Abstract
Genome-wide association studies recently identified 32 loci that associate with the age at menarche (AAM) in humans. Because the locus most robustly associated with AAM is in/near LIN28B, the goal of this study was to investigate how the Lin28 pathway might modulate pubertal timing by examining expression of Lin28b, and its homologue, Lin28a, across the pubertal transition in female mice. Quantitative reverse-transcriptase PCR data indicate that, prior to the onset of puberty, expression of both Lin28b and Lin28a decreases in the ovary, while expression of only Lin28a decreases in the hypothalamus; the expression of Lin28a increases after the onset of puberty in the pituitary. Immunohistochemistry in ovarian tissue verified that Lin28a protein levels decreased in parallel with gene expression. Although these data do not demonstrate cause and effect, they do suggest that decreased expression of Lin28a/Lin28b may facilitate the transition into puberty, consistent with previous data showing that overexpression of Lin28a in transgenic mice leads to delayed puberty. In addition, although Lin28b and/or Lin28a expression significantly decreased prior to puberty, neither Let-7a nor Let-7g miRNA levels changed significantly, raising the possibility that some effects of Lin28b and Lin28a within the hypothalamic-pituitary-gonadal (HPG) axis may be Let-7 miRNA independent. Subsequent studies, such as tissue and age specific modulation of Lin28b and Lin28a expression, could determine whether the expression patterns observed are responsible for modulating the onset of puberty and delineate further the role of this pathway in the HPG axis.
Keywords and topic category: puberty, Lin28a, Lin28b, hypothalamic-pituitary-gonadal axis
1. INTRODUCTION
Although the timing of puberty is highly heritable, the genes that control the onset of puberty in the general population have, until recently, been largely unknown [reviewed in - Gadjos et al., 2010; Towne et al., 2005]. Thirty-two loci that associate with variation in the age at menarche (AAM) have now been identified through genome wide association (GWA) studies. The most robustly associated locus is in/near the gene that encodes the RNA binding protein LIN28B [Elks et al., 2010; He et al., 2009; Ong et al., 2009; Perry et al., 2009; Sulem et al., 2009].
Two lines of evidence indicate that LIN28B may be the responsible gene at the GWA locus. First, prior to the GWA studies, lin28 had been shown to regulate developmental timing in C. elegans [Moss et al., 1997; Yang et al., 2003]. When lin28 is mutated and expression is decreased, progression through the larval stages is accelerated; conversely, when lin28 expression is increased, the nematode undergoes delayed development [Ambros and Horvitz, 1984; Moss et al., 1997; Yang and Moss, 2003]. Second, a relationship was found between the Lin28 pathway components and pubertal timing when it was examined in a fortuitously available transgenic mouse model that over-expressed Lin28a, a functional homologue of Lin28b [Zhu et al., 2010]. Vaginal opening (VO), time to first estrus, and age at first pregnancy were all delayed in the female mice carrying the transgene compared to littermate controls. These data provide evidence that the Lin28a/Lin28b pathway regulates the hypothalamic-pituitary-gonadal (HPG) axis and suggest that LIN28B is likely the gene at the GWA locus responsible for regulating AAM in humans.
How the Lin28a/Lin28b pathway regulates the HPG axis and modulates the timing of puberty is not known. However, the data from C. elegans and from the transgenic mice suggest that decreased expression would be associated with progression into puberty because over-expression of lin28 in C. elegans and of Lin28a in female mice both lead to delayed development.
To begin to assess where within the HPG axis the Lin28a/Lin28b pathway exerts effects and to determine whether levels of expression change across the pubertal transition, we assessed Lin28a and Lin28b expression in the hypothalamus, pituitary, and ovary of mice between prepubertal to postpubertal ages, ages 15 to 45 days, respectively. We also assessed expression of key members of the Let-7 miRNA family, a putative downstream mediator of Lin28a/Lin28b. To our knowledge, this is the first study to analyze the peripubertal expression of members of the LIN28B pathway, which has recently been linked to variation in AAM in humans, within the HPG axis.
2. MATERIAL AND METHODS
2.1 Animals
C57BL/6 (B6) mice used in this study were housed at the Toronto Centre for Phenogenomics (TCP) in a controlled lighting (12 h light/dark cycle) and temperature environment and were given ad libitum access to mouse chow (Tekland Standard Chow, Harland Laboratories, Mississauga, ON, CAN) and water, as described previously [Nathan et al., 2006]. The Animal Care Committee (ACC) at the TCP approved all experimental protocols.
2.2 Collection of tissues for quantitative reverse-transcriptase PCR (qRT-PCR) and immunohistochemistry (IHC)
On the day of collection, C57BL/6 female mice were euthanized with carbon dioxide (CO2) for 1 – 2 minutes followed by rapid decapitation.
For qRT-PCR, hypothalamic, pituitary, and ovarian tissue were collected at 15, 20, 25, 30, 35, 39, and 45 days of age from female C57BL/6 mice. Tissues collected at 35, 39, and 45 days were grouped and collectively analyzed as a ‘postpuberty’ time-point. For hypothalamic tissue, the whole brain was removed and placed ventral side up, where an en bloc dissection approach was taken with the following boundaries: rostral, posterior to the optic chiasm; lateral, the choroidal fissures; and caudal, the rostral pole of the mammillary bodies, and hypothalamic tissue was then excised via a 2 mm cut from the ventral side of the brain [Baker et al., 1983]. After collection, all tissues for qRT-PCR were either stored in RNAlater®, containing 10 % w/v sodium citrate tribasic dihydrate and 60 % w/v ammonium sulphate (Ambion, Foster City, CA, USA), or flash frozen in liquid nitrogen. For immunohistochemistry, hypothalamic, and ovarian tissue at 20 and 30 days of age were collected and stored in neutral-buffered formalin at room temperature for 24 hours, followed by long-term storage in 70% ethanol at 4°C. Pituitary tissue was collected at 15 and 20 days. Samples from the different ages were processed together.
2.3 RNA extraction
Total RNA extraction was performed using a modified guanidine isothiocyanate and silica membrane isolation procedure with Qiagen’s RNeasy® Mini Kit, AllPrep DNA/RNA Mini Kit, or miRNeasy Mini Kit (for Let-7a and Let-7g miRNA collection), following the manufacturer’s recommended protocols. RNA concentrations were quantified using ultraviolet-visible spectroscopy, and the integrity of total RNA was assessed using a denaturing formaldehyde agarose gel (1.2 % (w/v) agarose, 10 % (v/v) MOPS buffer, 3 % (v/v) formaldehyde, and 0.05 mg/mL ethidium bromide).
2.4 qRT-PCR for Lin28a and Lin28b
Total RNA from hypothalamic, pituitary, or ovarian tissue at each age was reverse transcribed into cDNA using the SuperScript® III Reverse Transcriptase First Strand System (Invitrogen, Carlsbad, CA, USA). After reverse transcription, quantification of Lin28a or Lin28b mRNA levels from 50 ng of hypothalamic and ovarian tissue, or 25 ng of pituitary tissue, was performed using threshold cycle (Ct) values compared to a standard curve of serially diluted hypothalamic or ovarian tissue. Ct is defined as the cycle where fluorescence in the reaction increased above a baseline threshold. Primers were designed to span exon-exon boundaries to ensure specificity, which was further ensured by dissociation curve analysis and sequencing (Lin28a F: 5′-AGGCGGTGGAGTTCACCTTTAAGA-3′, R: 5′-AGCTTGCATTCCTTGGCATGATGG-3′; Lin28b F: 5′-CAACATGGCCGAAGGCGGGG-3′, R: R 5′-CCCATGCGCACGTTGAACCA-3′; β-actin F: 5′-CCACACCCGCCACCAGTTCG-3′, R: 5′-TACAGCCCGGGGAGCATCGT-3′). A total volume of 20 μL was used, where each reaction contained 4 – 6.5 μL of reverse-transcribed sample or standard, 10 μL of SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), 600 nM of forward and reverse Lin28a or Lin28b primers, or 50 nM of forward and reverse β-actin primers. The real-time polymerase chain reaction consisted of a 10 minute initial denaturation step at 95°C, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 1 minute. A final dissociation curve analysis was also performed (95°C for 1 minute, 60°C for 30 seconds, and 95°C for 30 seconds) to ensure a single product was amplified.
2.5 qRT-PCR for Let-7a and Let-7g
Five (5) nanograms of total RNA were reverse transcribed from 15-, 20-, 25- and 30-day-old hypothalamic and pituitary tissues and 20-, 25-, and 30-day-old ovarian tissue using the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosytems), with specific reverse transcription (RT) primers designed to reverse transcribe Let-7a (Assay ID: 000377, Applied Biosystems), Let-7g (Assay ID: 002282, Applied Biosystems), and the housekeeping gene snoRNA142 (Assay ID: 001231, Applied Biosytems). For standard curves, 10 ng of RNA from 30-day-old ovarian tissue was reverse transcribed using Let-7a, Let-7g, and snoRNA142 RT primers, and was serially diluted from 1:10 to 1:40.
After reverse transcription, quantification of mature Let-7a or Let-7g was performed using the standard curve method [Chen et al., 2005]. A total volume of 20 μL was used, where each reaction contained 1.33 μL of reverse-transcribed sample or standard, 10 μL of TaqMan Universal PCR Master Mix no AmpErase® UNG (Applied Biosystems), and 1 μL of either 20x Let-7a (Assay ID: 000377, Applied Biosystems), Let-7g (Assay ID: 002282, Applied Biosystems), or housekeeping gene snoRNA142 (Assay ID: 001231, Applied Biosytems) TaqMan® MicroRNA Assay (Applied Biosytems). The real-time polymerase chain reaction consisted of an initial 10 minute denaturation step at 95°C, followed by 40 cycles of alternating denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 1 minute.
2.6 RT-PCR
A total of 50–80 ng of cDNA were loaded with 0.625 units of Taq polymerase (Fermentas, Burlington, ON, CAN), 0.4 μM of forward and reverse primers for Lin28a and Lin28b or 0.1 μMM of forward and reverse primers for β-actin, 0.4 mM of dNTP mix, 2 mM of MgCl2, and 10x Taq buffer (with (NH4)2S04 ) (Fermentas). The polymerase chain reaction consisted of an initial denaturation step at 95°C for 2 minutes, 40 cycles of denaturation (95°C for 1 minute), annealing (55°C for 1 minute) and extension (72°C for 90 seconds), followed by a final step at 72°C for 5 minutes.
The primers used to amplify targets are identical to those used for qRT-PCR analysis (see above). Amplified products were separated via polyacrylamide gel electrophoresis (100 mL 6 % acrylamide gel mixture, 100 μL TEMED, and 1000 μL 10 % APS) and visualized by ethidium bromide staining (1 mg/mL) followed by ultraviolet illumination.
2.7 Immunohistochemistry (IHC)
Immunohistochemical staining was performed by the Centre for Modeling Human Disease (CMHD) at the Toronto Centre for Phenogenomics. Tissues were incubated with polyclonal antibodies specifically designed to recognize murine Lin28a protein (1:100, Cell Signaling Technology, Beverly, MA, USA; Catalogue Number 3978), for 24 hours at 4°C, followed by incubation with secondary goat anti-rabbit biotinylated antibody (1:200, Vector labs, Burlingame, CA, USA, catalogue number: BA-1000) for 30 minutes at room temperature. Western blotting was used to verify the specificity of the polyclonal antibodies, where only one protein was recognized at approximately 26 kDa, corresponding to Lin28a. Tissues were then incubated with ABC reagent (1:50, Vector Labs, catalogue number PK-6100) and visualized with diaminobenzidine reagent. Samples were counterstained with haematoxylin. Two biological replicates at each time point from each tissue were assessed.
2.8 Statistical analysis
For qRT-PCR, all time points were compared with each other within each tissue, and differences were analyzed by one-way Analysis of Variance (ANOVA), followed by the Bonferroni Multiple Comparison Test. Data are expressed as mean ± standard error of the mean (SEM).
3. RESULTS
3.1 Expression of Lin28a and Lin28b in hypothalamic, pituitary, and ovarian tissue
Both Lin28a and Lin28b are expressed in hypothalamic, pituitary, and ovarian tissue (Figure 1A). However, the relative expression of these two genes differed among the tissues. Expression of Lin28b within hypothalamic tissue was approximately 30 times higher than Lin28a (Figure 1B), while expression of Lin28b within pituitary tissue was approximately 2 times higher than Lin28a. Conversely, in ovarian tissue, levels of Lin28a expression were approximately 60 times higher than Lin28b, which is in agreement with data from a recent study of expression within human ovarian tissue (El-Khairi et al., 2012).
Figure 1. Expression of Lin28a and Lin28b in hypothalamic, pituitary, and ovarian tissue.
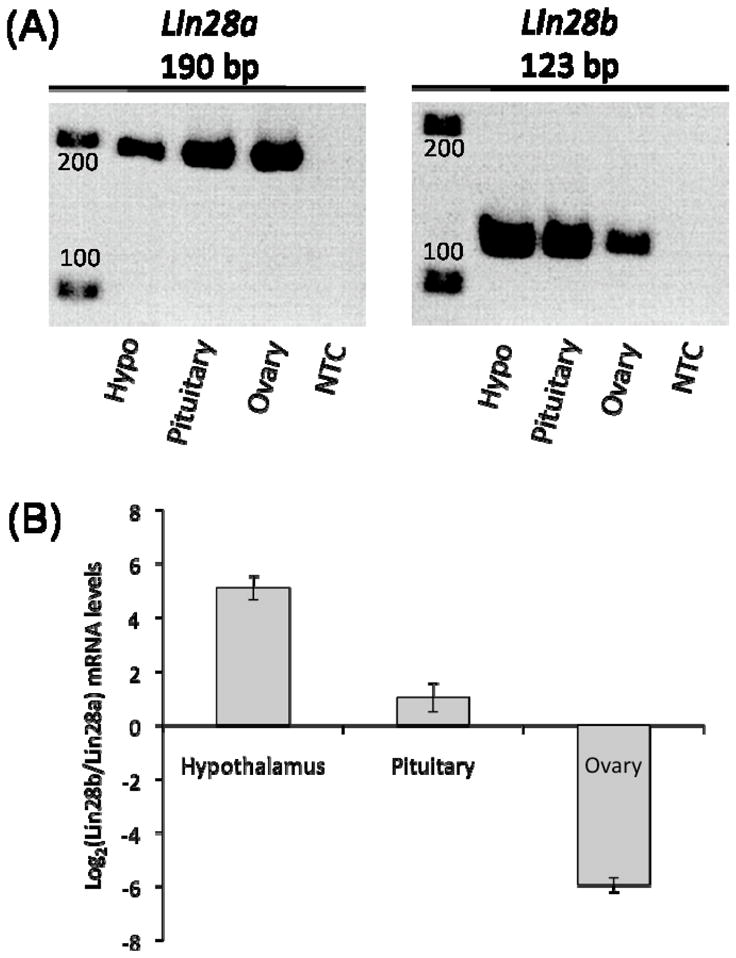
(A) RT-PCR of Lin28a (190 bp) and Lin28b (123 bp) mRNA in 25-day-old hypothalamic (hypo), pituitary, and ovarian tissue from C57BL/6 female mice. NTC = no template control. (B) Relative expression levels of Lin28a and Lin28b in different tissues at 25 days as determined by qRT-PCR, normalized to β-actin mRNA.
3.2 Expression of Lin28a and Lin28b across the pubertal transition in hypothalamic, pituitary, and ovarian tissue
Because increased expression of Lin28a has been shown to cause delayed puberty in mice [Zhu et al., 2010], it was hypothesized that a decrease in expression of Lin28b and/or Lin28a might facilitate the transition into puberty. Demonstrating a diminution in expression prior to the pubertal transition would support this hypothesis. Thus, levels of Lin28a and Lin28b expression in components of the HPG axis in female C57BL/6 mice were examined using qRT-PCR prior to the onset of puberty (at post natal days 15, 20, 25, 30) and postpubertally (between ages 35–45 days); the average age of pubertal onset (as assessed by vaginal opening) in our colony is 31.5 ± 2.4 days.
3.2.1 A decrease in Lin28a mRNA was observed in hypothalamic tissue prior to the pubertal transition
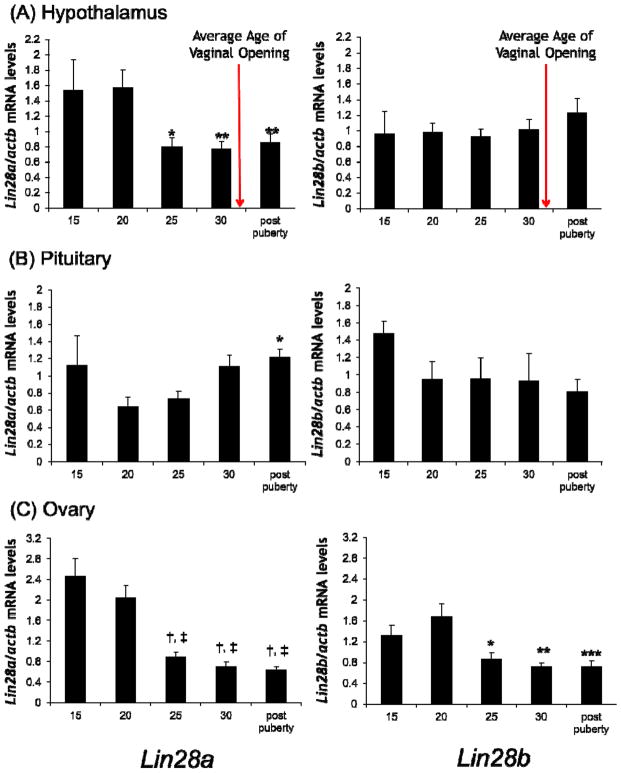
In hypothalamic tissue, a statistically significant decrease in Lin28a expression of 49%, 51%, and 46% was observed at 25 days, 30 days, and postpubertally, respectively, when compared to 20-day-old prepubertal tissue (Figure 2A). No change in Lin28b expression was noted.
Figure 2. Lin28a and Lin28b mRNA expression levels in hypothalamic, pituitary, and ovarian tissue across the pubertal transition.
qRT-PCR for Lin28a and Lin28b mRNA in tissues from prepubertal 15, 20, 25, 30 day old and postpubertal (35–45 days old) C57BL/6 female mice, normalized to β-actin in hypothalamus (A), pituitary (B), and ovary (C). Average age of vaginal opening in the mouse colony is indicated by the arrow (31.5 ± 2.4 days). Values are displayed as means ± SEM (n=5–21 mice for each time point). For Lin28a in hypothalamic tissue, *=p<0.05, and **=p<0.01, when compared to 20-day-old tissue. For Lin28a in pituitary tissue, *=p<0.05, when compared to 20-day-old tissue. For Lin28a in ovarian tissue, †=p<0.001, and ‡=p<0.001, when compared to 15 and 20-day-old tissue, respectively. For Lin28b in ovarian tissue, *=p<0.05, **=p<0.01, and ***=p<0.001, when compared to 20 day old tissue.
3.2.2 Both Lin28a and Lin28b mRNA levels decrease across the pubertal transition within ovarian tissue
Statistically significant decreases in both Lin28a and Lin28b expression were observed in ovarian tissue prior to the onset of puberty (Figure 2C). Specifically, 25-day-old, 30-day-old, and postpubertal mice had 64%, 72%, and 74% lower levels of Lin28a expression when compared to 15-day-old mice, and 56%, 66%, 69%, lower levels of Lin28a expression when compared to 20-day-old mice. Similarly, 25-day-old, 30-day-old, and postpubertal mice had 48%, 57%, and 57% lower levels of Lin28b, when compared to 20-day-old mice (p<0.05 for all).
3.2.3 Neither Lin28a nor Lin28b expression decreases across the pubertal transition within pituitary tissue
No statistically significant changes in mRNA levels were noted in the pituitary prior to the onset of puberty (Figure 2B), but there is a trend towards decreased levels of Lin28a and Lin28b mRNA between 15 and and 20 days. After 20 days, Lin28a mRNA levels tended to increase, with a statistically significant increase being seen between 20 days and the postpuberty timepoint (p<0.05). This increase postdates the onset of onset of puberty in our colony, and was not observed for Lin28b.
3.3 Protein levels within the ovary also decrease prior to the onset of puberty
Using immunohistochemistry (IHC), Lin28a protein levels were examined in hypothalamic, pituitary, and ovarian tissue (an antibody suitable for IHC is currently not available for Lin28b); time points were chosen according to the ages where decreases in expression were noted.
In the ovary, an overall decrease in Lin28a protein levels was observed from 20-day-old to 30-day-old tissue (see Figure 3), which parallels the observed decreased mRNA levels measured by qRT-PCR. Unlike the ovary, only diffuse low level IHC-staining, with no specific pattern nor apparent change over time, was observed in the pituitary and hypothalamic tissues (data not shown), indicating that only low levels of Lin28a protein are present in these tissues at these ages.
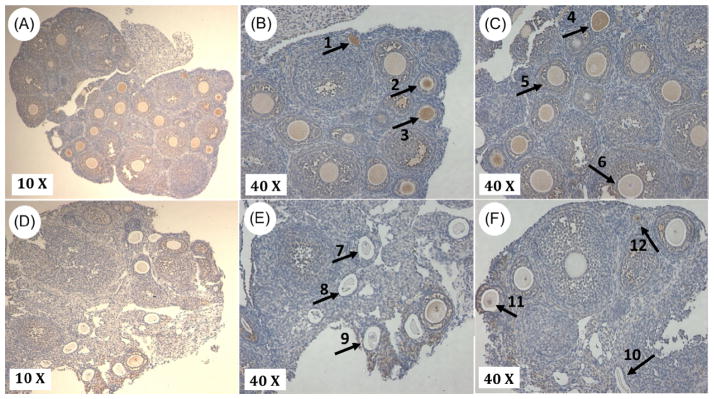
Figure 3. Lin28a protein levels decrease prior to the onset of puberty in ovarian tissue.
Immunohistochemical staining of Lin28A protein in 20 day (A–C) and 30 day (D–F) ovarian tissue. Lin28a protein expression is identified by diaminobenzidine staining. In 20-day-old tissue, Lin28a is detected in early developing follicles (arrow 1), early secondary follicles (arrows 2, 3, and 4), late secondary follicles and granulosa cells surrounding secondary follicles (arrow 5). In 30-day-old tissue, Lin28a protein is less evident, due to the abundance of atretic follicles (arrows 7, 8, 9 and 10). Lin28a still persists in the granulosa cells and oocytes of secondary follicles (arrow 11), and in early follicles (arrow 12). This pattern of Lin28a protein distribution was consistent in the two biological replicates that were examined
In 20-day-old tissue (Figure 3A, B and C), Lin28a protein was visible within oocytes of early developing follicles, and levels of Lin28a protein appear to decrease as follicles develop. In 30-day-old tissue, an overall reduction Lin28a protein level was observed when compared to 20-day-old tissue, and this was associated with an increase in more developed and atretic follicles.
3.4 Let-7a and Let-7g miRNA levels do not change in hypothalamic, pituitary, and ovarian tissues prior to the onset of puberty
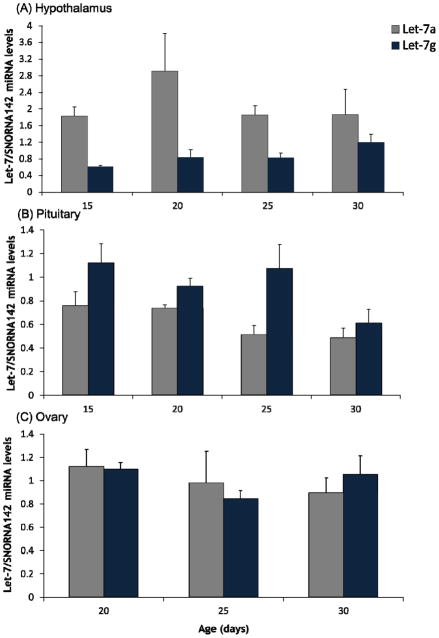
To investigate whether the expression of Let-7 miRNA changed across the time course, the miRNA expression levels of Let-7a and Let-7g were quantified using qRT-PCR. As with IHC studies, ages for investigation where chosen based on the timing of observed decreases in Lin28a and/or Lin28b expression. Hypothalamus was assessed at days 15, 20, 25, and 30, and ovary at days 20, 25, and 30 (Figure 4); to be complete, pituitary was also assessed at days 15, 20, 25, and 30 even though changes in Lin28a and Lin28b expression were not observed across the pubertal transition within the pituitary. Despite the observed decreases in Lin28b expression within the hypothalamus, and Lin28a and Lin28b mRNA within the ovary, there were no statistically significant changes observed in Let-7a and Let-7g miRNA levels between the younger versus older mice. No changes in Let-7a and Let-7g expression levels were noted in the pituitary either.
Figure 4. Expression of Let-7a and Let-7g miRNA in hypothalamic, pituitary, and ovarian tissue.
qRT-PCR for Let-7a and Let-7g miRNA in 15, 20, 25, and 30-day-old hypothalamic (A) and pituitary (B) tissue, and 20, 25, and 30-day-old ovarian (C) tissue, normalized to snoRNA142. Values are displayed as means ± SEM (n=3–7 for each time point).
4. DISCUSSION
Transgenic female mice that over-express Lin28a have delayed puberty [Zhu et al., 2010]. This finding is consistent with the earlier data from Moss et al. (1997) showing that in C. elegans, over-expression of lin28, an orthologue of Lin28a, through gain-of-function mutations, results in delayed development while lack of lin28 expression, through loss-of-function mutations, results in precocious development. The decreased Lin28a and Lin28b mRNA levels we observed prior to the onset of puberty in the ovary are consistent with these previous studies. We acknowledge that our data do not demonstrate cause and effect, and it remains possible that the decreases in expression that we noted are not related to the transition into puberty but simply represent part of a developmentally programmed diminution in Lin28a and Lin28b expression with age. However, in total, data from the transgenic mice, C. elegans, and the current study are all consistent and all support the working hypothesis that lowered levels of Lin28a and Lin28b expression may facilitate developmental transitions, while increased levels are associated with delayed development.
Within hypothalamic tissue, a significant decrease in Lin28a expression was observed at 25 days, 30 days, and postpubertally, when compared to 20-day-old female mice, while there was no significant decrease in Lin28b expression. Although these findings are consistent with the findings of Zhu et al. (2010) and suggest that lower levels of Lin28a expression may facilitate the transition into puberty, they also raise several questions that will warrant further investigation: what is the biological significance of steady as opposed to decreasing levels of Lin28b expression at these ages? Although IHC may not be quantitative enough to detect subtle changes in protein levels, does the lack of change in IHC staining for Lin28a in the hypothalamus negate the importance of the changes in gene expression? Moreover, it remains possible that decreases in Lin28a and/or Lin28b expression and protein levels will be observed in different strains of mice, at different time points (including in utero), or within particular nuclei or cell types within the hypothalamus and/or pituitary.
Our data do provide important new information and strongly suggest that changes of Lin28a/Lin28b expression in the ovary may play an important role in modulating maturation of the HPG axis. The most convincing and consistent changes in expression, for both Lin28a and Lin28b, prior to the onset of puberty were observed in the ovary. These data indicate that the ovary may be an important site where the Lin28 pathway exerts effects that modulate the timing of puberty. Although traditionally much attention has been focused on hypothalamic factors that modulate pubertal timing [Ojeda et al., 2006] and it is well accepted that the hypothalamus is the central site of regulation of the onset of puberty, it is also clear that gonadal products are also critical components of the network that modulates the onset of puberty [Mayer et al., 2010]. The hypothalamus receives modulating signals from the periphery (such as nutritional status and other inputs) and responds to them via a complex gene network/hierarchy that factors these inputs in when controlling the activation of GnRH neurons and onset of puberty [Ojeda et al., 2010]. Thus, it is possible that the Lin28a/Lin28b pathway influences the timing of puberty, at least in part, through indirect effects at the level of the ovary or perhaps even at other tissues in the periphery. For example, two groups have recently demonstrated that altering Lin28b expression in peripheral tissues, such as muscle, alters glucose metabolism and affects body size [Frost and Olson, 2012; Zhu et al., 2011, Zhu et al., 2010].
Importantly, the changes we observed in Lin28a gene expression were correlated with a decrease in protein levels in 30-day-old compared to 20-day-old ovaries. Moreover, Lin28a protein levels appear to be higher within early developing follicles (which are more prominent at 20 days) and lower as the follicles become more developed and/or atretic (which is more prominent in the 30-day-old samples), suggesting that the level of Lin28a protein is dependent on the stage of follicular development. Demonstrating that the alterations in expression that we observed modulate pubertal onset will require future experimentation. For example, one could envision using transgenic methodologies to modulate Lin28a and/or Lin28b expression in a tissue or even age-dependent manner and investigating whether changes of expression in the ovary (or other components of the HPG axis) alter pubertal timing. Such studies would also allow for investigation of whether the Lin28a/Lin28b pathway modulates aspects of reproductive endocrine function other than pubertal timing, such as follicle development, ovulation, and fertility.
Lin28 is known to repress generation of mature Let-7 miRNAs [Hagan et al., 2009; Heo et al., 2008; Piskounova et al., 2008; Viswanathan et al., 2008]. Thus, it is currently thought that the Lin28 pathway modulates pubertal onset by affecting levels of Let-7 miRNAs and, subsequently, affecting levels of expression of Let-7 targets. Based on this reasoning, it would follow that increased expression of Lin28a/Lin28b would lead to decreased Let-7 miRNA levels, which would then lead to increased expression of the Let-7 target genes. If these target genes usually restrain pubertal onset [Ojeda et al., 2010; Viswanathan and Daley, 2010; Zhu et al., 2010], this cascade of events would result in later pubertal onset. Indeed, this reasoning fits with the observation that increased Lin28a expression leads to later puberty [Zhu et al., 2010] and our finding of decreased Lin28a and Lin28b expression prior to the onset of puberty.
However, other data from the transgenic mice that overexpress Lin28a and the Let-7 data from the present studies raise the possibility that at least some of the Lin28a/Lin28b pathway effects in the HPG axis may be Let-7 independent. The transgenic mice displayed increased levels of Lin28a in the hypothalamus and ovary, but the expected decrease in Let-7a and Let-7g miRNA levels was not observed [Zhu et al, 2010]. Our current data show that, although Lin28a expression decreased in the hypothalamus and Lin28a and Lin28b expression decreased in the ovary, neither Let-7a nor Let-7g miRNA levels increased significantly across the time course. It is possible that subtle changes occurred that we did not detect or that changes in other Let-7 family members occurred that were not assessed by our assays. This last possibility seems unlikely, however, because Let-7a and Let-7g are prominent members of the Let-7 family, and it has been demonstrated that changes in these two miRNAs are reflective of changes within the whole family [Newman et al., 2008; Zhu et al., 2011].
That modulation of the timing of puberty in female mice may be Let-7 independent has important implications. Our data suggest that Lin28a and Lin28b’s effects may be mediated, at least in part, through a different miRNA pathway within the HPG axis [Balzer et al., 2010] or that Lin28a and Lin28b effects within the HPG axis may be miRNA independent. Indeed, a recent publication about regulation of metabolic pathways by the Lin28 system has demonstrated that the Lin28 pathway can affect phenotypes through both Let-7 dependent and independent actions [Zhu et al., 2011]. It is clear that investigation of the downstream mediators of Lin28a and Lin28b’s effects within the HPG axis will be an important area for further study. Determining whether modulation of pubertal timing is Let-7 dependent will require model systems, such as transgenic animals, where Let-7 levels can be increased or decreased and the effect on pubertal timing assessed.
An unexpected but interesting finding is that Lin28a, but not Lin28b, expression increased after puberty in the pituitary. The observed increase in Lin28a expression may indicate a role for Lin28a in adult neuroendocrine function and suggests that the role of the Lin28 pathway within the pituitary is an important area for further investigation.
In summary, we report what we believe are the first published data examining peripubertal expression of Lin28a, Lin28b, and Let-7 family members in the HPG axis. Consistent with the delayed developmental phenotype observed in C. elegans mutants over-expressing lin28 [Moss et al., 1997; Yang and Moss, 2003] and the delayed puberty observed in transgenic mice that over-express Lin28a [Zhu et al., 2010], our current findings demonstrate that Lin28a and Lin28b expression decrease across the pubertal transition. Moreover, our data raise the possibility that Lin28a and Lin28b expression in the ovary may be an important regulator of the HPG axis, affecting the timing of puberty and/or follicular development. Although this pathway clearly warrants further investigation in the hypothalamus and pituitary, our data point to the importance of gaining further understanding of how the Lin28 pathway modulates ovarian function and how that modulation may then regulate maturation of the HPG axis.
Highlights.
Expression of Lin28a and Lin28b decreases in the ovary prior to pubertal onset
Expression of Lin28a decreases in hypothalamus prior to the onset puberty
A decrease Lin28a protein levels mirrored the decline in Lin28a in ovarian tissue
Lin28 regulation of pubertal onset may occur at the ovarian and hypothalamic level
The effects of the Lin28 pathway may be mediated, in part, independently of Let-7
Acknowledgments
This work was supported by The Hospital for Sick Children and NIH grant (R01HD048960) to M.R.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ambros V, Horvitz HR. Heterochronic mutants of the nematode caenorhabditis elegans. Science. 1984;226(4673):409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 2.Baker H, Joh TH, Ruggiero DA, Reis DJ. Variations in number of dopamine neurons and tyrosine hydroxylase activity in hypothalamus of two mouse strains. J Neurosci. 1983;3(4):832–843. doi: 10.1523/JNEUROSCI.03-04-00832.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137(6):891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- 4.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbism M, Xu NL, Mahuvakar VR, Anderson MR, Lao KQ, Livak KJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Khairi R, Parnaik R, Duncan AJ, Lin L, Gerrelli D, Dattani MT, Conway GS, Achermann JC. Analysis of LIN28A in early human ovary development and as a candidate gene for primary ovarian insufficiency. Mol Cell Endocrinol. 2012;351(2):264–268. doi: 10.1016/j.mce.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, He C, Lunetta KL, Visser JA, Byrne EM, Cousminer DL, Gudbjartsson DF, Esko T, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42(12):1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gajdos ZK, Henderson KD, Hirschhorn JN, Palmert MR. Genetic determinants of pubertal timing in the general population. Mol Cell Endocrinol. 2010;324(1–2):21–29. doi: 10.1016/j.mce.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16(10):1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41(6):724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of Let-7 precursor MicroRNA. Mol Cell. 2008;32(2):276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Mayer C, Acosta-Martinez M, Dubois SL, Wolfe A, Radovick S, Boehm U, Levine JE. Timing and completion of puberty in female mice depend on estrogen receptor alpha-signaling in kisspeptin neurons. Proc Natl Acad Sci U S A. 2010;107(52):22693–22698. doi: 10.1073/pnas.1012406108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88(5):637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 13.Nathan BM, Hodges CA, Supelak PJ, Burrage LC, Nadeau JH, Palmert MR. A quantitative trait locus on chromosome 6 regulates the onset of puberty in mice. Endocrinology. 2006;147(11):5132–5138. doi: 10.1210/en.2006-0745. [DOI] [PubMed] [Google Scholar]
- 14.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14(8):1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ojeda SR, Dubay C, Lomniczi A, Kaidar G, Matagne V, Sandau US, Dissen GA. Gene networks and the neuroendocrine regulation of puberty. Mol Cell Endocrinol. 2010;324(1–2):3–11. doi: 10.1016/j.mce.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ojeda SR, Lomniczi A, Mastronardi C, Heger S, Roth C, Parent AS, Matagne V, Mungenast AE. Minireview: The neuroendocrine regulation of puberty: Is the time ripe for a systems biology approach? Endocrinology. 2006;147(3):1166–1174. doi: 10.1210/en.2005-1136. [DOI] [PubMed] [Google Scholar]
- 17.Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, Gillson CJ, Glaser B, et al. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009;41(6):729–733. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, Cherkas L, Eiriksdottir G, et al. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009;41(6):648–650. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem. 2008;283(31):21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 20.Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Alexandersen P, Feenstra B, Boyd HA, Aben KK, Verbeek AL, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet. 2009;41(6):734–738. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 21.Towne B, Czerwinski SA, Demerath EW, Blangero J, Roche AF, Siervogel RM. Heritability of age at menarche in girls from the fels longitudinal study. Am J Phys Anthropol. 2005;128(1):210–219. doi: 10.1002/ajpa.20106. [DOI] [PubMed] [Google Scholar]
- 22.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140(4):445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang DH, Moss EG. Temporally regulated expression of lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns. 2003;3(6):719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Grasemann C, Rinn JL, Lopez MF, Hirschhorn JN, Palmert MR, Daley GQ. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42(7):626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]