Abstract
Multisite phosphorylation modulates the function of regulatory proteins with complex signaling properties and outputs. The retinoblastoma tumor suppressor protein (Rb) is inactivated by Cyclin-dependent kinase (Cdk) phosphorylation in normal and cancer cell cycles, so understanding the molecular mechanisms and effects of Rb phosphorylation is imperative. Rb functions in diverse processes regulating proliferation, and it has been speculated that multisite phosphorylation might act as a code in which discrete phosphorylations control specific activities. The idea of an Rb phosphorylation code is evaluated here in light of recent studies of Rb structure and function. Rb inactivation is discussed with an emphasis on how multisite phosphorylation changes Rb structure and associations with protein partners.
Keywords: multisite phosphorylation, cell cycle regulation, Retinoblastoma protein, post-translational modifications, tumor suppressor protein, Cyclin-dependent kinases
Rb inactivation by multisite phosphorylation
The Rb tumor suppressor protein is inactivated in a number of diverse cancers [1]. Genetic lesions that lead to upregulated Cyclin-dependent kinase activity are more common than mutations to the Rb gene itself, reflecting the important function of Cdk in negatively regulating Rb and motivating intense research to develop therapeutic Cdk inhibitors that prevent Rb inactivation [2, 3]. Rb and its homologs p107 and p130, collectively known as the “pocket protein” family, were identified as cell cycle regulatory proteins and targets of oncogenic viral proteins that induce aberrant proliferation [4]. Like many other cell cycle Cdk substrates, pocket proteins are phosphorylated on multiple sites. Since the early observation of differentially phosphorylated forms of Rb, it is been proposed that phosphorylation events have distinct regulatory effects on Rb function [5]. Analogous to posttranslational modifications of histones and p53 [6, 7], this Rb phosphorylation code is an intriguing mechanism for integrating regulatory input signals and generating manifold responses.
Recent studies have reaffirmed the potential for an Rb code while revealing aspects of its mechanism and function. First, structural analysis has demonstrated that phosphorylation events induce unique conformations in Rb, each capable of making different protein interactions. These structural changes are a clear mechanism for transducing discrete phosphorylation signals into distinct functional outputs. Second, several mechanisms have been found that can independently control the phosphorylation state of individual sites; there is potential for code writing. Finally, new Rb tumor suppressor activities have been characterized that might utilize independent regulation, and clear predictions can now be made for how the code could separately turn off these different activities.
Rb structure and biochemical function
Rb was initially characterized as a regulator of transcription [8, 9]. It inhibits expression of genes under the control of E2F transcription factors by binding E2Fs and recruiting co-repressor proteins that modify histones and chromatin. More recently, Rb has been found to maintain chromosome structure and mediate protein degradation in mechanisms that are transcription-independent [10]. Despite its role in these diverse processes, the molecular function of Rb is consistent. Rb is an adaptor that assembles different protein and protein-DNA complexes for executing a wide-range of cellular control mechanisms. Indeed, nearly two hundred cellular proteins have been reported to associate with Rb [11]. Understanding how Rb is inactivated by phosphorylation, therefore, requires investigations of how Rb structure facilitates protein interactions and how phosphorylation modulates structure to regulate those interactions.
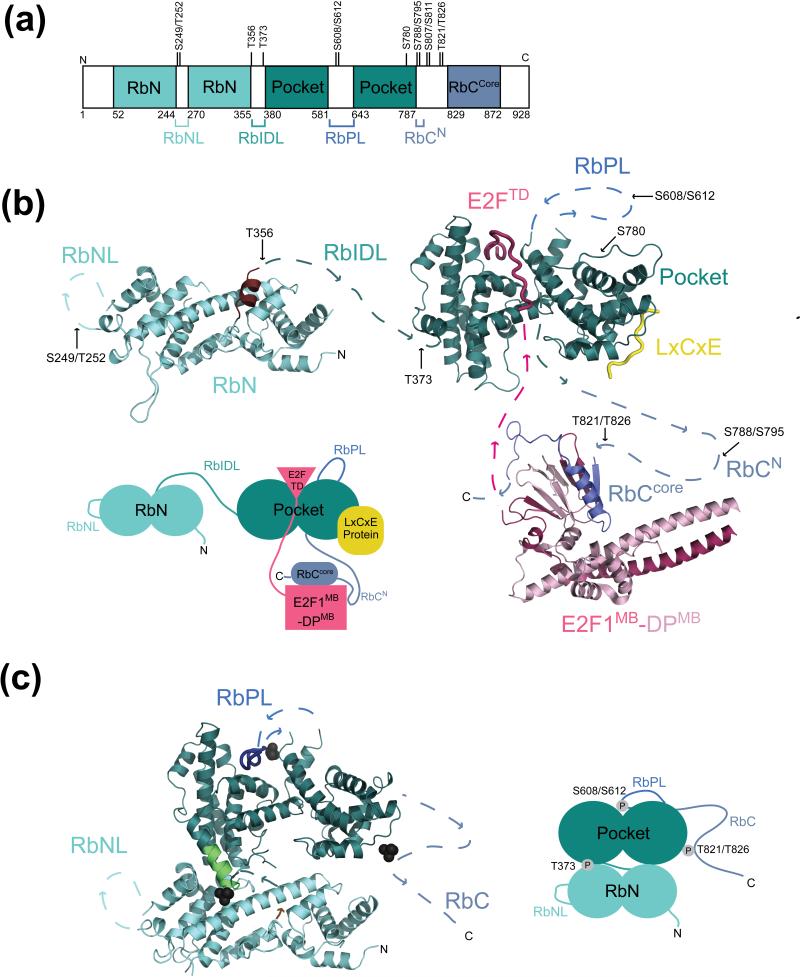
Rb contains two independently folded domains and a considerable amount (~33% of 928 amino acids) of intrinsically disordered primary sequence (Figure 1a). Both the structured N-terminal domain (RbN) and central pocket domain consist of two helical subdomains [12, 13] (Figure 1b). The disordered sequences include loops within RbN (here called RbNL) and the pocket (RbPL), and the C-terminal domain (RbC) (Figure 1a). In the unphosphorylated state, RbN and the pocket are flexibly tethered by an interdomain linker (RbIDL) (Figure 1b).
Figure 1.
Retinoblastoma protein (Rb) structure, phosphorylation sites, and protein interactions. (a) Domain organization and location of Cdk phosphorylation sites. The structured domains, which are colored, include the N-terminal domain (RbN) and the pocket domain. In contrast, several regions are intrinsically disordered including two large loops in RbN (RbNL) and the pocket (RbPL), an interdomain linker (RbIDL), and parts of the C-terminal domain (RbC). (b) Model of unphosphorylated Rb from crystal structures of individual domains and complexes with E2F and an LxCxE peptide (PDB codes: 2QDJ, 1GUX, 1N4M, and 2ACE). Unstructured loops and linkers connecting the structured domains are represented as broken lines. Approximate locations of phosphorylation sites are indicated. The Rb domains are colored as in (a); E2F is in pink, and the LxCxE peptide is in yellow. The C-terminal helix of RbN, which becomes disordered upon T356 phosphorylation is colored brown. (c) Model of phosphorylated Rb from crystal structures (PDB codes: 4ELJ and 4ELL). Only phosphates that are known to promote intramolecular interactions (T373, S608/S612, and T821/T826) are shown. T821/T826 induces binding of RbC to the pocket domain at the LxCxE site, although there is no high-resolution structure detailing this interaction. The N-terminal helix of the pocket domain that is nucleated by T373 phosphorylation is shown in green.
Rb interactions with other proteins have been mapped to several different surfaces, consistent with the idea that Rb utilizes multiple binding modes to assemble protein complexes. The best-characterized Rb interactions are with E2Fs. The pocket domain binds the E2F transactivation domain (E2FTD) in a cleft between the two helical subdomains (Figure 1b) [14, 15]. A second binding interface exists between RbC and the “marked box” domains of E2F and its obligate heterodimer partner DP (E2FMB-DPMB) [16]. Two regions of RbC, called RbCN and RbCcore, are responsible for this secondary interaction. As seen in the crystal structure of RbCcore-E2F1MB-DP1MB, RbCcore adopts a helix-turn-strand structure to bind the marked-box beta-sandwich domain [16] (Figure 1b). Although RbC-marked box binding has been observed between Rb and all E2Fs in vitro [16], the interaction in cells appears exclusive for E2F1 and mediates the specific E2F1 function of inducing apoptosis [17, 18].
The pocket domain contains a cleft within its second helical subdomain, which binds a linear ‘LxCxE’ sequence originally identified in viral oncoproteins [13, 19] (Figure 1b). Histone modifying enzymes, chromatin remodeling complexes, Cdh1 (of the anaphase promoting complex), the condensin II complex, and several other cellular proteins bind Rb in an LxCxE cleft-dependent manner [9, 11, 20-22]. There is no detailed structural data characterizing the particular nature of these protein interactions, and several of these proteins might bind Rb indirectly. Some of them contain an LxCxE motif in their sequence and are thought to bind similarly to the E7 protein from human papilloma virus [13]. However, biophysical data indicate the LxCxE residues are not sufficient for tight interaction [23], and some proteins that fail to bind an Rb with mutations in its LxCxE-biding cleft do not actually contain an ‘LxCxE’ sequence. These observations suggest that interactions at this site are likely more complex than commonly understood, and there could even be different, nonoverlapping modes of binding this pocket surface.
Regions of Rb outside the pocket domain mediate associations with several other proteins. RbN is responsible for binding several proteins, including the E1A-like inhibitor of differentiation (EID1), which inhibits Rb interactions with transcription-repressing chromatin factors, and origin replication complex proteins [12, 24]. A region of RbC C-terminal to RbCcore is the docking site for Cdks and the Rb-activating enzyme protein phosphatase 1 (PP1) [25-28].
Code translation: Rb phosphorylation induces diverse structures
The global conformational changes that occur in Rb upon phosphorylation are site-specific and remarkably diverse (Table 1 and Figure 1), providing a mechanism through which different phosphorylation events can code for different functional Rb outputs. Rb contains 13 conserved sites that are phosphorylated by Cdk in cycling cells (Figure 1a) [29]. These S/TP sites are located in disordered regions of the protein. Cdk phosphorylation typically promotes protein-protein interactions through creation of a phospho-epitope that becomes structured upon binding its target [30, 31]. In contrast, phosphorylation in Rb disrupts interactions with its binding partners, which creates a seeming paradox: how does phosphate addition in disordered protein regions inhibit protein interactions at the structured domains? Structural studies have answered this question, demonstrating that phosphorylation promotes interdomain interactions within Rb that render its structure incompatible with binding other proteins [16, 32, 33]. As commonly observed, phosphorylation promotes structure; however here, the disordered-to-ordered transitions are entirely intramolecular and result in intermolecular binding inhibition.
Table 1.
Summary of Cdk phosphorylation events in Rb
| Sites | Domain | Structural Effect | Biochemical Output |
|---|---|---|---|
| S249/T252 | RbN | Unknown | Inhibits protein interactions with RbN |
| T356 | RbIDL | C-terminal helix of RbN becomes disordered | Unknown |
| T373 | RbIDL | Nucleates N-terminal pocket helix to induce RbN-pocket association | Inhibits E2FTD and LxCxE binding to pocket domain |
| S608/S612 | RbL | RbL binds pocket | Inhibits E2FTD binding |
| S780 | Unknown | Unknown | |
| S788/S795 | RbC | Unknown | Inhibits RbC-E2F1MB-UPMB binding |
| S807/S811 | RbC | Unknown | Might prime phosphorylation at other sites |
| T821/T826 | RbC | Induces RbC binding to the pocket domain | Inhibits RbC-E2F1MB-DPMB binding and inhibits LxCxE binding to pocket domain. |
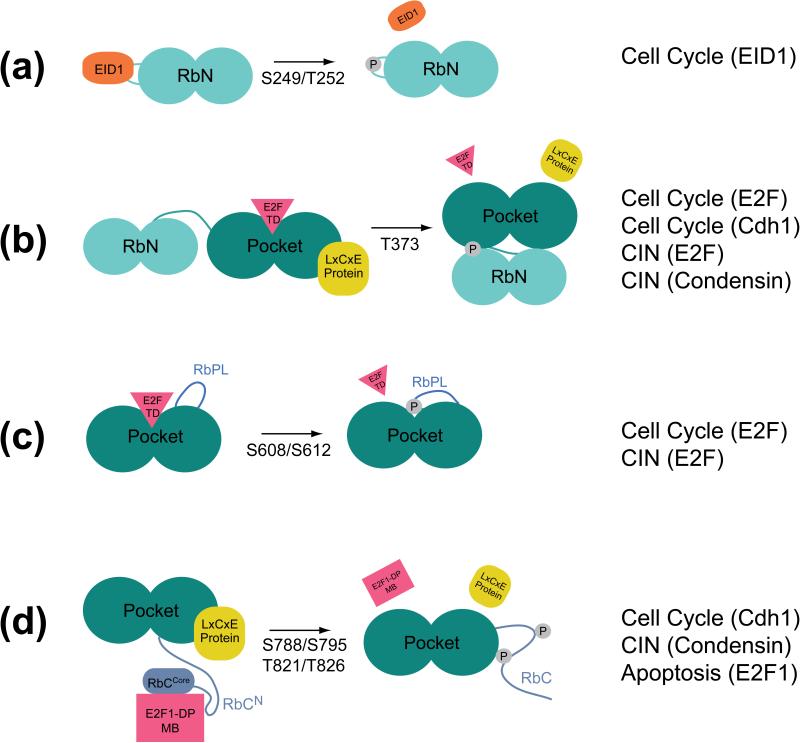
This mechanism was first demonstrated for Rb T821/T826 phosphorylation, which inhibits viral protein and histone deacetylase binding to the pocket domain [34, 35]. It was shown with purified proteins that phosphorylation of T821/T826 induces RbC binding to the pocket domain [16]. Phosphorylated RbC binding excludes LxCxE peptide binding, explaining how T821/T826 inhibits proteins that access the LxCxE cleft. T821/T826 immediately precedes the RbCcore sequence required for E2F1MB-DPMB binding, and the phosphorylation dependent RbC-pocket association is also incompatible with RbCcore binding to the marked-box domains (Figure 1b and 1c). S788/S795 phosphorylation in RbCN also inhibits Rb binding to E2F1MB-DPMB by excluding the interactions made in this region [16].
The association between the pocket domain and E2FTD is inhibited by S608/S612 and T373 phosphorylation [32, 33]. S608/S612 phosphorylation induces RbPL binding to the pocket domain (Figure 1c). A short sequence in RbPL binds the pocket as a helix in a manner that mimics and competes with E2FTD. The S608 phosphate stabilizes the RbPL-pocket association by capping the N-terminus of a pocket helix with hydrogen bonding and electrostatic interactions [32, 33]. T373 phosphorylation nucleates two additional turns in the most N-terminal pocket helix, creating a hydrophobic surface that binds a cleft in RbN (Figure 1c) [33]. This RbN-pocket docking allosterically disrupts the E2FTD binding cleft. Whereas T373 phosphorylation promotes helix formation, T356 phosphorylation appears to destabilize the C-terminal helix of RbN (Figure 1b and 1c) [33]. The function of this less common order-to-disorder transition in Rb is uncertain, as no protein interactions have yet been mapped to this region.
There are several possibilities for why two distinct and independent mechanisms exist for E2FTD inhibition. Rb pocket affinity for E2FTD is strong (Kd ~ 50 nM), and it could be that redundancy is needed for collective inhibition. Indeed, studies have shown that multiple phosphorylation events additively inhibit Rb-E2F binding and repression of E2F transcription [32, 36, 37]. However, there are interesting structural differences in how T373 (allosteric) and S608/S612 phosphorylation (competitive) inhibit E2FTD. The allosteric change to the E2FTD binding cleft is an efficient mechanism for release of bound E2F, whereas the competitive binding of RbPL is better suited for preventing the binding of free E2F. The interdomain interface promoted by T373 phosphorylation, but not S608/S612 phosphorylation, also inhibits binding at the LxCxE cleft in the pocket [33]. Notably, the RbN-pocket interface inhibits LxCxE peptide binding but not phosphorylated RbC (Figure 1c), suggesting that T373 and T821/T826 phosphorylation can inhibit distinct interactions made by the LxCxE binding surface.
The structural effects of phosphorylating other Cdk sites are less well understood. S259/T252 phosphorylation has no effect on E2F inhibition and does not influence the interdomain docking mechanism induced by T373 phosphorylation [32, 33]. S259/T252 phosphorylation inhibits binding of EID1 to RbN, and it was suggested that the effect of phosphorylation is electrostatic (Figure 2a) [12]. S780, implicated in cell cycle regulation [38], is located in a C-terminal tail of the pocket domain [13]. This extended peptide, which folds back to cover one helical subdomain, is not visible in all pocket crystal structures and may not be well ordered [19, 33]. One possibility is that phosphorylation modulates the association of this C-terminal element with the rest of rigid domain, which may regulate accessibility to an unknown binding surface. S807/S811 phosphorylation also does not directly inhibit E2F binding and does not contribute to the RbC-pocket association [16]. As described in the next section, an alternative function for S807/S811 might be in regulating phosphorylation at other sites.
Figure 2.
Specific phosphorylation events can inhibit protein interactions relevant to different Rb tumor suppressor functions. (a) S249/T252 phosphorylation inhibits EID1 binding through electrostatic repulsion. (b) T373 phosphorylation induces RbN-pocket docking and inhibits E2FTD and LxCxE protein binding. (c) S608/S612 phosphorylation inhibits E2FTD association by promoting RbPL binding to the pocket. (d) S788/S795 and T821/T826 phosphorylation inhibit E2F1MB-DPMB binding. T821/T826 promotes binding of RbC to the pocket and inhibits LxCxE protein binding.
Code writing: mechanisms for controlling Rb phosphorylation
There is considerable evidence that Rb can be selectively modified at discrete phosphorylation sites. Phosphorylation has been best characterized in the context of cell cycle progression, for which both Cdk4-CyclinD and Cdk2-CyclinE must phosphorylate Rb to induce S phase entry [5, 39]. Both kinases can phosphorylate the same S/TP consensus, but utilize unique docking sites in RbC. Cdk2-CyclinE binds a KxLxF sequence on Rb that is present in many Cdk substrates and inhibitors [25, 40]. Kinetic studies have implicated a region in the C-terminal ~50 residues of RbC as a necessary Cdk4-CyclinD docking site [27]. The recent crystal structures of Cdk4-CyclinD reveal that unlike in Cdk2, Cyclin binding and activation loop phosphorylation do not transform Cdk4 into its active conformation, suggesting that binding of Rb substrate or another protein might be important for stimulating the Cdk4 catalytic step [41, 42].
Consistent with these differences in enzymatic mechanisms, Cdk2 and Cdk4 phosphorylate Rb on only partially overlapping sets of sites [43, 44]. Past experiments to correlate phosphorylation events with particular kinases have been hampered by loss of enzyme specificity in in vitro experiments that utilize short peptides and in cell based experiments that rely on ectopic kinase expression [45]. Considering recent advances in mass spectrometry that enable quantitative analysis of phosphorylation content and the development of more selective small molecule kinase inhibitors, more accurate analysis of Cdk site specificity in Rb under endogenous conditions should now be feasible. With more sophisticated technologies for identifying phosphorylation sites, it will also be interesting to determine whether Rb is phosphorylated on nonconsensus Cdk sites; nonconsensus phosphorylation has been observed in other cell cycle regulatory proteins [46, 47].
Both Cdk2-CyclinE and Cdk4-CyclinD are required under endogenous cellular conditions to fully hyperphosphorylate Rb [5, 39, 43, 48]. Cdk4-CyclinD is activated earlier in G1 than Cdk2-CyclinE, and partial phosphorylation by Cdk4 is necessary for Cdk2 phosphorylation [39, 48]. Several lines of evidence suggest S807/S811 might function as Cdk4-activated priming sites. Cdk4 preferentially phosphorylates S807/S811 in vitro, and deletion of the Cdk4 docking site in RbC abrogates S807/S811 phosphorylation in cells [44, 49]. S807/S811 mutation results in loss of Rb phosphorylation levels beyond expected for mutation of only two sites, indicating an S807/S811 dependence for other phosphorylation events [36, 44]. K810 methylation, which directly inhibits S807/S811 phosphorylation, also results in reduced phosphorylation throughout Rb [50]. Together these results implicate a special role for S807/S811 in stimulating phosphorylation at other sites.
Although the enzymatic mechanism has not been explored for Rb, phosphate priming has been observed to stimulate Cdk and other kinases by promoting an enzyme-substrate association [47, 51]. That phosphorylation of S807/S811, unlike most other sites in Rb, does not seem to cause a structural change is consistent with the idea that priming at these sites promotes an intermolecular association to facilitate further phosphorylation. Another priming mechanism has recently been identified, in which phosphorylation of S608/S612 facilitates recruitment of the prolyl isomerase Pin1 and further phosphorylation of Rb [52].
Two phosphatases, PP1 and PP2A, dephosphorylate Rb at its Cdk sites and might also contribute to the production of distinct Rb-isoforms [53]. PP1 dephosphorylates Rb at mitotic exit by binding a sequence that overlaps with the Cdk2-Cyclin RxLxF docking site in RbC [26]. It was shown using phosphospecific antibodies that Cdk sites become dephosphorylated at different rates through mitotic exit into G1 [54]. Interestingly, sites that inhibit protein interactions at the LxCxE cleft (T373 and T826) are dephosphorylated first, whereas S608, which can prevent rebinding of E2FTD, is dephosphorylated most slowly. PP1 and PP2A also dephosphorylate Rb in response to different cellular stresses [53]. Although this dephosphorylation is usually global, in one case, hypoxia and DNA damage were observed to result specifically in T821 dephosphorylation [55]. T821 is not efficiently dephosphorylated at mitotic exit, indicating that it plays a role in Rb-mediated stress response, rather than normal cell cycle regulation [54, 55].
Several site-specific phosphorylation events have been observed outside the context of Cdk-dependent Rb inactivation in the cell cycle. For example, S795 is specifically phosphorylated in response to angiotensin II, a mitogenic agonist that signals through MAP kinase pathways [56]. In a recent study looking at the mechanism of HIV-induced neurotoxicity, factors secreted by HIV-infected macrophages specifically induce phosphorylation of S795 [57]. Some phosphorylation events are mediated by other kinases. For example, Aurora B phosphorylates Rb to prevent endoreplication after an aberrant mitosis [58]. In response to DNA damage, Chk2 phosphorylates S612, which stimulates E2F1 binding by an unknown mechanism [59]. S567 has been described as a Cdk site in the literature [35]. However, structural data indicate this site in the pocket is inaccessible in both the active and inactive (phosphorylated T373) conformations [13, 33], and it is likely not phosphorylated in normal, cycling cells [29]. Alternatively, S567 is phosphorylated by the MAP kinase p38 in response to genotoxic stress and promotes Rb degradation [60]. Together these examples demonstrate that specific phosphorylated Rb isoforms are generated in response to different signaling inputs. In the next section, the different functional outputs of Rb phosphorylation are considered.
Code output: inactivating Rb tumor suppression functions
Several Rb functions have been connected to its capacity for tumor suppression, each of which is mediated by interactions with different proteins. Therefore, considering how phosphorylation inhibits specific interaction surfaces in Rb provides a unique perspective for understanding how phosphorylation could inactivate tumor suppression function in specific contexts (Figure 2). The first and most extensively studied mechanism for Rb growth suppression is its negative regulation of the cell cycle through E2F repression [8]. Expression of Rb arrests proliferating cells in G1, and this arrest is overcome by Cdk phosphorylation. Many studies have examined the relative importance of specific Rb domains and phosphorylation sites in regulation of the cell cycle, E2F binding, and E2F transcription [32, 35-38, 61]. Sites in RbPL, RbIDL, and RbC all contribute to Rb inactivation in these assays, which is consistent with structural analyses demonstrating these phosphorylation events all induce conformational changes that inhibit binding of both E2F domains (Figures 1c, 2b, and 2c) [16, 32, 33]. Importantly, in assays of E2F reporter repression, RbN is required and T373 is the only site sufficient and necessary for complete Rb inactivation [36, 37, 61]. These observations point to the particular potency of the allosteric RbN-pocket docking mechanism for E2F transactivation domain release (Figure 2b).
An additional role for Rb in cell cycle regulation that is independent of E2F repression has been identified. Rb can induce G1 arrest through stabilizing the Cdk inhibitor p27 [20, 62, 63]. Rb promotes degradation of the p27 ubiquitin ligase Skp2 by recruiting Skp2 to its ubiquitin ligase Cdh1. The interaction site for Skp2 has been mapped to both the pocket domain and RbC, while Cdh1 binds the pocket domain. The role of phosphorylation in inactivating Rb stabilization of p27 has not yet been investigated in detail, but p27 is degraded upon progression into S phase when both Rb and p27 are phosphorylated, and Rb phosphorylation inhibits Cdh1 binding [20]. Mutations in the pocket LxCxE cleft also inhibit Cdh1 binding, suggesting that phosphorylation at T373 and T821/T826, both of which induce conformations that occlude the cleft, could inhibit the Rb-Cdh1 association (Figures 2b and 2d).
A role for Rb in maintaining chromosome structure during mitosis has been recently characterized and explains the chromosomal instability (CIN) and aneuploidy observed upon Rb loss [64]. This function has been connected in part to proper control of E2F transcription; misregulated expression of checkpoint and replication genes early in the cell cycle leads to chromosome errors later in division. Rb prevention of CIN has also been linked to its recruitment of factors involved in histone modifications and chromatin condensation [22, 65, 66]. Cells harboring an Rb mutant with specific defects in the LxCxE binding cleft (RbΔL) show chromosomal defects in mitosis [21, 65]. These defects result from failures in regulating pericentric heterochromatin formation through histone methylation and in recruiting the condensin II complex to chromatin. The fact that RbΔL cells properly regulate E2F confirms that protecting against CIN is at least in part an E2F-independent tumor suppressor activity. Accordingly, phosphorylation events that would inactivate Rb mitotic functions likely include events that inhibit both E2F repression and interactions at the LxCxE cleft (Figure 2b-d).
Rb has a cell cycle independent function in mediating apoptosis. E2F1 upregulates proapoptotic genes in response to genotoxic stress in proliferating cells, and in several studies Rb was shown to inhibit E2F1-induced apoptosis by repressing E2F gene expression [67, 68]. More consistent with the role of Rb as a tumor suppressor, it was recently found that Rb stimulates proapoptotic gene expression [69]. A phosphorylated Rb-E2F1 complex found at the promoters of actively transcribed genes following DNA damage depends on the E2F1-specific association with RbC [17, 18]. Interestingly, even in the absence of DNA damage, E2F1-Rb complexes have been observed during S phase when Rb is phosphorylated, however the specific sites have not been mapped [70, 71]. These observations support the notion that the RbC-E2F1 interaction is regulated independently of other Rb-E2F interactions that drive the normal cell cycle. Considering that the E2F1-specific interaction with RbC is inhibited by S788/S795 and T821/T826 phosphorylation (Figure 2d), it will be interesting to explore whether these sites specifically inactivate either the positive or negative apoptotic functions of Rb.
Is Rb an exceptional Cdk substrate?
Cdk phosphorylation has been well studied as a mechanism for controlling cell cycle events [72], but there are few structures of phosphorylated substrates that reveal the molecular mechanisms for how phosphorylation changes protein function. In the best-studied cases, phosphorylation creates a linear binding epitope that promotes a protein-protein interaction [30, 31]. Rb is the first example in which Cdk phosphorylation induces global structural changes. Analysis of Cdk phosphorylation site conservation in yeast indicates that for most sites, the location and sequence context of phosphorylation sites are not well conserved [73]. Such sites are suited for modulating multivalent protein interactions that rely on short epitopes. In contrast, Rb resembles a much smaller subset of proteins in which sites are in disordered regions but within conserved sequences near structured domains. It will be important to learn whether Rb is a rare or common example of a protein containing conserved sites that mediate specific conformational changes.
Cdk substrates are often phosphorylated many times, and this multisite phosphorylation is crucial for producing the proper signaling output [46, 47, 74-77]. Multisite phosphorylation influences signaling properties such as sensitivity and has been shown to be important for generating irreversible, switch-like cell cycle transitions [46, 74, 75, 77]. From these studies, a model for the function of multisite phosphorylation has been proposed in which sites are redundant in function and together shape signaling sensitivity towards a single output [74, 76, 77]. Outside of cell cycle regulation there are also several cases of multisite phosphorylation acting as a rheostat, incrementally tuning a single protein interaction affinity and corresponding signaling response [78, 79]. These mechanisms for modulating protein interactions by phosphorylation all rely on multivalency (redundant phosphorylated epitopes all bind the same site) or the tuning of bulk electrostatics. Strikingly, Rb does not behave in this way, as discrete phosphorylation events generate diverse structures with unique biochemical consequences. The observation that distinct conformations are capable of inhibiting different protein interactions is among the strongest indications that multisite phosphorylation of Rb codes for different outputs. Rb could be an exception among cell cycle regulatory proteins because it functions in other pathways, such as apoptosis, which are potentially subject to distinct regulation.
In the specific context of E2F repression for G1-S regulation, Rb might follow the paradigm that multiple sites together create sensitivity towards a single output. Several sites in Rb all inhibit E2F binding, which theory predicts could be utilized to generate an E2F activation response that is ultrasensitive to Cdk input [75]. Ultrasensitive responses, observed for the multiply phosphorylated Cdk regulators Sic1 and Wee1, are described by sharp changes in signaling output over a narrow range of input and are an important mechanism for generating irreversible cell cycle phase transitions. A recent study demonstrated that Rb-E2F is critical for an irreversible G1-S switch in response to growth factor signals [80], but further investigations are needed to clarify the role of multisite Rb phosphorylation in the mechanism.
Concluding Remarks
Several key observations support the notion of an Rb code in which distinct phosphorylation events modulate different Rb functions. Rb is a multifunctional protein, and studies of its inappropriate inactivation continue to reveal novel roles for Rb in controlling growth. There is evidence for selective phosphorylation in response to different signals, and there are enzymatic mechanisms capable of generating specific phosphoisoforms. Perhaps most importantly, phosphorylation events each generate unique structures that inhibit different protein binding sites.
Many questions remain, particularly about the cellular contexts in which specific sites are phosphorylated and what complexes are specifically disrupted by these events. Even considering only a particular context such as cell cycle regulation, it is uncertain whether different functions such as E2F regulation and p27 stabilization are in fact independently regulated or whether they are coordinately inactivated by hyperphosphorylation. Another manner of posing this question is to ask whether different subpopulations of Rb phosphoisoforms simultaneously regulate different functions in a cell or whether phosphorylation incites a uniform response to a specific signal. Rb protection from CIN is lost when Rb is genetically inactivated, but it is unclear why and whether this activity is regulated by phosphorylation in normal cells. To address these questions, future studies must identify more precisely the phosphorylation state of Rb throughout the cell cycle and in response to upstream signals and correlate specific phosphorylation events with the inactivation of each Rb tumor suppressor activity. Assigning specific inactivating functions to phosphorylation sites is an important remaining goal for understanding the complexity of Rb regulation and ultimately for designing therapeutics that can counter Rb inactivation in tumors with a greater degree of specificity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone A, et al. Inhibitors of cell cycle kinases: recent advances and future prospects as cancer therapeutics. Crit Rev Oncog. 2012;17:175–198. doi: 10.1615/critrevoncog.v17.i2.40. [DOI] [PubMed] [Google Scholar]
- 4.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 5.Mittnacht S. Control of pRB phosphorylation. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 6.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 9.Brehm A, Kouzarides T. Retinoblastoma protein meets chromatin. Trends Biochem Sci. 1999;24:142–145. doi: 10.1016/s0968-0004(99)01368-7. [DOI] [PubMed] [Google Scholar]
- 10.Henley SA, Dick FA. The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 2012;7:10. doi: 10.1186/1747-1028-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- 12.Hassler M, et al. Crystal structure of the retinoblastoma protein N domain provides insight into tumor suppression, ligand interaction, and holoprotein architecture. Mol Cell. 2007;28:371–385. doi: 10.1016/j.molcel.2007.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JO, et al. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 14.Lee C, et al. Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor. Genes Dev. 2002;16:3199–3212. doi: 10.1101/gad.1046102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao B, et al. Crystal structure of the retinoblastoma tumor suppressor protein bound to E2F and the molecular basis of its regulation. Proc Natl Acad Sci U S A. 2003;100:2363–2368. doi: 10.1073/pnas.0436813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubin SM, et al. Structure of the Rb C-terminal domain bound to E2F1-DP1: a mechanism for phosphorylation-induced E2F release. Cell. 2005;123:1093–1106. doi: 10.1016/j.cell.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 17.Julian LM, et al. Characterization of an E2F1-specific binding domain in pRB and its implications for apoptotic regulation. Oncogene. 2008;27:1572–1579. doi: 10.1038/sj.onc.1210803. [DOI] [PubMed] [Google Scholar]
- 18.Dick FA, Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol Cell. 2003;12:639–649. doi: 10.1016/s1097-2765(03)00344-7. [DOI] [PubMed] [Google Scholar]
- 19.Kim HY, et al. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 2001;20:295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binne UK, et al. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007;9:225–232. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- 21.Isaac CE, et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol Cell Biol. 2006;26:3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longworth MS, et al. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22:1011–1024. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh M, et al. Molecular determinants for the complex formation between the retinoblastoma protein and LXCXE sequences. J Biol Chem. 2005;280:37868–37876. doi: 10.1074/jbc.M504877200. [DOI] [PubMed] [Google Scholar]
- 24.Ahlander J, et al. The N-terminal domain of the Drosophila retinoblastoma protein Rbf1 interacts with ORC and associates with chromatin in an E2F independent manner. PLoS One. 2008;3:e2831. doi: 10.1371/journal.pone.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams PD, et al. Retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin-cdk complexes. Mol Cell Biol. 1999;19:1068–1080. doi: 10.1128/mcb.19.2.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirschi A, et al. An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein. Nat Struct Mol Biol. 2010;17:1051–1057. doi: 10.1038/nsmb.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan W, et al. A cyclin D1/cyclin-dependent kinase 4 binding site within the C domain of the retinoblastoma protein. Cancer Res. 2001;61:2885–2891. [PubMed] [Google Scholar]
- 28.Tamrakar S, Ludlow JW. The carboxyl-terminal region of the retinoblastoma protein binds non-competitively to protein phosphatase type 1alpha and inhibits catalytic activity. J Biol Chem. 2000;275:27784–27789. doi: 10.1074/jbc.M004542200. [DOI] [PubMed] [Google Scholar]
- 29.Lees JA, et al. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J. 1991;10:4279–4290. doi: 10.1002/j.1460-2075.1991.tb05006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao B, et al. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 31.Orlicky S, et al. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 32.Burke JR, et al. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem. 2010;285:16286–16293. doi: 10.1074/jbc.M110.108167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burke JR, et al. Structures of inactive retinoblastoma protein reveal multiple mechanisms for cell cycle control. Genes Dev. 2012;26:1156–1166. doi: 10.1101/gad.189837.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knudsen ES, Wang JY. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J Biol Chem. 1996;271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- 35.Harbour JW, et al. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999;98:859–869. doi: 10.1016/s0092-8674(00)81519-6. [DOI] [PubMed] [Google Scholar]
- 36.Brown VD, et al. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol Cell Biol. 1999;19:3246–3256. doi: 10.1128/mcb.19.5.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knudsen ES, Wang JY. Dual mechanisms for the inhibition of E2F binding to RB by cyclin-dependent kinase-mediated RB phosphorylation. Mol Cell Biol. 1997;17:5771–5783. doi: 10.1128/mcb.17.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chew YP, et al. pRB phosphorylation mutants reveal role of pRB in regulating S phase completion by a mechanism independent of E2F. Oncogene. 1998;17:2177–2186. doi: 10.1038/sj.onc.1202443. [DOI] [PubMed] [Google Scholar]
- 39.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe ED, et al. Specificity determinants of recruitment peptides bound to phospho-CDK2/cyclin A. Biochemistry. 2002;41:15625–15634. doi: 10.1021/bi0268910. [DOI] [PubMed] [Google Scholar]
- 41.Day PJ, et al. Crystal structure of human CDK4 in complex with a D-type cyclin. Proc Natl Acad Sci U S A. 2009;106:4166–4170. doi: 10.1073/pnas.0809645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takaki T, et al. The structure of CDK4/cyclin D3 has implications for models of CDK activation. Proc Natl Acad Sci U S A. 2009;106:4171–4176. doi: 10.1073/pnas.0809674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmitz NM, et al. Limited redundancy in phosphorylation of retinoblastoma tumor suppressor protein by cyclin-dependent kinases in acute lymphoblastic leukemia. Am J Pathol. 2006;169:1074–1079. doi: 10.2353/ajpath.2006.051137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem. 1997;272:12738–12746. doi: 10.1074/jbc.272.19.12738. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD. Regulation of the retinoblastoma tumor suppressor protein by cyclin/cdks. Biochim Biophys Acta. 2001;1471:M123–133. doi: 10.1016/s0304-419x(01)00019-1. [DOI] [PubMed] [Google Scholar]
- 46.Harvey SL, et al. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 2005;122:407–420. doi: 10.1016/j.cell.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 47.Koivomagi M, et al. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011;480:128–131. doi: 10.1038/nature10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ezhevsky SA, et al. Hypo-phosphorylation of the retinoblastoma protein (pRb) by cyclin D:Cdk4/6 complexes results in active pRb. Proc Natl Acad Sci U S A. 1997;94:10699–10704. doi: 10.1073/pnas.94.20.10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Driscoll B, et al. Discovery of a regulatory motif that controls the exposure of specific upstream cyclin-dependent kinase sites that determine both conformation and growth suppressing activity of pRb. J Biol Chem. 1999;274:9463–9471. doi: 10.1074/jbc.274.14.9463. [DOI] [PubMed] [Google Scholar]
- 50.Carr SM, et al. Interplay between lysine methylation and Cdk phosphorylation in growth control by the retinoblastoma protein. EMBO J. 2011;30:317–327. doi: 10.1038/emboj.2010.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma HT, Poon RY. How protein kinases co-ordinate mitosis in animal cells. Biochem J. 2011;435:17–31. doi: 10.1042/BJ20100284. [DOI] [PubMed] [Google Scholar]
- 52.Rizzolio F, et al. Retinoblastoma tumor-suppressor protein phosphorylation and inactivation depend on direct interaction with Pin1. Cell Death Differ. 2012;19:1152–1161. doi: 10.1038/cdd.2011.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolupaeva V, Janssens V. PP1 and PP2A phosphatases - cooperating partners in modulating retinoblastoma protein activation. FEBS J. 2012 doi: 10.1111/j.1742-4658.2012.08511.x. [DOI] [PubMed] [Google Scholar]
- 54.Rubin E, et al. Site-specific and temporally-regulated retinoblastoma protein dephosphorylation by protein phosphatase type 1. Oncogene. 2001;20:3776–3785. doi: 10.1038/sj.onc.1204518. [DOI] [PubMed] [Google Scholar]
- 55.Lentine B, et al. Dephosphorylation of threonine-821 of the retinoblastoma tumor suppressor protein (Rb) is required for apoptosis induced by UV and Cdk inhibition. Cell Cycle. 2012;11 doi: 10.4161/cc.21693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garnovskaya MN, et al. Mitogen-induced rapid phosphorylation of serine 795 of the retinoblastoma gene product in vascular smooth muscle cells involves ERK activation. J Biol Chem. 2004;279:24899–24905. doi: 10.1074/jbc.M311622200. [DOI] [PubMed] [Google Scholar]
- 57.Akay C, et al. Site-specific hyperphosphorylation of pRb in HIV-induced neurotoxicity. Mol Cell Neurosci. 2011;47:154–165. doi: 10.1016/j.mcn.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nair JS, et al. Aurora B kinase regulates the postmitotic endoreduplication checkpoint via phosphorylation of the retinoblastoma protein at serine 780. Mol Biol Cell. 2009;20:2218–2228. doi: 10.1091/mbc.E08-08-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue Y, et al. Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. EMBO J. 2007;26:2083–2093. doi: 10.1038/sj.emboj.7601652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delston RB, et al. p38 phosphorylates Rb on Ser567 by a novel, cell cycle-independent mechanism that triggers Rb-Hdm2 interaction and apoptosis. Oncogene. 2011;30:588–599. doi: 10.1038/onc.2010.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lents NH, et al. Reverse mutational analysis reveals threonine-373 as a potentially sufficient phosphorylation site for inactivation of the retinoblastoma tumor suppressor protein (pRB). Cell Cycle. 2006;5:1699–1707. doi: 10.4161/cc.5.15.3126. [DOI] [PubMed] [Google Scholar]
- 62.Ji P, et al. An Rb-Skp2-p27 pathway mediates acute cell cycle inhibition by Rb and is retained in a partial-penetrance Rb mutant. Mol Cell. 2004;16:47–58. doi: 10.1016/j.molcel.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 63.Wang H, et al. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/- mice. Nat Genet. 2010;42:83–88. doi: 10.1038/ng.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Manning AL, Dyson NJ. pRB, a tumor suppressor with a stabilizing presence. Trends Cell Biol. 2011;21:433–441. doi: 10.1016/j.tcb.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Coschi CH, et al. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010;24:1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manning AL, et al. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010;24:1364–1376. doi: 10.1101/gad.1917310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hallstrom TC, Nevins JR. Balancing the decision of cell proliferation and cell fate. Cell Cycle. 2009;8:532–535. doi: 10.4161/cc.8.4.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–657. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ianari A, et al. Proapoptotic function of the retinoblastoma tumor suppressor protein. Cancer Cell. 2009;15:184–194. doi: 10.1016/j.ccr.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cecchini MJ, Dick FA. The biochemical basis of CDK phosphorylation-independent regulation of E2F1 by the retinoblastoma protein. Biochem J. 2011;434:297–308. doi: 10.1042/BJ20101210. [DOI] [PubMed] [Google Scholar]
- 71.Wells J, et al. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sites during S phase. Oncogene. 2003;22:1445–1460. doi: 10.1038/sj.onc.1206264. [DOI] [PubMed] [Google Scholar]
- 72.Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 73.Holt LJ, et al. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nash P, et al. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- 75.Salazar C, Hofer T. Multisite protein phosphorylation--from molecular mechanisms to kinetic models. FEBS J. 2009;276:3177–3198. doi: 10.1111/j.1742-4658.2009.07027.x. [DOI] [PubMed] [Google Scholar]
- 76.Strickfaden SC, et al. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128:519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trunnell NB, et al. Ultrasensitivity in the Regulation of Cdc25C by Cdk1. Mol Cell. 2011;41:263–274. doi: 10.1016/j.molcel.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee CW, et al. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc Natl Acad Sci U S A. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pufall MA, et al. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 80.Yao G, et al. A bistable Rb-E2F switch underlies the restriction point. Nat Cell Biol. 2008;10:476–482. doi: 10.1038/ncb1711. [DOI] [PubMed] [Google Scholar]