Abstract
Genetic cross is a powerful tool for studying malaria genes contributing to drug resistance, parasite development, and pathogenesis. Cloning and identification of recombinant progeny (RP) is laborious and expensive, especially when a large proportion of progeny derived from self-fertilization are present in the uncloned progeny of a genetic cross. Since the frequency of cross-fertilization affects the number of recombinant progeny in a genetic cross, it is important to optimize the procedure of a genetic cross to maximize the cross-fertilization. Here we investigated the factors that might influence the chances of obtaining RP from a genetic cross and showed that different Plasmodium yoelii strains/subspecies/clones had unique abilities in producing oocysts in a mosquito midgut. When a genetic cross is performed between two parents producing different numbers of functional gametocytes, the ratio of parental parasites must be adjusted to improve the chance of obtaining RP. An optimized parental ratio could be established based on oocyst counts from single infection of each parent before crossing experiments, which may reflect the efficiency of gametocyte production and/or fertilization. The timing of progeny cloning is also important; cloning of genetic cross progeny from mice directly infected with sporozoites (vs. frozen blood after needle passage) at a time when parasitemia is low (usually <1%) could improve the chance of obtaining RP. This study provides an optimized protocol for efficiently cloning RPs from a genetic cross of malaria parasites.
Keywords: Malaria, genetic cross, rodent, parasite cloning, microsatellite
1. Introduction
Genetic crosses of malaria parasites have played an important role in studying drug resistance (Beez, et al., 2010; Carlton, et al., 1998; Culleton, et al., 2005; Ferdig, et al., 2004; Nguitragool, et al., 2011; Patel, et al., 2010; Sa, et al., 2009; Su, et al., 1997; Vaidya et al., 1995 Wang, et al., 1997; Wellems, et al., 1990; Yuan, et al., 2011; Yuan, et al., 2009), parasite growth and pathology (Li, et al., 2011; Pattaradilokrat, et al., 2009; Reilly Ayala, et al., 2010); gene expression regulation (Gonzales, et al., 2008), strain specific immunity (Martinelli, et al., 2005a; Pattaradilokrat, et al., 2007), and invasion (Hayton, et al., 2008). Performing a genetic cross and subsequent cloning of progeny are generally labor intensive and expensive, particularly for crosses of human malaria parasites that requires non-human primates. The rodent malaria parasite Plasmodium yoelii comprises three subspecies (Plasmodium yoelii yoelii, Plasmodium yoelii nigeriensis, and Plasmodium yoelii killicki) that share many traits with human malaria parasites and, hence, are widely used to study host immune responses and the genetic basis of parasite phenotypes. Although technically it is easier to perform a genetic cross using rodent malaria parasites, cloning individual progeny requires large numbers of rodents because of the lack of an in vitro system for culturing the parasites. It is therefore important to maximize the efficiency of obtaining independent recombinant progeny (IRP) from a genetic cross.
The general procedure for performing a genetic cross in rodent malarias includes: 1) crossing two parasites with different phenotypes in an Anopheles (An.) mosquito and transmitting the parasites to mice to obtain the blood forms of the parasites; 2) cloning the products of the genetic cross from mouse blood and; 3) genotyping clonal parasites to identify IRP. Theoretically, the greatest proportion of RP (50% of the total progeny clones) will likely occur when an equal number of gametocytes from each parent are transmitted through mosquitoes and fertilized in a non-biased fashion. However, in practice (and in our observations), the percentage of RPs obtained from a genetic cross is often lower than 10% of total parasites cloned (Li, et al., 2011). Indeed, the numbers of IRPs obtained from several malaria crosses have been small, generally 35 or fewer, due to the low efficiency of obtaining RP, and the time and resources to clone a large number of progeny (Carlton, et al., 1998; Hayton, et al., 2008; Martinelli, et al., 2005b; Su, et al., 1997; Vaidya, et al., 1995; Wellems, et al., 1990). Many factors can affect the chance of cloning RPs such as the ability to produce functional gametocytes or the timing of cloning progeny after a cross. A rodent malaria parasite that has been through multiple needle passages in laboratory mice often have reduced or lost the ability to produce gametocytes, oocysts, or sporozoites (Janse, et al., 1992), likely due to accumulation of mutations during asexual passages in mice. A genetic cross involving one parent that produces more infective gametocytes than the other may lead to high rates of self-fertilization of the dominant parent. A second important factor is the timing of cloning or isolating RPs from uncloned products of a genetic cross, especially when a genetic cross is performed between parasite strains with different blood stage growth rates in mice (Li, et al., 2011; Otsuki, et al., 2009; Pattaradilokrat, et al., 2009). Cloning progeny at an early stage when asexual parasites appear in the bloodstream may increase the likelihood of obtaining fast-growing progeny. On the other hand, cloning progeny at a late infection, when the slow-growing parasites appear, may result in isolation of large numbers of duplicated parasite clones after multiple asexual replications. It is therefore worthwhile to investigate parasite population dynamics after a genetic cross so that an optimal time for cloning RPs can be determined.
Here we investigated the parameters that might affect the efficiency of obtaining an RP from a genetic cross of P. yoelii. We performed several genetic crosses, including using a parasite that produced green-fluorescent oocysts as a means to determine the rate of self- and cross-fertilizations, and showed that it was necessary to adjust the ratio of parental parasites used in a cross if the parents had different ability in producing functional gametocytes (as reflected in oocyst count). We found that cloning progeny from a sporozoite-initiated infection, at a time when parasitemia was below <1%, could improve the probability of obtaining IRPs, compared with cloning progeny from frozen blood samples of high parasitemia. A better understanding of parasite population dynamics from a genetic cross can help determine the time of parasite cloning and improve the chances of obtaining IRPs for genetic studies.
2. Materials and Methods
2.1 Parasites, mice, and mosquitoes
All parasites used in this study have been described previously (Fu, et al., 2009; Li, et al., 2011). Parasite BY265 was a gift from the Chinese Academy of Military Science. The parasite was originally obtained by Y. Boulard and I. Landau from a Thamnomys rutilans in 1969 and described by I. Landau in 1992 (Landau, 1992). BY265G was a parasite we generated previously by inserting a copy of the gene encoding a green fluorescent protein (GFP) from the jellyfish Aequorea victoria into the genome of BY265 parasite (Fu, et al., 2009). Briefly, plasmid vector PL10017 with tgdhfr/ts selectable marker for targeted integration into the C- or D-small subunit rRNA loci deposited by A.P. Waters was obtained form Malaria Research and Reference Reagent Center (MR4) for parasite transformation. The plasmid was electroporated into BY265 parasites, and parasites were selected and cloned as described (Janse, et al., 2006; van Spaendonk, et al., 2001). N67, NSM, 17XL, and 17XNL are abbreviated names for P. y. nigeriensis N67, P. y. nigeriensis NSM, P. y. yoelii 17XL, and P. y. yoelii 17XNL, respectively. A colony of An. stephensi mosquitoes maintained in the laboratory of the Third Military Medical University in Chongqing, China was used for completion of the life cycle of genetic crosses. Mosquitoes for transmission of parasites were maintained at 23°C, 70–80% relative humidity, and fed with 5% sugar solution. Kunming (KM) outbred mice or BALB/c inbred mice were purchased from the Institute of Laboratory Animals of the Third Military Medical University and were used in genetic crosses and parasite passages. All animal procedures were performed in accordance with the approved protocols at the Third Military Medical University or Xiamen University in China.
2.2 Mouse infection, blood smears, and parasitemia
To initiate a blood-stage infection, 100 μl of donor mouse blood containing approximately 1 × 106 infected red blood cells (iRBCs) was injected intraperitoneally (i.p.) into a recipient mouse. To monitor parasites in the infected mouse, thin blood smears of tail blood were prepared, stained with Giemsa’s stain, and examined daily for parasitemia. Approximately 1,000 RBCs per smear were examined to estimate parasitemia, and 30–40 microscopic fields with approximately 1,000 iRBC were counted per slide for calculating the percentage of RBCs infected with gametocytes (gametocytemia).
2.3 Mosquito infection
Infected mice with gametocytemia ≥0.5% or at a fixed time after injection of parasites were anesthetized and placed onto a pot of 50–60 female, 5- to 7-day-old An. stephensi mosquitoes. Thirty min after the feeding, the mice were removed and immediately euthanized; unfed or partially engorged mosquitoes were removed from the cages. The mosquitoes were dissected on day 3-23 (different days on different experiments) after the blood meal, and mosquito midguts were examined under a light microscope for oocysts.
2.4 Genetic crosses and parasite cloning
Genetic crosses were performed according to procedures described (Li, et al., 2011; Pattaradilokrat, et al., 2011). Progeny from the crosses were cloned after injecting 100 μl of phosphate buffered saline (PBS) diluted parasite solution containing ~1.0 infected into a mouse. Generally, 50–200 mice were injected for each cloning experiment, given the resources of animal housing and manpower available.
2.5 Preparation of P. yoelii genomic DNA and Microsatellite (MS) typing
A 50 μl volume of mouse tail blood in 1 ml of heparinized PBS was used to extract genomic DNA. Blood cells were pelleted and lyzed with 0.5% saponin in PBS on ice for 5 min and washed three times with PBS to remove hemoglobin. The lysate was then diluted in 50 μl of water, boiled at 95°C for 12 min, and centrifuged to remove cell debris. Parasite genomic DNA in aqueous phase (supernatant) was then transferred to nuclease-free tubes and stored at −20°C prior to quantitative MS typing. PCR primers for MS markers Py2354 Py1618, Py2673, Py1428, Py1404, Py2311, Py1352, Py2000, Py2857, Py2469 and Py1318 and amplification conditions for MS typing were as described previously (Li, et al., 2009). PCR products were analyzed on 2% agarose gels, stained with ethidium-bromide, and visualized under UV transillumination.
3. Results and Discussion
3.1 Different numbers of oocysts produced by various P. yoelii parasites
It is well known that malaria parasites often loose (or reduce) the ability to produce gametocytes or to infect mosquitos after an extended period of time in culture (Plasmodium falciparum) or through needle passages in mice (rodent malaria) (Day, et al., 1993; Janse, et al., 1992). Before performing a genetic cross, it is often necessary to make sure that both parental parasites can complete a sporogonic developmental cycle in mosquitoes. Here we fed six P. yoelii strains/subspecies (N67, NSM, 17XL, 17XNL, BY265, and BY265G) to An. stephensi mosquitoes to investigate the transmissibility of these parasites. Mice were fed to mosquitoes 96 h (optimal time for most P. yoelii (Pattaradilokrat, et al., 2011)) after injection of 1×106 iRBCs, and infected mosquitoes were dissected 8 days post infection (p.i.). Feeding results showed that these parasites indeed produced different numbers of oocysts in the mosquito midgut, with the parasite 17XNL producing an average of approximately 1.2 oocysts and N67/NSM producing an average of 250.5/235.7 oocysts per midgut (Table 1). The BY265 parasite produced an average of 15.4 oocysts per midgut when mosquitoes were fed with parasites 96 h post infection; however, much higher counts (average 133.5) were obtained when parasites of 110 h (optimal time for BY265) were used, likely due to slightly slower gametocyte maturation rate. BY265G, a parasite derived from BY265, had an average of 27.2 oocysts per midgut when fed on 110-hour parasites (Table 1). These results showed that different P. yoelii strains could produce different numbers of functional gametocytes or oocysts, which can be influenced by the feeding time.
Table 1.
Average oocyst counts of six different Plasmodium yoelii strains or subspecies
| Parasite | N67 | NSM | 17XL | 17XNL | BY265 | BY265* | BY265G | BY265G* |
|---|---|---|---|---|---|---|---|---|
| No. mos | 30 | 30 | 30 | 30 | 30 | 30 | 24 | 30 |
| Ave. oocyst | 250.5 | 235.7 | 161.5 | 1.2 | 15.4 | 133.5 | 18.6 | 27.2 |
| STD | 127.8 | 149.9 | 109.4 | 2.3 | 21.3 | 80.1 | 22.4 | 54.9 |
Note: N67, Plasmodium yoelii nigeriensis N67; NSM, P. y. nigeriensis NSM; 17XL, P. y. yoelii 17XL; 17XNL, P. y. yoelii 17XNL; BY265, P. y. yoelii BY265; BY265G, P. y. yoelii BY265G, green fluorescent; No. mos, number of mosquitoes dissected; Avg. oocyst, average number of oocysts; SD, standard deviation. All mosquitoes were fed when gametocytemia was 0.5% or higher. All the feedings were performed 96 h after injection of 1×106 parasites into mice, except those indicated with “*” that were fed at 110 h. The mosquitoes dissected were from at least two separate experiments.
3.2 Parental ratios and variation in recombination rate in genetic crosses
A successful genetic cross will allow isolation of as many independent recombinant progeny (IRP) as possible after cloning and genotyping. Typically, cloning of blood stage malaria parasites from uncloned progeny of a genetic cross requires injections of single parasites into a large number of laboratory mice. It is therefore preferable to generate a population of the uncloned progeny that contains a good proportion of IRPs (instead of parental clones derived from self-fertilization) to improve the chance of identifying IRP.
A parental strain that produces more functional gametocytes tends to undergo self-fertilization at higher rates than the other strain that produces only few functional gametocytes. Crossing two parasites with different abilities in producing functional gametocytes (or at a time favoring one parent) may result in parental-type progeny mostly coming from self-fertilization of the strain that produce more gametocytes and/or more functional oocysts. One solution to compensate for differences in gametocyte production or efficiency of fertilization is to use a larger number of the parent that produces fewer gametocytes in a cross so the parental mixture will contain similar numbers of functional gametocytes from the parents. In practice, it is difficult to determine whether a gametocyte is functional (mature) or not in a blood smear, and to estimate the numbers of hybrid oocysts in mosquitoes. Indeed, it has been shown in both rodent and human malaria parasite infections that gametocytemia do not always correlate with parasite infectivity to mosquitoes (Gadsby, et al., 2009; Gouagna, et al., 2003). We therefore decided to evaluate whether we could use oocyst counts from single-strain infections to guide the parental proportion in a genetic cross and use the ratios of oocysts expressing GFP (sans genotyping) and those without GFP to estimate the frequency of cross-fertilization. In a perfect situation when the numbers of gametocytes of the two parents are equal, half of the total oocysts will originate from self-fertilization (25% of which will be oocysts expressing GFP and 25% will be oocysts without GFP) and half will be hybrids derived from cross-fertilization, with some oocysts containing sporozoites that inherit the chromosome(s) inserted with the GFP plasmid construct.
We performed several genetic crosses with different parental ratios between the fluorescent BY265G parasite and non-fluorescent N67 or NSM parasites 96 hours after injection of parasites into mice. N67 and NSM were parasites of the same genetic background (Li, et al., 2011). Because the N67 and NSM parasites produce approximately 10 times more oocysts than the BY256G parasite, we compared oocyst counts and the ratios of green oocysts and non-green oocysts from genetic crosses with parental ratios of 1:1, 5:1, 10:1, and 20:1 (BY265G:N67), respectively. The results from the experiments showed that a parental ratio of 10:1 (BY265G:N67) produced the largest number of fluorescent oocysts or the best ratios of fluorescent/non-fluorescent oocysts The RFOs were averages from ratios of individual mosquitoes examined in crosses involving N67, with ~5-fold increase in fluorescent oocyst counts compared with those of parental BY265G (Table 2 and Fig. 1). The increase in the numbers of fluorescent oocysts suggests higher frequencies of fertilization events either between fluorescent BY265G and non-fluorescent N67 or BY265G self-fertilizations at 10:1 ratio. One explanation for the increased numbers of green oocysts is that more gametocytes of BY265G than N67 were transmitted to mosquitoes, resulting in more self-fertilized BY265G oocysts. However, this cannot explain the lower green oocyst counts at 20:1 parental ratio (Table 2). Another explanation is that recombinant green oocysts have developmental advantage over self-fertilized green oocysts; e.g., more green recombinant oocysts survived after fertilization. Recombinant oocysts might gain some important genetic factors from N67 that helped their development in mosquito.
Table 2.
Oocyst counts from genetic crosses of a single parent (self-fertilized) or two parents with different parental ratios
| Ratio | No. mos | No. FO (SD) | No. NFO (SD) | RFO |
|---|---|---|---|---|
| BY265G | 46 | 20.4(15.2) | 0 | |
| N67 | 41 | 234.6(86.1) | ||
| 1:1 | 32 | 19.8(11.0) | 178.3(60.1) | 0.2# |
| 5:1 | 33 | 75.4(55.0) | 91.2(66.0) | 1.0*# |
| 10:1 | 32 | 96.8(58.4) | 42.6(28.8) | 2.7* |
| 20:1 | 33 | 42.8(29.3) | 46.7(35.6) | 1.2*# |
| NSM | 45 | 0 | 243.8(93.9) | |
| 1:1 | 34 | 14.8(10.2) | 198.0(77.8) | 0.1# |
| 5:1 | 34 | 22.4 (16.5) | 132.9 (51.4) | 0.2*# |
| 10:1 | 40 | 123.6 (68.1) | 81.0 (44.9) | 1.7* |
| 20:1 | 32 | 54.9(39.2) | 54.6(29.5) | 1.0*# |
Note: The ratios are BY265G:N67 or NSM; No. mos, number of mosquitoes dissected; No. FO, number of fluorescent oocysts; No. NFO, number of non-fluorescent oocysts; RFO, ratio of fluorescent oocysts over non-fluorescent oocysts; SD, standard deviation; the relatively large standard deviations were likely due to the differences in the amount of blood taken by individual mosquitoes. *, P<0.01 compared with the ratios from 1:1 crosses (unpaired t-test), and #, P<0.01 compared with ratios of 10:1 crosses (unpaired t-test). Mosquitoes were dissected on day 7 after feeding infected mice.
Fig. 1.
Relationship of the ratios of parental parasites and oocyst counts in genetic crosses. A) Green fluorescent parasite (GFP) -BY265G- producing few oocysts; BY265G was crossed with non-fluorescent N67 (B–D) or with NSM (E) parasites at 5:1 (B), 20:1 (C), and 10:1 (D and E) ratios. (F) A ratio of 10:1 consistently produced more green oocysts from day 7 to day 14; standard deviation bars were from at least 10 mosquitoes. Crossing BY265G with N67 or NSM can greatly improve the efficiency of green oocyst production by BY265G, suggesting gain of genetic elements from the N67 or NSM during the crosses. In this case, the majority of green oocysts should be recombinant containing BY265 genetic background with the gene expressing GFP and genes that are necessary for producing large numbers of oocysts.
We then repeated the experiments replacing N67 with NSM (essentially the same parasites) with the same parental ratios. Again, the ratios of 5:1, 10:1, and 20:1 (all BY265G:NSM) produced significantly more fluorescent oocysts (or RPs) than the cross using a 1:1 ratio. These results suggest that adjusting the parental ratio can indeed change or improve the chance of fertilization of the parent, either by itself or with the other parent that produces fewer oocysts. A ratio>1 of fluorescent/non-fluorescent oocysts seen in the group of BY265G: N67/NSM (10:1) suggests recombinant progeny among the fluorescent oocysts (Table 2).
3.3 Use of PCR to estimate proportion of parental alleles and parasite cloning time
Another factor that may influence the chance of obtaining RPs from a genetic cross is the timing of parasite cloning after infecting a mouse with sporozoites. After injection of sporozoites into a mouse, the parasite replicates in hepatocytes, producing thousands of merozoites that are released into the bloodstream and eventually invade RBCs. After invading RBCs, the parasites will grow and replicate again. It is therefore reasonable to clone parasites when parasitemia is low. If we clone the parasites when parasitemia is too high (>1%), there will likely be more parasites with the same genotype due to mitotic replication. Another complicating factor is that different parasite strains grow at different rates in mouse blood; the fast grower may appear in the bloodstream in the first few days after initial infection. To determine when is the best time to clone progeny so as to maximize the chance of obtaining RPs, we injected mice with different amounts of iRBCs (5 × 106 and 1 × 107 in frozen blood samples; the real numbers of parasites were lower after thawing due to lysis of RBCs) and monitored parasitemia up to 72 h (Table 3). Parasites could be seen in blood smears at around 30–35 h after injection, and parasitemia increased 6 to 10 fold between 48 to 72 h. These results suggested that delaying parasite cloning for 24 h could lead to ~10-fold increase in redundant clones.
Table 3.
Parasitemia at different hours after injection of Plasmodium yoelii parasites into BALB/c mice
| Parasitemia at different hours post infection (%) |
||||||
|---|---|---|---|---|---|---|
| Mouse | Parasite Inj | 24 h | 36 h | 48 h | 60 h | 72 h |
| 1 | 200 μl | 0 | 0.05 | 0.9 | 2.5 | 5.3 |
| 2 | 200 μl | 0 | 0.05 | 0.6 | 1.9 | 6.1 |
| 3 | 100 μl | 0 | 0.01 | 0.5 | 1.8 | 6.9 |
| 4 | 100 μl | 0 | 0.01 | 0.3 | 1.0 | 3.2 |
Note: Parasite Inj, the amount of parasites injected; a 200 μl inoculum contained approximately 1×107 frozen infected red blood cells (iRBCs). The actual numbers of infective parasites could be lower due to lysis of iRBCs after thawing.
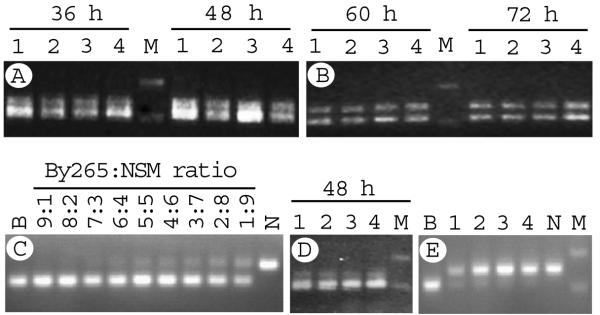
To estimate the ratios of parental parasites during infection, we first used an MS marker (Py2354) that produces products with large size differences (so that PCR products could be separated on an agarose gel) between BY265 (113 bp) and NSM/N67 (154 bp) to estimate the ratios of parental genotypes in the progeny mixtures. Two bands were produced from blood samples of the mice infected with progeny mixture of a genetic cross after separation of PCR products on agarose gels, suggesting mixed infections with both parents or RP (Fig. 2). Band intensity shifted slightly from stronger BY265 (smaller) bands at 36 h to approximately equal intensity of the two bands of BY265 and NSM at 72 h (Fig. 2A and 2B). Considering the higher efficiency in amplifying the smaller products (Fig. 2C), the shift to equal band intensity suggests more parasites carrying NSM genotype at 60 and 72 h, likely due to faster growth rate of NSM.
Fig. 2.
PCR products amplified from parasite mixtures collected at different time points for two NSM×BY265 genetic crosses. Microsatellite Py2354 on chromosome 12 produced an 113-bp product from BY265 and a 154-bp product from NSM. (A) Parasite samples collected from four individual mice at 36 h and 48 h from the first cross, respectively. M, molecular weight marker. (B) The same samples collected from the same cross at 60 h and 72 h, respectively. (C) Products from mixtures of different DNA ratios. B was from Plasmodium yoelii yoelii BY265 DNA, and N was from Plasmodium yoelii nigeriensis NSM DNA. (D) Parasites collected at 48 h from a second cross. Parasites from mouse number 1 (lane 1) were used in the second cloning experiments (Table 4). (E) PCR products showing allelic proportions when parasite mixtures were cloned at 120 h (1, parasitemia 0.74%) and 144 h (3, parasitemia 6.22%) after injection of frozen infected red blood cells into a mouse, respectively. Samples 2 and 4 were from parasite mixtures of 132 and 156 h (not cloned), respectively. B and N are BY265 and NSM DNA controls. M, molecular weight marker.
To further determine the proportion of parental genotypes in blood samples after crossing, we performed limiting dilution to clone individual progeny from two mice that had parasite mixtures producing different intensities of PCR bands (Fig. 2 and Table 4). In the first cloning experiment, each of 50 mice was injected with 1.0 iRBC in 100 μl PBS diluted from the mouse producing similar intensity of DNA bands for both parental genotypes when parasitemia was ~0.3% (Fig. 2A, lane 4 of 48 h). Only six mice were infected, and no RPs (five with NSM genotype and one with mixed genotypes) were obtained after typing the DNA samples with 10 MSs (Table 4). The results were consistent with the PCR band intensities suggesting more NSM parasites in the mixture. In a second cloning experiment, we injected a parasite mixture with stronger intensity of the BY265 band (Fig. 2D, lane 1) and cloned the progeny similarly. Nine RPs from 12 infected mice—including four IRP—were obtained even though the initial parasitemia was slightly higher than the first cloning experiment (Table 4). After adjusting for the difference in the number of infected mice (6 vs. 12), the results from experiment 2 were still better. To avoid variation between cloning from different mice, we also cloned progeny from a mouse injected with infected blood of another cross 24 h apart at parasitemia of 0.74% and 6.22%, respectively (Table 4). PCR typing suggested the progeny mixture was heavily biased toward NSM (Figure 2E). Again, the group cloned at low parasitemia had five recombinant progeny (16.7%), whereas the group cloned at high parasitemia had no recombinant progeny. Although the results were from relatively small-scale cloning experiments, the data indicate that cloning at a time with relatively equal parental genotypes or low parasitemia is crucial. It is also necessary to consider PCR amplification bias when using relative DNA band intensity from MSs to determine the timing of parasite cloning.
Table 4.
Numbers of infected mice and recombinant progeny from genetic crosses cloned at different ratios of parental genotypes or parasitemia.
| Number(%) |
||||||||
|---|---|---|---|---|---|---|---|---|
| Experiments | Parasitemia | No. Mice | No. Inf. Mice | NSM(%) | BY265(%) | MG(%) | RP(%) | IRP(%) |
| 1 | 0.27% | 50 | 6 | 5 (83.3) | 0(0) | 1(16.7) | 0(0) | 0(0) |
| 2 | 0.57% | 46 | 12 | 2(16.7) | 0(0) | 1(8.3) | 9(75.0) | 4(33.0) |
| 3 | 0.74% | 51 | 30 | 20(66.7) | 0(0) | 5(16.7) | 5(16.7) | 3 (10.0) |
| 4 | 6.22 % | 50 | 29 | 27(93.1) | 0(0) | 2(6.9) | 0(0) | 0(0) |
Note: Experiment 1 and 2 were cloned based on different intensity of PCR bands on agarose gels. Parasite mixture in experiment 1 produced similar band intensity for both parental alleles (Fig. 2A, lane 4), whereas experiment 2 had a stronger band for BY265G (Fig. 2D, lane 1). Experiment 3 and 4 were cloned (at 1.3 infected red blood cells per mouse) from a mouse injected with infected blood from another cross at different parasitemia. . Recombinant progeny were identified using 10 microsatellite loci (Py1618, Py2673, Py1428, Py1404, Py2311, Py1352, Py2000, Py2857, Py2469, Py1318) as described previously (Li et al., 2011). No. Mice, numbers of mice used in the cloning experiments; No. Inf. Mice, numbers of infected mice; NSM(%), numbers and percentage of progeny with NSM type; BY265(%), number and percentage of BY265 type; MG(%), numbers and percentage of mixed genotype; RP(%), number and percentage of recombinant progeny; IRP(%), numbers and percentage of independent recombinant progeny.
3.4 Cloning from mice infected with frozen iRBC or sporozoites
Cloning from mice infected with frozen iRBCs of a cross progeny is more convenient than from mice infected with sporozoites, because infection with sporozoites requires performing a cross each time that may also introduce variations during individual crossing experiments; however, cloning from frozen iRBCs requires additional propagation of a parasite mixture, which leads to more redundant progeny, particularly from fast-growers. To investigate how a single passage in mice before cloning would affect the chance of obtaining IRPs, we compared cloned progeny from two independent crosses of direct sporozoite infection with those from frozen mixtures of iRBCs of the same crosses (Table 5). More RPs, including IRPs, were obtained from cloning experiments with mice infected with sporozoites directly (even after adjusting for the numbers of infected mice), suggesting that a passage in mice before cloning could greatly increase numbers of progeny with the same genotypes and that the practice of parasite cloning from propagated iRBCs should be avoided if possible.
Table 5.
Numbers of infected mice and recombinant progeny cloned from mice infected with frozen blood samples or direct sporozoite infection
| Para. Source | NM Inf. with: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| (Parental ratio) | Parasitemia | NM | NIM | NSM | BY265 | NMG | NRP | NIRP | %NIRP |
| Blood (10:1) | 0.68% | 60 | 10 | 7 | 0 | 2 | 1 | 1 | 10 |
| Blood (10:1) | 0.96% | 100 | 13 | 12 | 1 | 0 | 0 | 0 | 0 |
| Sporozoite (10:1) | 0.30% | 102 | 42 | 26 | 0 | 5 | 11 | 10 | 23.8 |
| Sporozoite (10:1) | 1.28% | 94 | 19 | 3 | 0 | 1 | 15 | 13 | 68.4 |
Note: Para. Source (parental ratio), parasites used in infecting the mice for subsequent cloning. Ratios in parentheses are those of BY265:NSM; Parasitemia, parasitemia when the parasites were cloned; NM, numbers of mice; NIM, numbers of infected mice; NM Inf. with, number of mice infected with; NSM, Plasmodium yoelii nigeriensis NSM; BY265, Plasmodium y. yoelii BY265; NMG, numbers of mixed genotypes; NRP, numbers of recombinant progeny; NIRP, numbers of independent recombinant progeny; %NIRP, percentage of infected mice having independent recombinant progeny.
3.5 Conclusions
Many factors can affect the outcome of a malaria genetic cross; two major factors that can influence cloning RPs are 1) the difference in growth rate of asexual stages in the mouse and 2) the variation in the parents’ ability to produce functional gametocytes or oocysts, which in turn affect the frequency of RP production. The chance of cloning IRPs is dependent on the numbers of cross-fertilization events that are influenced by the numbers or the ratio of gametes from each parent. The parental ratio for a cross can be estimated by counting the numbers of oocysts from each parent; e.g., using a parental ratio inverse of their oocyst count ratio in mosquito. To avoid cloning redundant progeny, it is preferable to perform parasite cloning at low parasitemia (<1%) and use mice infected with sporozoites rather than blood containing cross progeny. MSs can be used to estimate a DNA mixture from a cross, but amplification efficiency (and bias) of each parental allele need to be calibrated using DNA mixtures of known parental ratios.
-
▶
Parameters for improving the efficiency of cloning recombinant progeny from 510 genetic crosses of malaria parasites
-
▶
Differences in parasite growth rate, gametocyte/oocsyst counts, time of cloning are important
-
▶
Cloning progeny from sporozoite infection are encouraged
Acknowledgements
This work was funded by the Fundamental Research Funds for the Central Universities (2011121031); by “Project 111” sponsored by the State Bureau of Foreign Experts and Ministry of Education of China (B06016); by a 973 National Basic Research Program of China #2007CB513103; and by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health. We thank NIAID intramural editor Brenda Rae Marshall and Cindy Clark of NIH Library Writing Center for assistance in manuscript editing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beez D, Sanchez CP, Stein WD, Lanzer M. Genetic predisposition favors the acquisition of stable artemisinin resistance in malaria parasites. Antimicrobial Agents and Chemotherapy. 2010;55:50–55. doi: 10.1128/AAC.00916-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J, Mackinnon M, Walliker D. A chloroquine resistance locus in the rodent malaria parasite Plasmodium chabaudi. Molecular and Biochemical Parasitology. 1998;93:57–72. doi: 10.1016/s0166-6851(98)00021-8. [DOI] [PubMed] [Google Scholar]
- Culleton R, Martinelli A, Hunt P, Carter R. Linkage group selection: rapid gene discovery in malaria parasites. Genome Research. 2005;15:92–97. doi: 10.1101/gr.2866205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day KP, Karamalis F, Thompson J, Barnes DA, Peterson C, Brown H, Brown GV, Kemp DJ. Genes necessary for expression of a virulence determinant and for transmission of Plasmodium falciparum are located on a 0.3-megabase region of chromosome 9. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8292–8296. doi: 10.1073/pnas.90.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su X.-z., Wellems TE. Dissecting the loci of low-level quinine resistance in malaria parasites. Molecular Microbiology. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Fu Y, Ding Y, Zhou TL, Chen JD, Peng XH, Xu WY. Recombinant Plasmodium yoelii expressing green fluorescent protein in erythrocytic and mosquito stages. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2009;27:488–491. [PubMed] [Google Scholar]
- Gadsby N, Lawrence R, Carter R. A study on pathogenicity and mosquito transmission success in the rodent malaria parasite Plasmodium chabaudi adami. International Journal for Parasitology. 2009;39:347–354. doi: 10.1016/j.ijpara.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Gonzales JM, Patel JJ, Ponmee N, Jiang L, Tan A, Maher SP, Wuchty S, Rathod PK, Ferdig MT. Regulatory hotspots in the malaria parasite genome dictate transcriptional variation. PLoS Biol. 2008;6:e238. doi: 10.1371/journal.pbio.0060238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouagna LC, Okech BA, Kabiru EW, Killeen GF, Obare P, Ombonya S, Bier JC, Knols BG, Githure JI, Yan G. Infectivity of Plasmodium falciparum gametocytes in patients attending rural health centres in western Kenya. East African Medical Journal. 2003;80:627–634. doi: 10.4314/eamj.v80i12.8779. [DOI] [PubMed] [Google Scholar]
- Hayton K, Gaur D, Liu A, Takahashi J, Henschen B, Singh S, Lambert L, Furuya T, Bouttenot R, Doll M, Nawaz F, Mu J, Jiang L, Miller LH, Wellems TE. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe. 2008;4:40–51. doi: 10.1016/j.chom.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse CJ, Franke-Fayard B, Mair GR, Ramesar J, Thiel C, Engelmann S, Matuschewski K, van Gemert GJ, Sauerwein RW, Waters AP. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Molecular and Biochemical Parasitology. 2006;145:60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Janse CJ, Ramesar J, van den Berg FM, Mons B. Plasmodium berghei: in vivo generation and selection of karyotype mutants and non-gametocyte producer mutants. Experimental Parasitology. 1992;74:1–10. doi: 10.1016/0014-4894(92)90133-u. [DOI] [PubMed] [Google Scholar]
- Landau I. Ph.D. thesis. National Museum of Natural History; Paris, France: 1992. Plasmodium parasites from Thamnomys rutilans from La Maboke. A zoological study. [Google Scholar]
- Li J, Pattaradilokrat S, Zhu F, Jiang H, Liu S, Hong L, Fu Y, Koo L, Xu W, Pan W, Carlton JM, Kaneko O, Carter R, Wootton JC, Su XZ. Linkage maps from multiple genetic crosses and loci linked to growth-related virulent phenotype in Plasmodium yoelii. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E374–382. doi: 10.1073/pnas.1102261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Y, Liu S, Hong L, Sullivan M, McCutchan TF, Carlton JM, Su XZ. Hundreds of microsatellites for genotyping Plasmodium yoelii parasites. Molecular and Biochemical Parasitology. 2009;166:153–158. doi: 10.1016/j.molbiopara.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli A, Cheesman S, Hunt P, Culleton R, Raza A, Mackinnon M, Carter R. A genetic approach to the de novo identification of targets of strain-specific immunity in malaria parasites. Proceedings of the National Academy of Sciences of the United States of America. 2005a;102:814–819. doi: 10.1073/pnas.0405097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli A, Hunt P, Fawcett R, Cravo PV, Walliker D, Carter R. An AFLP-based genetic linkage map of Plasmodium chabaudi chabaudi. Malar Journal. 2005b;4:11. doi: 10.1186/1475-2875-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguitragool W, Bokhari AA, Pillai AD, Rayavara K, Sharma P, Turpin B, Aravind L, Desai SA. Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell. 2011;145:665–677. doi: 10.1016/j.cell.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki H, Kaneko O, Thongkukiatkul A, Tachibana M, Iriko H, Takeo S, Tsuboi T, Torii M. Single amino acid substitution in Plasmodium yoelii erythrocyte ligand determines its localization and controls parasite virulence. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7167–7172. doi: 10.1073/pnas.0811313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JJ, Thacker D, Tan JC, Pleeter P, Checkley L, Gonzales JM, Deng B, Roepe PD, Cooper RA, Ferdig MT. Chloroquine susceptibility and reversibility in a Plasmodium falciparum genetic cross. Molecular Microbiology. 2010;78:770–787. doi: 10.1111/j.1365-2958.2010.07366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattaradilokrat S, Cheesman SJ, Carter R. Linkage group selection: towards identifying genes controlling strain specific protective immunity in malaria. PLoS ONE. 2007;2:e857. doi: 10.1371/journal.pone.0000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattaradilokrat S, Culleton RL, Cheesman SJ, Carter R. Gene encoding erythrocyte binding ligand linked to blood stage multiplication rate phenotype in Plasmodium yoelii yoelii. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7161–7166. doi: 10.1073/pnas.0811430106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattaradilokrat S, Li J, Su X.-z. Protocol for production of a genetic cross of the rodent malaria parasites. Journal of Visualized Experiments. 2011:1–6. doi: 10.3791/2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly Ayala HB, Wacker MA, Siwo G, Ferdig MT. Quantitative trait loci mapping reveals candidate pathways regulating cell cycle duration in Plasmodium falciparum. BMC Genomics. 2010;11:577. doi: 10.1186/1471-2164-11-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.-z., Kirkman LA, Fujioka H, Wellems TE. Complex polymorphisms in an approximately 330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- Vaidya AB, Muratova O, Guinet F, Keister D, Wellems TE, Kaslow DC. A genetic locus on Plasmodium falciparum chromosome 12 linked to a defect in mosquito-infectivity and male gametogenesis. Molecular and Biochemical Parasitology. 1995;69:65–71. doi: 10.1016/0166-6851(94)00199-w. [DOI] [PubMed] [Google Scholar]
- van Spaendonk RM, Ramesar J, van Wigcheren A, Eling W, Beetsma AL, van Gemert GJ, Hooghof J, Janse CJ, Waters AP. Functional equivalence of structurally distinct ribosomes in the malaria parasite, Plasmodium berghei. Journal of Biological Chemistry. 2001;276:22638–22647. doi: 10.1074/jbc.M101234200. [DOI] [PubMed] [Google Scholar]
- Wang P, Read M, Sims PF, Hyde JE. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Molecular Microbiology. 1997;23:979–986. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- Wellems TE, Panton LJ, Gluzman IY, do Rosario VE, Gwadz RW, Walker-Jonah A, Krogstad DJ. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- Yuan J, Cheng KC, Johnson RL, Huang R, Pattaradilokrat S, Liu A, Guha R, Fidock DA, Inglese J, Wellems TE, Austin CP, Su XZ. Chemical genomic profiling for antimalarial therapies, response signatures, and molecular targets. Science. 2011;333:724–729. doi: 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Johnson RL, Huang R, Wichterman J, Jiang H, Hayton K, Fidock DA, Wellems TE, Inglese J, Austin CP, Su XZ. Genetic mapping of targets mediating differential chemical phenotypes in Plasmodium falciparum. Nature Chemical Biology. 2009;5:765–771. doi: 10.1038/nchembio.215. [DOI] [PMC free article] [PubMed] [Google Scholar]