Abstract
Background
Peanut-allergic subjects have highly stable pathologic antibody repertoires to the immunodominant B cell epitopes of the major peanut allergens Ara h 1-3.
Objective
We used a peptide microarray technique to analyze the effect of treatment with peanut oral immunotherapy (OIT) on such repertoires.
Methods
Measurements of total peanut-specific IgE (psIgE) and psIgG4 were made with CAP-FEIA. We analyzed sera from 22 OIT subjects and 6 controls and measured serum specific IgE and IgG4 binding to epitopes of Ara h 1-3 using a high-throughput peptide microarray technique. Antibody affinity was measured using a competitive peptide microarray as previously described.
Results
At baseline, psIgE and psIgG4 diversity were similar between subjects and controls, and there was broad variation in epitope recognition. After a median 41 months of OIT, polyclonal psIgG4 increased from a median 0.3 mcg/mL (IQR 0.1-0.43) at baseline to 10.5 mcg/mL (3.95-45.48) (p<0.0001) and included de novo specificities. PsIgE was reduced from a median baseline of 85.45 kUA/L (23.05-101.0) to 7.75 kUA/L (2.58-30.55) (p<0.0001). Affinity was unaffected. Although the psIgE repertoire contracted in most OIT-treated subjects, several subjects generated new IgE specificities even as the total psIgE decreased. Global epitope-specific shifts from IgE to IgG4 binding occurred, including at an informative epitope of Ara h 2.
Conclusion
OIT differentially alters Ara h 1-3 binding patterns. These changes are variable between subjects, not observed in controls, and include a progressive polyclonal increase in IgG4, with concurrent reduction in IgE amount and diversity.
Keywords: peanut allergy, oral immunotherapy, IgE, IgG4, peptide microarray, epitope, B cell, antibody affinity
INTRODUCTION
Peanut allergy is a serious immunologic disorder characterized by the production of highly pathogenic IgE antibodies that can mediate life-threatening anaphylaxis. No treatment is currently available to modify the natural history of the disease, although several experimental interventions are currently in clinical trials. One such intervention, oral immunotherapy (OIT), appears to induce a state of altered state of clinical reactivity called desensitization1-3. Mechanistically, desensitization is complex and may involve multiple mechanisms, including suppression of effector cells and allergen-specific Th2 responses, possibly via induction of regulatory T cells; and alterations in the amount of circulating allergen-specific antibody4. Specifically, peanut-specific IgE levels rise early in OIT, when administered allergen doses are low, but subsequently plateau and then begin to decrease, continuing to fall below pre-treatment values. This progressive fall in peanut-specific IgE levels is similar to the trend seen in patients who naturally outgrow peanut allergy. Consistent with other forms of immunotherapy, the repeated oral administration of allergen also leads to increased production of specific IgG, IgG4 and an inhibitory IgG-dependent serum factor which inhibits the binding of allergen to IgE1. Thus, previous studies have established that OIT affects the quantity of allergen-specific antibodies, but it is not known if the diversity, specificity, and affinity of such antibody repertoires change during treatment.
The potential of any food allergy treatment to alter these repertoires may be critical, given that antibody binding patterns have been shown to associate with peanut allergy phenotypes5-7. Specifically, IgE recognition of certain immunodominant regions within the major peanut allergens Ara h 1-3, as well as broad IgE epitope specificity overall, correlates with persistent disease and more severe reactions. These antibody repertoires appear durable and remain stable over time, although the longest interval studied so far was 20 months7,8. It is important to note that these data were generated from studies of patients receiving standard of care treatment with allergen avoidance.
Here we use a peptide microarray system to examine for the first time the longitudinal antibody repertoires of subjects undergoing treatment with OIT. We show that in subjects on OIT but not elimination diets, the amplitude, specificity and diversity of the peanut-specific IgE and IgG4 response are altered in a complex. These alterations in binding patterns are individualized, occur broadly across the major peanut allergens and affect informative epitopes within immunodominant regions.
METHODS
Subject Recruitment and Selection
Peanut-allergic subjects aged 1 to 16 years were recruited from the Allergy and Immunology clinics at Arkansas Children’s Hospital and Duke University Medical Center or surrounding community physician offices. These subjects were enrolled in an IRB-approved open-label trial of oral peanut immunotherapy. Protocol details and interim results of this study have been published previously1. Serum samples at baseline and throughout the study were collected and frozen at −20°C. Twenty-two subjects for whom multiple time points were available were included in this study. Six control samples, managed with diet restriction alone, were obtained from an IRB-approved biorepository of untreated peanut-allergic subjects recruited from the clinic of one of the investigators (A.W.B.).
Conventional Measurement of IgE and IgG4
Peanut-specific IgE and IgG4 levels were measured in serum samples in the laboratory of one of the investigators (A.W.B.) using the ImmunoCAP 100 instrument (Phadia AB, Uppsala SE) according to the manufacturer’s instructions.
Peptide Microarray Analyses
De-identified serum samples were sent to Mount Sinai for microarray analysis. A library of peptides, consisting of 15 amino acids overlapping by 12 (3-offset), corresponding to the primary sequences of Ara h 1, Ara h 2 and Ara h 3 was commercially synthesized by JPT Peptide Technologies (Berlin, Germany). Peptides were printed in two sets of triplicates onto Arrayit® SuperEpoxy glass slides (Arrayit Corporation, Sunnyvale, CA) as previously described9.
Immunolabeling was performed as previously described with some modifications10,11. In brief, the slides were blocked with 400μl of 1% human serum albumin (HSA) in phosphate-buffered saline containing 0.05% Tween 20 (PBS-T) for 60 minutes at room temperature, followed by incubation with 250μl of patient serum diluted 1:5 in PBS-T/HSA for 24 hours at 4°C. Slides were then washed with PBS-T and incubated for 24 hours at 4°C with a cocktail of several monoclonal antibodies including three biotinylated monoclonal anti-human IgE antibodies: one from Invitrogen (Carlsbad, CA, USA) diluted 1:250, one from BD Biosciences Pharmingen (San Jose, CA, USA) diluted 1:250, and one as a gift from Phadia (Uppsala, Sweden), biotinylated in our laboratory and diluted 1:1000, and one monoclonal anti-human IgG4-FITC (Southern Biotech, Birmingham, AL, USA) diluted 1:1000 in PBS-T/HSA. Slides were then incubated for 3 hours at 31°C with a cocktail of Anti-Biotin-Dendrimer_Oyster 550 (350) (Genisphere) and Anti-FITC_Dendrimer_Oyster 650 (350) (Genisphere, Hatfield, PA) in Dendrimer Buffer (Genisphere), both at 0.6μg/ml with the addition of 0.02μg/ml of salmon sperm DNA (Invitrogen), followed by wash with PBS-T, 15 mM Tris, 0.1X PBS, and 0.05X PBS. Slides were centrifuged dried and scanned using a ScanArray®Gx (PerkinElmer, Waltham, MA). Images were saved as TIF format.
Fluorescence signal of each spot was digitized with the ScanArray Express Microarray Analysis System (PerkinElmer), exported as comma-delimited (CSV) files and transformed into robust Z-scores, as previously described12. An individual peptide is considered positive if its Z-score is >3, meaning that the signal was above the background with p value less than 0.003.
A competition assay was performed by immunolabeling the slide as described above with an additional incubation using 1 mg/ml of peanut extract at 16°C for 1 hour after serum incubation and before application of the 2nd antibody. Peanut extract was prepared from commercially available roasted peanut as described previously13. Protein concentration was determined using BCA protein assays (Pierce, Rockport, IL)
Statistical Analyses
Descriptive statistics, including medians, means, and measures of variance, were tabulated for all study parameters. Nonparametric methods, including two-sample Wilcoxon tests, Spearman correlation, and simple linear regression models, were used to analyze results (Prism 5 for Windows, GraphPad, La Jolla CA). Wilcoxon signed rank pair-wise comparisons were utilized where appropriate for pre-post values obtained from the same subject. A p-value < 0.05 was considered significant. Pooled IgE and IgG4 binding peptides/regions were identified using TileMap, a tool for tiling array analysis,14 based on a hierarchical empirical Bayes model with moving average method. As hundreds of peptides were analyzed simultaneously, false discovery rate (FDR), was calculated to adjust for multiple comparisons.
RESULTS
The reduction in psIgE during OIT is polyclonal and may preferentially affect IgE that binds with high intensity
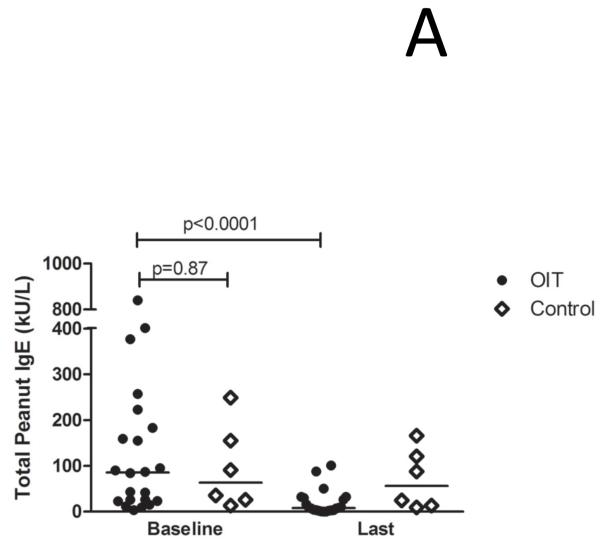
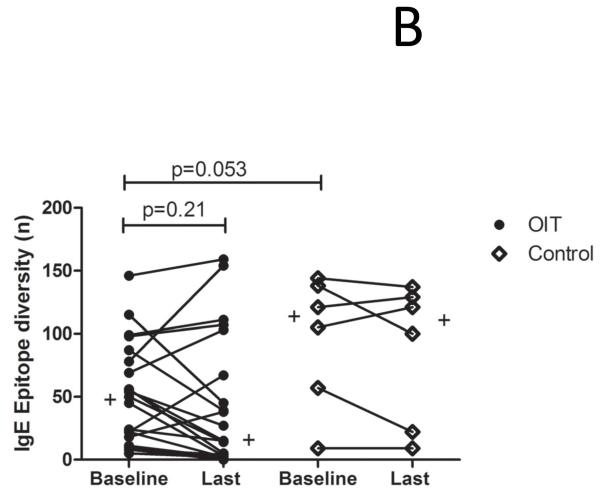
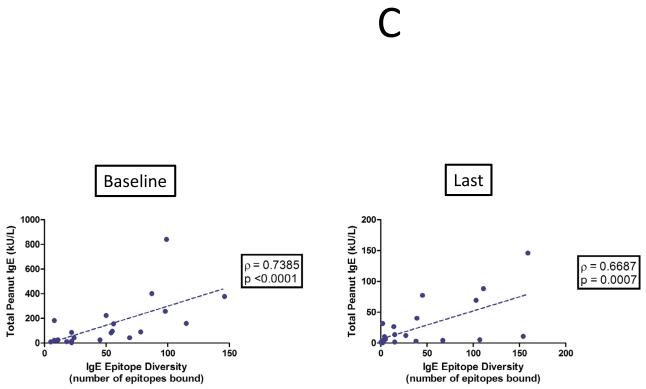
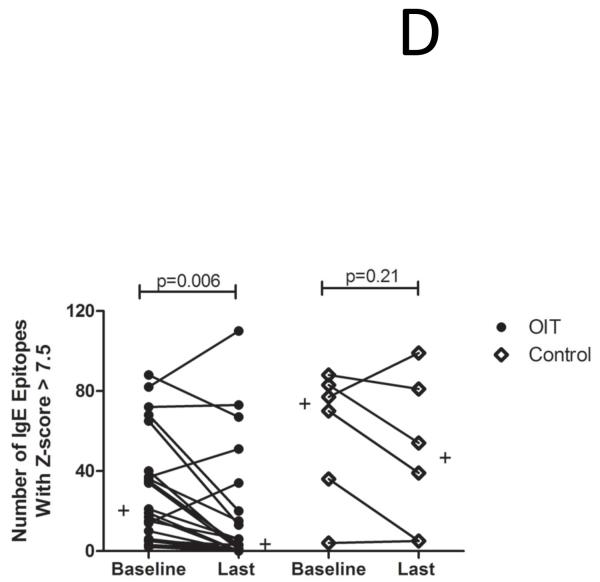
The demographics of the study and control populations are shown in Table E1 (Online Repository). Whereas psIgE levels remain stable in untreated controls, they decrease from a median of 85.5 kUA/L at baseline to 7.8 kUA/L after a median of 41 months (range 24-53 mo) of OIT treatment (p<0.0001) (Figure 1A). To account for differences in age between the study populations, we performed a nonparametric analysis of pre-post differences in peanut-specific IgE using median regression while adjusting for baseline age. In the control group, there was no significant difference in pre-post peanut-specific IgE levels when controlling for age (p=0.77), whereas in OIT subjects the pre-post difference remained significant even after adjustment (p=0.002). Although the total production of psIgE as measured by CAP-FEIA is reduced while on OIT, the median number of total Ara h 1-3 epitopes bound by IgE did not change over time in either group (Figure 1B), suggesting that the quantitative reduction in psIgE levels observed during OIT is broad in scope and not due to the elimination of a few specific B cell clones. Although the control group appeared to have a more diverse IgE repertoire, this difference did not achieve statistical significance. PsIgE concentration and repertoire diversity were highly correlated at baseline (correlation coefficient ρ= 0.7385 [p<0.0001]) and remained so at 41 months (ρ=0.6687 [p=0.0007]), further confirming the overall preservation of IgE epitope diversity while total psIgE changes during OIT (Figure 1C). Since we did not see significant numbers of peptides in which IgE binding converted from positive to negative (i.e., a Z-score < 3), we next assessed whether OIT could reduce high-intensity binding on the array. When we focused on areas of Ara h 1-3 that bound IgE with a Z-score > 7.5 at baseline, we observed a reduction in the number of such epitopes bound over time, from a median of 20 at baseline, which decreased to 3 (p=0.006) after treatment. (Figure 1D).
Figure 1.
OIT alters the peanut-specific immunoglobulin response in quantity and intensity. In these figures, each data point represents one subject. Lines or plus signs indicate medians. (A) Peanut-specific IgE, as measured by conventional CAP-FEIA, decreases from a median of 85.5 kUA/L at baseline to 7.8 kUA/L after OIT treatment (p<0.0001). (B) The overall IgE repertoire remains stable even as psIgE decreases. All array spots with a Z-score > 3 were considered positive. (C) IgE level and epitope diversity are strongly correlated at baseline and at the time of the last measurement, after a median of 41 months of treatment. (D) Fewer peanut peptides are bound by IgE with high intensity over the course of OIT treatment.
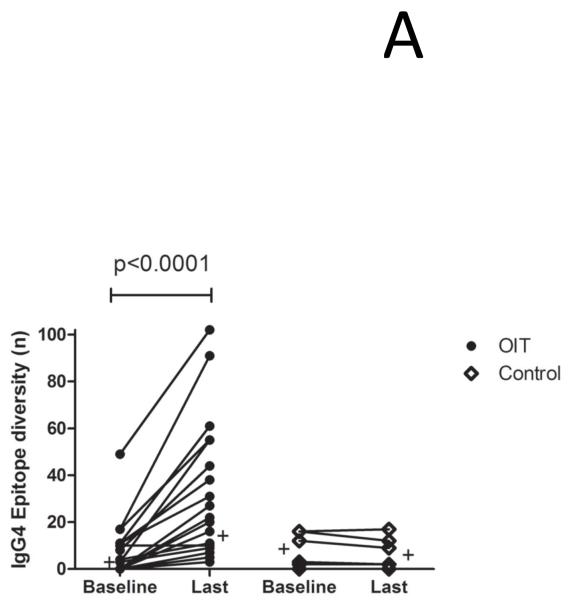
OIT induces a polyclonal expansion of peanut specific IgG4 production which includes novel specificities
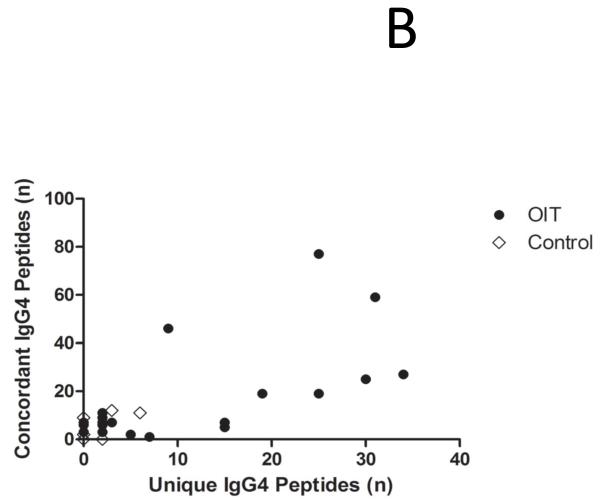
As shown in Figure 2A, a small repertoire of psIgG4 is detectable at baseline in both groups, but only in treated subjects does the production of psIgG4 diversify over time, increasing from a median of 1.5 array spots at baseline to 18 after a median treatment length of 41 months (p<0.0001). This is concurrent with a progressive rise in the total amount of psIgG4 produced, from a median 0.3 mcg/mL (IQR 0.1-0.43) at baseline to 10.5 mcg/mL (3.95-45.48) (p<0.0001) (Figure E1 in the Online Repository). In order to understand whether the IgG4 that emerges during OIT has a similar specificity as the original sensitization profile, we compared the binding patterns of IgG4 at the last measurement to the IgE binding patterns at baseline. Peptides were defined as new if IgE did not bind them at baseline, and concordant if they were. As shown in Figure 2B, the IgG4 repertoire in untreated controls generally does not expand to include new specificities. In contrast, in the serum of 8/22 (36%) OIT subjects, the number of de novo peptides recognized by the IgG4 repertoire expands by > 10. At the last timepoint measured, IgG4 of the 22 OIT subjects recognized 584 total peptides (mean 26.54 per subject), of which 228 (39%) were new, compared to 45 total among 6 controls (mean 7.5 per subject), of which 11 were new (24.4%) [p=0.056,Fisher’s exact test].
Figure 2.
OIT increases the production of broadly diverse IgG4 antibodies, some of which have de novo specificity. (A) A polyclonal expansion of IgG4 production occurs while on OIT but not an allergen elimination diet. Plus signs indicate median values. (B) The specificity of the IgG4 response after treatment includes both peptides recognized before OIT and new specificities. Each circle or diamond represents one subject.
The binding affinity of peanut-specific IgE and IgG4 is stable over time and largely unaffected by OIT
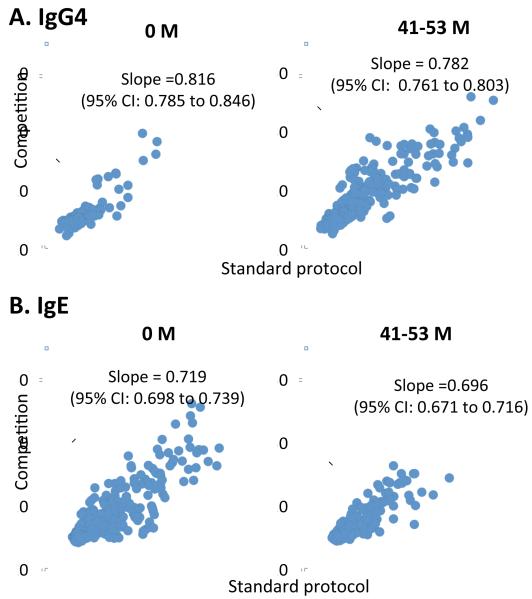
Because high-intensity IgE binding appeared to be preferentially reduced over time, paralleled by an emerging polyclonal IgG4 response, we considered the possibility that alterations in antibody affinity may occur through affinity maturation during OIT. To investigate this, we performed competition experiments in a subset of subjects who had detectable IgE binding at both baseline and at the last time point. As shown in Figure 3, compared with the standard protocol, antibody binding to peptides was slightly reduced when peanut extract was added as a competitor. The overall decrease of IgE binding achieved a statistically significant difference. However, the competition effect was weak for both IgE and IgG4 binding, and no obvious difference between time points could be observed.
Figure 3.
Scatter plot comparing IgG4 (A) and IgE (B) binding to the array between the standard protocol (x-axis) and the competition assay (y-axis) from baseline and last time points of 5 subjects. Each spot represents antibody binding (represented in Z-score) to one peanut peptide on the array.
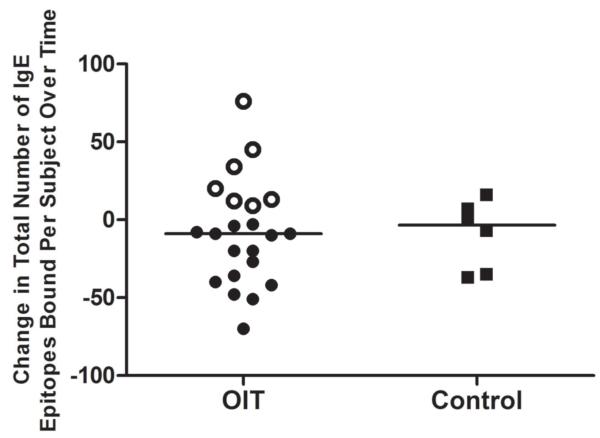
Diversity of the psIgE repertoire varies by individual during OIT and may change independently of the psIgE level
To further assess OIT’s effect on the IgE repertoire, we investigated individual variation by calculating the difference in the number of Ara h 1-3 epitopes bound by individual subjects’ IgE at baseline and after a median of 41 months on OIT. As demonstrated in Figure 4, the number of epitopes bound by IgE was reduced in 15/22 (68%) of subjects, indicating a tapering of the antibody repertoire over time. However, an increase in the diversity of IgE binding was observed in 7/22 (32%). Interestingly, in all of these seven subjects, as the IgE repertoire was expanding, the total psIgE decreased, from a median of 90.3 kUA/L (IQR, 15-377 kUA/L) at baseline to 9.1 kUA/L (4.5-88 kUA/L) after median 41 months (data not shown). In contrast, repertoires and total IgE levels of controls remained stable. These data suggest that new B cell clones could emerge and produce novel epitope-specific IgE during the course of OIT, even as the overall peanut IgE response is being suppressed.
Figure 4.
The peanut IgE repertoire contracts in most but not all subjects treated with OIT. The number of array spots bound by IgE was compared from baseline to the last available measurement for each subject.
OIT induces shifts in global binding patterns involving informative epitopes, with varying degrees of concordance
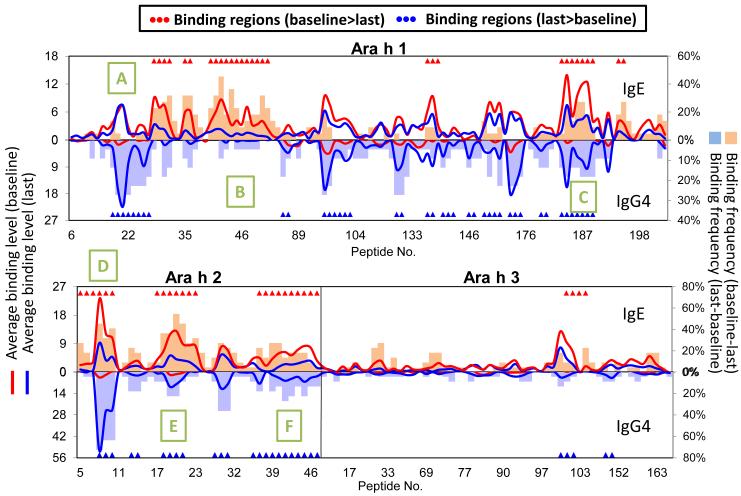
In order to assess global epitope binding patterns and to observe any possible relationship between epitope-specific IgE and IgG4 production, we constructed epitope maps of binding patterns for all subjects in the study. As previously reported, we observed recognition of immunodominant regions and significant variation in binding patterns between individual subjects (not shown). The frequencies of antibody binding across the primary sequence of Ara h 1-3, as well as the average z-score for each peptide, and the changes over time in each parameter, are represented in Figure 5 for all treated subjects. For these subjects, ten IgE binding regions consisting of 57 peptides (Ara h 1: 30 peptides/6 regions, Ara h 2: 23 peptides/3 regions, Ara h 3: 4 peptides/1 region) were identified using TileMap as having significantly greater IgE binding (p<0.01, FDR<0.1) at baseline than at last time point measured. Similarly, eighteen IgG4 binding regions consisting of 69 peptides (Ara h 1: 41 peptides/11 regions, Ara h 2: 23 peptides/5 regions, Ara h 3: 5 peptides/2 region) were identified as having significantly greater IgG4 binding (p<0.01, FDR<0.1) at the last time point than at baseline. No region was found to have significantly greater IgE binding at the last time point than baseline, or conversely, significantly greater IgG4 binding at baseline than at the last time point. Interestingly, reduction in IgE binding was discordant with specific IgG4 production in two regions of Ara h 1, as indicated by the green boxes labeled A and B in Figure 5. However, in another immunodominant region of the protein (Box C), the reduction in IgE binding was inversely associated with IgG4 recognition. Similarly, the changes in Ara h 2 binding patterns (Boxes D-F) was much more concordant, including importantly at epitopes previously identified to be “informative (Box D).” Binding patterns did not significantly change over time in controls (Figure E2 in the Online Repository).
Figure 5.
Comparison of IgE and IgG4 binding of peanut OIT treated patients to peptides of Ara h 1-3 between baseline and last time point. The x-axis shows the overlapping peptides. The left y-axis shows the average IgE and IgG4 binding level (represented as Z-scores) to each peptide. The right y-axis shows the difference in the percentage of patients showing positive binding to each peptide between time points. Binding for IgE is indicated above the x-axis line while binding for IgG4 is below the line. Regions/peptides with significantly (p<0.01, FDR<0.1, identified using TileMap) higher binding at baseline than last time point are indicated with red triangles, while regions with significantly higher binding at last time point than baseline are indicated with blue triangles. Green boxes labeled A-F indicate regions where varied concordance of IgE and IgG4 binding is of interest.
DISCUSSION
Although several studies have now examined in detail the epitope-specific antibody repertoires of untreated food-allergic patients, few have had longitudinal designs, and no prior studies have investigated whether oral immunotherapy can change the specificity and diversity of such repertoires. To address this knowledge gap, we utilized a high-throughput peptide microarray system12 to investigate the longitudinal effects of peanut OIT on the specificity and diversity of the antibody repertoire directed against the three major peanut allergens Ara h 1-3.
As was previously reported6, we observed marked variation in IgE and IgG4 binding patterns between individuals. We confirmed the finding that peanut-allergic patients on allergen elimination diets have highly stable antibody repertoires by studying untreated controls over an average follow-up duration of 58 months (not shown), far longer than the 20 months previously reported.7 However, in contrast to these controls, we show that the antibody repertoire undergoes dynamic and individualized changes during OIT. In our previous studies of peanut OIT, we have shown that the overall quantity of peanut-specific IgE is reduced on treatment, while the concentration of peanut-specific IgG4 is concurrently increased, and that over time a serum factor (presumed to be IgG4) emerges that can functionally inhibit IgE binding in vitro1. These data were consistent with the hypothesis that functionally inhibitory “blocking” antibodies of the IgG class develop during immunotherapy15,16. Some of the findings of the current study support this hypothesis, e.g., a progressive isotype shift during OIT in some subjects, in which a reduction in IgE binding occurs at the same epitopes that IgG4 binding gradually increases. Importantly, we observed this shift towards IgG4 predominance occurring especially within Ara h 2 and involving critical “informative” epitopes previously shown to be predictive of natural tolerance acquisition5. Studies utilizing SPOTS membranes5, immunoblotting8,17, and component-resolved techniques18 have all previously demonstrated the importance of IgE binding to Ara h 2 in peanut allergic patients, as compared to controls that outgrew peanut allergy or alternatively were sensitized to peanut but not allergic.
However, several additional findings from this study suggest that the antibody response to OIT is more complex than simply the gradual induction of epitope-identical blocking antibodies of a different isotype. First, the IgG4 response was broadly expanded in a polyclonal fashion during OIT, including induction of novel specificities. At certain peptides, this IgG4 production had no apparent impact on IgE binding. In addition, the clonality of the IgE repertoire was generally preserved, even while total psIgE levels fell, suggesting that the diversity and the amplitude of the allergen-specific antibody responses to OIT are differentially regulated. Indeed, we unexpectedly also observed a concomitant expansion of the IgE repertoire in roughly one-third of our subjects, even while an overall reduction in peanut-specific IgE levels occurred in each of them. The induction of de novo specificities in both the IgE and IgG4 compartments had been previously shown with the aid of recombinant test antigens in a study of specific subcutaneous immunotherapy with grass pollen19. It has been proposed that low-affinity IgE with novel specificities may emerge early in the course of specific immunotherapy, when doses are low; and that because these clones bind allergen with less relative avidity than antibody produced by hypermutated, long-lived memory B or plasma cells, they are not likely to be clinically deleterious20. Remarkably in our study, the IgE repertoire expansion occurred over a period of almost four years, and not just during the early phases of treatment. This paradoxical expansion of IgE diversity concurrent to the reduction in amplitude of allergen-specific IgE production has never been shown in a study of specific immunotherapy. Finally, using peanut extract, we were unable to out-compete IgE or IgG4 binding to target epitopes in the serum of subjects treated with OIT for several years, suggesting that the affinity of these antibodies is and remains high. This may be unique to peanut allergy and this form of therapy, due to the use of crude whole antigen preparations for treatment, the complex process of food preparation, digestion, and absorption, and/or differences in the immunogenic potential of food antigens in more severe forms of food allergy. A previous study of subjects with milk allergy similarly found that patients remaining allergic to unheated milk had a substantial fraction of high affinity antibodies, when compared to those tolerant of heated milk or nonallergic controls21.
Several previous studies have used similar methodologies to analyze the epitope binding patterns of patients allergic to peanut6,7, milk21-23, shrimp11, and lentil24. Collectively, these studies have established (1) the individuality of epitope binding patterns among patients; (2) the association between the diversity of the IgE repertoire and the persistence and severity of food allergy; and (3) the long term stability of IgE and IgG4 binding patterns in peanut-allergic patients treated with elimination diets. Wang et al21 extended these observations to include the antibody responses of those patients who can tolerate a heated milk product, which is considered to be a form of partial tolerance, and included affinity studies. However, all of these studies were observational, and most were conducted in patients practicing allergen avoidance, utilizing cross-sectional designs. A recent prospective study23 followed milk-allergic patients over time, and analyzed the evolution of antibody responses occurring as milk allergy was outgrown. Compared to those with persistent milk allergy, patients who recover spontaneously experience a longitudinal reduction in the diversity of the IgE repertoire, and a decrease in intensity of IgE binding that is concurrent with an increase in IgG4 binding intensity at similar epitopes. We observed similar changes in some but not all of our subjects on OIT, suggesting that while the serologic response to OIT may share some overlap with natural tolerance induction, it may have its own unique features. In future analyses, we will investigate the relationship of particular binding patterns to clinical responses using oral food challenge outcomes. Intriguingly, Savilahti et al23 used mathematical modeling to show that a signature that considered the IgE and IgG4 binding to fifteen regions across four main milk allergens at the time of diagnosis could predict whether milk allergy would resolve or persist, suggesting that this type of approach may have merit in tolerance evaluation.
We have previously used CAP-FEIA™ to show that peanut OIT changes the amplitude of the antibody response to peanut, and others have used peptide microarrays to study IgE and IgG4 responses in food-allergic children on elimination diets. This paper extends these previous studies by demonstrating for the first time that in subjects undergoing peanut OIT, but not those on elimination diets, the amplitude, specificity, and diversity of the antibody response to the major peanut allergens Ara h 1-3 is modified over time. These changes occur without evident effect on the affinity of the repertoire. Although the changes in binding patterns are individualized, they tend to affect the previously shown immunodominant regions, including at epitopes within Ara h 2 previously shown to be informative for naturally occurring tolerance. Future study will be necessary to further elucidate how these findings contribute to clinical tolerance.
Supplementary Material
Key Messages.
Allergen avoidance does not change the highly stable peanut-specific antibody repertoires of peanut-allergic patients.
In contrast, oral immunotherapy induces dynamic and individualized changes in IgE and IgG4 binding patterns with a minimal effect on antibody affinity.
Some of these changes are consistent with those seen in other forms of specific immunotherapy or natural tolerance.
Acknowledgements
We wish to thank the subjects and their families. We acknowledge Xiaohong Yue, Huamei Zhang, Anne Hiegel, Suzanne Carlisle, Henry Beresford, Meredith Ross, and Nikolas and Jan Kamilaris for their contributions to these studies. Brian Smith, MD MPH MHS provided invaluable statistical expertise. We would also like to thank the Duke Clinical Research Unit for providing the clinic space and staff support necessary to conduct our study.
Source of Funding: NIH-NIAID
Abbreviations Used
- IgE
immunoglobulin E
- IgG4
immunoglobulin G4
- IQR
25-75% interquartile range
- OIT
oral immunotherapy
- psIgE
peanut-specific immunoglobulin E
- Th2
T-helper-2 type response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Jones SM, Pons L, Roberts JL, Scurlock AM, Perry TT, Kulis M, et al. Clinical efficacy and immune regulation with peanut oral immunotherapy. J Allergy Clin Immunol. 2009 Aug;124(2):292–300. doi: 10.1016/j.jaci.2009.05.022. 300.e1-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark AT, Islam S, King Y, Deighton J, Anagnostou K, Ewan PW. Successful oral tolerance induction in severe peanut allergy. Allergy. 2009 Aug;64(8):1218–20. doi: 10.1111/j.1398-9995.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 3.Blumchen K, Ulbricht H, Staden U, Dobberstein K, Beschorner J, de Oliveira LC, et al. Oral peanut immunotherapy in children with peanut anaphylaxis. J Allergy Clin Immunol. 2010 Jul;126(1):83–91.e1. doi: 10.1016/j.jaci.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 4.Nowak-Węgrzyn A, Sampson HA. Future therapies for food allergies. J Allergy Clin Immunol. 2011 Mar;127(3):558–73. doi: 10.1016/j.jaci.2010.12.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyer K, Ellman-Grunther L, Järvinen KM, Wood RA, Hourihane J, Sampson HA. Measurement of peptide-specific IgE as an additional tool in identifying patients with clinical reactivity to peanuts. J Allergy Clin Immunol. 2003;112(1):202–207. doi: 10.1067/mai.2003.1621. [DOI] [PubMed] [Google Scholar]
- 6.Shreffler WG, Beyer K, Chu TH, Burks AW, Sampson HA. Microarray immunoassay: association of clinical history, in vitro IgE function, and heterogeneity of allergenic peanut epitopes. J Allergy Clin Immunol. 2004;113(4):776–82. doi: 10.1016/j.jaci.2003.12.588. [DOI] [PubMed] [Google Scholar]
- 7.Flinterman AE, Knol EF, Lencer D, Bardina L, den Hartog Jager CF, Lin J, et al. Peanut epitopes for IgE and IgG4 in peanut-sensitized children in relation to severity of peanut allergy. J Allergy Clin Immunol. 2008;121(3):737–43. doi: 10.1016/j.jaci.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 8.Flinterman AE, van Hoffen E, den Hartog Jager CF, Koppelman S, Pasmans SG, Hoekstra MO, et al. Children with peanut allergy recognize predominantly Ara h2 and Ara h6, which remains stable over time. Clin Exp Allergy. 2007 Aug;37(8):1221–8. doi: 10.1111/j.1365-2222.2007.02764.x. [DOI] [PubMed] [Google Scholar]
- 9.Flinterman AE, Knol EF, Lencer DA, Bardina L, Jager C, Lin J, et al. Peanut epitopes for IgE and IgG4 in peanut-sensitized children in relation to severity of peanut allergy. J Allergy Clin Immunol. 2008;121:737–43. doi: 10.1016/j.jaci.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Bardina L, Bruni FM, Shreffler WG, Wang J, Sampson HA. Development of a reliable and sensitive peptide microarray for large-scale epitope mapping of milk allergens. J Allergy Clin Immunol. 2008;121:949. doi: 10.1016/j.jaci.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayuso R, Garcia S, Lin J, Fu Z, Ibáñez MD, Carrillo T, et al. Greater epitope recognition of shrimp allergens by children than by adults suggests that shrimp sensitization decreases with age. J Allergy Clin Immunol. 2010 Jun;125(6):1286–1293.e3. doi: 10.1016/j.jaci.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Lin J, Bardina L, Shreffler WG, Andreae DA, Ge Y, Wang J, et al. Development of a novel peptide microarray for large-scale epitope mapping of food allergens. J Allergy Clin Immunol. 2009 Aug;124(2):315–22. doi: 10.1016/j.jaci.2009.05.024. 322.e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beyer K, Bardina L, Grishina G, Sampson HA. Identification of sesame seed allergens by 2-dimensional proteomics and Edman sequencing: seed storage proteins as common food allergens. J Allergy Clin Immunol. 2002 Jul;110(1):154–9. doi: 10.1067/mai.2002.125487. [DOI] [PubMed] [Google Scholar]
- 14.Ji HK, Wong WH. TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–36. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- 15.Wachholz PA, Durham SR. Mechanisms of immunotherapy: IgG revisited. Curr Opin Allergy Clin Immunol. 2004 Aug;4(4):313–8. doi: 10.1097/01.all.0000136753.35948.c0. [DOI] [PubMed] [Google Scholar]
- 16.Uermösi C, Beerli RR, Bauer M, Manolova V, Dietmeier K, Buser RB, et al. Mechanisms of allergen-specific desensitization. J Allergy Clin Immunol. 2010 Aug;126(2):375–83. doi: 10.1016/j.jaci.2010.05.040. Epub 2010 Jul 10. [DOI] [PubMed] [Google Scholar]
- 17.Koppelman SJ, Wensing M, Ertmann M, Knulst AC, Knol EF. Relevance of Ara h1, Ara h2 and Ara h3 in peanut-allergic patients, as determined by immunoglobulin E Western blotting, basophil-histamine release and intracutaneous testing: Ara h2 is the most important peanut allergen. Clin Exp Allergy. 2004 Apr;34(4):583–90. doi: 10.1111/j.1365-2222.2004.1923.x. [DOI] [PubMed] [Google Scholar]
- 18.Nicolaou N, Poorafshar M, Murray C, Simpson A, Winell H, Kerry G, et al. Allergy or tolerance in children sensitized to peanut: prevalence and differentiation using component-resolved diagnostics. J Allergy Clin Immunol. 2010 Jan;125(1):191–7e1-13. doi: 10.1016/j.jaci.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Ball T, Sperr WR, Valent P, Lidholm J, Spitzauer S, Ebner C, Kraft D, Valenta R. Induction of antibody responses to new B cell epitopes indicates vaccination character of allergen immunotherapy. Eur J Immunol. 1999 Jun;29(6):2026–36. doi: 10.1002/(SICI)1521-4141(199906)29:06<2026::AID-IMMU2026>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Shamji MH, James LK, Durham SR. Serum immunologic markers for monitoring allergen-specific immunotherapy. Immunol Allergy Clin North Am. 2011 May;31(2):311–23. doi: 10.1016/j.iac.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Lin J, Bardina L, Goldis M, Nowak-Wegrzyn A, Shreffler WG, et al. Correlation of IgE/IgG4 milk epitopes and affinity of milk-specific IgE antibodies with different phenotypes of clinical milk allergy. J Allergy Clin Immunol. 2010 Mar;125(3):695–702. doi: 10.1016/j.jaci.2009.12.017. 702.e1-702.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerecedo I, Zamora J, Shreffler WG, Lin J, Bardina L, Dieguez MC, et al. Mapping of the IgE and IgG4 sequential epitopes of milk allergens with a peptide microarray-based immunoassay. J Allergy Clin Immunol. 2008 Sep;122(3):589–94. doi: 10.1016/j.jaci.2008.06.040. [DOI] [PubMed] [Google Scholar]
- 23.Savilahti EM, Rantanen V, Lin JS, Karinen S, Saarinen KM, Goldis M, et al. Early recovery from cow’s milk allergy is associated with decreasing IgE and increasing IgG4 binding to cow’s milk epitopes. J Allergy Clin Immunol. 2010 Jun;125(6):1315–1321.e9. doi: 10.1016/j.jaci.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vereda A, Andreae DA, Lin J, Shreffler WG, Ibañez MD, Cuesta-Herranz J, et al. Identification of IgE sequential epitopes of lentil (Len c 1) by means of peptide microarray immunoassay. J Allergy Clin Immunol. 2010 Sep;126(3):596–601.e1. doi: 10.1016/j.jaci.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.