Abstract
Translational control of gene expression is instrumental in the regulation of eukaryotic cellular form and function. Neurons in particular rely on this form of control as their numerous synaptic connections need to be independently modulated in an input-specific manner. Brain cytoplasmic (BC) RNAs implement translational control at neuronal synapses. BC RNAs regulate protein synthesis by interacting with eIF4 translation initiation factors. Recent evidence suggests that such regulation is required to control synaptic strength, and that dysregulation of local protein synthesis precipitates neuronal hyperexcitability and a propensity for epileptogenic responses. A similar phenotype results from lack of fragile X mental retardation protein (FMRP), indicating that BC RNAs and FMRP use overlapping and convergent modes of action in neuronal translational regulation.
Key terms: local protein synthesis, regulatory RNAs, synapses, translational control
Translational control in neurons
In eukaryotes, the cellular regulation of gene expression unfolds on several levels, including transcriptional control in the nucleus and translational control in the cytoplasm. In large and complex cell types in particular, translational control of gene expression [1] plays an important role in the temporal-spatial management of local protein reservoirs in subcellular microdomains. The requirements for local control are especially demanding in nerve cells. Neurons make large numbers of synaptic connections with each other, often more than 1000 per cell. These synapses need to be independently maintained and regulated in a manner that is specific and responsive to the physiological input that each of them receives. For this reason, local control of translation is key to synaptic function and plasticity.
Protein-synthetic machinery (i.e. ribosomes, translation factors, tRNAs, etc.) is actively present at postsynaptic sites, and increasing numbers of mRNAs have been reported to be selectively delivered to such sites [2–4]. Dendritic mRNAs are diverse, encoding synapto-dendritic proteins such as cytoskeletal components, receptors, and kinases, among others [3, 4]. The intimate juxtaposition of mRNAs and translational machinery at the synapse necessitates effective mechanisms to ensure that translation of such mRNAs is tightly controlled. Inappropriate or premature synthesis of local proteins can result in excessive strengthening of synapses, which in turn can cause neuronal hyperexcitability and, at the network level, a propensity for epileptogenic responses. To maintain an appropriately balanced neuronal excitability, it is therefore critically important that translational regulators are in place at the synapse to implement temporal-spatial control of local protein synthesis. Several such regulators have now been identified, prominent among them the fragile X mental retardation protein (FMRP, see glossary), micoRNAs (miRNAs), and BC (brain cytoplasmic) RNAs.
This review discusses the significance of translational control in brain function, and it highlights recent advances in our understanding of regulatory BC RNAs in such mechanisms. We do not cover neuronal miRNAs as they have been discussed extensively in recent years [5–10].
FMRP
Functional absence of FMRP, the protein product of the FMR1 gene, causes the fragile X syndrome (FXS) [11, 12]. Lack of FMRP is typically the consequence of CGG triplet repeat expansions in the 5′ UTR of the FMR1 gene. Transcriptional silencing is induced once the repeat length exceeds 200 units [11, 13]. FXS is characterized by intellectual disability, often in conjunction with epileptic and autistic phenotypes [11, 13].
FMRP is a negative regulator of translation although this function is subject to modulation by phosphorylation. How FMRP represses translation has been a subject of debate. While some reports associate FMRP with the initiation phase of translation [14–18], data to indicate that FMRP targets translating polyribosomes [19–22] are consistent with a regulatory role at a later step, such as elongation or termination. This notion is also supported by recent data showing that FMRP binds to protein-coding regions of target mRNAs and stalls translocating ribosomes [23]. In view of the combined evidence, it cannot be ruled out that FMRP employs multi-modal repression mechanisms, possibly targeting translation via initiation, via postinitiation steps (e.g. elongation), or via combinations of these.
FMRP has been reported to interact physically with BC1 RNA and to control translation of FMRP target mRNAs via this interaction [15, 24]. However, these claims could not be independently corroborated by other groups [25, 26]. A further report [27] found that FMRP bound BC1 RNA in vitro with an estimated equilibrium dissociation constant of KD = 25 nM but stated that FMRP bound tRNA with the same affinity, and that similar results were obtained with single-stranded DNA oligodeoxynucleotides of 56–98 nt in length. It would appear that the observed in vitro interactions between FMRP and BC1 RNA are entirely unspecific.
Using high-throughput sequencing of RNAs isolated by crosslinking immunoprecipitation (HITS-CLIP), Darnell et al. [23] identified 842 FMRP target transcripts, among them some previously reported dendritic mRNAs (e.g. CaMKIIα mRNA, MAP2 mRNA) as well as well as novel FMRP targets that encode proteins involved in synaptic function and plasticity. Notably, the list of FMRP target transcripts also includes a substantial number of autism candidate gene products, supporting a functional link between loss of FMRP and autistic manifestations [23].
The collective evidence has identified FMRP as a reversible repressor of translation that targets a large number of neuronal mRNAs. Although the mechanism of action continues to be debated, the data suggest that translational control by FMRP is an important determinant of brain function.
BC RNAs
It has become increasingly clear over the past decade that molecular control of cellular form and function is often performed by regulatory RNAs [10, 28–30]. Protein-coding genes make up less than 2% of the human genome [30, 31]. By contrast, a major part of the rest of the genome is transcribed in various cell types [28, 32]. During evolution, non-protein-coding elements have steadily increased their presence in genomes, accounting for about 10% of genomic content in some prokaryotes but more than 90% in mammals (Figure 1) [33]. At the same time, protein-coding genomic content has remained rather constant at a level below 40 Mb (Figure 1). It is assumed that many non-protein-coding elements are transcribed into RNAs with regulatory functions, and that the need for such cellular regulators has increased with growing organismal complexity during phylogenetic development [33].
Figure 1.
Ratios of non-protein-coding DNA and megabases of protein-coding regions per haploid genome across species. (a) Ratios of bases in non-protein-coding regions vs. total genomic DNA per sequenced genome. Prokaryotes are in black, unicellular eukaryotes in gray, organisms known to be both uni- and multicellular, depending on lifecycle, in light blue, basal multicellular organisms in blue, plants in green, nematodes in purple, arthropods in orange, chordates in yellow, vertebrates in red. (b) Amount (in megabases) of protein-coding regions per genome for species ranked by fraction of non-protein-coding DNA in (a). Adapted from Taft et al. [33], with permission.
The mammalian central nervous system is unsurpassed in terms of complexity, and neuronal regulatory RNAs are being identified at an accelerating pace. Here we focus on neuronal regulatory RNAs that are in the small RNA category, i.e. in a size range of up to 400 nucleotides. We define 400 nucleotides as the cutoff between small (or short) RNAs and long RNAs (i.e. sRNAs and lRNAs, respectively) [10]. This definition is consistent with established terminology (see glossary) such as snRNA for small nuclear RNA (e.g. 7SK RNA), snoRNA for small nucleolar RNA (e.g. U3 RNA), and scRNA for small cytoplasmic RNA (e.g. SRP RNA), with the three examples given each measuring about 300 nucleotides in size.
BC RNAs comprise a small family of scRNAs. Prototypical BC1 RNA is a 152-nucleotide scRNA that is expressed in rodent neurons where it is located in synapto-dendritic domains [29, 34]. A second, shorter species (BC2 RNA, about 100 nucleotides) has been detected in rodent brains but has not been functionally characterized [34, 35]. BC200 RNA, a 200-nucleotide scRNA expressed in primate nervous systems [36, 37], is an analog rather than an ortholog (see glossary) of rodent BC1 RNA. The BC1 RNA and BC200 RNA genes were each generated by retroposition (Box 1) but are of different evolutionary pedigree: whereas the former is a phylogenetic descendant of tRNAAla, the latter derives from SRP RNA [38–40]. To the extent this has been examined, mature BC RNAs are collinear with their genes and do not undergo co-or posttranscriptional modification [38, 40, 41].
Box 1. Retroposition.
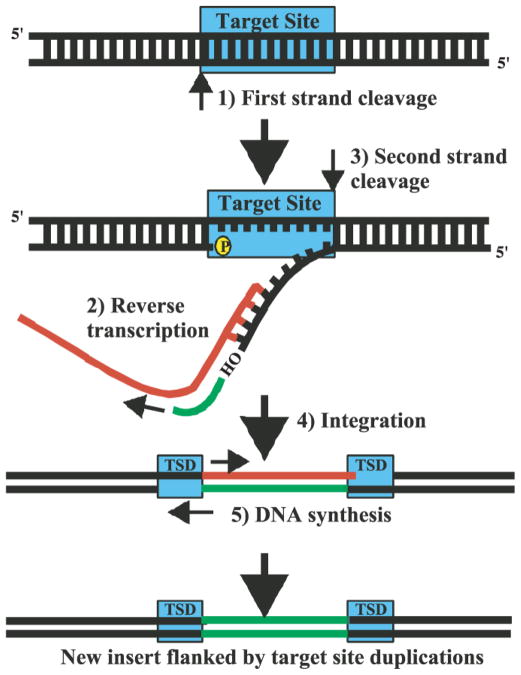
Retroposition (also known as retrotransposition) is an RNA-based mechanism that in the course of phylogenetic development has had a profound impact in shaping eukaryotic genomes [82–85]. In a retroposition event, a cellular transcript is reverse-transcribed into multiple DNA elements that are in turn inserted into genomic DNA (Figure I). Whereas somatic retroposition has been implicated in organismal development and disease, heritable retroposition requires that these mechanisms occur in the germline [82]. A typical retroposition event proceeds through the stages of DNA cleavage at the genomic locus of integration, annealing of a template RNA molecule, reverse transcription, and gap closure with associated target site duplication (Figure I) [84]. Especially with RNA molecules that constitute efficient templates for reverse transcription, the number of insertions can be high, and it is estimated that about 45% of the human genome has been generated by retroposition events [83]. Because of mutational neutralization, more ancient retroposons are difficult to detect. Nevertheless, this process is ancient, and it has been estimated [86] and recently confirmed [87] that about two thirds (and by extrapolation almost the entire human genome [86]) has been generated by RNA-to-DNA conversion.
The retroposition mechanism has the potential to generate large numbers of copies, partial copies, or variant copies of existing genes, that are inserted into novel genomic environments. The insertion environment is therefore a crucial determinant of functional outcome. Retroposition events can simply result in insertion mutations, in which case they can be detrimental and cause genetic disorders [82]. However, retroposition can also initiate genomic innovation and set the stage for the evolution of novel regulatory RNAs [88]. This scenario is particularly relevant whenever the retroposition event originates with an RNA polymerase III (Pol III) transcript. RNA Pol III genes carry their promoters in the coding regions. As a result, Pol III retroposons are often transcriptionally competent, and their transcripts can therefore be recruited into novel functional roles. The expression competence of Pol III retrogenes will also depend on integration site flanking (mostly upstream) sequences that can act as additional promoter elements. It is noted in this context that information sufficient for the tissue-specific expression of BC1 RNA is contained within a 1.4 kb upstream region [89].
Because the generation of regulatory RNA genes via retroposition is an ongoing process in mammalian evolution, it appears likely that retrogenes often arise in specific orders or genera, thus bestowing selection advantages and promoting phylogenetic diversification.
Figure I.
The retroposition mechanism. The sequence is initiated with a nicking of the first (bottom) DNA strand by an endonuclease (step 1). The new 3′ end serves as a primer for reverse transcription of a template RNA (red, step 2) [84]. Second strand cleavage (step 3) occurs during or after reverse transcription. After integration (step 4), the hybrid RNA is degraded, and second strand synthesis ensues (step 5). Newly synthesized DNA strands are in green. Target site duplications (TSDs) are hallmarks of retroposons. They occur because target site cleavage produces staggered ends, and the resulting small gaps are filled in on both sides of the insertion, giving rise to short flanking duplications. Adapted from Kazazian et al. [84], with permission.
BC RNAs are thus regulatory RNAs that, in the course of mammalian phylogenetic development, have been recruited to operate in increasingly complex neural systems. They regulate neuronal gene expression through translational control.
BC RNAs in translational control mechanisms
BC RNAs are negative regulators of translation. The first evidence about the mechanism of translational control (see glossary) by BC RNAs was obtained by differential internal ribosome entry site analysis coupled with sucrose density gradient centrifugation. This work established that BC1 RNA inhibits translation initiation (see glossary) by repressing assembly of 48S initiation complexes [42]. Three factors that operate in the translation initiation pathway are targeted by BC1 RNA: eukaryotic initiation factors (eIFs) 4A and 4B and poly(A) binding protein (PABP) [42–44]. Repression is mainly achieved through interactions with eIFs 4A and 4B [44]. eIF4A is an ATP-dependent RNA helicase that unwinds double-stranded content in the 5′ untranslated regions (UTRs) of mRNAs, enabling those mRNAs to recruit small ribosomal subunits to their start codons [45]. eIF4B is a multifunctional factor that stimulates the helicase activity of eIF4A and also promotes recruitment of the small ribosomal subunit by interacting with 18S rRNA and eIF3 [45, 46]. Thus, BC1 RNA targets a rate-limiting step in eukaryotic translation initiation, the recruitment of the small ribosomal subunit to the mRNA.
Rodent BC1 RNA and primate BC200 RNA have indistinguishable translational repression competencies and mechanisms [44]. Despite the distinct phylogenetic pedigrees of the two RNAs, they share important common structural features. These include a tripartite structure with a 5′ stem-loop domain, a central A-rich region, and a 3 stem-loop domain [34, 36]. The BC1 and BC200 3′ stem-loop domains feature noncanonical C-loop motifs (Box 2) that are identical in both RNAs [44]. It is through this motif, and the adjacent A-rich region, that BC1 and BC200 RNAs interact with eIF4B and eIF4A, respectively, and exert their translational repression competence [44].
Box 2. Noncanonical RNA motifs.
Noncanonical nucleotide interactions are characterized by types of hydrogen bonds that are different from those seen in canonical (i.e. Watson-Crick, WC) interactions. In its interactions with other nucleotides, a nucleotide base can use any of its three ‘edges’: Watson-Crick (WC), Hoogsteen (H), and Sugar-Edge (SE) [90]. All of the 12 theoretically possible interaction types are naturally occurring in RNA (e.g. trans-H/SE, cis-WC/SE, and others), in contrast to DNA which is typically restricted to canonical cis-WC/WC. The geometry of noncanonical interactions and pairings, especially those involving H and/or SE hydrogen bonding, is often such that they force RNA stem-loops into distinct 3D architectural conformations that serve as recognition codes for transacting proteins [58]. Thus, except for strictly protein-coding regions, it is often the 3D structure of architectural RNA motifs, rather than nucleotide sequence per se, that is subject to natural selection in evolution [59, 91].
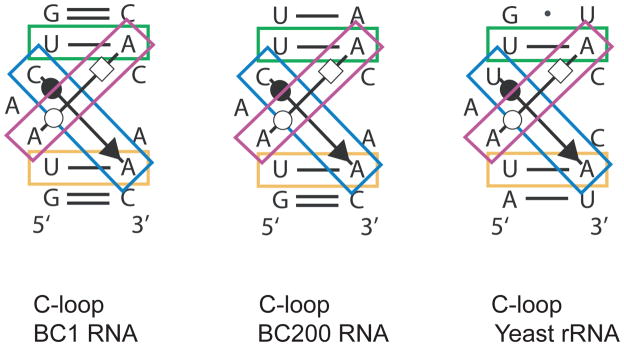
Numerous types of noncanonical RNA motifs have been identified [59]. C-loop motifs are important for BC RNA translational repression competence (Figure I). These motifs feature standard cis-WC/WC pairs as well as noncanonical trans-WC/H and cis-WC/SE interactions [44, 58]. A second type of noncanonical RNA motifs is used by BC RNAs to encode spatial information for dendritic targeting [62]. These motifs, known as GA motifs, feature noncanonical purine•purine interactions in which tandem G•A/A•G pairs engage in trans-H/SE interactions. GA motifs are dimorphic in that they can assume a straight or a kinked conformation. The latter conformation, also known as kink-turn or K-turn, is prevalent at high divalent cation concentrations and/or when engaged with cognate binding proteins [58, 62].
Figure I.
Noncanonical C-loop motifs. BC RNA C-loop motifs are aligned with a previously described eukaryotic C-loop motif [58]. Canonical and non-canonical nucleotide interactions are indicated. Base pairings are typified using the Leontis and Westhof scheme of symbolic representations [58, 90] as follows: solid symbols, cis pairing; open symbols, trans pairing; lines, cis-WC/WC (green and yellow boxes); solid circle-arrow, cis-WC/SE (blue box); open circle-square, trans-WC/H (purple box). Adapted from Eom et al. [44], with permission. Copyright © American Society for Microbiology.
Role of metabotropic glutamate receptor (mGluR) signaling in translational control
In the mGluR theory of fragile X mental retardation, Bear and colleagues [12] proposed that FMRP functions as a repressor in translation pathways that are activated via stimulation of group I mGluRs (see glossary). Thus, FMRP would act as a downstream brake that balances translational output upon activation of upstream group I mGluR signaling. This concept has since received significant experimental support. Group I mGluR-dependent protein synthesis [47] is defective in Fmr1 knockout (KO) animals [48, 49]. Lack of FMRP results in neuronal hyperexcitability, manifesting in epileptogenic responses that require signaling through the group I mGluR – MEK-ERK (mitogen-activated protein kinase kinase/extracellular signal-regulated kinase) pathway [50]. Correspondingly, elevated basal protein synthesis in Fmr1 KO animals depends on MEK-ERK signaling, but not Akt-mTOR signaling [51]. These data support the notion that FMRP represses translation that operates downstream from group I mGluR and MEK-ERK pathway signaling. In addition, however, FMRP has also been implicated in upstream signaling events. Interactions between mGluRs and Homer, a scaffolding protein that links signaling molecules in postsynaptic densities, are reduced in Fmr1 KO mice, and the consequential downregulation of mGluR activation of PI3K-mTOR activity was proposed to decrease mGluR-stimulated translation [52].
In many respects, the phenotypic consequences of a lack of BC1 RNA are very similar to those of a lack of FMRP. BC1 KO animals exhibit neuronal hyperexcitability, as evidenced by a predisposition for prolonged epileptiform discharges, exaggerated cortical oscillations in the gamma frequency range, and a susceptibility to convulsive seizures upon auditory stimulation [53]. These phenotypic manifestations require activation of group I mGluRs and MEK-ERK signaling, indicating that BC1 RNA targets translation pathways that depend on such signaling. The close correspondence between the BC1 KO and the Fmr1 KO phenotypes prompted the generation of double KO (dKO) animals that lack both BC1 RNA and FMRP. These animals exhibit the same phenotypic manifestations as the single KO (sKO) animals, however in significantly more severe forms [54]. Hyperexcitability is exacerbated in dKO neurons, as evidenced by epileptiform discharges that materialize earlier and occur with higher frequency and longer duration, as compared with respective sKO neurons. Spatial learning, which is only mildly affected in BC1 and Fmr1 sKO animals [54, 55], is severely impaired in dKO animals [54]. The dKO phenotypic features indicate that the contributions of the BC1 and FMRP translation repression mechanisms are modular-additive.
The close phenotypic resemblance of the respective animal models strongly suggests that the two repression systems cooperate functionally, although not physically, to generate significantly overlapping translational output profiles. Recent work has identified a comprehensive set of FMRP target mRNAs [23]; analogous work with BC1 RNA should establish the extent of overlap. A possible scenario to explain their functional convergence is that BC RNAs and FMRP act in series along the translation pathway (Figure 2). It is expected, however, that the functional convergence of the BC RNA and FMRP translational control mechanisms is less than complete because it is possible that (i) the respective complements of target mRNAs overlap only partially, and that (ii) FMRP is multifunctional and performs roles in addition to repression of translation [52, 56]. An in-depth understanding of the nature and extent of functional convergence might ultimately also provide a basis for designing substitution approaches, for example remedying a lack of FMRP by increasing BC RNA expression.
Figure 2. Translational control by BC RNAs and FMRP.
Activation of group I mGluRs (metabotropic glutamate receptors) couples to the translational machinery via the MEK-ERK (mitogen-activated protein kinase kinase/extracellular signal-regulated kinase) pathway. This translational stimulation is counteracted by two translational repression systems acting in series. BC RNAs (here represented by BC1 RNA) repress assembly of 48S initiation complexes [42, 44] and thus negatively regulate translation that is activated by signaling via the group I mGluR – MEK-ERK pathway [53]. Analogously, FMRP represses translation that is stimulated via this pathway [12, 50]. The asterisks associated with the two red inhibition bars indicate that published evidence has implicated FMRP in the repression of initiation or postinitiation steps, or both. Earlier evidence [14] suggested a role of FMRP in inhibiting formation of 80S ribosomes, but more recent data [23] support a role in stalling elongating ribosomes. The combined actions of translational stimulation and repression are assumed to ensure an appropriate excitability balance at the synapse. Dark green arrows indicate activation, red T-shaped bars indicate inhibition, and light green arrows indicate steps in a pathway.
RNA transport
The targeted delivery of RNAs (including mRNAs, tRNAs, and regulatory RNAs) to synapto-dendritic domains is a prerequisite for local protein synthetic competence and a determinant of its specificity. The number of mRNAs that are locally available at a given synapse is physically limited [57], and the makeup of this mRNA reservoir is a key underlying factor in the plastic modulation of synaptic strength. It is for this reason important that we understand the mechanisms of dendritic RNA transport, and the nature of the spatial codes that RNAs use to specify their intracellular destination sites.
Several RNA localization pathways have been described in eukaryotic cells, including the Staufen, the zipcode binding protein (ZBP), and the heterogeneous nuclear ribonucleoprotein (hnRNP, see glossary) A2 pathways. In neurons, dendritically targeted RNAs include mRNAs and regulatory RNAs such as miRNAs and BC RNAs (Figure 3). However, common principles in neuronal RNA transport have been slow to emerge. Contributing to the problem has been our limited understanding of functional RNA architecture.
Figure 3. RNA transport in dendrites.
The hnRNP (heterogeneous nuclear ribonucleoprotein) A2 and ZBP1 (zipcode binding protein 1) pathways represent two of the major RNA targeting mechanisms in neurons. hnRNP A2 (blue spheres) interacts with GA-type noncanonical motifs, as exemplified by those in the 5′ domain of BC1 and BC200 RNAs, and in the 3′ UTR of PKMζ mRNA [62]. At the synapse, BC RNAs repress translation (red T-shaped bar) by preventing 48S complex formation [42, 44], thus providing a counterbalance to translational stimulation through group I mGluR activation (dark green arrow; center, right) [53]. hnRNP A2 also interacts with expanded CGG repeat stem-loop structures in the 5′ UTR of premutation FMR1 mRNA (FMR1 mRNA*) [69, 73]. Premutation FMR1 mRNA can compete with GA motif RNAs for access to hnRNP A2, which could lead to impairments in their dendritic delivery [62]. ZBP1 (orange spheres) interacts with the β-actin mRNA zipcode, promoting dendritic delivery of the mRNA while at the same time inhibiting its translation [3]. It is possible that at the synapse, phosphorylation of ZBP1 by Src kinase (bottom, right) causes it to release β-actin mRNA, permitting translation [3]. Black arrows indicate movement and association, dark green arrow indicates stimulation, red T-shaped bars indicate inhibition.
In recent years, major advances in the field of RNA biology have led to a growing appreciation of the significance of RNA architectural motifs in RNA functionality [58–60]. Specifically, the collective work highlights the role of noncanonical nucleotide interactions as structural determinants of RNA motifs (Box 2) [58, 59]. Noncanonical motifs of the GA type (see glossary) are requisite for the dendritic delivery of BC RNAs [61, 62]. BC1 and BC200 RNAs contain analogous GA motifs in their 5′ stem-loop domains. An unpaired U residue (U22, located directly adjacent to the GA motif) is also required for the dendritic delivery of BC1 RNA [61]. The BC RNA transport motifs interact with hnRNP A2 [62], a previously identified RNA transport factor (Figure 3) [63]. Noncanonical motif features are essential as conversion to standard Watson-Crick (WC) pairing abolishes both hnRNP A2 binding and dendritic delivery [62]. hnRNP A2 has also been shown to bind to an A2 response element (A2RE), an RNA element that is found in various localized mRNAs [63, 64] and that is contained within the GA motif structure. Structural data will be needed to establish the precise contributions of single-stranded, double-stranded, and noncanonical content that underlie recognition by hnRNP A2. A second potential transport factor, Purα, binds to the transport-specifying 5′ domains of BC1 and BC200 RNAs [65, 66], but its role in dendritic delivery of BC RNAs remains to be established.
As mentioned before, CGG repeats in excess of 200 units in the 5′ UTR of the FMR1 gene (known as full mutation) trigger transcriptional silencing, thus causing FXS. In contrast, CGG repeats that have expanded into a rage of about 55 to 200 units (known as premutation) are transcribed as part of FMR1 mRNA. They cause various disorders including the fragile X-associated tremor/ataxia syndrome (FXTAS) which is characterized by cognitive decline and motor disorders at advanced age [13]. The premutation, alleles of which are carried by approximately 1:250–810 males and 1:130–250 females [67], also causes neurodevelopmental deficits including cognitive impairment, seizure activity, autism-spectrum disorder (ASD), and symptoms that resemble mild fragile X syndrome [13]. Premutation FMR1 mRNA is expressed at significantly elevated levels [68], and it is assumed that expanded repeats give rise to RNA toxicity via a dominant gain-of-function mechanism [13]. CGG repeat RNA interacts with cellular factors, and it is likely that premutation FMR1 mRNA sequesters specific factors and makes them unavailable to perform their normal functions. Several CGG-interacting factors have been identified [13, 69], among them hnRNP A2 which is colocalized with premutation FMR1 mRNA in cellular inclusions that are characteristic of FXTAS [70].
CGG repeats form stable stem-loops that feature noncanonical G•G pairs [71, 72]. Given that hnRNP A2 binds to the noncanonical targeting motif of BC RNAs [62] and to CGG repeat RNA [69, 73], it was conceivable that these two types of RNA might engage in a molecular competition for the common cellular resource hnRNP A2. Such competition was indeed observed. CGG repeat RNA, used at repeat lengths and intracellular concentrations that corresponded to those in human premutation cells [68], compete directly with BC1 RNA for binding to hnRNP A2, resulting in reduced dendritic transport [62]. The data suggest that impairments in intracellular RNA localization mechanisms can result in disease. In addition, other cellular factors (including Purα, Sam68) have been identified as interacting with expanded CGG repeat RNA [69, 74, 75]. CGG premutation phenotypic manifestations might thus result from competitive interactions of expanded CGG repeat RNA with multiple cellular target factors.
The BC1 and BC200 RNA genes trace their origins to retroposition events (Box 1), and both genes in turn have given rise to retroposed elements in the rodent and primate genomes, respectively [38, 40, 76]. In the well-studied case of BC1 retroposition, so-called ID elements have been disseminated in rodent genomes. These elements are derived from, and are hence identical or similar to, the 5′ BC1 domain [76]. It was therefore conceivable that some of these elements, when inserted into a host mRNA gene and transcribed in neurons, would confer dendritic targeting competence to that mRNA. Recent evidence indicates that this is indeed the case. Buckley et al. [77] reported that ID element retroposons can reside in introns that are retained in cytoplasmic transcripts, and that such ID elements confer dendritic targeting competence to their host mRNAs. This work underscores the impact of retroposition in the genomic dissemination of RNA targeting codes during the course of phylogenetic development.
As a caveat, it has been observed that the 5′ BC1 domain as well as ID elements, as part of reporter RNAs expressed in transgenic animals, failed to reach dendritic domains [78]. It is noted, however, that chronic overexpression of GA motif RNAs, as is the case in conventional transgenic animals, can elicit anti-dsRNA cellular defense mechanisms [79, 80]. As part of anti-viral response strategies, such mechanisms are specifically triggered by GA motifs containing tandem noncanonical G•A pairs [81]. Cellular responses directed against GA motif dsRNA are mediated by activation of PKR which binds directly to the motif [79–81].
The work discussed in this section again underscores the significance of noncanonical RNA motifs. The decoding of such motifs by cognate protein factors is key to RNA functionality, and we posit that compromised decoding can cause cellular impairments.
Concluding remarks
A fundamental conundrum in neurobiology is posed by the following consideration. Whereas the long-term plastic modulation of synaptic strength requires changes in gene expression, the nucleus is unlikely to “know” which specific synapses require modulation at any given time. It is increasingly recognized that one solution to this problem is the translational control of gene expression at local synaptic sites. This mechanism provides neurons with the capability to synthesize synapto-dendritic proteins in a manner that is specifically reactive to the input a given synapse receives [4]. However, it is imperative for a cell to impose strict control over local protein synthetic capacity because excessive or premature translation of mRNAs available at a synapse may cause inappropriate synaptic strengthening. Regulatory BC RNAs are representative mediators of such local control.
Mammalian genomes devote a significant percentage of their coding capacity to regulatory RNA genes. It has been suggested that such a high level of regulatory power was necessitated by an ever growing complexity of eukaryotic systems, as evidenced by the ultimate example of a complex system, the human brain [33]. The question arises why regulatory functions have increasingly been assumed by RNAs, rather than proteins, in eukaryotic cells [31]. RNAs, possibly the most ancient of all bio-macromolecules, have undergone a long and continuing co-evolution with RNA-interacting proteins. In the course of this evolution, the number of RNA genes has significantly expanded as a result of genomic retroposition events, and transcripts of such genes have become eligible for functional recruitment (Box 1) [82–85].
With the beginning of the mammalian radiation about 65 million years ago, BC1 and BC200 RNAs were independently recruited into their novel functional roles in the rodent and primate lineages, respectively [85]. The mechanistic mode of action of these two RNAs appears to be identical; that is, translational repression via interaction with eIFs 4A and 4B [44]. An identical C-loop motif in the 3′ domains of these RNAs is part of the structural basis that likely underlies their functional selection by eIF proteins. Thus, the two RNAs were independently recruited because a need had arisen for the local regulation of protein synthesis in neurons, and because they were uniquely suited to address this need. Similarly, the 5′ domains of BC1 and BC200 RNAs contain highly similar GA motif structures that serve as dendritic targeting codes [62]. Again, despite their separate phylogenetic ancestry, the two RNAs have acquired convergent functional motif content that allows them to interact with identical protein factors, in this case to mediate selective delivery to synapto-dendritic domains. The rapid and adaptive evolution of RNA genes thus satisfies regulatory needs that arise from increasing systems complexities.
The reliance on multiple regulators in the administration of complex systems, although clearly vital, is not without risks. Who controls the regulators? Out-of-control regulators could easily wreak havoc on the system they are supposed to govern. Such problems might be caused, for instance, by inappropriate expression of regulators outside of their functional contexts (e.g. in an inappropriate cell type). The consequences of dysregulated translation could include inadequate growth control and malignant potential. In future research (Box 3), aberrant control of regulatory RNAs is likely to be increasingly identified as causing cellular dysfunction and disease.
Box 3. Outstanding questions.
How does FMRP target the translation mechanism? The mode of action used by FMRP in the regulation of translation is not well understood. FMRP might target the initiation phase of translation, the elongation or termination phase, or a combination thereof. In either case, mechanisms of interaction with translation machinery components need to be elucidated.
What are the mRNAs that are subject to regulation by BC RNAs? Target mRNAs that controlled by BC RNAs are only now being identified. The degree of overlap between this complement and FMRP target mRNAs remains to be established.
What is the human equivalent of the epileptogenic phenotype observed in model animals? Lack or dysfunction of neuronal translational repressors seems to precipitate hyperexcitability and epileptogenic responses. However, for BC RNAs this has not yet been demonstrated with human subjects.
Can inadequate RNA transport cause disease? Impaired neuronal RNA transport can be expected to impact regulation of synaptic function. However, mechanics and specificities of transport mechanisms are only vaguely understood, as are phenotypic consequences and disease manifestation of dysfunctional RNA transport.
Acknowledgments
We regret that space limitations did not permit inclusion of all recent literature relevant to the subject area, and we apologize to those authors whose work could not be directly referenced. Work in the authors’ laboratory was in part supported by National Institutes of Health grants NS046769 and DA026110 (to HT).
Glossary
- Fragile X mental retardation protein (FMRP)
a translational regulator. Functional absence of FMRP causes fragile X syndrome (FXS), the most common form of inherited intellectual disability
- GA motifs
architectural RNA motifs with a core of noncanonical purine•purine base pairs. This core typically comprises tandem G•A/A•G pairs that interact using trans-Hoogsteen/Sugar Edge hydrogen bonds. See also Box 2
- Heterogeneous nuclear ribonucleoproteins (hnRNPs)
a family of nucleo-cytoplasmic RNA binding proteins. hnRNPs play various roles in RNA metabolism, including transcription, RNA processing, RNA transport, and translation
- Metabotropic glutamate receptors (mGluRs)
a group of glutamate receptors that, upon binding of an agonist, activate cellular signaling pathways. mGluR-activated pathways couple to the regulation of translational mechanisms
- Orthologs
genes in different species that evolved from a common ancestor in phylogenetic development. Orthologous gene products typically retain functional equivalence
- snRNAs
snoRNAs, scRNAs, small nuclear, nucleolar, or cytoplasmic RNAs. These RNAs often perform regulatory or structural functions in their respective cellular domains
- Translational control
mechanisms to ensure that proteins are synthesized only when needed and where needed. Translational control mechanisms typically but not exclusively target the initiation phase
- Translation initiation
the first phase of the translation pathway. During initiation, the small ribosomal subunit is recruited to the mRNA (usually to the 5′ end) and is joined at the initiator codon by the large ribosomal subunit. In eukaryotes, translation initiation is mediated by various eukaryotic initiation factors (eIFs)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mathews MB, et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 2.Tiedge H, Brosius J. Translational machinery in dendrites of hippocampal neurons in culture. J Neurosci. 1996;16:7171–7181. doi: 10.1523/JNEUROSCI.16-22-07171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle M, Kiebler MA. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011;30:3540–3552. doi: 10.1038/emboj.2011.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Job C, Eberwine J. Localization and translation of mRNA in dendrites and axons. Nat Rev Neurosci. 2001;2:889–898. doi: 10.1038/35104069. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, et al. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosik KS, Krichevsky AM. The elegance of the microRNAs: a neuronal perspective. Neuron. 2005;47:779–782. doi: 10.1016/j.neuron.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 8.Muddashetty RS, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012;11:189–200. doi: 10.1016/S1474-4422(11)70286-1. [DOI] [PubMed] [Google Scholar]
- 11.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bear MF, et al. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Hagerman R, et al. Fragile X and autism: Intertwined at the molecular level leading to targeted treatments. Mol Autism. 2010;1:12. doi: 10.1186/2040-2392-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laggerbauer B, et al. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 15.Zalfa F, et al. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 16.Monzo K, et al. Fragile X mental retardation protein controls trailer hitch expression and cleavage furrow formation in Drosophila embryos. Proc Natl Acad Sci USA. 2006;103:18160–18165. doi: 10.1073/pnas.0606508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Napoli I, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 18.Lacoux C, et al. BC1-FMRP interaction is modulated by 2′-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res. 2012;40:4086–4096. doi: 10.1093/nar/gkr1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiler IJ, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceman S, et al. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet. 2003;12:3295–3305. doi: 10.1093/hmg/ddg350. [DOI] [PubMed] [Google Scholar]
- 21.Khandjian EW, et al. Biochemical evidence for the association of fragile X mental retardation protein with brain polyribosomal ribonucleoparticles. Proc Natl Acad Sci USA. 2004;101:13357–13362. doi: 10.1073/pnas.0405398101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stefani G, et al. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zalfa F, et al. FMRP binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA binding motif. J Biol Chem. 2005;280:33403–33410. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- 25.Iacoangeli A, et al. On BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci USA. 2008;105:734–739. doi: 10.1073/pnas.0710991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schütt J, et al. Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. J Biol Chem. 2009;284:25479–25487. doi: 10.1074/jbc.M109.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gabus C, et al. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amaral PP, et al. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 29.Cao X, et al. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- 30.Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- 31.Mattick JS. RNA regulation: a new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 32.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taft RJ, et al. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 34.DeChiara TM, Brosius J. Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc Natl Acad Sci USA. 1987;84:2624–2628. doi: 10.1073/pnas.84.9.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, et al. Transcription and processing of the rodent ID repeat family in germline and somatic cells. Nucleic Acids Res. 1995;23:2245–2251. doi: 10.1093/nar/23.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiedge H, et al. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci. 1993;13:2382–2390. doi: 10.1523/JNEUROSCI.13-06-02382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skryabin BV, et al. The BC200 RNA gene and its neural expression are conserved in Anthropoidea (Primates) J Mol Evol. 1998;47:677–685. doi: 10.1007/pl00006426. [DOI] [PubMed] [Google Scholar]
- 38.Martignetti JA, Brosius J. BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc Natl Acad Sci USA. 1993;90:11563–11567. doi: 10.1073/pnas.90.24.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozhdestvensky TS, et al. Neuronal BC1 RNA structure: evolutionary conversion of a tRNAAla domain into an extended stem-loop structure. RNA. 2001;7:722–730. doi: 10.1017/s1355838201002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martignetti JA, Brosius J. Neural BC1 RNA as an evolutionary marker: guinea pig remains a rodent. Proc Natl Acad Sci USA. 1993;90:9698–9702. doi: 10.1073/pnas.90.20.9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozhdestvensky TS, et al. Isolation and posttranscriptional modification analysis of native BC1 RNA from mouse brain. RNA Biol. 2007;4:11–15. doi: 10.4161/rna.4.1.4306. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, et al. Dendritic BC1 RNA: functional role in regulation of translation initiation. J Neurosci. 2002;22:10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondrashov AV, et al. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP) J Mol Biol. 2005;353:88–103. doi: 10.1016/j.jmb.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 44.Eom T, et al. Dual nature of translational control by regulatory BC RNAs. Mol Cell Biol. 2011;31:4538–4549. doi: 10.1128/MCB.05885-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pestova T, et al. The mechanism of translation initiation in eukaryotes. In: Mathews MB, et al., editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 46.Méthot N, et al. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA recognition motif. RNA. 1996;2:38–50. [PMC free article] [PubMed] [Google Scholar]
- 47.Huber KM, et al. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 48.Weiler IJ, et al. Fragile X mental retardation protein is necessary for neurotransmitter-activated protein translation at synapses. Proc Natl Acad Sci USA. 2004;101:17504–17509. doi: 10.1073/pnas.0407533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waung MW, Huber KM. Protein translation in synaptic plasticity: mGluR-LTD, Fragile X. Curr Opin Neurobiol. 2009;19:319–326. doi: 10.1016/j.conb.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chuang SC, et al. Prolonged epileptiform discharges induced by altered group I metabotropic glutamate receptor-mediated synaptic responses in hippocampal slices of a fragile X mouse model. J Neurosci. 2005;25:8048–8055. doi: 10.1523/JNEUROSCI.1777-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osterweil EK, et al. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30:15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ronesi JA, Huber KM. Homer interactions are necessary for metabotropic glutamate receptor-induced long-term depression and translational activation. J Neurosci. 2008;28:543–547. doi: 10.1523/JNEUROSCI.5019-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong J, et al. BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J Neurosci. 2009;29:9977–9986. doi: 10.1523/JNEUROSCI.3893-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong J, et al. Regulatory BC1 RNA and the fragile X mental retardation protein: convergent functionality in brain. PLoS One. 2010;5:e15509. doi: 10.1371/journal.pone.0015509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lewejohann L, et al. Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav Brain Res. 2004;154:273–289. doi: 10.1016/j.bbr.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 56.Dictenberg JB, et al. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schuman EM, et al. Synaptic regulation of translation of dendritic mRNAs. J Neurosci. 2006;26:7143–7146. doi: 10.1523/JNEUROSCI.1796-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lescoute A, et al. Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leontis NB, et al. The building blocks and motifs of RNA architecture. Curr Opin Struct Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grandin K, editor. The Nobel Prizes 2009. Nobel Foundation; 2010. [Google Scholar]
- 61.Muslimov IA, et al. Spatial codes in dendritic BC1 RNA. J Cell Biol. 2006;175:427–439. doi: 10.1083/jcb.200607008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muslimov IA, et al. Spatial code recognition in neuronal RNA targeting: Role of RNA–hnRNP A2 interactions. J Cell Biol. 2011;194:441–457. doi: 10.1083/jcb.201010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith R. Moving molecules: mRNA trafficking in mammalian oligodendrocytes and neurons. Neuroscientist. 2004;10:495–500. doi: 10.1177/1073858404266759. [DOI] [PubMed] [Google Scholar]
- 64.Gao Y, et al. Multiplexed dendritic targeting of α calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell. 2008;19:2311–2327. doi: 10.1091/mbc.E07-09-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson EM, et al. Role of Purα in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res. 2006;83:929–943. doi: 10.1002/jnr.20806. [DOI] [PubMed] [Google Scholar]
- 66.Ohashi S, et al. The single-stranded DNA- and RNA-binding proteins pur α and pur β link BC1 RNA to microtubules through binding to the dendrite-targeting RNA motifs. J Neurochem. 2000;75:1781–1790. doi: 10.1046/j.1471-4159.2000.0751781.x. [DOI] [PubMed] [Google Scholar]
- 67.Cao Z, et al. Clustered burst firing in FMR1 premutation hippocampal neurons: amelioration with allopregnanolone. Hum Mol Genet. 2012;21:2923–2935. doi: 10.1093/hmg/dds118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tassone F, et al. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swanson MS, Orr HT. Fragile X tremor/ataxia syndrome: blame the messenger! Neuron. 2007;55:535–537. doi: 10.1016/j.neuron.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 70.Iwahashi CK, et al. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 71.Zumwalt M, et al. Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol. 2007;4:93–100. doi: 10.4161/rna.4.2.5039. [DOI] [PubMed] [Google Scholar]
- 72.Napierala M, et al. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 2005;33:451–463. doi: 10.1093/nar/gki186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sofola OA, et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sellier C, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin P, et al. Pur α binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim J, et al. Rodent BC1 RNA gene as a master for ID element amplification. Proc Natl Acad Sci USA. 1994;91:3607–3611. doi: 10.1073/pnas.91.9.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buckley PT, et al. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011;69:877–884. doi: 10.1016/j.neuron.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khanam T, et al. Can ID repetitive elements serve as cis-acting dendritic targeting elements? An in vivo study. PLoS One. 2007;2:e961. doi: 10.1371/journal.pone.0000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pe’ery T, Mathews MB. Viral translation strategies and host defense mechanisms. In: Sonenberg N, et al., editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; 2000. pp. 400–424. [Google Scholar]
- 80.Kaufman RJ. The double-stranded RNA-activated protein kinase PKR. In: Sonenberg N, et al., editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; 2000. pp. 503–527. [Google Scholar]
- 81.Bevilacqua PC, et al. Binding of the protein kinase PKR to RNAs with secondary structure defects: role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry. 1998;37:6303–6316. doi: 10.1021/bi980113j. [DOI] [PubMed] [Google Scholar]
- 82.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Herbert A. The four Rs of RNA-directed evolution. Nat Genet. 2004;36:19–25. doi: 10.1038/ng1275. [DOI] [PubMed] [Google Scholar]
- 84.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 85.Brosius J. Echoes from the past--are we still in an RNP world? Cytogenet Genome Res. 2005;110:8–24. doi: 10.1159/000084934. [DOI] [PubMed] [Google Scholar]
- 86.Brosius J. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica. 1999;107:209–238. [PubMed] [Google Scholar]
- 87.De Koning AP, et al. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brosius J. Retroposons - seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- 89.Martignetti JA, Brosius J. BC1 RNA: transcriptional analysis of a neural cell-specific RNA polymerase III transcript. Mol Cell Biol. 1995;15:1642–1650. doi: 10.1128/mcb.15.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leontis NB, et al. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pang KC, et al. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]