Abstract
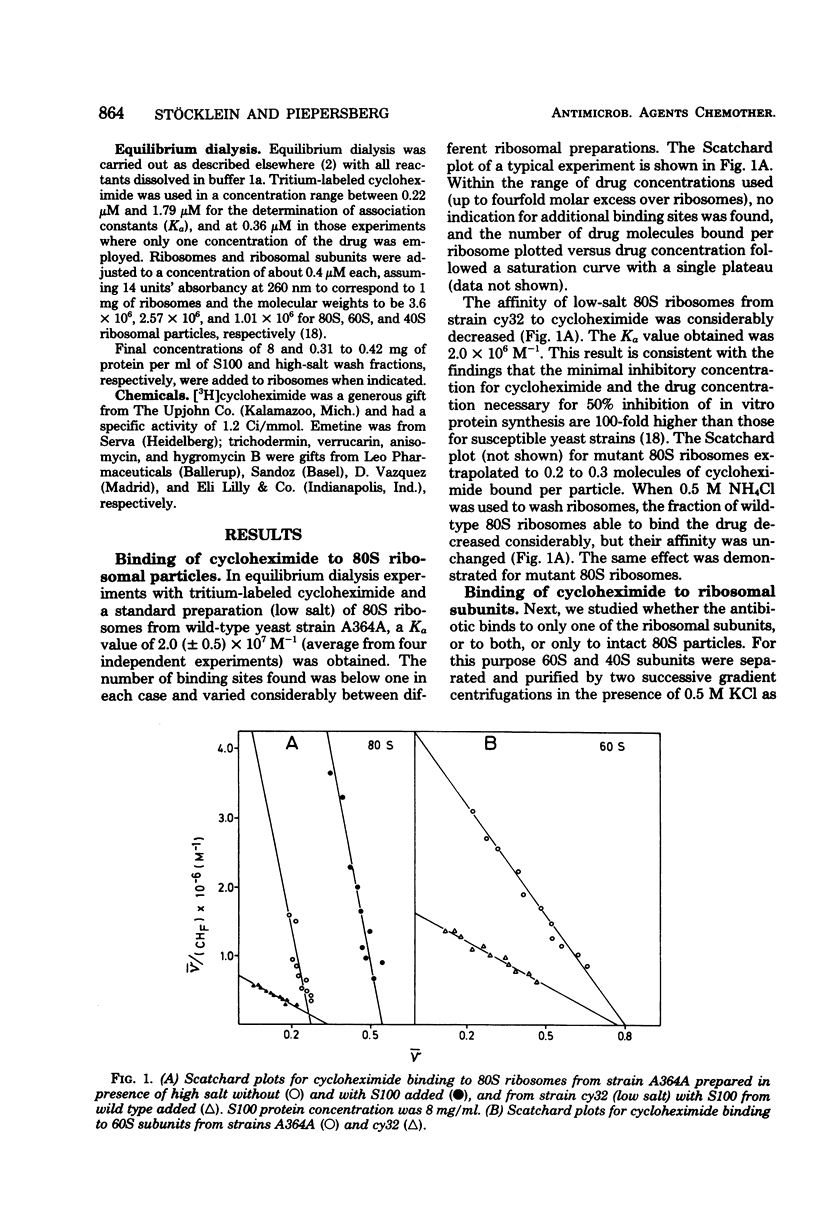
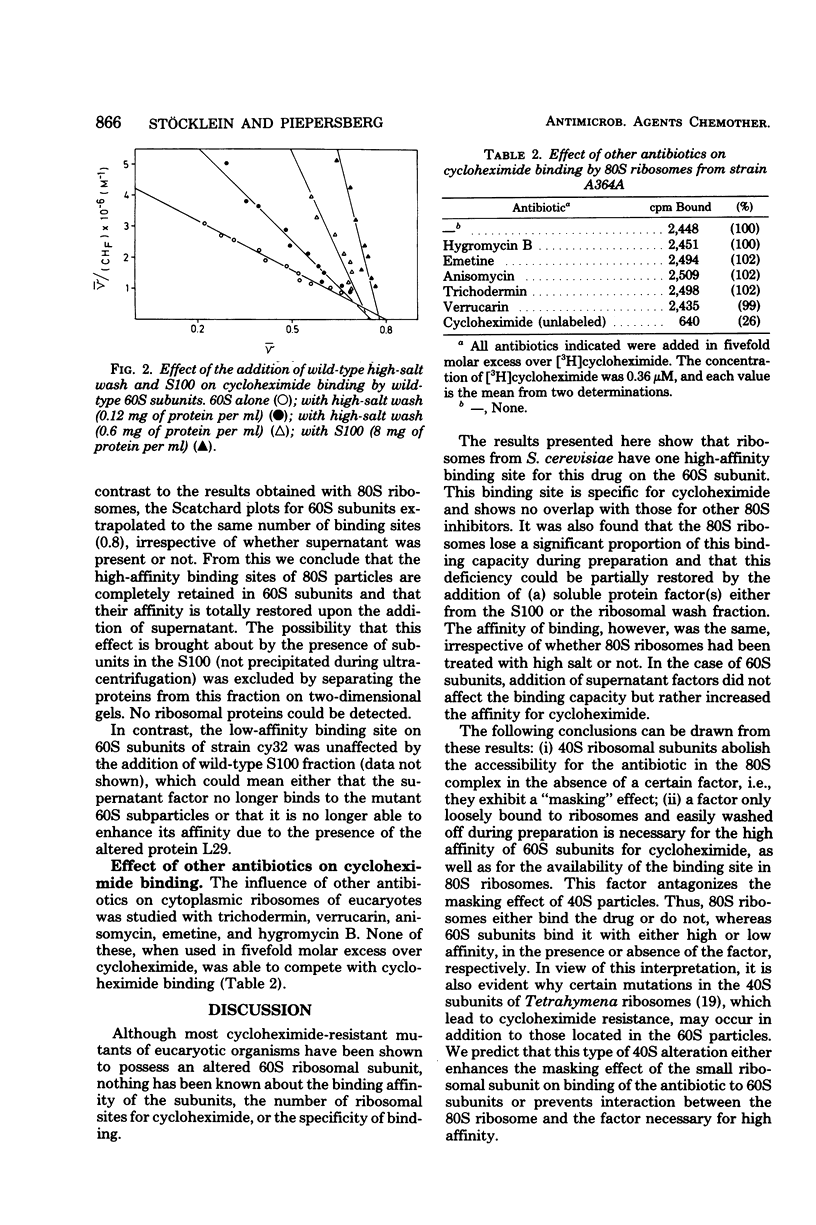
Cycloheximide bound to cytoplasmic (80S) ribosomes of the yeast Saccharomyces cerevisiae with an association constant (Ka) of 2.0 (+/- 0.5) x 10(7) M-1. The number of binding sites found per ribosome was between 0.4 and 0.6; it was reduced by high-salt treatment of ribosomes 60S particles prepared in the presence of high salt had a lower affinity (Ka: 5.5 [+/- 0.5] x 10(6) M-1) than did 80S ribosomes, but a greater proportion of particles (0.8) were able to bind. No specific binding to 40S subunits was observed. The addition of supernatant fractions (S100, high-salt wash fraction) increased the number of binding sites found per 80S ribosome up to 0.8, leaving the association constant unchanged. In contrast, the affinity of 60S subunits was enhanced to a Ka value of 3.5 x 10(-7) M-1 by the addition of supernatant fractions, whereas the number of binding sites stayed constant. A model to explain these facts is proposed. 80S ribosomes, as well as 60S subunits of strain cy32, which is highly resistant to cycloheximide and altered in ribosomal protein L29 (18), showed a drastically reduced affinity for the drug (Ka values of 2.0 x 10(6) M-1).
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Begueret J., Perrot M., Crouzet M. Ribosomal proteins in the fungus Podospora anserina: evidence for an electrophoretically altered 60S protein in a cycloheximide resistant mutant. Mol Gen Genet. 1977 Nov 14;156(2):141–144. doi: 10.1007/BF00283486. [DOI] [PubMed] [Google Scholar]
- Böck A., Petzet A., Piepersberg W. Ribosomal ambiguity (ram) mutations facilitate diyhydrostreptomycin binding to ribosomes. FEBS Lett. 1979 Aug 15;104(2):317–321. doi: 10.1016/0014-5793(79)80842-x. [DOI] [PubMed] [Google Scholar]
- Coddington A., Fluri R. Characterisation of the ribosomal proteins from Schizosaccharomyces pombe by two-dimensional polyacrylamide gel electrophoresis: demonstration that a cycloheximide resistant strain, cyh1, has an altered 60S ribosomal protein. Mol Gen Genet. 1977 Dec 14;158(1):93–100. doi: 10.1007/BF00455123. [DOI] [PubMed] [Google Scholar]
- Cooper D., Banthorpe D. V., Wilkie D. Modified ribosomes conferring resistance to cycloheximide in mutants of Saccharomyces cerevisiae. J Mol Biol. 1967 Jun 14;26(2):347–350. doi: 10.1016/0022-2836(67)90302-6. [DOI] [PubMed] [Google Scholar]
- Cooper T. G., Bossinger J. Selective inhibition of protein synthesis initiation in Saccharomyces cerevisiae by low concentrations of cycloheximide. J Biol Chem. 1976 Nov 25;251(22):7278–7280. [PubMed] [Google Scholar]
- Haugli F. B., Dove W. F., Jimenez A. Genetics and biochemistry of cycloheximide resistance in Physarum polycephalum. Mol Gen Genet. 1972;118(2):97–107. doi: 10.1007/BF00267081. [DOI] [PubMed] [Google Scholar]
- Mazelis A. G., Petermann M. L. Physical-chemical properties of stable yeast ribosomes and ribosomal subunits. Biochim Biophys Acta. 1973 Jun 8;312(1):111–121. doi: 10.1016/0005-2787(73)90056-7. [DOI] [PubMed] [Google Scholar]
- Oleinick N. L. Initiation and elongation of protein synthesis in growing cells: differential inhibition by cycloheximide and emetine. Arch Biochem Biophys. 1977 Jul;182(1):171–180. doi: 10.1016/0003-9861(77)90296-x. [DOI] [PubMed] [Google Scholar]
- Pongratz M., Klingmüller W. Role of ribosomes in cycloheximide resistance of Neurospora mutants. Mol Gen Genet. 1973 Aug 28;124(4):359–363. doi: 10.1007/BF00267664. [DOI] [PubMed] [Google Scholar]
- Pöche H., Junghahn I., Geissler E., Bielka H. Cycloheximide resistance in Chinese hamster cells. III. Characterization of cell-free protein synthesis by polysomes. Mol Gen Genet. 1975;138(2):173–177. [PubMed] [Google Scholar]
- Pöche H., Zakrzewski S., Nierhaus K. H. Resistance against cycloheximide in cell lines from Chinese hamster and human cells is conferred by the large subunit of cytoplasmic ribosomes. Mol Gen Genet. 1979 Sep;175(2):181–185. doi: 10.1007/BF00425534. [DOI] [PubMed] [Google Scholar]
- Rao S. S., Grollman A. P. Cycloheximide resistance in yeast: a property of the 60s ribosomal subunit. Biochem Biophys Res Commun. 1967 Dec 15;29(5):696–704. doi: 10.1016/0006-291x(67)90273-2. [DOI] [PubMed] [Google Scholar]
- Skogerson L., Wakatama E. A ribosome-dependent GTPase from yeast distinct from elongation factor 2. Proc Natl Acad Sci U S A. 1976 Jan;73(1):73–76. doi: 10.1073/pnas.73.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasundaran U., Skogerson L. A cycloheximide sensitivity factor from yeast required for N-acetylphenylalanylpuromycin formation. Biochemistry. 1976 Nov 2;15(22):4760–4764. doi: 10.1021/bi00667a002. [DOI] [PubMed] [Google Scholar]
- Sutton C. A., Ares M., Jr, Hallberg R. L. Cycloheximide resistance can be mediated through either ribosomal subunit. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3158–3162. doi: 10.1073/pnas.75.7.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]



