs1047699 and rs1063193) and a single SNP was selected for PRH1 (rs10492098).
The addition of these three SNPs upstream of TAS2R13 slightly redefined the LD structure we observed between the genotyped SNPs. After inclusion of these SNPs, we observed four LD blocks. Three of the LD blocks were the same as that defined in Figure 1. A new LD block was formed that consisted of two of the three SNPs that were introduced in this later analysis (rs1047699 and rs1063193). These SNPs showed low to moderate LD with rs1015443 (r 2 = 0.10 and 0.65, respectively; Figure 2). The other SNP included in this LD analysis, rs10492098, was in tight LD with rs1015443 (r 2 = 0.96; Figure 2). rs10492098 was also significantly associated with the measures PERDAY and SIXORMORE, though the p-values of its association were higher than that observed for rs1015443 (Table 3). When taken together, these data suggest that variation in the bitter taste receptor gene TAS2R13 is principally responsible for the association observed in our head and neck cancer cohort.
Figure 2 .
Pairwise LD (r 2) among the 9 SNPs located both upstream and downstream of rs1015443 on chromosome 12 and 3 additional SNPs located directly upstream of TAS2R13. r 2 values × 100 are indicated within squares, with darker shades indicating higher r 2 values.
Discussion
Our results indicate that genetic variation in the gene TAS2R13 is associated with a self-reported increase in alcohol consumption as assessed using two measures from the AUDIT: PERDAY and SIXORMORE. We also observed a significant association between genetic variation in the gene TAS2R38 and a measure of alcohol consumption (DRINKSCONTAININGALCOHOL). Other measures of alcohol consumption have been associated with variation in TAS2R38. Variation in responses to the measure Maxdrinks (i.e., responses to the question: what is the largest number of drinks you have ever had in a 24-h period?) was significantly associated with genetic variation in the gene TAS2R38 (Wang et al. 2007) in an African-American cohort. Hayes JE et al. (2011) reported associations with genetic variation in TAS2R38 and a measure of the frequency of alcohol intake in a convenience sample of 96 healthy adults subjects. Thus, our results are consistent with previous studies that have reported that variation in bitter taste receptor genes is associated with alcohol consumption.
The AUDIT is an easily administered, widely used and referenced screening tool for the assessment of alcohol use. However, this instrument does not provide information on lifetime alcohol use/problems. Specifically, subjects who are lifetime abstainers are collapsed together with subjects who may be alcohol dependent but are abstaining from alcohol use. If such information were incorporated into our analyses, these data would likely have strengthened our results and, potentially, led to the discovery of other genotype–phenotype associations.
Taste abnormalities have been observed in patients with head and neck cancer, and it has been postulated that these defects are caused by treatment-based radiation exposure (Mirza et al. 2008). It is possible that any genetic or environmental factors that predominate in head and neck cancer patients may also allow the impact of TAS2Rs on alcohol intake to be more easily observed. More research is needed to elucidate any putative factors that may be influencing the relationship between genetic variation in TAS2R genes and alcohol intake. Nevertheless, our results provide new evidence supporting the link between gustatory functioning and the ingestion of alcoholic beverages.
Participants who were homozygous for the major allele of rs1015443 consumed alcoholic beverages less frequently relative to subjects that were heterozygotes or those subjects that were homozygous for the minor allele. It is unknown whether the amino acid change from asparagine (in receptors encoded by genes that possess the minor allele of rs1015443) to serine in those that possess the major allele would influence the functioning of TAS2R13. Based on the influence of other TAS2Rs on alcohol intake (i.e., alleles that decrease the sensitivity of a TAS2R receptor lead to increased alcohol consumption [Wang et al. 2007; Hayes et al. 2008]), we would predict that the minor allele of rs1015443 impacts negatively on the functioning of TAS2R13. The natural ligand or ligands for TAS2R13 are unknown. However, the synthetic compounds diphenidol and denatonium benzoate are known TAS2R13 agonists (Meyerhof et al. 2010). Experiments using diphenidol and/or denatonium benzoate designed to determine the impact of variation at rs1015443 will be needed to test this prediction.
The analysis of LD surrounding TAS2R13 revealed only a single other candidate locus that could be responsible for the association signal emanating from this genomic region. The SNP rs10492098, located in PRH1, is in extremely tight LD with rs1015443. PRH1 encodes a member of the proline rich protein (PRP) family of peptides. These peptides are thought to play a role in oral mucosal defense by their involvement in maintaining calcium concentrations, as well as in retarding the aggregation capacity of microorganisms, thereby impacting their ability to colonize on tissue surfaces (Moreno et al. 1982; Gibbons et al. 1988; Ligtenberg et al. 1992; Fung et al. 2004). However, surprisingly, salivary levels of some of these peptides have been implicated in the modulation of bitter taste perception (Cabras et al. 2012). It should be noted that rs10492098 is on the borderline of Hardy Weinberg equilibrium in the sample (Table 2, HWE p value = 0.009). This fact presents a significant potential confound in the interpretation of the results based on data derived from the genotyping of this allele and, as such, adds to our confidence that rs1015443 is the true causal allele. Moreover, the observed P-values of the associations for rs10492098 were higher, and in one case, an order of magnitude higher, than that observed for rs1015443 (Table 3). Indeed, considering these factors, in addition to our a priori hypothesis regarding the impact of variation in bitter taste receptor genes on alcohol consumption, we conclude that rs1015443, located in the gene TAS2R13, is principally responsible for the association signal emanating from this genomic region. That said, based on the proximity of these two loci and the fact that in any population they are likely to be in tight LD, it will be exceedingly difficult to disentangle the influence of each locus on alcohol consumption using genotype/phenotype association testing alone. Additional in vivo experiments (e.g., Roudnitzky et al. 2011), and possibly the use of animal models, will be needed to determine the individual effects of these two genes on taste-related behavior.
It is well known that cigarette use is confounded with alcohol consumption (Bottoni et al. 1997; Koh et al. 2005), and some studies suggest that variation in bitter taste receptors may impact on cigarette usage (Cannon et al. 2005; Mangold et al. 2008). Although none of the SNPs in our head and neck cancer cohort was directly associated with smoking status (data not shown), we did control for cigarette use in our analyses. Thus, the use of cigarettes by our head and neck cancer patients does not appear to account for the observed associations between bitter taste receptor variants and alcohol consumption. Nevertheless, a more systematic evaluation of the relationship between bitter taste perception and its influence on the use of both cigarette and alcohol seems warranted.
In this report, we hypothesized that T2R receptor function influences alcohol consumption via mediation of bitter taste perception. However, it is also possible that bitter taste receptors affect alcohol intake via their expression and function in nonoral tissues. In addition to their expression in the oral–nasal cavity where they act as taste receptors, it is now well known that taste receptors, including T2Rs, are expressed in tissues all over the body, where they appear to act as general chemoreceptors (Behrens and Meyerhof, 2010, 2011). For example, T2Rs are expressed in tissues of the gastrointestinal tract (GI), where they appear to play a role in gastrointestinal hormone secretion as well as in the nutrient dependent regulation of metabolism (Sternini 2007; Kaji et al. 2009; Dotson et al. 2010; Janssen et al. 2011; Jeon et al. 2011). Thus, it is possible that it is the extraoral expression and function of T2Rs that is impacting on alcohol consumption. For example, it is known that plasma levels of certain gastrointestinal hormones, which are known to be affected by taste receptor expression and function in the GI tract, are correlated with alcohol intake (Yeomans et al. 2003; Pravdova and Fickova 2006; Leggio et al. 2011).
In summary, alcohol intake is a complex trait influenced by numerous genes. We have observed an association between genetic variation in a bitter taste receptor gene and the consumption of alcohol. As alcohol consumption is a risk factor in developing head and neck cancer, taste perception may represent one of the many pathways that contribute to the development of these cancers of the oral cavity.
FUNDING
This research was partially funded by the National Cancer Institute [CA111593] and the National Institute of Dental and Craniofacial Research [U54DEO19261] awarded to H.L.L.
Acknowledgements
We thank Michelle Burch and Kaitlin Healy for technical assistance and are grateful to the subjects who participated in this study.
References
- Anantharaman D, Marron M, Lagiou P, Samoli E, Ahrens W, Pohlabeln H, Slamova A,, Schejbalova M, Merletti F, Richiardi L, et al. 2011. Population attributable risk of tobacco and alcohol for upper aerodigestive tract cancer Oral Oncol 47:725– 731 [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. 2005. Haploview: analysis and visualization of LD and haplotype maps Bioinformatics 21:263– 265 [DOI] [PubMed] [Google Scholar]
- Basson M, Bartoshuk L, Dichello S, Panzini L, Weiffenbach J, Duffy V. 2005. Association between 6-n-propylthiouracil (PROP) bitterness and colonic neoplasms Dig Dis Sci 50:483– 489 [DOI] [PubMed] [Google Scholar]
- Behrens M, Meyerhof W. 2010. Oral and extraoral bitter taste receptors Results Probl Cell Differ 52: 87– 99 [DOI] [PubMed] [Google Scholar]
- Behrens M, Meyerhof W. 2011. Gustatory and extragustatory functions of mammalian taste receptors Physiol Behav 105:4– 13 [DOI] [PubMed] [Google Scholar]
- Bender R, Lange S. 2001. Adjusting for multiple testing—when and how? J Clin Epidemiol 54:343– 349 [DOI] [PubMed] [Google Scholar]
- Blizard DA. 2007. Sweet and bitter taste of ethanol in C57BL/6J and DBA2/J mouse strains Behav Genet 37:146– 159 [DOI] [PubMed] [Google Scholar]
- Bottoni A, Cannella C, Del Balzo V. 1997. Lifestyle and dietary differences in smokers and non-smokers from an Italian employee population Public Health 111:161– 164 [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. 2005. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception Curr Biol 15:322– 327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabras T, Melis M, Castagnola M, Padiglia A, Tepper BJ, Messana I, Tomassini Barbarossa I. 2012. Responsiveness to 6-n-propylthiouracil (PROP) is associated with salivary levels of two specific basic proline-rich proteins in humans PLoS One. 7:e30962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon D, Baker T, Piper M, Scholand M, Lawrence D, Drayna D, McMahon W, Villegas G, Caton T, Coon H, et al. 2005. Associations between phenylthiocarbamide gene polymorphisms and cigarette smoking Nicotine Tob Res 7:853– 858 [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. 2000. T2Rs function as bitter taste receptors Cell 100:703– 711 [DOI] [PubMed] [Google Scholar]
- Di Lorenzo PM, Kiefer SW, Rice AG, Garcia J. 1986. Neural and behavioral responsivity to ethyl alcohol as a tastant Alcohol 3:55– 61 [DOI] [PubMed] [Google Scholar]
- Dinehart M, Hayes J, Bartoshuk L, Lanier S, Duffy V. 2006. Bitter taste markers explain variability in vegetable sweetness, bitterness, and intake Physiol Behav. 87:304– 313 [DOI] [PubMed] [Google Scholar]
- Dotson C, Vigues S, Steinle N, Munger S. 2010. T1R and T2R receptors: the modulation of incretin hormones and potential targets for the treatment of type 2 diabetes mellitus Curr Opin Investig Drugs 11:447– 454 [PMC free article] [PubMed] [Google Scholar]
- Drewnowski A, Henderson S, Hann C, Berg W, Ruffin M. 2000. Genetic taste markers and preferences for vegetables and fruit of female breast care patients J Am Diet Assoc 100:191– 197 [DOI] [PubMed] [Google Scholar]
- Drewnowski A, Henderson S, Levine A, Hann C. 1999. Taste and food preferences as predictors of dietary practices in young women Public Health Nutr 2:513– 519 [DOI] [PubMed] [Google Scholar]
- Drewnowski A, Henderson S, Shore A. 1997. Taste responses to naringin, a flavonoid, and the acceptance of grapefruit juice are related to genetic sensitivity to 6-n-propylthiouracil Am J Clin Nutr 66:391– 397 [DOI] [PubMed] [Google Scholar]
- Drewnowski A, Henderson S, Shore A, Barratt-Fornell A. 1998. Sensory responses to 6-n-propylthiouracil (PROP) or sucrose solutions and food preferences in young women Ann NY Acad Sci 855:797– 801 [DOI] [PubMed] [Google Scholar]
- Duffy VB, Davidson AC, Kidd JR, Kidd KK, Speed WC, Pakstis AJ, Reed DR, Snyder DJ, Bartoshuk LM. 2004. Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake Alcohol Clin Exp Res 28:1629– 1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy VB, Hayes JE, Davidson AC, Kidd JR, Kidd KK, Bartoshuk LM. 2010. Vegetable intake in college-aged adults is explained by oral sensory phenotypes and TAS2R38 genotype Chemosens Percept 3:137– 148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney E, O'Brien S, Scannell A, Markey A, Gibney ER. 2011. Genetic variation in taste perception: does it have a role in healthy eating? Proc Nutr Soc 70:135– 143 [DOI] [PubMed] [Google Scholar]
- Fischer R, Griffin F, England S, Garn SM. 1961. Taste thresholds and food dislikes Nature 191:1328 [DOI] [PubMed] [Google Scholar]
- Fung KY, Morris C, Sathe S, Sack R, Duncan MW. 2004. Characterization of the in vivo forms of lacrimal-specific proline-rich proteins in human tear fluid Proteomics 4:3953– 3959 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. 2002. The structure of haplotype blocks in the human genome Science 296:2225– 2229 [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Hay DI, Cisar JO, Clark WB. 1988. Adsorbed salivary proline-rich protein 1 and statherin: receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces Infect Immun 56:2990– 2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville EV, Kaplan AR. 1965. Food preference and sensitivity of taste for bitter compounds Nature 205:851– 853 [Google Scholar]
- Groome PA, Rohland SL, Hall SF, Irish J, Mackillop WJ, O'Sullivan B. 2011. A population-based study of factors associated with early versus late stage oral cavity cancer diagnoses Oral Oncol 47:642–– 647 [DOI] [PubMed] [Google Scholar]
- Hamer D, Sirota L. 2000. Beware the chopsticks gene Mol Psychiatry 5:11– 13 [DOI] [PubMed] [Google Scholar]
- Hayes JE, Bartoshuk L, Kidd J, Duffy V. 2008. Supertasting and PROP bitterness depends on more than the TAS2R38 gene Chem Senses 33:255– 265 [DOI] [PubMed] [Google Scholar]
- Hayes JE, Wallace MR, Knopik VS, Herbstman DM, Bartoshuk LM, Duffy VB. 2011. Allelic variation in TAS2R bitter receptor genes associates with variation in sensations from and ingestive behaviors toward common bitter beverages in adults Chem Senses 36:311– 319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes RB, Bravo-Otero E, Kleinman DV, Brown LM, Fraumeni JF, Harty LC, Winn DM. 1999. Tobacco and alcohol use and oral cancer in Puerto Rico Cancer Causes Control 10:27– 33 [DOI] [PubMed] [Google Scholar]
- Hinrichs AL, Wang JC, Bufe B, Kwon JM, Budde J, Allen R, Bertelsen S, Evans W, Dick D, Rice J, et al. 2006. Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence Am J Hum Genet 78:103– 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Hapmap Consortium 2005. A haplotype map of the human genome Nature 437:1299– 1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intranuovo LR, Powers AS. 1998. The perceived bitterness of beer and 6-n-propylthiouracil (PROP) taste sensitivity Ann NY Acad Sci 855:813– 815 [DOI] [PubMed] [Google Scholar]
- Janssen S, Laermans J, Verhulst PJ, Thijs T, Tack J, Depoortere I. 2011. Bitter taste receptors and α-gustducin regulate the secretion of ghrelin with functional effects on food intake and gastric emptying Proc Natl Acad Sci USA 108:2094– 2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon TI, Seo YK, Osborne TF. 2011. Gut bitter taste receptor signalling induces ABCB1 through a mechanism involving CCK Biochem J 438:33– 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzsa-Latta M, Krondl M, Coleman P. 1990. Use and perceived attributes of cruciferous vegetables in terms of genetically-mediated taste sensitivity Appetite 15:127– 134 [DOI] [PubMed] [Google Scholar]
- Kaji I, Karaki S, Fukami Y, Terasaki M, Kuwahara A. 2009. Secretory effects of a luminal bitter tastant and expressions of bitter taste receptors, T2Rs, in the human and rat large intestine Am J Physiol Gastrointest Liver Physiol 296:G971– 981 [DOI] [PubMed] [Google Scholar]
- Kiefer SW, Lawrence GJ. 1988. The sweet-bitter taste of alcohol: aversion generalization to various sweet-quinine mixtures in the rat Chem. Senses 13:633– 641 [Google Scholar]
- Kiefer SW, Mahadevan RS. 1993. The taste of alcohol for rats as revealed by aversion generalization tests Chem. Senses 18:509– 522 [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. 2003. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide Science 299:1221– 1225 [DOI] [PubMed] [Google Scholar]
- Kim UK, Wooding S, Ricci D, Jorde L, Drayna D. 2005. Worldwide haplotype diversity and coding sequence variation at human bitter taste receptor loci Hum Mutat 26:199– 204 [DOI] [PubMed] [Google Scholar]
- Koh WP, Yuan JM, Sun CL, Lee HP, Yu MC. 2005. Middle-aged and older Chinese men and women in Singapore who smoke have less healthy diets and lifestyles than nonsmokers J Nutr 135:2473– 2477 [DOI] [PubMed] [Google Scholar]
- Lambert R, Sauvaget C, de Camargo Cancela M, Sankaranarayanan R. 2011. Epidemiology of cancer from the oral cavity and oropharynx Eur J Gastroenterol Hepatol 23:633– 641 [DOI] [PubMed] [Google Scholar]
- Lanier SA, Hayes JE, Duffy VB. 2005. Sweet and bitter tastes of alcoholic beverages mediate alcohol intake in of-age undergraduates Physiol Behav 83:821– 831 [DOI] [PubMed] [Google Scholar]
- Lawrence GJ, Kiefer SW. 1987. Generalization of specific taste aversions to alcohol in the rat. Chem Senses 12:591– 599 [Google Scholar]
- Leggio L, Addolorato G, Cippitelli A, Jerlhag E, Kampov-Polevoy AB, Swift RM. 2011. Role of feeding-related pathways in alcohol dependence: A focus on sweet preference, NPY, and ghrelin Alcohol Clin Exp Res 35:194– 202 [DOI] [PubMed] [Google Scholar]
- Ligtenberg AJ, Walgreen-Weterings E, Veerman EC, de Soet JJ, de Graaff J, Amerongen AV. 1992. Influence of saliva on aggregation and adherence of Streptococcus gordonii HG 222 Infect Immun 60:3878– 3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan HL, Fillingim RB, Bartoshuk LM, Sandow P, Tomar SL, Werning JW, Mendenhall WM. 2010. Smoking status and pain level among head and neck cancer patients J Pain 11:528– 534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold JE, Payne TJ, Ma JZ, Chen G, Li MD. 2008. Bitter taste receptor gene polymorphisms are an important factor in the development of nicotine dependence in African Americans J Med Genet 45:578– 582 [DOI] [PubMed] [Google Scholar]
- Meyerhof W. 2005. Elucidation of mammalian bitter taste Rev Physiol Biochem Pharmacol 154:37– 72 [DOI] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. 2010. The molecular receptive ranges of human TAS2R bitter taste receptors Chem Senses 35:157– 170 [DOI] [PubMed] [Google Scholar]
- Mirza N, Machtay M, Devine PA, Troxel A, Abboud SK, Doty RL. 2008. Gustatory impairment in patients undergoing head and neck irradiation Laryngoscope 118:24– 31 [DOI] [PubMed] [Google Scholar]
- Moreno EC, Kresak M, Hay DI. 1982. Adsorption thermodynamics of acidic proline-rich human salivary proteins onto calcium apatites. J Biol Chem 257:2981– 2989 [PubMed] [Google Scholar]
- Mueller K, Hoon M, Erlenbach I, Chandrashekar J, Zuker C, Ryba N. 2005. The receptors and coding logic for bitter taste Nature 434:225– 229 [DOI] [PubMed] [Google Scholar]
- Perneger TV. 1998. What's wrong with Bonferroni adjustments BMJ 316:1236– 1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pravdova E, Fickova M. 2006. Alcohol intake modulates hormonal activity of adipose tissue Endocr Regul 40:91– 104 [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies Nat Genet 38:904– 909 [DOI] [PubMed] [Google Scholar]
- Roudnitzky N, Bufe B, Thalmann S, Kuhn C, Gunn HC, Xing C, Crider BP, Behrens M, Meyerhof W, Wooding SP. 2011. Genomic, genetic and functional dissection of bitter taste responses to artificial sweeteners Hum Mol Genet 20:3437– 3449 [DOI] [PubMed] [Google Scholar]
- Sacerdote C, Guarrera S, Smith GD, Grioni S, Krogh V, Masala G, Mattiello A, Palli D, Panico S, Tumino R, et al. 2007. Lactase persistence and bitter taste response: instrumental variables and mendelian randomization in epidemiologic studies of dietary factors and cancer risk Am J Epidemiol 166:576– 581 [DOI] [PubMed] [Google Scholar]
- Saunders JB, Aasland OG, Babor TF, de la Fuente JR, Grant M. 1993. Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons with Harmful Alcohol Consumption—II. Addiction 88:791– 804 [DOI] [PubMed] [Google Scholar]
- Sternini C. 2007. Taste receptors in the gastrointestinal tract. IV. Functional implications of bitter taste receptors in gastrointestinal chemosensing. Am J Physiol Gastrointest Liver Physiol 292:G457– G461 [DOI] [PubMed] [Google Scholar]
- Tepper BJ, Williams TZ, Burgess JR, Antalis CJ, Mattes RD. 2009. Genetic variation in bitter taste and plasma markers of anti-oxidant status in college women Int J Food Sci Nutr 60(Suppl 2):35– 45 [DOI] [PubMed] [Google Scholar]
- Wang J, Hinrichs A, Bertelsen S, Stock H, Budde J, Dick D, Bucholz K, Rice J, Saccone N, Edenberg H, et al. 2007. Functional variants in TAS2R38 and TAS2R16 influence alcohol consumption in high-risk families of African-American origin Alcohol Clin Exp Res 31:209– 215 [DOI] [PubMed] [Google Scholar]
- Yeomans MR, Caton S, Hetherington MM. 2003. Alcohol and food intake Curr Opin Clin Nutr Metab Care 6:639– 644 [DOI] [PubMed] [Google Scholar]
- Zygogianni AG, Kyrgias G, Karakitsos P, Psyrri A, Kouvaris J, Kelekis N, Kouloulias V. 2011. Oral squamous cell cancer: early detection and the role of alcohol and smoking Head Neck Oncol 3:2 [DOI] [PMC free article] [PubMed] [Google Scholar]