Abstract
Many changes must occur to the RNA polymerase II (pol II) transcription complex as it makes the transition from initiation into transcript elongation. During this intermediate phase of transcription, contact with initiation factors is lost and stable association with the nascent transcript is established. These changes collectively comprise promoter clearance. Once the transcript elongation complex has reached a point where its properties are indistinguishable from those of complexes with much longer transcripts, promoter clearance is complete. The clearance process for pol II consists of a number of steps and it extends for a surprisingly long distance downstream of transcription start.
Keywords: Transcript initiation, Abortive initiation, Transcript elongation, TFIIB, TFIIH
1. Introduction and terminology
There is not a well-established terminology to describe the conversion of pol II from an initiation complex to a fully committed transcript elongation complex. The terms “promoter clearance” and “promoter escape” have both been used. I will refer to the initial stage of transcription as promoter clearance in this review. Designating this process as promoter escape could imply that the transcription machinery has completely disengaged from the promoter region. As will be discussed in the last section, recent work on the three dimensional organization of actively transcribed chromatin suggests that pol II may retain promoter contacts during transcript elongation. Thus, promoter clearance seems the more appropriate designation.
Promoter clearance has been most extensively studied with mammalian pol II and general transcription factors, using templates containing a TATA box. The following discussion will focus primarily on results with these systems. In the interests of brevity, the details of the transcription systems will not be described but it should be noted that the transcription complexes were assembled in a variety of ways, with extracts or with purified or recombinant factors. Structural studies of pol II and early transcription complexes have used yeast RNA polymerase and general factors. Relevant findings from these studies will also be described. However, there are fundamental differences between the initial stages of transcription for metazoans and budding yeast, as will be discussed below.
2. The preinitiation complex and the initiation of transcription
The preinitation complex (PIC) is the molecular assembly through which pol II can locate and utilize a promoter. On double-stranded templates bearing a TATA box, pol II PIC formation requires, at minimum, five general transcription factors: the TATA box binding protein TBP, TFIIB, TFIIF, TFIIE and TFIIH [1–3]. In the complete PIC, pol II is encompassed by many interactions. The B-finger/reader segment of TFIIB occupies a channel within pol II that will ultimately provide the exit path of the nascent RNA. TFIIB also contacts other portions of pol II as well as the upstream promoter DNA [4,5]. Portions of TFIIH interact with DNA downstream of transcription start [6,7]. TFIIF and TFIIE bind to polymerase on either side of the central cleft in which the template will ultimately reside [8,9]. All of these interactions must presumably be lost through the clearance process. The release of the general transcription factors may serve to reveal interaction surfaces on pol II which can be utilized by other components during transcript elongation (see, for example, [8,10]).
Once assembly of the PIC is complete, pol II cannot initiate transcription until the template strands are separated. Uniquely among the DNA-dependent RNA polymerases, pol II does not drive template opening itself. Open complex formation requires ATP and the XPB subunit of TFIIH [11,12]- see also [13]. Unwinding is not driven by an intercalation mechanism; instead, TFIIH apparently acts by rotating downstream DNA while retaining a fixed upstream contact within the complex [6]. The initial transcription bubble extends only from -9 to -2 relative to transcription start (Fig. 1A); the unpaired region does not expand to encompass the start site until NTPs are added to support formation of the first bond in the RNA [14].
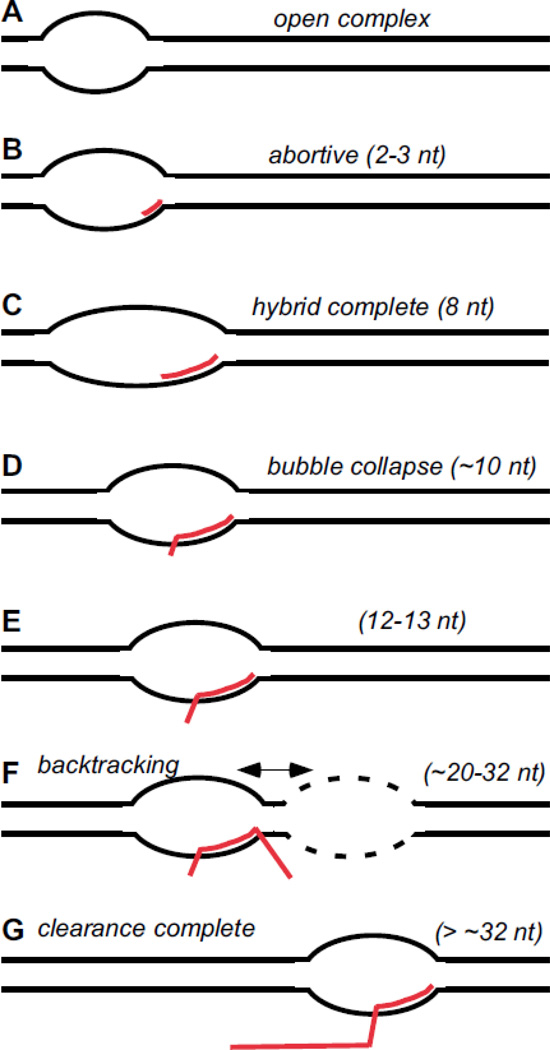
Fig. 1. Important stages in the progress of pol II to promoter clearance.
The downstream progress of the pol II transcription bubble from the initial open complex to the completely cleared elongation complex is shown schematically. RNA is in red and the transcript length for that stage is indicated. This diagram is based on results obtained in mammalian systems with TATA box promoters. Only a very limited number of different promoters and initially transcribed regions have been investigated. (A) The location of the upstream edge of the bubble is determined by distance from the TATA element [15,16]; the initiation site remains double-stranded [14]. (B) Initiation is abortive and transcription can proceed for many rounds if RNA synthesis is limited to 2–3 nt [14,21,22]. (C) The RNA-DNA hybrid reaches its mature length and the structure of the hybrid is normal at this point [35,36]. The upstream edge of the bubble remains in its initial location as the leading edge advances [14,16]. Loss of transcripts through the abortive pathway is considerably reduced [14,27]. Transcript slippage within repeated template segments falls off abruptly just downstream of this point [34]. Nascent RNA should begin to clash with the finger/reader segment of TFIIB just upstream of this point [44]. (D) At this point, roughly the upstream half of bubble closes abruptly as the unpaired region extends to ~18 bases [14,16]. For a promoter with the canonical spacing of the TATA box and transcription start, this occurs at a transcript length of 9–10 nt [16]. TFIIH is no longer required for effective elongation [16]. Loss of an upstream contact of TFIIB with the template has been proposed to occur here [5]. (E) TFIIB is destabilized within the advancing complex [22]. (F) As elongation continues, the 5’ end of the nascent RNA emerges from within pol II at ~17 nt [54,55] . Complexes paused over about the next 15 bases have a strong tendency to backtrack. The exact location of the onset of backtracking, and the extent to which particular complexes backtrack, is a function of promoter sequence, but all backtracked complexes apparently retreat to a common template location, essentially the same as that in stage E [57]. Downstream of ~+23, there is no measurable tendency for complexes to slip during transcription of repeated template segments [34]. (G) Clearance is apparently complete at this point, with no general tendency to backtrack.
The downstream progress of the bubble is a central part of the clearance process. This will be discussed further below, but it is important to note two critical features of the bubble: the position of its upstream edge is fixed relative to the TATA box, not to transcription start [15,16], and the upstream edge remains in place during the earliest stages of transcript elongation [14,16]. Thus, the pol II transcription bubble stretches in the downstream direction immediately after initiation, rather than simply translocating downstream with the polymerase.
3. Abortive versus productive transcription: synthesis of the first 10 nt of the transcript
All RNA polymerases must cope with the fact that immediately after transcription initiates, the RNA-DNA hybrid is too short to be stable. The nascent transcript can be released from the advancing transcription complex, resulting in abortive initiation. Loss of the transcript will be driven by the tendency of template and nontemplate strands to reanneal. For bacterial RNA polymerase, transcription passes through many cycles of abortive initiation in which short RNAs, typically up to about 10 nt, are repeatedly synthesized and released [17,18]. The polymerase remains at the promoter during this process and only infrequently escapes into productive transcript elongation. For example, a study of transcript initiation at three E. coli promoters showed that 7, 32 and 165 abortive transcripts were made for each full length RNA [19]. The promoter that was least effective in escaping abortive initiation supported the synthesis of abortive products as long as 15 nt [19]. Abortive initiation is not an artifact of the in vitro assay since the aborted RNAs can be detected in cells [20]. The example of bacterial RNA polymerase emphasizes the inherent difficulty of converting the initiation-competent polymerase into a processive elongation complex.
When pol II open complexes are provided only with the substrates necessary to make the first bond in a transcript (Fig. 1B), the polymerase will engage in multiple rounds of RNA synthesis without leaving the template [14,21,22]. The addition of NTPs which allow transcription beyond the first bond strongly reduces the abortive production of single bond transcripts [14,21,23]. Pol II can be forced to pause during the earliest stages of transcript elongation by leaving one NTP out of the reaction mix or simply by using very low levels of the NTPs (i.e., ~ 1 µM). Complexes which pause during the synthesis of about the first 10 bases of the nascent RNA are unstable and prone to disassociate [24,25]. When pol II transcription reactions were performed with nonlimiting (200 µM) NTP levels (similar to typical NTP levels in mammalian cells- see [26]), no short RNAs other than one or two bond products failed to chase into longer transcripts, indicating that cycling abortive production of 4–10 nt transcripts did not occur at significant levels under these conditions [27]. However, evidence for low levels of abortive synthesis of RNAs as long as 9 nt was presented in another study [14]. In neither case were the abortive products in high excess over full-length RNAs, in contrast to the behavior of bacterial RNA polymerase in similar reactions.
Given the underlying structural similarity of prokaryotic RNA polymerase and pol II [28], it may be surprising that pol II has a much lower tendency to abortively initiate. This difference probably arises because pol II is assisted in the early stages of transcript elongation by TFIIH. Efficient extension of nascent RNA to about +10 is significantly diminished if the XPB helicase is absent or inhibited [13,29–31]. The helicase presumably counters the tendency of the template and nontemplate strands to reanneal and expel the short nascent RNA. The importance of this assistance can be demonstrated in a particularly striking way using templates which are mismatched over the region upstream of transcription start that normally melts at open complex formation. Transcription of these bubble templates allows the early stages of transcript elongation to be studied with or without TFIIH. On such templates, very little RNA synthesis can proceed past +15 without TFIIH and ATP [16,30].
The RNA-DNA hybrid in pol II early transcription complexes grows until it is 8 bp long (Fig. 1C). Downstream of this point the 5’ end of the transcript separates from the template strand and enters the RNA exit channel of the polymerase [32]. Even though there is minimal loss of transcript during the synthesis of the first ten nt of RNA by pol II when NTPs are not limiting, the transcript is not as stably associated with the transcription complex during this early stage as it is later in elongation. This is particularly evident from the ability of transcripts from repetitive segments of the initially transcribed region to slip upstream and reanneal with the DNA, leading to the synthesis of RNAs longer than predicted from the template sequence [27,33]. This lateral instability of the hybrid falls off by 10-fold once the RNA-DNA hybrid has reached its mature length [34]. Recent crystallographic studies demonstrate that the structure of the RNA-DNA hybrid in pol II complexes with less than 8 nt RNAs is not the same as the structure expected in the final elongation complex [35,36]. Results from these studies are not in complete agreement, but it is clear that between 6 and 8 nt must be added to the growing RNA chain before the hybrid structure reaches its mature form. The sequence of the template is probably critical in determining the exact transition point into the final hybrid structure [35].
4. The bubble collapse transition
The completion of the RNA-DNA hybrid and entry of the RNA into the exit channel does not mark the end of the clearance process. A detailed investigation of transcript slippage with promoter variants differing in their TATA to +1 distances revealed that the transition to a stable complex is not driven primarily by the length of the transcript [16]. As noted above, the upstream edge of the transcription bubble remains at its initial location during the early stages of transcript elongation, resulting in continuous lengthening of the bubble as the RNA is extended. Once the bubble reaches a length of 17–18 bases (Fig. 1D), the upstream segment abruptly closes [14,16]. This leaves only about 10 bases unpaired, a bubble size typical of the mature transcript elongation complex. Once this “bubble collapse” transition occurs, the transcription machinery takes on many of the characteristics of an elongation complex, regardless of the length of the transcript. In particular, bubble collapse marks the end of the requirement for TFIIH assistance for effective elongation [16]. Since the location of the upstream edge of the initial bubble is fixed relative to the TATA element and not to transcription start, bubble collapse can occur at different transcript lengths depending on the sequence of the promoter. Almost all mammalian TATA box promoters have a TATA to + 1 spacing of 28 to 33 bp [37], with the most typical distance being 30 or 31 bp. For a promoter with this canonical spacing of TATA and transcription start, bubble collapse occurs upon synthesis of a 9–10 nt transcript [16].
It was suggested that the energy expended to generate the overly-long initial transcription bubble is utilized during collapse to remodel the transcription complex and thus complete promoter clearance [16]. This idea is reminiscent of the “stressed intermediate” model of the prokaryotic RNA polymerase early elongation complex [38]. Single molecule studies with bacterial RNA polymerase support a “DNA scrunching” model, in which polymerase remains essentially stationary on the template just after initiation while drawing in downstream DNA during RNA synthesis [39,40]. The accumulated strain from scrunching drives abortive initiation. This strain is eventually expended to break promoter contacts and drive clearance, apparently analogous to the bubble expansion and ultimate collapse just described for pol II. However, recent work with T7 RNA polymerase questions the idea that the stressed state actually drives abortive initiation, at least for the single-subunit polymerase [41]. In any event, the analogy between the prokaryotic and eukaryotic cases is limited, in that pol II clearance is uniquely assisted by a helicase.
The changes in the pol II complex that accompany bubble collapse (acquisition of overall complex stability, fall-off of slippage, loss of requirement for the XPB helicase) suggested that collapse could correspond to the completion of promoter clearance by pol II. If bubble collapse does mark the end of clearance it should also correspond to the loss of contacts between pol II and the transcript initiation factors. Of particular interest in the context of bubble collapse is the fate of TFIIB. An initial study showed that TFIIB is released from the complex shortly after the start of transcription [42]. More recent experiments using a different promoter agreed with the earlier observations [43]. Structural studies of the pol II-TFIIB complex have led to differing predictions on the point at which TFIIB should be destabilized during transcript elongation. The initial presence of a TFIIB domain within the RNA exit channel suggested that TFIIB should be displaced by the advancing nascent RNA, beginning with 6 or 7 nt transcripts [44]. Other studies identified an important contact of the linker region of TFIIB with the upstream single-stranded segment of the transcription bubble. This contact should be lost, and thus TFIIB-pol II interaction should be destabilized, once the upstream section of the transcription bubble reanneals during bubble collapse [5]. This question was recently addressed by directly assaying for loss of TFIIB from pol II early elongation complexes advanced in single nt increments [22]. A slight loss of TFIIB was observed as pol II advanced from +6 to +7, but TFIIB remained relatively stably associated with the transcription complex until the nascent RNA was 13 nt long (Fig. 1E). This is well past the predicted initial collision point of TFIIB with the nascent RNA in the RNA exit channel, and crucially it is also 2 bp downstream of the point at which bubble collapse occurs on the promoter used [16,22]. The somewhat surprising retention of TFIIB beyond the point of bubble collapse suggests that additional important contacts govern the association of TFIIB in the transcription complex after initiation. At least some of these contacts must involve TFIIF. TFIIB remains in human pol II complexes as far downstream as stage E in Fig. 1 when transcription complex assembly is performed such that the PICs retain TFIIF. However, when PICs lose TFIIF following PICs assembly, most of their TFIIB is lost upon open complex formation [22]. This is consistent with the recent observation, made with yeast pol II and factors, that addition of TFIIF to bubble template complexes with pol II and TFIIB re-orients TFIIB [45].
During promoter clearance, the initiation-specific interactions of pol II with the general transcript factors other than TFIIB should also be lost. TBP remains upstream on the TATA box as pol advances into elongation [42]. TFIIE is required for PIC formation but it is not stably retained within the pol II PIC, at least not for PICs assembled with the minimal general transcription factor set [46]. TFIIE binds to a site on the pol II clamp domain which is also the binding site for the Spt5 subunit of the DSIF transcript elongation factor [8,10]. Loss of TFIIE may be therefore important in the subsequent loading of DSIF, similar to the analogous situation with archaeal TFE and Spt5 [47]. TFIIH loss from the advancing transcription complex was placed downstream of +30 in an earlier study [42], but as noted above TFIIH is not required for efficient transcript elongation once pol II has passed through the bubble collapse transition ([16]; see also [7]).
TFIIF is unique among the general transcript initiation factors in that it can also associate with pol II throughout transcription and thereby stimulate the rate of transcript elongation [48–51]. TFIIF is transiently lost from the transcription complex after initiation [42]. TFIIF binds to paused pol II elongation complexes [52], but it does not stably associate with pol II during active transcription [48]. Even though TFIIF is required for assembly of the pol II PIC, TFIIF can under some conditions be removed from the PIC before initiation [22]. The direct comparison of transcription with PICs that retained or lacked TFIIF showed no difference in the ability of those complexes to pass through the bubble collapse transition, indicating that TFIIF does not have a major influence on this phase of transcription [22]. An earlier study had proposed an important role for TFIIF in pol II promoter clearance [53]. The apparent discrepancy with the more recent work may be based on the different choice of template types: bubble templates were used for the Yan et al. study [53] versus the double-stranded templates used by Čabart et al. [22].
5. Transitions downstream of bubble collapse; the completion of promoter clearance
If bubble collapse marks the end of promoter clearance, the properties of the transcription complex should not change significantly downstream of collapse. The next important physical change after bubble collapse is the filling of the RNA exit channel by the nascent RNA. At this point the complex should have acquired all of the stabilizing effects from the transcript-exit channel interactions. RNA emerges from pol II at a transcript length of about 17 nt [54,55]. However, the properties of the complex continue to change during elongation, through the channel-filling process and ultimately well downstream of that point. For example, transcript slippage was evident in pol II complexes with 15-mer and 21-mer RNAs when the polymerase was forced to pause over a repetitive template segment [34]. Slippage was completely absent after pausing over the same repetitive element only when poi II had synthesized RNAs 23 nt or longer [34]. Surprisingly, complexes paused from +17/+18 to as far downstream as +32 (Fig. 1F) have a very strong tendency to backtrack and in some cases to arrest, depending on the template sequence [55–57]. This is in contrast to mature pol II elongation complexes which do not backtrack when paused except at a very small subset of sequences that cause arrest [58]. All pol II complexes in the +17/+32 range which backtrack retreat to a similar location, such that only about 12–13 bases remain upstream of the active site as judged by the length of transcript cleavage products released when these complexes are treated with TFIIS ([57]; compare Figs. 1E and 1F).
It is not known why pol II complexes with 17–32 nt transcripts backtrack, nor is it understood why complexes with 12–13 bases upstream of the active site after backtracking are especially stable. It is possible that secondary structure in the emerging RNA is important in blocking reverse threading of the transcript, thereby preventing backtracking once the RNA is of sufficient length. However, in the examples studied, no strong secondary structures could be predicted in the newly emerged transcripts for the complexes that no longer backtracked [57]. More importantly, hybridizing short DNA oligonucleotides to the transcripts at positions well upstream of the point of emergence of the RNA from the polymerase strongly increased backtracking and arrest [55], inconsistent with a model in which immediate self-association of the transcript is important to stabilize the transcription complex against backtracking.
In the experiments just cited, the transcription complexes were detergent-rinsed and thus lacked any additional factors (such as the capping machinery) which could have bound to the emerging transcript and inhibited reverse threading of the RNA. With that caveat in mind, once pol II has extended the transcript beyond roughly +30, it has acquired all of the characteristics of an elongation complex: it has no general tendency to backtrack, no measurable ability to slip when transcribing a dinucleotide repeat that drives slippage effectively in promoter-proximal complexes, and no residual interactions with the general initiation factors (Fig. 1G). Thus, as judged by these properties, promoter clearance is complete for pol II by about 30 bases downstream of transcription start.
6. Challenges for the future
While the clearance pathway just described has been reasonably well established for mammalian pol II at a TATA box promoter, it is important to acknowledge the limitations of this analysis. Only a small minority of pol II promoters in mammalian cells contain TATA boxes and many mammalian promoters do not contain any known promoter sequence motifs (recently reviewed in [37,59,60]). It is not yet known whether the canonical set of general initiation factors identified for TATA box promoters are either necessary or sufficient for initiation at non-TATA promoters (see in particular [61]). In the absence of a TATA element to anchor the upstream end of the initial transcription bubble, it is not clear how the extension and ultimate collapse of the bubble would contribute to the clearance process.
The propagation of the initial transcription bubble in budding yeast is especially perplexing. Recent studies indicate that most S. cerevisiae pol II promoters do contain a functional TATA box, even though matches to the TATA consensus are not always evident [62]. In addition, the initial transcription bubble in yeast is apparently anchored at its upstream end to TATA, as in mammalian cells [15]. However, transcription in yeast can start over a wide range of permissible distances downstream from TATA: from 40 to 120 bp [45,63], in contrast to the narrow 28–33 bp window in mammalian cells [37]. This makes it difficult to envision how expansion of the transcription bubble to a single maximum length would drive the clearance process in yeast, as hypothesized for mammalian pol II transcription [16]. Recent results from Fishburn and Hahn [45] show that the minimal yeast transcriptional machinery (TBP and TFIIB, in addition to pol II) can utilize the normal initiation sites for the yeast HIS4 gene, which are located ~80 bp downstream of the HIS4 TATA element, when those sites are located within a short preformed bubble. Thus, it is not necessary to have an exceptionally long bubble within the open complex for yeast pol II to initiate transcription. Work from Ponticelli and colleagues suggests that yeast pol II itself selects the transcript start site as it scans downstream from the initial, more TATA-proximal bubble position (see [64] and [65] for more detailed discussion).
The clearance pathway in Fig. 1 is primarily based on results obtained with the minimal transcription machinery necessary to direct initiation. During the clearance process in the nucleus, pol II transcription complexes will be modified and supplemented by the addition of many factors necessary to support effective elongation. The acquisition of these modifications and factors, as well as the loading of additional factors involved in RNA processing, are reviewed in several of the accompanying papers in this issue. (See especially: Handa, Price, Hartzog, Eick, Conaway, Reese-??)
One point in particular is worth noting. Regulatory factors, often in conjunction with the Mediator coactivator complex, occupy the DNA upstream of the promoter of active genes (see [66] for a recent review). Association of pol II with Mediator and other factors provides additional stabilizing interactions for the transcription complex just after initiation, which could act to suppress abortive initiation. This stands in contrast to the case in prokaryotes where, as noted above, abortive production of short RNAs during the initial stages of transcription is evident both in vitro and in vivo. Interestingly, Mediator subunits associate not only with the promoter region but also with the bodies of actively transcribed genes, as determined by ChIP assays ([67]; see also [68]). Based on these observations and the fact that Mediator can bind in a specific way to pol II [69], it is tempting to speculate that the transcription complex remains in contact with promoter-bound Mediator throughout the transcription process. This is a central feature of the “transcription factory” concept, in which polymerase and associated factors remain in a fixed location while the template is threaded through this complex (recently reviewed in [70]). In this model, pol Il presumably transitions into a stable elongation complex through a clearance path similar to that described above, but the polymerase also maintains continued association with the promoter through interactions with Mediator and other factors. Thus, in this sense pol II may never actually “escape” from the promoter. A mechanism for retaining some promoter contacts for pol II throughout the transcription cycle would allow polymerase to rapidly and effectively reinitiate transcription after termination. This is consistent with reports on the retention of subsets of the initiation factors at the promoter after initiation in yeast [71] and the connection of termination and initiation through looping interactions mediated in part by TFIIB [72,73].
Highlights.
Pol II must break promoter and factor interactions to enter transcript elongation
Abortive initiation is not extensive for pol II
Clearance is driven by TFIIH and collapse of the initial transcription bubble
Pol II early elongation complexes backtrack, even after RNA exits the polymerase
Clearance is incomplete until the pol II elongation complex is fully stabilized
Acknowledgements
Work from our laboratory discussed in this review was supported by grant GM 29487 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 2.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 3.Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA Polymerase II-TFIIB Complex and the Transcription Initiation Mechanism. Science. 2010;327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 6.Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1421. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- 7.Wang XX, Spangler L, Dvir A. Promoter escape by RNA polymerase II - Downstream promoter DNA is required during multiple steps of early transcription. J. Biol. Chem. 2003;278:10250–10256. doi: 10.1074/jbc.M210848200. [DOI] [PubMed] [Google Scholar]
- 8.Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichner J, Chen HT, Warfield L, Hahn S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 2010;29:706–716. doi: 10.1038/emboj.2009.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30:1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 12.Coin F, Bergmann E, Tremeau-Bravard A, Egly JM. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 1999;18:1357–1366. doi: 10.1093/emboj/18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin YC, Choi WS, Gralla JD. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat. Struct. Mol. Biol. 2005;12:603–607. doi: 10.1038/nsmb949. [DOI] [PubMed] [Google Scholar]
- 14.Holstege FCP, Fiedler U, Timmers HTM. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997;16:7468–7480. doi: 10.1093/emboj/16.24.7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giardina C, Lis JT. DNA Melting on Yeast RNA Polymerase-II Promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 16.Pal M, Ponticelli AS, Luse DS. The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell. 2005;19:101–110. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 17.Carpousis AJ, Gralla JD. Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. J. Mol. Biol. 1985;183:165–177. doi: 10.1016/0022-2836(85)90210-4. [DOI] [PubMed] [Google Scholar]
- 18.Carpousis AJ, Gralla JD. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 19.Hsu LLM, Vo NV, Kane CM, Chamberlin MJ. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 1. RNA chain initiation, abortive initiation, and promoter escape at three bacteriophage promoters. Biochemistry. 2003;42:3777–3786. doi: 10.1021/bi026954e. [DOI] [PubMed] [Google Scholar]
- 20.Goldman SR, Ebright RH, Nickels BE. Direct Detection of Abortive RNA Transcripts in Vivo. Science. 2009;324:927–928. doi: 10.1126/science.1169237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luse DS, Jacob GA. Abortive initiation by RNA polymerase II in vitro at the Adenovirus 2 major late promoter. J. Biol. Chem. 1987;262:14990–14997. [PubMed] [Google Scholar]
- 22.Čabart P, Újvári A, Pal M, Luse DS. TFIIF is not required for initiation by RNA polymerase II but it is essential to stabilize TFIIB in early transcription complexes. Proc. Natl. Acad. Sci. USA. 2011;108:15786–15791. doi: 10.1073/pnas.1104591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y, Yan M, Gralla JD. Abortive initiation and first bond formation at an activated adenovirus E4 promoter. J. Biol. Chem. 1995;270:27332–27338. doi: 10.1074/jbc.270.45.27332. [DOI] [PubMed] [Google Scholar]
- 24.Coppola JA, Luse DS. Purification and characterization of ternary complexes containing accurately initiated RNA polymerase II and less than 20 nucleotides of RNA. J. Mol. Biol. 1984;178:415–437. doi: 10.1016/0022-2836(84)90151-7. [DOI] [PubMed] [Google Scholar]
- 25.Cai H, Luse DS. Transcription initiation by RNA polymerase II in vitro. Properties of preinitiation, initiation and elongation complexes. J. Biol. Chem. 1987;262:298–304. [PubMed] [Google Scholar]
- 26.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 27.Pal M, Luse DS. Strong natural pausing by RNA polymerase II within 10 bases of transcription start may result in repeated slippage and reextension of the nascent RNA. Mol. Cell. Biol. 2002;22:30–40. doi: 10.1128/MCB.22.1.30-40.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebright RH. RNA polymerase: Structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 29.Dvir A, Conaway RC, Conaway JW. Promoter escape by RNA polymerase II - A role for an ATP cofactor in suppression of arrest by polymerase at promoter- proximal sites. J. Biol. Chem. 1996;271:23352–23356. doi: 10.1074/jbc.271.38.23352. [DOI] [PubMed] [Google Scholar]
- 30.Dvir A, Conaway RC, Conaway JW. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc. Natl. Acad. Sci. USA. 1997;94:9006–9010. doi: 10.1073/pnas.94.17.9006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreland RJ, Tirode F, Yan Q, Conaway W, Egly JM, Conaway RC. A role for the TFIIH XPB DNA helicase in promoter escape by RNA polymerase II. J. Biol. Chem. 1999;274:22127–22130. doi: 10.1074/jbc.274.32.22127. [DOI] [PubMed] [Google Scholar]
- 32.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: Separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 33.Gilman B, Drullinger LF, Kugel JF, Goodrich JA. TATA-binding Protein and Transcription Factor IIB Induce Transcript Slipping during Early Transcription by RNA Polymerase II. J. Biol. Chem. 2009;284:9093–9098. doi: 10.1074/jbc.M900019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pal M, Luse DS. The initiation-elongation transition: Lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at+8/+9 and absent by+23. Proc. Natl. Acad. Sci. USA. 2003;100:5700–5705. doi: 10.1073/pnas.1037057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung ACM, Sainsbury S, Cramer P. Structural basis of initial RNA polymerase II transcription. EMBO J. 2011;30:4755–4763. doi: 10.1038/emboj.2011.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD. Initiation Complex Structure and Promoter Proofreading. Science. 2011;333:633–637. doi: 10.1126/science.1206629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, Sandelin A. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biology. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straney DC, Crothers DM. A stressed intermediate in the formation of stably initiated RNA chains at the Escherichia coli lac UV5 promoter. J. Mol. Biol. 1987;193:267–278. doi: 10.1016/0022-2836(87)90218-x. [DOI] [PubMed] [Google Scholar]
- 39.Revyakin A, Liu CY, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vahia AV, Martin CT. Direct Tests of the Energetic Basis of Abortive Cycling in Transcription. Biochemistry. 2011;50:7015–7022. doi: 10.1021/bi200620q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 43.Tran K, Gralla JD. Control of the timing of promoter escape and RNA catalysis by the transcription factor IIB fingertip. J. Biol. Chem. 2008;283:15665–15671. doi: 10.1074/jbc.M801439200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science. 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 45.Fishburn J, Hahn S. Architecture of the Yeast RNA Polymerase II Open Complex and Regulation of Activity by TFIIF. Mol. Cell. Biol. 2012;32:12–25. doi: 10.1128/MCB.06242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Čabart P, Luse DS. Inactivated RNA Polymerase II Open Complexes Can Be Reactivated with TFIIE. J. Biol. Chem. 2012;287:961–967. doi: 10.1074/jbc.M111.297572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grohmann D, Nagy J, Chakraborty A, Klose D, Fielden D, Ebright RH, Michaelis J, Werner F. The Initiation Factor TFE and the Elongation Factor Spt4/5 Compete for the RNAP Clamp during Transcription Initiation and Elongation. Mol. Cell. 2011;43:263–274. doi: 10.1016/j.molcel.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price DH, Sluder AE, Greenleaf AL. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell. Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Izban MG, Luse DS. Factor-Stimulated RNA Polymerase-II Transcribes at Physiological Elongation Rates on Naked DNA But Very Poorly on Chromatin Templates. J. Biol. Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 50.Kephart DD, Wang BQ, Burton ZF, Price DH. Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. J. Biol. Chem. 1994;269:13536–13543. [PubMed] [Google Scholar]
- 51.Lei L, Ren DL, Finkelstein A, Burton ZF. Functions of the N- and C-terminal domains of human RAP74 in transcriptional initiation, elongation, and recycling of RNA polymerase II. Mol. Cell. Biol. 1998;18:2130–2142. doi: 10.1128/mcb.18.4.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng B, Price DH. Analysis of factor interactions with RNA polymerase II elongation complexes using a new electrophoretic mobility shift assay - art. no. e135. Nucl. Acids Res. 2008;36:E135. doi: 10.1093/nar/gkn630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan Q, Moreland RJ, Conaway JW, Conaway RC. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J. Biol. Chem. 1999;274:35668–35675. doi: 10.1074/jbc.274.50.35668. [DOI] [PubMed] [Google Scholar]
- 54.Gu WG, Wind M, Reines D. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc. Natl. Acad. Sci. USA. 1996;93:6935–6940. doi: 10.1073/pnas.93.14.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Újvári A, Pal M, Luse DS. RNA polymerase II transcription complexes may become arrested if the nascent RNA is shortened to less than 50 nucleotides. J. Biol. Chem. 2002;277:32527–32537. doi: 10.1074/jbc.M201145200. [DOI] [PubMed] [Google Scholar]
- 56.Samkurashvili I, Luse DS. Structural changes in the RNA polymerase II transcription complex during transition from initiation to elongation. Mol. Cell. Biol. 1998;18:5343–5354. doi: 10.1128/mcb.18.9.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pal M, McKean D, Luse DS. Promoter clearance by RNA polymerase II is an extended, multistep process strongly affected by sequence. Mol. Cell. Biol. 2001;21:5815–5825. doi: 10.1128/MCB.21.17.5815-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samkurashvili I, Luse DS. Translocation and transcriptional arrest during transcript elongation by RNA polymerase II. J. Biol. Chem. 1996;271:23495–23505. doi: 10.1074/jbc.271.38.23495. [DOI] [PubMed] [Google Scholar]
- 59.Juven-Gershon T, Hsu J-Y, Theisen JWM, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr. Opin. Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis BA, Sims RJ, Lane WS, Reinberg D. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol. Cell. 2005;18:471–481. doi: 10.1016/j.molcel.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 62.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuehner JN, Brow DA. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J. Biol. Chem. 2006;281:14119–14128. doi: 10.1074/jbc.M601937200. [DOI] [PubMed] [Google Scholar]
- 64.Khaperskyy DA, Ammerman ML, Majovski RC, Ponticelli AS. Functions of Saccharomyces cerevisiae TFIIF during Transcription Start Site Utilization. Mol. Cell. Biol. 2008;28:3757–3766. doi: 10.1128/MCB.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang C, Ponticelli AS. Evidence that RNA polymerase II and not TFIIB is responsible for the difference in transcription initiation patterns between Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucl. Acids Res. 2012 doi: 10.1093/nar/gks323. published online Apr. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, Young RA, Price DH. Functional Association of Gdown1 with RNA Polymerase II Poised on Human Genes. Mol. Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernecky C, Grob P, Ebmeier CC, Nogales E, Taatjes DJ. Molecular Architecture of the Human Mediator-RNA Polymerase II–TFIIF Assembly. PLoS Biol. 2011;9:e1000603. doi: 10.1371/journal.pbio.1000603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papantonis A, Cook PR. Fixing the model for transcription: The DNA moves, not the polymerase. Transcription. 2011;2:41–44. doi: 10.4161/trns.2.1.14275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 72.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol. Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 73.El Kaderi B, Medler S, Raghunayakula S, Ansari A. Gene Looping Is Conferred by Activator-dependent Interaction of Transcription Initiation and Termination Machineries. J. Biol. Chem. 2009;284:25015–25025. doi: 10.1074/jbc.M109.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]