Abstract
Double-stranded break (DSB) repair during meiotic recombination in yeast Saccharomyces cerevisiae leads to the formation of heteroduplex DNA, a hybrid DNA molecule composed of single strands from two homologous chromosomes. Differences in sequence between the strands within heteroduplex DNA generate mismatches or large unpaired loops that are substrates for repair. At least two pathways function to repair large loops that form within heteroduplex DNA: the RAD1-dependent large loop repair (LLR) pathway and another as yet uncharacterized RAD1-independent LLR pathway. Repair of large loops during meiotic recombination is especially important for the genomic stability of the repetitive DNA sequences known as minisatellites. Minisatellite DNA tracts are generally stable during mitotic cell divisions but frequently alter in length during meiosis. Using a yeast minisatellite system in which the human minisatellite associated with the HRAS1 proto-oncogene has been inserted into the recombination hotspot region upstream of HIS4 in S. cerevisiae, our lab previously showed that the RAD1-dependent LLR pathway controls minisatellite length expansions, but not contractions. Here we show that minisatellite length expansions are controlled by the products of the CSM3 and TOF1 genes, while contractions are controlled by MRC1. By examining meiotic segregation patterns in yeast strains heterozygous for the 26bp his4-lopd insert, we found that deleting CSM3 caused a loss of LLR activity similar to that seen in a RAD1 mutant. Double mutant analysis revealed that failure to repair loops is exacerbated upon deleting both RAD1 and CSM3 - specifically the type of repair that fills in loops, which would generate minisatellite length expansions. A model for minisatellite length alteration based on these results is presented.
Keywords: Meiosis, Recombination, DNA repair, Gene conversion
1. Introduction
Meiotic recombination is an elaborate process that functions to repair a programmed DNA double-stranded break (DSB) during meiosis, using the homologous chromosome as the template (Hunter, 2006; Roeder, 1997). During recombination, 3′ single-stranded DNA from the broken chromosome invades the DNA duplex of the homologue, forming a heteroduplex molecule composed of DNA strands from each of the chromosomes. If a difference exists in the DNA sequences that make up the heteroduplex, a mismatch or a loop will form that is a substrate for repair (Figure 1) (Alani et al., 1994; Fogel et al., 1981; Jensen et al., 2005; Kearney et al., 2001; Kirkpatrick, 1999).
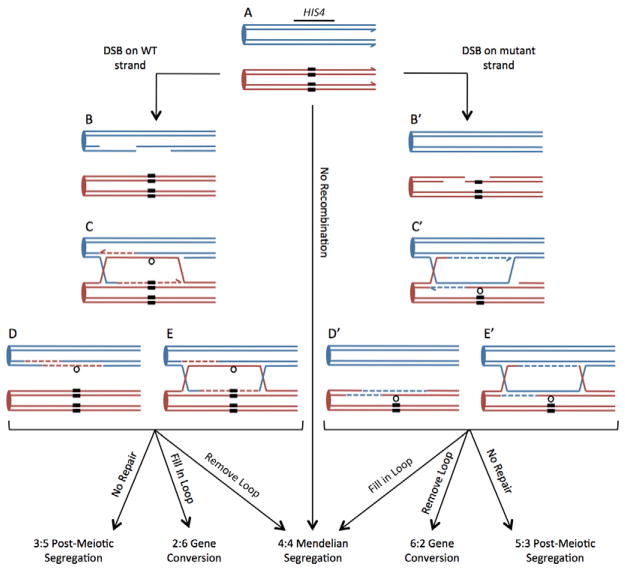
Figure 1.
DNA Repair events during meiotic recombination. Two chromosomes containing the HIS4 locus are shown following DNA replication in early meiosis. One homolog is in blue, the other in red, and both strands of each sister chromatid are depicted as thin lines. The solid black boxes on the red chromosome indicate a region of inserted DNA. Double-strand breaks (DSBs) initiate recombination on either the wild-type (WT) chromosome (left side) or the mutant chromosome (right side). The breaks are processed to generate single-stranded ends (B and B′), which invade the homolog to form heteroduplex DNA (C and C′). Mismatches that occur in the heteroduplex DNA are shown as loops. Repair synthesis is indicated by dashed lines. The mismatched loops in heteroduplex DNA can be resolved by either removing the loop or nicking the strand opposite the loop and filling it in via repair synthesis, leading to either a gene conversion event or a restoration of normal Mendelian segregation. Failure to recognize or repair the mismatched loop will generate a post-meiotic segregation event.
In yeast Saccharomyces cerevisiae, there are several possible fates for a loop-generating mismatch after formation. Depending on which strand initiates repair, repair of the loop can either restore normal Mendelian segregation or generate a gene conversion (GC) (Kirkpatrick, 1999). If the loop is not detected or repaired, one of the four spores will receive the duplex DNA molecule containing the mismatch. When this spore germinates and goes through the first round of DNA synthesis, the two strands comprising the mismatched region will each serve as a template, producing daughter DNA molecules with different sequences in the mismatched region. The result will be a spore colony with a sectored phenotype that can easily be detected if the mismatched region is within an auxotrophic marker; this type of segregation is known as a post-meiotic segregation (PMS) (Alani et al., 1994; Fogel and Hurst, 1967; Fogel et al., 1981; Kirkpatrick, 1999; Petes et al., 1991). The efficiency with which a mismatch is detected and repaired during meiotic recombination is reflected in the percent of GC and PMS tetrads (Alani et al., 1994; Fogel et al., 1981; Jensen et al., 2005; Kearney et al., 2001; Kirkpatrick, 1999; Kirkpatrick and Petes, 1997).
In the AS4/AS13 Saccharomyces cerevisiae strain background used in this study, the HIS4 locus exhibits a very high level of recombination activity during meiosis (Detloff et al., 1992; Jauert et al., 2002; Kearney et al., 2001; Kirkpatrick and Petes, 1997; Kirkpatrick et al., 1999; Nag and Petes, 1993; Nag et al., 1989; White et al., 1993). This hotspot recombination activity leads to a high level of mismatch formation in diploids heterozygous for HIS4 alleles. Research using strains that are heterozygous for his4-lopd, a 26 bp non-palindromic insertion in HIS4, has shown that at least two pathways function to repair large loop mismatches during meiosis. The first large loop repair (LLR) pathway, consisting of RAD1, RAD10, MSH2, and MSH3, can repair loops no less than 16 bp and up to 5.6 kb in size (Jensen et al., 2005; Kearney et al., 2001; Kirkpatrick and Petes, 1997). Although no genes have been identified, studies indicate that a second RAD1-independent large loop repair pathway exists, as significant large loop repair is still seen when the RAD1-dependent LLR pathway is eliminated (Kearney et al., 2001; Kirkpatrick and Petes, 1997).
One of the goals of our lab has been to identify other genes involved in meiotic large loop repair. This is especially important for characterizing the meiotic stability of repetitive DNA sequences such as minisatellite DNA. Minisatellites are composed of tandemly repeated units that are 15 to 100 bp in length. Our lab utilizes the minisatellite found in the 3′ region of the human proto-oncogene HRAS1, which is composed of 28 bp units that are repeated in tandem (Capon et al., 1983; Cohen et al., 1987). Studies have shown that the HRAS1 minisatellite is generally stable during mitosis but frequently alters in length during meiosis (Jauert et al., 2002; Jeffreys et al., 1998). Meiotic alterations generate new HRAS1 minisatellite alleles that can enhance the expression of the HRAS1 gene and predispose cells to becoming malignant (Krontiris, 1995; Krontiris et al., 1993; Krontiris et al., 1985; Spandidos and Holmes, 1987; Trepicchio and Krontiris, 1992). By inserting the HRAS1 minisatellite into the yeast genome upstream of HIS4, our lab has been able to identify factors that influence meiotic minisatellite stability (Jauert et al., 2002; Jauert and Kirkpatrick, 2005; Jensen et al., 2005). Previously, our lab has shown that the RAD1-dependent LLR pathway controls the expansions, but not the contractions, of the common HRAS1-A1 minisatellite allele, and thereby plays a direct role in the meiotic stability of the HRAS1 minisatellite (Jauert et al., 2002).
In this study we demonstrate that CSM3 plays a role in the meiotic stability of the HRAS1 minisatellite in yeast. CSM3 (chromosome segregation in meiosis) was first identified in a screen for genes that are necessary for wild-type meiosis in S. cerevisiae (Rabitsch et al., 2001). The loss of CSM3 resulted in a “mild chromosome missegregation” phenotype and a loss of spore viability, but the functional role of CSM3 during meiosis remains poorly understood. Since then research has focused on the role of CSM3 during S phase in mitotically dividing cells. Studies have shown that the products of the CSM3, MRC1, and TOF1 genes form a heterotrimeric complex that interacts directly with the replisome, specifically the MCM helicase, during S phase (Bando et al., 2009; Calzada et al., 2005; Nedelcheva et al., 2005). The CSM3/MRC1/TOF1 complex is not required for DNA synthesis, but instead serves as a mediator of replication fork progression. In the event of DNA damage or replicative stress, DNA synthesis pauses leading to the accumulation of RPA bound single-stranded DNA regions, the activation of the replication checkpoint, and phosphorylation of Mrc1 by Mec1. The Csm3/Mrc1/Tof1 complex stabilizes the MCM helicase and prevents further DNA unwinding, effectively keeping the replisome intact and stabilizing the replication fork (Bando et al., 2009; Calzada et al., 2005; Nedelcheva et al., 2005). Additionally, Mrc1 and Tof1, and presumably Csm3, have been shown to play a role in replication fork progression recovery (Tourriere et al., 2005).
In this study we analyzed CSM3, MRC1, and TOF1 for a role in meiotic minisatellite stability. We further characterized the role of CSM3 in large loop repair by analyzing the loss of CSM3 in strains heterozygous for the his4-lopd allele and compared the results to a RAD1 mutant. Finally, we analyzed CSM3 RAD1 double mutants in a heterozygous his4-lopd diploid to determine if they are in the same LLR pathway, or if CSM3 is the first gene to be identified that is part of the RAD1-independent LLR pathway. We present a model for the activities of the proteins based on their known enzymatic roles during mitotic DNA repair and our observations of the effects on meiotic recombination upon deletion of these genes.
2. Materials and Methods
2.1 Strain construction
All strains are derived from the haploid strains AS4 (MATα trp1-1 tyr7-1 arg4-17 ade6 ura3) and AS13 (MATα leu2-3 ura3 ade6 rme1) and are isogenic except for alterations introduced by transformation (Stapleton and Petes, 1992). The his4-A1 minisatellite allele was inserted into PD63 and PD57 derived strains at the site of the deleted promoter sequences upstream of HIS4 (Jauert et al., 2002), as shown in Figure 2. The his4-H10 minisatellite allele was derived from the his4-A1 allele as a naturally occurring tract reduction during meiosis (Jauert and Kirkpatrick, 2005). The his4-lopc and his4-lopd alleles, 26 bp insertions into the coding region of HIS4, were inserted into the AS13-derived haploids (Jauert et al., 2002; Jensen et al., 2005). Candidate gene deletions were constructed with one-step transformation by replacing the ORF of the respective gene with the KANMX4 cassette encoding geneticin resistance (Jauert et al., 2005). RAD1 deletions were previously constructed by two-step transformation (Kirkpatrick and Petes, 1997).
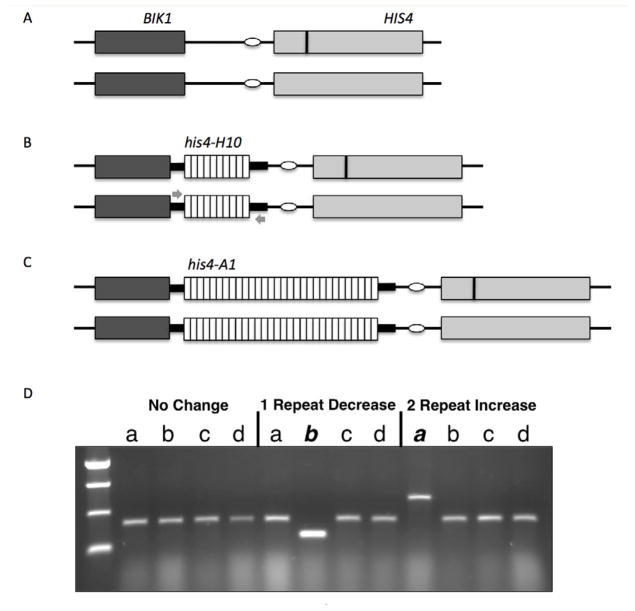
Figure 2.
Diagram of the HIS4 locus of chromosome III in diploid strains. A) The HIS4-BIK1 locus, shown in a diploid strain bearing wildtype HIS4 promoter sequences is heterozygous for either the his4-lopd or his4-lopc insert (dark vertical bar) that disrupts the HIS4 coding sequence. B) A diploid strain isogenic to (A) but bearing the his4-H10 minisatellite insert (white boxes) upstream of HIS4 replacing the endogenous promoter sequence. The TATA box (white circle) is left intact. Small grey arrows represent the primers used in PCR reactions to amplify the repetitive DNA tract. C) A diploid strain bearing the his4-A1 minisatellite insert (white boxes) upstream of HIS4, replacing the same endogenous promoter sequences as in (B). D) A representative gel showing minisatellite tract alterations in spore colonies from DTK751. Three complete tetrads are shown, with each spore colony labeled a – d. PCR was conducted as described in Materials and Methods, using the primers indicated in (B) above. The first tetrad had no spores that contained an altered his4-H10 tract. The second tetrad had a spore (b) that lost a repeat unit, while the third tetrad had a spore (a) that gained two repeat units.
2.2 Strains generated
All haploid strains are listed in Table 1. Diploid strains were constructed by the following crosses: DNY27k (DNY24 × AS4), DTK314 (DTK288 × DTK305), DTK508 (DTK506 × DTK507), DTK751 (DTK608 × DTK750), DTK802 (DTK669 × DTK796), DTK1459 (DTK1451 × DTK1452), DTK1460 (DTK1454 × DTK1453), DTK1480 (DTK1470 × DTK1469), DTK1487 (DTK1462 × DTK1461), DTK1567 (DTK1566 × DTKDTK1565), DTK1667 (DTK1659 × DTK1660), DTK1684 (DTK1679 × DTK1680), DTK1685 (DTK1681 × DTK1682), DTK1724 (DTK1661 × DTK1662), DTK1731 (DTK221 × DTK225), DTK1785 (DTK1782 × DTK1783), DTK1887 (DTK1886 × DTK1885)
Table 1.
Haploid Strains
| Strain | HIS4 promoter | HIS4 coding sequence | Relevant Mutation | Parental Strain | Source |
|---|---|---|---|---|---|
| Mata: | |||||
| AS4 | WT | WT | WT | Stapleton and Petes 1991 | |
| PD63 | his4-Δ52 | WT | WT | AS4 | Detloff, White and Petes 1992 |
| DTK225 | WT | WT | rad1Δ | AS4 | Kirkpatrick and Petes 1997 |
| DTK288 | his4-A1 | WT | WT | PD63 | Jauert et al. 2002 |
| DTK506 | his4-A1 | WT | rad1Δ | DTK288 | Jauert et al. 2002 |
| DTK750 | his4-H10 | WT | WT | PD63 | Jauert and Kirkpatrick 2005 |
| DTK796 | his4-H10 | WT | rad1Δ | DTK750 | Jauert and Kirkpatrick 2005 |
| DTK1452 | his4-H10 | WT | csm3Δ | DTK750 | this study |
| DTK1454 | his4-H10 | WT | slx1Δ | DTK750 | this study |
| DTK1462 | his4-H10 | WT | slx4Δ | DTK750 | this study |
| DTK1470 | his4-H10 | WT | ubx7Δ | DTK750 | this study |
| DTK1566 | his4-H10 | WT | mlh3Δ | DTK750 | this study |
| DTK1659 | WT | WT | csm3Δ | AS4 | this study |
| DTK1662 | his4-A1 | WT | csm3Δ | DTK288 | this study |
| DTK1680 | his4-H10 | WT | mrc1Δ | DTK750 | this study |
| DTK1682 | his4-H10 | WT | tof1Δ | DTK750 | this study |
| DTK1783 | WT | WT | rad1Δ csm3Δ | DTK225 | this study |
| DTK1885 | WT | WT | mrc1Δ | AS4 | this study |
| Matα: | |||||
| AS13 | WT | WT | WT | Stapleton and Petes 1991 | |
| DNY24 | WT | his4-lopd | WT | AS13 | Nag, White and Petes 1989 |
| PD57 | his4-Δ52 | WT | WT | AS13 | Detloff, White and Petes 1992 |
| PD80 | his4-Δ52 | his4-lopc | WT | PD57 | Detloff, White and Petes1992 |
| DTK221 | WT | his4-lopd | rad1Δ | DNY24 | Kirkpatrick and Petes 1997 |
| DTK294 | his4-A1 | WT | WT | PD57 | Jauert et al. 2002 |
| DTK305 | his4-A1 | his4-lopc | WT | DTK294 | Jauert et al. 2002 |
| DTK507 | his4-A1 | his4-lopc | rad1Δ | DTK305 | Jauert et al. 2002 |
| DTK608 | his4-H10 | his4-lopc | WT | PD80 | Jauert and Kirkpatrick 2005 |
| DTK669 | his4-H10 | his4-lopc | rad1Δ | DTK608 | Jauert and Kirkpatrick 2005 |
| DTK1451 | his4-H10 | his4-lopc | csm3Δ | DTK608 | this study |
| DTK1453 | his4-H10 | his4-lopc | slx1Δ | DTK608 | this study |
| DTK1461 | his4-H10 | his4-lopc | slx4Δ | DTK608 | this study |
| DTK1469 | his4-H10 | his4-lopc | ubx7Δ | DTK608 | this study |
| DTK1565 | his4-H10 | his4-lopc | mlh3Δ | DTK608 | this study |
| DTK1660 | WT | his4-lopd | csm3Δ | DNY24 | this study |
| DTK1661 | his4-A1 | his4-lopc | csm3Δ | DTK305 | this study |
| DTK1679 | his4-H10 | his4-lopc | mrc1Δ | DTK608 | this study |
| DTK1681 | his4-H10 | his4-lopc | tof1Δ | DTK608 | this study |
| DTK1782 | WT | his4-lopd | rad1Δ csm3Δ | DTK221 | this study |
| DTK1886 | WT | his4-lopd | mrc1Δ | DNY24 | this study |
2.3 Media, meiotic protocols and minisatellite tract-length analysis
Standard media were used (Guthrie and Fink, 1991; Jauert et al., 2002). Diploid sporulation was performed using standard protocols (Jauert et al., 2002). All sporulating strains were incubated at 18°C for three days. Whole-cell PCR was performed on spore colonies to analyze the minisatellite tract length in four-spore tetrads. The following 50 μl reaction was used to amplify the his4-H10 allele: 5 μl 10X PCR buffer (Sigma P-2317), 7.5 μl 25 mM MgCl2 (Sigma M-8787), 7.5 μl of 1.25 mM dNTPs, 1.25 μl each of 10 mM Ras 3′ and Ras 5′ primers (Jauert et al., 2002), 2 μl Taq polymerase, and 19.5 μl of H2O for a total of 44 μl, which was added to 6 μl of H2O containing DNA. Reagents were identical but the following volumes were changed for the his4-A1 allele: 3.75 μl 25 mM MgCl2, 8.3 μl of 1.25 mM dNTPs, and 22.45 μl of H2O. PCR was performed in a Hybaid PCR Express machine using the following paramaters: 1 cycle of 94°C for 1 min; 30 cycles of 92°C for 1 min, 58°C for 45 s, and 72°C for either 1 min (for the his4-H10 allele) or 2 min (for the his4-A1 allele); 72°C for 10 min; and then held at 15°C. At least 200 tetrads were examined per strain.
2.4 HIS4 marker segregation analysis
Marker segregation is described using the nomenclature of the eight-spored fungi - a tetrad exhibiting a Mendelian segregation event is described as 4 His+: 4 His−, while deviations from Mendelian segregation (aberrant segregation) events include gene conversions (6:2, 2:6, 0:8 and 8:0) and post-meiotic segregation (PMS) events (5:3, 3:5, aberrant 2:6, aberrant 6:2, deviant 3:5, deviant 5:3, deviant 4:4, 1:7 and 7:1). At least 300 tetrads were evaluated for segregation and crossover patterns. In order to distinguish meiotic events from mitotic alterations, only four viable-spore tetrads were analyzed.
2.5 Mathematical analyses
Aberrant segregation and minisatellite tract-length alteration comparisons were done with Instat 1.12 (GraphPad) for Macintosh, using the Fisher’s Exact version of the chi-square test. Results are considered statistically significant if p ≤ 0.05. Crossover frequencies were calculated using the Perkins equation (Perkins, 1949). To compare crossover frequencies, the Fisher’s Exact test was performed using the number of nonrecombinant (PD − NPD + ½ (T − 2NPD)) and recombinant (PD − NPD + ½(T − 2NPD)) tetrads (Chua and Roeder, 1997).
3. Results
3.1 Loss of CSM3, MRC1, or TOF1 affects minisatellite stability during meiosis
We took a candidate gene approach to identify genes that have a role in minisatellite stability during meiosis, focusing on genes whose products are known to play a role in some aspect of DNA repair and/or recombination during meiosis. SLX1, SLX4, and MLH3 are involved in crossing over (Fekairi et al., 2009; Munoz et al., 2009; Santucci-Darmanin et al., 2002; Svendsen et al., 2009; Wang et al., 1999), UBX7 has been implicated in minisatellite instability in stationary phase yeast cells (Alver and Kirkpatrick, unpublished results), and CSM3, MRC1 and TOF1 form a complex that acts at the replication fork (Bando et al., 2009; Calzada et al., 2005; Nedelcheva et al., 2005). We used a yeast minisatellite model system developed by our lab, in which an HRAS1 minisatellite allele is inserted upstream of the HIS4 gene on chromosome III (Jauert et al., 2002). The primary minisatellite allele used in this study is the his4-H10 allele (Figure 2), which contains ten and a half repeats derived from the most common human HRAS1 allele A1 (Jauert and Kirkpatrick, 2005). In diploid strains homozygous for the his4-H10 insert and a deletion of the candidate gene of interest we used PCR to analyze the length of the minisatellite tracts in four-spore tetrads, to determine the total number of tetrads with an altered minisatellite allele after meiosis.
The wild-type his4-H10 strain had a total meiotic alteration frequency of 22% (46 altered alleles in 205 tetrads - Table 2). The most common minisatellite alteration during meiosis was a minisatellite length contraction in a single spore (16%), followed by tetrads with a minisatellite length expansion in a single spore (5%), while tetrads with two, three, or four spores with an altered minisatellite were observed at 0–1% (Table 2).
Table 2.
Distribution of minisatellite length alterations in various strain backgrounds
| Strain | Relevant mutation | HIS4 promoter | Total tetrads | % (No.) of tetrads with alteration | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No alteration | Single spore expansion | Single spore contraction | Two spore alterations | Three spore alterations | Four spore alterations | Total alterations | ||||
| DTK751 | + | his4-H10 | 205 | 78 (159) | 5 (10) | 16 (32) | 1 (2) | 1 (2) | 0 (0) | 22 (46) |
| DTK802 | rad1Δ | his4-H10 | 263 | 86 (226) | 1 (3)* | 11 (29) | 1 (3) | 0.4 (1) | 0.4 (1) | 14 (37)* |
| DTK1459 | csm3Δ | his4-H10 | 237 | 87 (207) | 1 (3)* | 11 (25) | 0.4 (1) | 0.4 (1) | 0 (0) | 13 (30)* |
| DTK1684 | mrc1Δ | his4-H10 | 255 | 85 (218) | 4 (11) | 8 (21)* | 0.4 (1) | 1 (2) | 1 (2) | 15 (37)* |
| DTK1685 | tof1Δ | his4-H10 | 224 | 85 (191) | 2 (5) | 11 (24) | 1 (3) | 0.4 (1) | 0 (0) | 15 (33)* |
| DTK1460 | slx1Δ | his4-H10 | 204 | 78 (159) | 2 (4) | 15 (31) | 2 (4) | 3 (6) | 0 (0) | 22 (45) |
| DTK1487 | slx4Δ | his4-H10 | 222 | 83 (184) | 2 (5) | 13 (28) | 1 (2) | 1 (2) | 0.5 (1) | 17 (38) |
| DTK1480 | ubx7Δ | his4-H10 | 222 | 84 (186) | 2 (4) | 11 (25) | 2 (4) | 1 (2) | 0.5 (1) | 16 (36) |
| DTK1567 | mlh3Δ | his4-H10 | 220 | 77 (169) | 2 (5) | 17 (38) | 2 (5) | 1 (3) | 0 (0) | 23 (51) |
| DTK314a | + | his4-A1 | 213 | 55 (117) | 9 (19) | 19 (41) | 10 (22) | 6 (13) | 0.5 (1) | 45 (95) |
| DTK508a | rad1Δ | his4-A1 | 275 | 52 (142) | 3 (9) | 24 (67) | 16 (44) | 4 (11) | 1 (2) | 48 (133) |
| DTK1724 | csm3Δ | his4-A1 | 206 | 57 (117) | 8 (17) | 22 (45) | 8 (16) | 4 (9) | 1 (2) | 43 (89) |
Asterisks indicates statistically significant difference compared to wild-type strain: DTK751 vs: DTK802 p = 0.0207; DTK1459 p = 0.0078; DTK1684 p = 0.0377; DTK1685 p = 0.0460.
Four of the candidate genes we analyzed (SLX1, SLX4, UBX7, and MLH3) had no statistically significant effect on the meiotic stability of the his4-H10 minisatellite (Table 2). We included SLX1, SLX4, and MLH3 in this study because they have each been previously determined to have a role in promoting meiotic crossing over in yeast and in humans (Fekairi et al., 2009; Munoz et al., 2009; Santucci-Darmanin et al., 2002; Svendsen et al., 2009; Wang et al., 1999), and in the case of SLX1 and SLX4 also have a role in maintaining the stability of the repetitive ribosomal DNA (rDNA) loci in S. cerevisiae and S. pombe (Coulon et al., 2004; Fricke and Brill, 2003; Mullen et al., 2001). While UBX7 has no role in meiotic crossing over, it has been implicated in minisatellite stability during stationary phase in yeast (Alver and Kirkpatrick, unpublished). Loss of these candidate genes had no significant effect on the pattern or the total percent of minisatellite length alterations (Table 2).
Our lab has previously shown that RAD1 plays a direct role in the meiotic stability of the larger HRAS1-A1 allele (Jauert et al., 2002). To determine if the loss of RAD1 also has an effect on the shorter A1-derived H10 allele, we examined minisatellite length alteration in a RAD1 mutant. We found that the total percent of tetrads with an altered minisatellite significantly decreased from 22% to 14% (p = 0.021, Table 2). Specifically, we observed a significant decrease in the percent of tetrads with a minisatellite length expansion (from 5% to 1%, p = 0.021), and an insignificant decrease in tetrads with a length contraction (from 16% to 11%, p = 0.17), while the percent of tetrads with minisatellite alterations in two, three, or four spores remained at 1% or less (Table 2). These data demonstrate that RAD1 functions in the meiotic stability of the his4-H10 allele, primarily in the type of repair that generates expansions, in agreement with our prior work with the HRAS1-A1 allele.
As our lab recently identified CSM3 in a forward screen for genes that have a significant effect on minisatellite stability during stationary phase in S. cerevisiae (B. Alver and D. Kirkpatrick, unpublished data), we examined CSM3 mutants for a meiotic minisatellite stability phenotype. In a CSM3 mutant, the total percent of tetrads with an altered minisatellite significantly decreased from 22% to 13% (p = 0.008, Table 2). Similar to the RAD1 mutant, we observed a significant decrease in the percent of tetrads with a length expansion (to 1%, p = 0.044) and a small but not significant decrease in length contractions (to 11%, p = 0.12), while the percent of tetrads with two, three or four altered spores remained at 1% or less (Table 2).
This result prompted us to analyze his4-H10 stability in strains bearing deletions in the CSM3 interacting partners MRC1 and TOF1. The his4-H10 mrc1Δ and tof1Δ strains also exhibited significant decreases in the total percent of minisatellite alterations, to 15% each (p = 0.038 and p = 0.046, respectively, Table 2). Similar to CSM3, a TOF1 mutation decreased the percent of tetrads with a minisatellite length expansion (to 2%, p = 0.19) and moderately decreased length contractions (to 11%, p = 0.15), although neither of these changes was significant (Table 2). In contrast, the MRC1 mutant exhibited a significant loss of minisatellite length contractions (to 8%, p = 0.018), about half that of a wild-type strain, and very little change in length expansions (Table 2). These results demonstrate that CSM3, MRC1, and TOF1 play roles in the meiotic stability of the HRAS1 minisatellite in yeast; CSM3 and TOF1 mutants have a loss of LLR activity similar to a RAD1 mutant, while MRC1 mutants exhibit a different spectrum of meiotic alterations, possibly indicating a different LLR activity (Table 2).
3.2 Loss of CSM3, MRC1, or TOF1 has no effect on HIS4 segregation and crossovers
As part of our analysis on minisatellite stability during meiosis, we measured the level of meiotic recombination at the HIS4 locus. Each diploid strain is heterozygous for the HIS4 gene, containing one wild-type allele and a mutant his4-lopc allele that bears a twenty-six base pair palindromic insertion within the HIS4 coding region that forms a stem-loop structure when incorporated into heteroduplex DNA (Nag et al., 1989). Utilizing an insert that is not easily repaired gives a better indication of the level of recombination events initiating at the HIS4 locus because it prevents large loop repair that restores Mendelian segregation (Figure 1). By following the segregation pattern of the HIS4/his4-lopc alleles we can measure the level of recombination at the HIS4 locus. A high frequency of aberrant segregation indicates a high level of recombination.
In a wild-type his4-H10 strain, the majority of tetrads exhibited 4:4 Mendelian segregation (53%, Table 3). Of the tetrads with an aberrant segregation, most exhibited a 3:5 or 5:3 PMS event (14% and 16%, respectively), while a smaller percent exhibited a 2:6 or 6:2 GC event (4% and 5%, respectively - Table 3). Deleting RAD1, CSM3, MRC1, or TOF1 in his4-H10 strains heterozygous for his4-lopc did not cause a significant deviation from the wild-type pattern of segregation, or in the overall percent of aberrant segregation (from 47% to 52% (rad1Δ), 50% (csm3Δ), 47% (mrc1Δ), and 48% (tof1Δ), respectively, Table 3).
Table 3.
Meiotic segregation patterns and crossover frequencies of diploid strains heterozygous for his4-lopc
| Strain | Relevant mutation | HIS4 promoter | Total tetrads | % Ab segb | Percent (No.) of tetrads with HIS4 segregaton patterna | HIS4-LEU2 distance (cM)a | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4:4 | 2:6 | 6:2 | 3:5 | 5:3 | Ab 4:4 | Otherd | ||||||
| DTK751 | WT | his4-H10 | 866 | 47 | 53 (455) | 4 (38) | 5 (47) | 14 (121) | 16 (137) | 3 (30) | 5 (46) | 32 |
| DTK802 | rad1Δ | his4-H10 | 311 | 52 | 48 (150) | 2 (6) | 7 (22) | 14 (43) | 18 (56) | 5 (16) | 6 (18) | 34 |
| DTK1459 | csm3Δ | his4-H10 | 318 | 50 | 50 (158) | 5 (15) | 3 (9) | 16 (52) | 19 (60) | 3 (9) | 5 (15) | 29 |
| DTK1684 | mrc1Δ | his4-H10 | 361 | 47 | 53 (190) | 4 (14) | 6 (21) | 11 (41) | 15 (55) | 1 (5) | 10 (35) | 35 |
| DTK1685 | tof1Δ | his4-H10 | 345 | 48 | 52 (178) | 2 (8) | 7 (25) | 15 (51) | 16 (55) | 2 (8) | 6 (20) | 33 |
| DTK1460 | slx1Δ | his4-H10 | 389 | 55 | 45 (175) | 3 (13) | 4 (16) | 19 (75) | 19 (74) | 4 (15) | 5 (21) | 34 |
| DTK1487 | slx4Δ | his4-H10 | 345 | 50 | 50 (173) | 2 (8) | 6 (20) | 17 (59) | 16 (54) | 4 (13) | 5 (18) | 28 |
| DTK1480 | ubx7Δ | his4-H10 | 381 | 50 | 50 (189) | 3 (10) | 6 (21) | 13 (50) | 21 (79) | 4 (17) | 4 (15) | 32 |
| DTK1567 | mlh3Δ | his4-H10 | 357 | 55 | 45 (161) | 4 (14) | 8 (27) | 14 (51) | 18 (64) | 5 (18) | 6 (22) | 35 |
| DTK314c | WT | his4-A1 | 733 | 47 | 53 (392) | 2 (17) | 9 (64) | 15 (109) | 15 (108) | 2 (16) | 4 (27) | 30 |
| DTK508c | rad1Δ | his4-A1 | 487 | 43 | 57 (279) | 5 (24) | 6 (28) | 13 (66) | 12 (56) | 5 (22) | 2 (12) | 25 |
| DTK1724 | csm3Δ | his4-A1 | 323 | 37 | 63 (205) | 5 (15) | 8 (26) | 7 (24) | 11 (37) | 3 (10) | 2 (6) | 33 |
See Materials and Methods.
Ab seg, aberrant segregation.
Includes: aberrant 2:6, ab 6:2, deviant 3:5, dev 5:3, dev 4:4, 1:7, 7:1, 0:8, and 8:0. WT = wild-type
We measured the level of intergenic crossing over occurring in the his4-H10 minisatellite-spanning interval by examining linkage between the HIS4/his4-lopc and LEU2/leu2-3 alleles. A wild-type his4-H10 strain had a map distance of 32 cM (Table 3). Deleting RAD1, CSM3, MRC1, or TOF1 did not significantly affect the level of crossovers in the HIS4-LEU2 interval (to 34, 29, 35, and 33 cM, respectively, Table 3).
3.3 Effect of the CSM3 mutation on his4-A1 minisatellite allele
Our lab has previously shown that RAD1 plays a direct role in the meiotic stability of the his4-A1 allele (Jauert et al., 2002). Because we identified CSM3 as having a role in the meiotic stability of the shorter his4-H10 allele, we analyzed the effect of deleting CSM3 on his4-A1 stability. A wild-type his4-A1 strain (Figure 2) exhibited minisatellites alterations in 45% of all tetrads (Table 2), with 19% of all tetrads exhibiting a minisatellite length decrease and 9% exhibiting a length increase (Table 2). These data demonstrate that the longer his4-A1 allele is significantly more unstable than the shorter his4-H10 derivative. (45% alteration versus 22%)
The his4-A1 rad1Δ strain showed a significant drop in the percent of tetrads that exhibited a minisatellite length expansion in a single spore, although the frequency of tetrads with a minisatellite alteration did not significantly differ from the wild-type strain (Jauert et al., 2002). In contrast, the his4-A1 csm3Δ strain did not show any deviation in the pattern of minisatellite alterations and the overall frequency of tract alterations was the same as in the wild-type strain (Table 2). Thus, loss of CSM3 appears to specifically affect the shorter his4-H10 tract, unlike RAD1 loss which affects both tracts. The reason behind this tract length difference for CSM3 is not known; it is possible that longer tract lengths adopt structures during alteration that are not Csm3 substrates. Neither the CSM3 nor the RAD1 mutation altered the level of his4-lopc aberrant segregation or the level of intergenic recombination between HIS4 and LEU2 in the his4-A1 strain (Table 3).
3.4 CSM3 and RAD1 mutations affect his4-lopd repair
The experiments above focused on minisatellite stability evaluation. To characterize the role of CSM3 in meiotic large loop repair (LLR) and compare it to the LLR activity of RAD1, we examined the meiotic segregation pattern of diploid strains heterozygous for the his4-lopd insertion. The his4-lopd insert is a 26 bp non-palindromic insertion within the HIS4 coding region that has been used extensively in examining large loop repair pathways during meiotic recombination (Jauert et al., 2002; Jensen et al., 2005; Kearney et al., 2001; Kirkpatrick and Petes, 1997). When incorporated into heteroduplex DNA, the his4-lopd allele forms a large 26 bp loop that is frequently repaired.
A wild-type strain exhibited his4-lopd aberrant segregation in 28% of tetrads, comprised of 21% GC events and 7% PMS events (Table 4). Within the class of tetrads with aberrant segregation of HIS4 alleles, 24% exhibited PMS (Table 4). These data indicate that the 26 bp loop formed by the his4-lopd insert is efficiently recognized and repaired when incorporated into heteroduplex DNA. Deleting either CSM3 or RAD1 did not significantly alter the total percent of his4-lopd aberrant segregation (27%; p = 0.75 and 30%; p = 0.5, respectively), however both mutations decreased the percent of GC events (from 21% to 15 and 17% respectively; Table 4)) and nearly doubled the percent of PMS events (from 7% to 12 and 14%; Table 4). This shift from GC to PMS events reflects a loss of repair of the 26 bp loop formed by the his4-lopd insert when incorporated into heteroduplex DNA (Figure 1). Both mutations increased the percent of 3:5 and 5:3 tetrads and reduced the percent of 2:6 tetrads, while the percent of 6:2 tetrads did not significantly change (Table 4). These data suggest that CSM3 and RAD1 are used primarily in the type of repair that fills-in loops when either the wild-type or his4-lopd strand initiates recombination (Figure 1), in agreement with previous data on meiotic large loop repair in RAD1 mutants (Jauert et al., 2002; Kearney et al., 2001; Kirkpatrick and Petes, 1997).
Table 4.
Meiotic segregation patterns and crossover frequencies of WT promoter strains heterozygous for his4-lopd
| Strain | Relevantmutation | Total tetrads | Percent (No.) of tetrads with HIS4 segregaton patterna | % Ab Seg | % GC | % PMS | % PMS/Ab Seg | HIS4-LEU2 distance (cM)a | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4:4 | 2:6 | 6:2 | 3:5 | 5:3 | Ab 4:4 | Otherb | ||||||||
| DNY27 | WT | 407 | 72 (293) | 11 (45) | 9 (35) | 1 (6) | 4 (17) | 0.5 (2) | 2 (9) | 28 | 21 | 7 | 24 | 28 |
| DTK1667 | csm3Δ | 357 | 73 (261) | 6 (22) | 8 (27) | 3 (12) | 8 (29) | 0 (0) | 2 (6) | 27 | 15 | 12 | 44 | 30 |
| DTK1731 | rad1Δ | 326 | 70 (227) | 4 (14) | 12 (38) | 5 (15) | 8 (26) | 0.6 (2) | 1 (4) | 30 | 17 | 14 | 46 | 29 |
| DTK1785 | csm3Δ rad1Δ | 320 | 66 (211) | 3 (10) | 9 (28) | 3 (10) | 16 (52) | 2 (5) | 1 (4) | 34 | 12 | 22 | 64 | 23 |
| DTK1887 | mrc1Δ | 568 | 78 (444) | 7 (38) | 7 (42) | 2 (12) | 4 (25) | 0.2 (1) | 1 (6) | 22 | 15 | 7 | 33 | 27 |
See Materials and Methods.
Includes: aberrant 2:6, ab 6:2, deviant 3:5, dev 5:3, dev 4:4, 1:7, 7:1, 0:8, and 8:0. WT = wild-type.
We also examined the effect of MRC1 loss on meiotic repair of the his4-lopd mismatch. Unlike the rad1Δ and csm3Δ strains, the mrc1Δ strain exhibited significantly lower levels of aberrant segregation (p = 0.03), with reductions in both classes of gene conversion tetrads (to 15% from 21% GC in the WT strain), although post-meiotic segregation tetrads were not affected (Table 4). This decrease in GC led to an apparent increase in the PMS/Ab seg ratio (Table 4), but for different reasons than the increases seen in the rad1Δ and csm3Δ strains. In addition, the mrc1Δ strain had a significantly lower level of spore viability (74%) compared to the other strains (i.e. csm3Δ = 82%, p < 0.0001). The mrc1Δ strain also showed the largest drop in spore viability in the his4-H10 his4-lopc strain set (Table 5). Finally, the individual isolates of mrc1Δ had a large amount of variability in sporulation frequency, viability and recombination frequency from plate to plate; these phenotypes were not seen with the other mutants we examined. The spore viability pattern indicated random spore death, rather than defects in meiosis I or II chromosome segregation (Table 5). These data suggest that MRC1 is required in at least a subset of gene conversion events, and lack of MRC1 function leads to a loss of those spores.
Table 5.
Meiotic spore viability patterns
| Strain | Relevant mutation | HIS4 promoter | HIS4 mutation | Total spore viability % | Spore viability patterna | ||||
|---|---|---|---|---|---|---|---|---|---|
| 4:0 | 3:1 | 2:2 | 1:3 | 0:4 | |||||
| DTK751 | WT | his4-H10 | his4-lopc | 90 | 866 | 188 | 101 | 18 | 9 |
| DTK802 | rad1Δ | his4-H10 | his4-lopc | 87 | 311 | 119 | 49 | 9 | 4 |
| DTK1459 | csm3Δ | his4-H10 | his4-lopc | 80 | 318 | 148 | 81 | 26 | 26 |
| DTK1684 | mrc1Δ | his4-H10 | his4-lopc | 67 | 361 | 253 | 231 | 108 | 76 |
| DTK1685 | tof1Δ | his4-H10 | his4-lopc | 73 | 345 | 214 | 192 | 80 | 25 |
| DTK1460 | slx1Δ | his4-H10 | his4-lopc | 86 | 389 | 86 | 36 | 9 | 33 |
| DTK1487 | slx4Δ | his4-H10 | his4-lopc | 85 | 345 | 110 | 58 | 12 | 15 |
| DTK1480 | ubx7Δ | his4-H10 | his4-lopc | 92 | 381 | 55 | 25 | 8 | 4 |
| DTK1567 | mlh3Δ | his4-H10 | his4-lopc | 80 | 357 | 129 | 84 | 35 | 27 |
| DTK314 | WT | his4-A1 | his4-lopc | 80 | 733 | 369 | 238 | 62 | 31 |
| DTK508 | rad1Δ | his4-A1 | his4-lopc | 77 | 487 | 357 | 172 | 77 | 29 |
| DTK1724 | csm3Δ | his4-A1 | his4-lopc | 62 | 323 | 366 | 350 | 192 | 87 |
| DNY27 | WT | WT | his4-lopd | 90 | 407 | 89 | 41 | 9 | 5 |
| DTK1667 | csm3Δ | WT | his4-lopd | 82 | 357 | 181 | 94 | 25 | 10 |
| DTK1731 | rad1Δ | WT | his4-lopd | 83 | 326 | 186 | 92 | 17 | 4 |
| DTK1785 | csm3Δ rad1Δ | WT | his4-lopd | 79 | 320 | 233 | 110 | 36 | 13 |
| DTK1887 | mrc1Δ | WT | his4-lopd | 74 | 568 | 411 | 291 | 94 | 48 |
live spore:dead spore. WT = wild-type
3.5 CSM3 RAD1 double mutant analysis
Components of the RAD1-dependent LLR pathway were previously identified in the his4-lopd background; these studies indicated the existence of at least one other meiotic LLR pathway that is independent of RAD1 (Kearney et al., 2001; Kirkpatrick and Petes, 1997). We analyzed HIS4 segregation in a csm3Δ rad1Δ strain heterozygous for his4-lopd to determine if both genes function in the same meiotic LLR pathway.
A csm3Δ rad1Δ strain exhibited his4-lopd aberrant segregation in 34% of tetrads, which is slightly but not significantly increased compared to wild-type (p = 0.09) and single mutant strains (p = 0.06 and p = 0.35, Table 4). Compared to a wild-type strain, the percent of GC events decreased by almost half and the percent of PMS events nearly tripled (Table 4). These data reveal that the CSM3 RAD1 double mutant has an even greater loss of large loop repair activity than either single mutant. Specifically, the change in the percent of 2:6, 6:2, and 3:5 tetrads is similar to either single mutant; the greatest difference is the increased percent of 5:3 tetrads, which is twice that of either single mutant and quadruple that observed in a wild-type strain (Table 4). This is reflected in the increased percent of aberrant segregation tetrads that exhibited a post-meiotic segregation (64%; Table 4), which is significantly higher than either single mutant. However, the increase seen in the double mutant is not a fully additive effect, as might be expected if CSM3 and RAD1 were in two completely separable LLR pathways. These results suggest that CSM3 and RAD1 play distinct but overlapping roles in the type of LLR activity that fills-in loops.
4. Discussion
4.1 RAD1, CSM3, TOF1, and MRC1 control minisatellite stability during meiosis
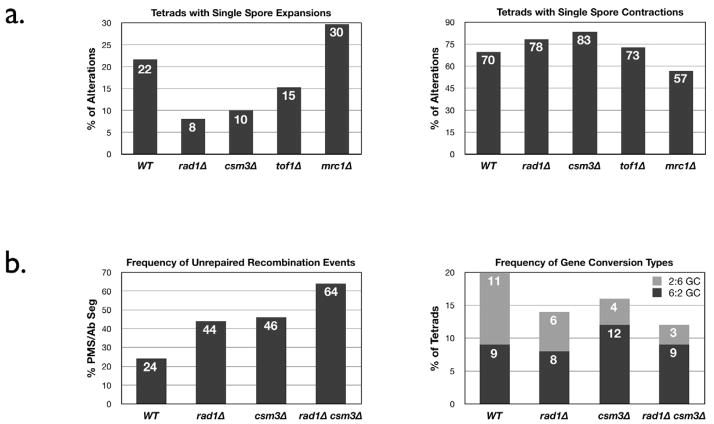
At least two pathways exist that function to repair large loops that form within heteroduplex DNA during meiotic recombination: the RAD1-dependent large loop repair (LLR) pathway and an uncharacterized RAD1-independent LLR pathway (Jauert et al., 2002; Kearney et al., 2001; Kirkpatrick and Petes, 1997). Our lab has previously shown that the RAD1-dependent LLR pathway controls the expansion, but not the contraction, of the HRAS1-A1 allele when inserted upstream of the HIS4 gene in yeast, and that these alterations are specific to meiosis (Jauert et al., 2002). In this study we identified three more factors that control meiotic minisatellite stability: CSM3, MRC1, and TOF1. Deleting each of these genes significantly decreases the frequency of meiotic minisatellite alterations. RAD1, CSM3, and TOF1 deletions significantly reduced expansion of the HRAS1 minisatellite tract length, while deletion of MRC1 significantly reduced the frequency of length contractions (Table 2). In Figure 3a, the frequency of alterations is expressed as a percentage of the total number of alterations seen in the strain. Loss of RAD1, CSM3 or TOF1 leads to a significant drop in observed expansions, while contractions are slightly increased. The RAD1 results match previous research on its role in minisatellite stability (Jauert et al., 2002; Kearney et al., 2001; Kirkpatrick and Petes, 1997), and suggests that CSM3 and TOF1 have a similar function in minisatellite stability. In contrast, MRC1 appears to act in a different manner, influencing tract contraction rather than expansion. Loss of MRC1 function decreases contractions from 70% of events to 57%, while expansions are mildly increased – a reversal of the rad1Δ/csm3Δ/tof1Δ pattern. MRC1 is the first gene we have identified that influences this aspect of minisatellite stability/
Figure 3.
A) Frequency of Meiotic Minisatellite Tract Alterations. Each bar represents the percentage of tetrads exhibiting a tract expansion or a contraction in a single spore for that particular mutant strain. Deletion of RAD1, CSM3 or TOF1 leads to a loss of expansions, while loss of MRC1 leads to a loss of contractions. B) Frequency of Meiotic his4-lopd Recombination Events. The left bar graph shows the frequency of unrepaired recombination events (PMS) to total aberrant segregation events as a percentage, demonstrating that deletion of RAD1 or CSM3 has equivalent effect on repair, while a double mutant exhibits significantly poorer repair. The right graph shows the frequencies of 2:6 and 6:2 gene conversion events in the mutant strains. 2:6 GC events are reduced in the mutants, indicating a loss of loop fill-in events (see Figure 1). Both graphs use data from Table 4.
4.2 An expanded model for minisatellite alterations during meiosis
We previously proposed a model for meiotic minisatellite alterations that involved misalignment of repeat units during strand invasion (Jauert et al., 2002). In this model, a DSB forms adjacent to the minisatellite, and strand resection generates 3′ single-stranded DNA which then invades the homologue, creating a region of heteroduplex DNA that includes the minisatellite. The repetitive minisatellite DNA can misalign, causing one or more of the repeat units to loop out of the heteroduplex region. Each repeat unit of the HRAS1 minisatellite is 28bp in length, so a one-repeat misalignment would generate a 28bp loop. The size of this loop is well within the functional range of the RAD1-dependent LLR pathway (Jensen et al., 2005).
The RAD1-dependent LLR pathway involves the RAD1, RAD10, MSH2, and MSH3 genes (Kearney et al., 2001; Kirkpatrick and Petes, 1997). We previously proposed a model, based on our research and the known mitotic activities of each of these gene products, in which the RAD1 and RAD10 endonucleases generate a nick in the strand opposite of the loop, with the MSH2 and MSH3 proteins serving as loop-recognition factors (Jauert et al., 2002; Jensen et al., 2005). The cleaved DNA strand is filled in by DNA synthesis, generating a minisatellite length expansion (Jauert et al., 2002; Jensen et al., 2005).
This model can be expanded to incorporate CSM3, MRC1, and TOF1. During mitotic growth, Csm3, Mrc1 and Tof1 form a heterotrimeric complex that interacts directly with the MCM helicase of the replisome, serving as a mediator of replication fork progression and DNA repair (Bando et al., 2009; Calzada et al., 2005; Nedelcheva et al., 2005). During meiotic recombination, DNA synthesis is necessary to repair the DSB using the homologue as template, and utilizes DNA replication factors such as POL30 and POL3 (Maloisel et al., 2004; Stone et al., 2008). CSM3, MRC1, and TOF1 genetically interact with POL30 and POL3 (Collins et al., 2007; Tong et al., 2004), with the MRX complex (MRE11/RAD50/XRS2) required for formation and processing of meiotic DSBs (Borde, 2007), and with RAD51, a strand exchange factor (Collins et al., 2007; Hunter, 2006; Pan et al., 2006; Tong et al., 2004). These data along with our results suggest that the CSM3/MRC1/TOF1 complex forms part of the DNA synthesis machinery involved in meiotic recombination, and is likely recruited to the heteroduplex intermediate.
We propose that the CSM3/MRC1/TOF1 complex localizes to heteroduplex DNA, and when a minisatellite length misalignment generates a large loop, the complex functions as a central player that mediates how the loop is repaired. Our data argue that CSM3 and TOF1 are involved in the type of repair that fills in loops (generating minisatellite length expansions), possibly by directly interacting with a helicase that opens the DNA duplex up around the region of the loop, providing easier access for the Rad1/10 endonuclease when it cleaves the strand opposite the loop. Candidate helicases include RAD3 and RAD5; CSM3 and TOF1 (but not MRC1) have been shown to genetically interact with both (Moriel-Carretero and Aguilera, 2010; Pan et al., 2006; Tong et al., 2004). Csm3 and Tof1 co-depend on each other for proper localization (Bando et al., 2009); this is likely why both mutants exhibit similar LLR phenotypes.
Intriguingly, our minisatellite data suggest that Mrc1 plays a role in the type of repair that removes the loop to generate minisatellite length contractions. In the event of replication stress during mitotic growth, Mrc1 (but not Csm3 or Tof1) is phosphorylated and recruits the sensor kinase Rad53 (Tanaka and Russell, 2001). It is possible that Mrc1 is also phosphorylated in the event of minisatellite misalignment and large loop formation, and transmits a signal to recruit an endonuclease to cleave the loop and generate a minisatellite length contraction (Figure 4). A candidate endonuclease is MUS81, which interacts genetically with MRC1 but not CSM3 or TOF1 (Collins et al., 2007). Unfortunately, MUS81 mutants do not sporulate in our AS4/AS13 strain background, preventing us from determining the role of MUS81 in minisatellite stability (unpublished data).
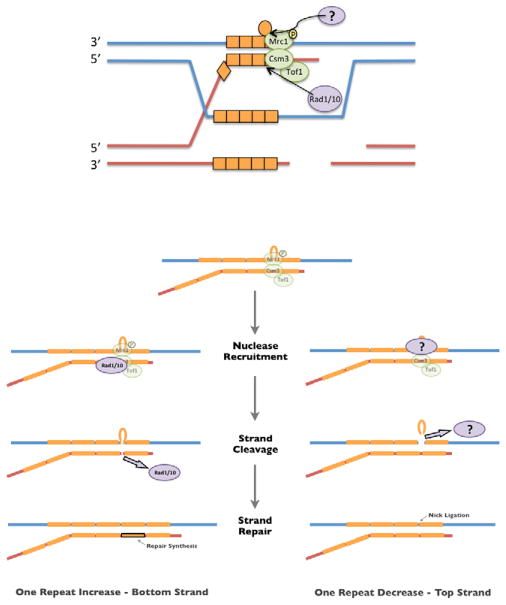
Figure 4.
A model for minisatellite length alterations during meiosis. An overview is shown at the top, with a detailed model for alterations at the bottom. During strand invasion and heteroduplex formation, a misalignment can occur within the repetitive minisatellite DNA (orange boxes). Extrusion of one of the repeat units will form a large 28bp loop; other misalignments are possible but not shown. Detection and repair of the loop can generate a minisatellite length expansion (left) or a contraction (right). Our data suggest that RAD1, RAD10, CSM3, and TOF1 function in large loop repair that generates length expansions. Our model depicts Rad1/10, directed by Csm3/Tof1, nicking the strand opposite the loop (red strand). The resulting gap will be filled in by DNA repair synthesis. Mrc1 functions in large loop removal (blue strand), which will generate a length contraction by recruiting an unknown endonuclease (possibly Mus81/Mms4) in response to the phosphorylation state of Mrc1.
While several studies have shown that MRC1, TOF1, and CSM3 have overlapping roles in DNA replication checkpoint activation (Bando et al., 2009; Calzada et al., 2005; Nedelcheva et al., 2005), our research is not the first to have observed a difference in function between MRC1 and CSM3/TOF1. Studies have shown that Mrc1 depends on Csm3-Tof1 for proper localization but the reverse is not true (Bando et al., 2009), and that Csm3-Tof1 but not Mrc1 is required for replication fork pausing at protein barriers (Calzada et al., 2005; Hodgson et al., 2007). Our data suggest a central role for the Csm3/Tof1/Mrc1 complex in discriminating between differing modes of repair during minisatellite tract alterations.
4.3 Analysis of CSM3 LLR activity in his4-lopd strains
CSM3 or RAD1 deletion in strains heterozygous for the his4-lopd insert lead to a nearly identical shift from GC to PMS tetrads in both strains (Table 4). Both mutants exhibited an increase in 3:5 and 5:3 tetrads and a decrease in 2:6 tetrads GC events (Figure 3b), while the total percent of HIS4 aberrant segregation remained unchanged (Table 4). These data can be explained by a loss of a specific type of large loop repair that fills in the loop (which would be accomplished by nicking the strand opposite the loop and filling in the gap with DNA synthesis), rather than repair that initiates with loop removal. This type of loop repair is consistent with the effect on minisatellite alterations observed in these mutants (Table 2). As shown in Figure 1, loss of fill-in loop repair would shift some of the 2:6 GC tetrads to 3:5 PMS tetrads when the DSB forms on the WT strand, and would shift the restored 4:4 tetrads to 5:3 tetrads when the DSB forms on the mutant strand. The expected result would be an increase in 3:5 and 5:3 tetrads, a loss of 2:6 tetrads, and no effect on 6:2 tetrads, which is what was observed in the RAD1 and CSM3 mutants (Table 4).
A double mutant analysis revealed that deleting both CSM3 and RAD1 exacerbated the phenotypes observed in either single mutant; the percent of PMS tetrads is significantly increased compared to either single mutant (Table 4, Figure 3b). This suggests that the CSM3 and RAD1 gene products may not have completely overlapping functions in LLR loop fill-in reactions.
4.4 Broader implications on minisatellite stability and human disease
This research has identified more factors that function in meiotic large loop repair. CSM3 appears to play a role in the type of large loop repair that fills-in loops, similar to the already characterized RAD1-dependent LLR pathway (Jensen et al., 2005; Kearney et al., 2001; Kirkpatrick and Petes, 1997). In contrast, MRC1 appears to be the first factor identified to function in the type of large loop repair that remove loops. The identification of large loop repair components is important for our understanding of factors that play a role in minisatellite stability. Minisatellite alterations during meiosis generate novel minisatellite alleles that are often associated with various types of human disease, as demonstrated by the correlation between human HRAS1 minisatellite alleles and cancer (Krontiris, 1995; Krontiris et al., 1993; Krontiris et al., 1985; Tamimi et al., 2003; Trepicchio and Krontiris, 1992). Furthering our understanding of how large loops are recognized and repaired during meiotic recombination will allow us to better understand the predisposition and initiation of human diseases associated with minisatellite DNA.
Highlights.
We find that Csm3, Mrc1 and Tof1 affect minisatellite stability during yeast meiosis
Minisatellite expansions are influenced by Csm3 and Tof1and contractions by Mrc1
Csm3 also affects large loop repair during gene conversion recombination
Csm3 acts in an overlapping pathway with Rad1/10 during gene conversion
We propose that Csm3/Mrc1/Tof1 mediates direction of repair during recombination
Acknowledgments
We thank Peter Jauert for help in strain construction and dissection of tetrads. This work was supported by a grant to David T. Kirkpatrick from the National Institutes of Health (5R01-GM-072598).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alani E, et al. Interaction between Mismatch Repair and Genetic Recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando M, et al. Csm3, Tof1, and Mrc1 Form a Heterotrimeric Mediator Complex That Associates with DNA Replication Forks. Journal of Biological Chemistry. 2009;284:34355–34365. doi: 10.1074/jbc.M109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Research. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- Calzada A, et al. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes & Development. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capon DJ, et al. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature. 1983;302:33–37. doi: 10.1038/302033a0. [DOI] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes & Development. 1997;11:1786–1800. doi: 10.1101/gad.11.14.1786. [DOI] [PubMed] [Google Scholar]
- Cohen JB, et al. A Repetitive Sequence Element 3′ of the human c-Ha-ras1 Gene Has Enhancer Activity. J Cell Physiol Suppl. 1987;5:75–81. doi: 10.1002/jcp.1041330415. [DOI] [PubMed] [Google Scholar]
- Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Coulon S, et al. Slx1-Slx4 Are Subunits of a Structure-specific Endonuclease That Maintains Ribosomal DNA in Fission Yeast. Molecular Biology of the Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detloff P, et al. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics. 1992;132:113–123. doi: 10.1093/genetics/132.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekairi S, et al. Human SLX4 is a Holliday Junction Resolvase Subunit that Binds Multiple DNA Repair/Recombination Endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S, Hurst DD. Meiotic gene conversion in yeast tetrads and the theory of recombination. Genetics. 1967;57 doi: 10.1093/genetics/57.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel S, et al. Mechanisms of meiotic gene conversion, or “Wanderings on a foreign strand”. In: Strathern JN, et al., editors. The Molecular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1981. pp. 289–393. [Google Scholar]
- Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes & Development. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Academic Press, Inc; San Diego, Calif: 1991. p. 194. [Google Scholar]
- Hodgson B, et al. Mrc1 and Tof1 Regulate DNA Replication Forks in Different Ways during Normal S Phase. Molecular Biology of the Cell. 2007;18:3894–3902. doi: 10.1091/mbc.E07-05-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic Recombination. Topics in Current Genetics. 2006;17:381–442. [Google Scholar]
- Jauert PA, et al. RAD1 Controls the Meiotic Expansion of the Human HRAS1 Minisatellite in Saccharomyces cerevisiae. Molecular and Cellular Biology. 2002;22:953–964. doi: 10.1128/MCB.22.3.953-964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauert PA, et al. A novel yeast genomic DNA library on a geneticin-resistance vector. Yeast. 2005;22:653–657. doi: 10.1002/yea.1250. [DOI] [PubMed] [Google Scholar]
- Jauert PA, Kirkpatrick DT. Length and Sequence Heterozygosity Differentially Affect HRAS1 Minisatellite Stability During Meiosis in Yeast. Genetics. 2005;170:601–612. doi: 10.1534/genetics.104.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, et al. Repeat instability at human minisatellites arising from meiotic recombination. EMBO J. 1998;17:4147–4157. doi: 10.1093/emboj/17.14.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LE, et al. The Large Loop Repair and Mismatch Repair Pathways of Saccharomyces cerevisiae Act on Distinct Substrates During Meiosis. Genetics. 2005;170:1033–1043. doi: 10.1534/genetics.104.033670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney HM, et al. Meiotic Recombination Involving Heterozygous Large Insertions in Saccharomyces cerevisiae: Formation and Repair of Large, Unpaired DNA Loops. Genetics. 2001;158:1457–1476. doi: 10.1093/genetics/158.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DT. Roles of the DNA mismatch repair and nucleotide excision repair proteins during meiosis. Cell Mol Life Sci. 1999;55:437–449. doi: 10.1007/s000180050300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DT, Petes TD. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature. 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DT, et al. Control of Meiotic Recombination and Gene Expression in Yeast by a Simple Repetitive DNA Sequence That Excludes Nucleosomes. Molecular and Cellular Biology. 1999;19:7661–7671. doi: 10.1128/mcb.19.11.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krontiris TG. Minisatellites and Human Disease. Science. 1995;269:1682–1683. doi: 10.1126/science.7569893. [DOI] [PubMed] [Google Scholar]
- Krontiris TG, et al. An association between cancer risk and mutations in the HRAS1 minisatellite locus. N Engl J Med. 1993;329:517–523. doi: 10.1056/NEJM199308193290801. [DOI] [PubMed] [Google Scholar]
- Krontiris TG, et al. Unique allelic restriction fragments of the human Ha-ras locus in leukocyte and tumour DNAs of cancer patients. Nature. 1985;313:369–374. doi: 10.1038/313369a0. [DOI] [PubMed] [Google Scholar]
- Maloisel L, et al. A Role for DNA Polymerase delta in Gene Conversion and Crossing Over During Meiosis in Saccharomyces cerevisiae. Genetics. 2004;167:1133–1142. doi: 10.1534/genetics.104.026260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel-Carretero M, Aguilera A. A Postincision-Deficient TFIIH Causes Replication Fork Breakage and Uncovers Alternative Rad51- or Pol32-Mediated Restart Mechanisms. Molecular Cell. 2010;37:690–701. doi: 10.1016/j.molcel.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Mullen JR, et al. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, et al. Coordination of Structure-Specific Nucleases by Human SLX4/BTBD12 is Required for DNA Repair. Molecular Cell. 2009;35:116–127. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- Nag DK, Petes TD. Physical Detection of Heteroduplexes during Meiotic Recombination in the Yeast Saccharomyces cerevisiae. Molecular and Cellular Biology. 1993;13:2324–2331. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag DK, et al. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:95–98. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- Nedelcheva MN, et al. Uncoupling of Unwinding from DNA synthesis Implies Regulation of the MCM helicase by Tof1/Mrc1/Csm3 Checkpoint Complex. Journal of Molecular Biology. 2005;347:509–521. doi: 10.1016/j.jmb.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Pan X, et al. A DNA Integrity Network in the Yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Perkins DD. Biochemical mutants in the smut fungus Ustilago maydis. Genetics. 1949;34:607–626. doi: 10.1093/genetics/34.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD, et al. Recombination in Yeast. In: Broach JR, et al., editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis, and Energetics. Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- Rabitsch KP, et al. A screen for genes required for meiosis and spore formation based on whole-genome expression. Current Biology. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- Roeder GS. Meiotic chromosomes: it takes two to tango. Genes & Development. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- Santucci-Darmanin S, et al. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Human Molecular Genetics. 2002;11:1697–1706. doi: 10.1093/hmg/11.15.1697. [DOI] [PubMed] [Google Scholar]
- Spandidos DA, Holmes L. Transcriptional enhancer activity in the variable tandem repeat DNA sequence downstream of the human Ha-ras1 gene. FEBS Lett. 1987;218:41–46. doi: 10.1016/0014-5793(87)81014-1. [DOI] [PubMed] [Google Scholar]
- Stapleton A, Petes TD. The Tn3 Beta-Lactamase Gene Acts as a Hotspot for Meiotic Recombination in Yeast. Genetics. 1992;127:39–51. doi: 10.1093/genetics/127.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JE, et al. Role of Proliferating Cell Nuclear Antigen Interactions in the Mismatch Repair-Dependent Processing of Mitotic and Meiotic Recombination Intermediates in Yeast. Genetics. 2008;178:1221–1236. doi: 10.1534/genetics.107.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, et al. Mammalian BTBD12/SLX4 Assembles A Holliday Junction Resolvase and is Required for DNA Repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamimi RM, et al. The HRAS1 Variable Number of Tandem Repeats and Risk of Breast Cancer. Cancer Epid Biomark. 2003;12:1528–1530. [PubMed] [Google Scholar]
- Tanaka K, Russell P. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nature Cell Biology. 2001;3:966–972. doi: 10.1038/ncb1101-966. [DOI] [PubMed] [Google Scholar]
- Tong AHY, et al. Global Mapping of the Yeast Genetic Interaction Network. Science. 2004;303:808–812. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Tourriere H, et al. Mrc1 and Tof1 Promote Replication Fork Progression and Recovery Independently of Rad53. Molecular Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- Trepicchio WL, Krontiris TG. Members of the rel/NF-kB family of transcriptional regulatory proteins bind the HRAS1 minisatellite DNA sequence. Nucleic Acids Res. 1992;20:2427–2434. doi: 10.1093/nar/20.10.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T-F, et al. Functional specificity of MutL homologs in yeast: Evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. NAS. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, et al. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. PNAS. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]