Abstract
Deletion of the core clock gene, Bmal1, ablates circadian rhythms and accelerates aging, leading to cognitive deficits and tissue atrophy (e.g., skeletal muscle) (Kondratov et al., 2006, Kondratova et al, 2010). Although normal aging has been shown to attenuate Bmal1 expression in the master circadian pacemaker in the suprachiasmatic nucleus (SCN), relatively little is known about age-related changes in Bmal1 expression in other tissues, where Bmal1 may have multiple functions. This study tested the hypothesis that aging reduces Bmal1 expression in extra-SCN oscillators including brain substrates for memory and in skeletal muscle. Brains and gastrocnemius muscles were collected from young (3–5 months) and old hamsters (17–21 months) euthanized at four times of day. Bmal1 mRNA expression was determined by conducting in situ hybridization on brain sections or real-time PCR on muscle samples. The results showed age-related attenuation of Bmal1 expression in many brain regions, and included loss of diurnal rhythms in the hippocampal CA2 and CA3 subfields, but no change in muscle. In situ hybridization for Per2 mRNA was also conducted and showed age-related reduction of diurnal rhythm amplitude selectively in the hippocampal CA1 and DG subfields. In conclusion, aging has tissue-dependent effects on Bmal1 expression in extra-SCN oscillators. These finding on normal aging will provide a reference for comparing potential changes in Bmal1 and Per2 expression in age-related pathologies. In conjunction with previous reports, the results suggest the possibility that attenuation of clock gene expression in some brain regions (the hippocampus, cingulate cortex and SCN) may contribute to age-related cognitive deficits.
Keywords: hippocampus, cortex, suprachiasmatic nucleus, Bmal1, Per2, clock genes
1. Introduction
The circadian timing system regulates the expression of a variety of physiological and behavioral processes, ensuring the optimal coordination of these processes with each other and with the ambient environment. Aging deleteriously affects the expression of circadian rhythms (see (Duncan, 2007; Monk, 2005) for review). The most consistent and prominent age-related change is an attenuation of the amplitude and robustness of rhythms; in some cases fragmentation or loss of rhythms occurs. With increased age, altered phase relationship of circadian rhythms to the daily lighting cycle as well as desynchrony of various rhythms within individuals have also been observed. Age-related deficits in circadian timekeeping may contribute to age-related changes in physiological and behavioral functions such as memory. In rodents, a variety of conditions or treatments that ablate circadian rhythms also induce memory deficits resembling those that occur spontaneously during aging (Devan et al., 2001a; Loh, D. H. et al, ; Ma et al., 2007; Ruby et al., 2008). In middle-aged hamsters, deficits in spatial and contextual memory in the absence of changes in motivation or sensory or motor function were correlated with fragmentation of circadian activity rhythms as well as apparent reduction of overall activity (Antoniadis et al., 2000; Devan et al., 2001b). An association between circadian rhythms and memory has also been observed in humans. For example, decreased amplitude and robustness of circadian rhythms is a risk factor for development of mild cognitive impairment or dementia in older women (Tranah et al., 2011).
Circadian rhythms are driven by molecular oscillations consisting of transcriptional-translational feedback loops [for review, see (Ko and Takahashi, 2006)]. The components of the positive feedback loop are the Bmal1 and Clock genes, whose products form heterodimers that induce transcription of other circadian genes, e.g., the Period and Cryptochrome genes, whose products inhibit Bmal1:Clock–mediated transcription, acting as negative feedback loop regulators. Bmal1: Clock heterodimers also stimulate the transcription of many clock-controlled genes that are not part of the circadian clock mechanism. Genetic deletion of Bmal1 (but not Clock) ablates circadian behavioral rhythms, indicating that Bmal1 is an essential, non-redundant component of the molecular circadian clock (Bunger et al., 2000). Interestingly, Bmal1 knock-out mice also exhibit an early aging phenotype, characterized by greatly reduced longevity, tissue atrophy (e.g., muscle and bone), and short- and long-term memory deficits appearing by four months of age (Kondratov et al., 2006; Kondratova et al., 2010). Molecular circadian oscillators are expressed not only in the SCN, the site of the master mammalian circadian pacemaker, but also in many other regions, including the hippocampus, an important substrate for learning and memory, and peripheral tissues such as muscle (Guo et al., 2005; Jilg et al., 2010; Wang, L. M. et al, ; Yoo et al., 2004).
Because genetic deletion of Bmal1, a critical regulator of circadian rhythms, induces aging-like changes in cognition and muscle function, elucidating the effect of natural aging on Bmal1 expression will help clarify the role of this gene in the normal aging process and provide a foundation for interpreting potential changes in Bmal1 expression in pathological conditions of aging. Previous studies have shown that aging decreases Bmal1 expression in the SCN in old Syrian hamsters exposed to constant darkness (Kolker et al., 2003). Whether aging also attenuates SCN Bmal1 expression during exposure to a lighting cycle, a more common environmental condition during the lifespan, and whether aging attenuates Bmal1 expression in other brain regions or peripheral tissues, has not been reported. Because genetic deletion of Bmal1 induces aging changes including memory deficits, the current study tested the hypothesis that natural aging is associated with decreased expression of Bmal1 mRNA in two structures that are essential for learning and memory, the hippocampus and the cingulate cortex, as well as in the SCN, the master circadian pacemaker that synchronizes other oscillators through its output pathways. In order to determine if potential effects of aging on Bmal1 expression are selective for these regions, other brain regions were also investigated. In view of previous findings that genetic deletion of Per2 also deleteriously affects memory (Wang, L. M. et al, ), the effect of aging on Per2 expression in the hippocampus and other brain regions was also explored. Furthermore, since Bmal1 deletion is associated with abnormalities in skeletal muscle structure and function (Andrews et al., 2010) and aging is associated with decreased motor activity (wheel running) (Duncan et al., 2010), the hypothesis that aging decreases Bmal1 expression in this peripheral tissue was also tested. Syrian hamsters, which exhibit age-related changes in circadian rhythms, running, memory, and SCN gene expression (Antoniadis et al., 2000; Duncan et al., 2001; Duncan et al., 2010; Kolker et al., 2003), were the subjects of these studies.
2. Results
Bmal1 expression in brain
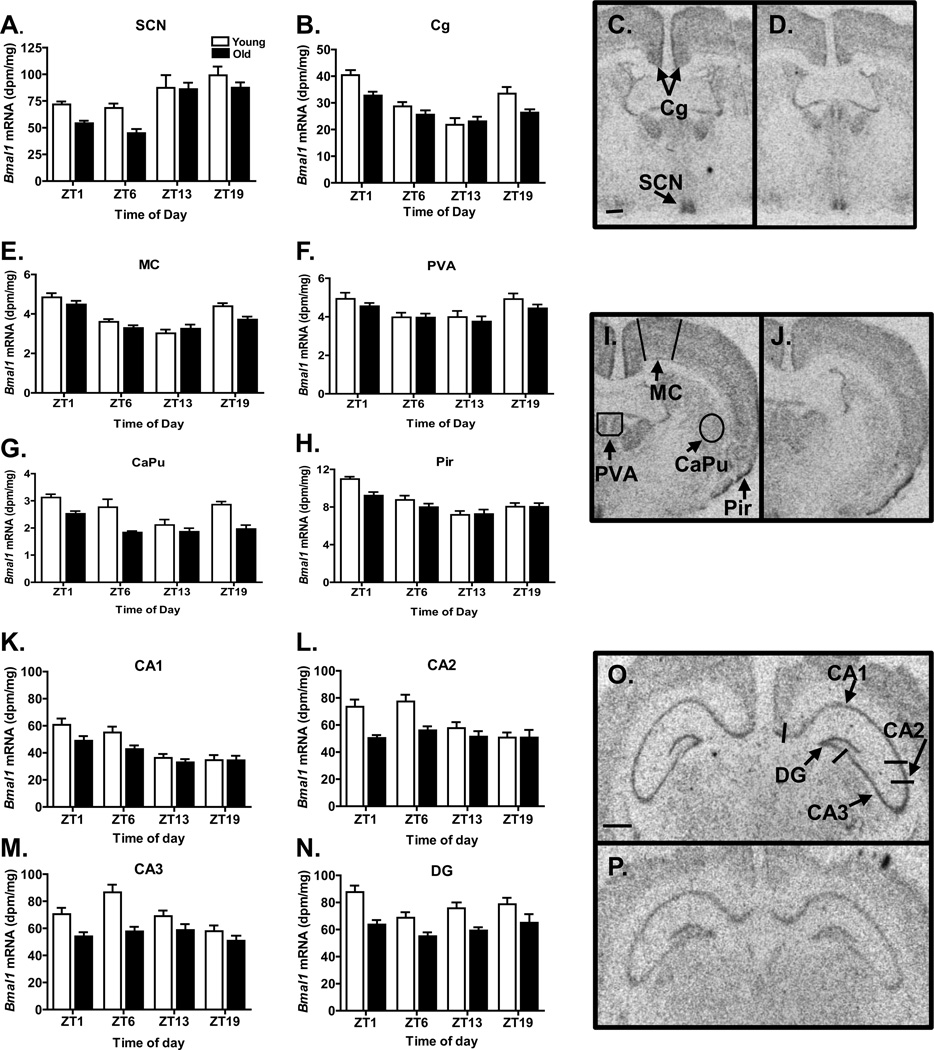
Bmal1 expression exhibited time of day variations in all regions examined (SCN, P<0.0001; hippocampal regions CA1, P<0.00001, CA2, P<0.005, CA3, P<0.005; DG, P<0.02, cingulate cortex, P<0.0001; motor cortex, P<0.0001; piriform cortex, P<0.0001; caudate putamen, P<0.0001; paraventricular thalamic nucleus, anterior (PVA), P<0.0005) (Figures 1 and 2), as expected. In all of these brain regions except the PVA, old animals exhibited decreased Bmal1 expression (SCN, P<0.005; CA1, P<0.01; CA2, P<0.0001; CA3, P<0.0001; DG, P<0.0001, cingulate cortex, P<0.002; motor cortex, P<0.05; piriform cortex, P<l0.05; caudate putamen, P<0.0001; PVA, P=0.119). An interaction between the main effects of time of day and aging on Bmal1 expression was observed in the CA2 (P<0.05), and a trend towards an interaction effect was found in the CA3 (P=0.069), the cingulate cortex (P=0.061), and the piriform cortex (P=0.095) but not in other regions. To further elucidate the age-related differences in rhythms exhibiting a significant or borderline significant time of day by age interaction effect, cosinor analysis was conducted (Table 1). The cosinor analysis also suggested that Bmal1 expression in the CA3 was arrhythmic in the old hamsters (P=0.30) but robustly rhythmic in the young hamsters (P=0.0008). Cosinor analysis of the Bmal1 expression rhythms in the cingulate cortex and piriform cortex yielded significant fits for both young and old, and indicated age-related decreases in amplitude of ~50% and small age-related advances in the acrophases (0.71 h and 0.90 h, respectively).
Figure 1.
Bmal1 expression in selected brain regions. Bars in the graphs represent the mean + S.E.M. level of expression for each age group (N=8–12/group); open bars indicate young animals and filled bars indicate old animals. In each brain region (indicated on the top of each panel), Bmal1 expression varied significantly across the day, and was significantly lower in old animals, except in the PVA. Autoradiograms depicting Bmal1 expression in representative young (C, I, O) and old (D, J, P) hamsters euthanized at ZT1. Scale bar at bottom of C represents 500 microns; D,I and J are the same magnification as C. Scale bar at bottom of O represents 1500 microns; P is the same magnification as O. CaPu, caudate putamen; CG, cingulate cortex; DG, dentate gyrus; MC, motor cortex; Pir, piriform cortex; PVA, paraventricular thalamic nucleus, anterior; SCN, suprachiasmatic nucleus.
Figure 2.
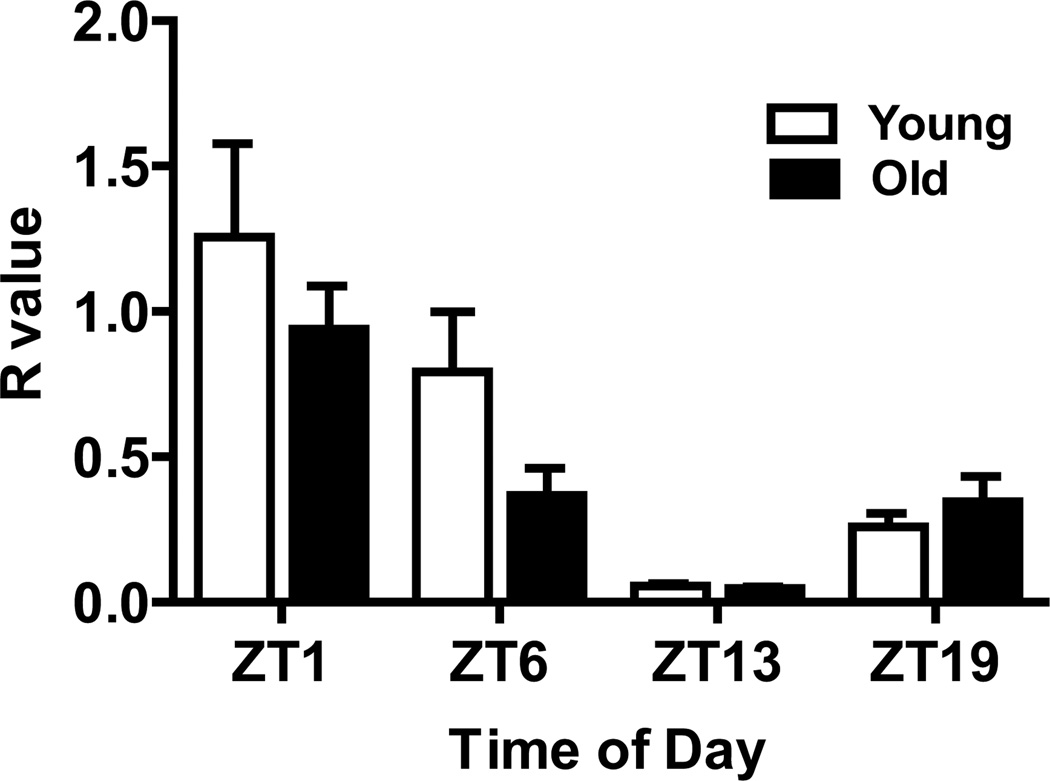
Bmal1 expression in the gastrocnemius muscle. Bars represent the mean + S.E.M. for each group; N=8/group. Bmal1 expression varied significantly with time of day, but was not affected by aging, nor was there an interaction of age and time.
Table 1.
Amplitudes and acrophases of clock gene rhythms revealed with cosinor analysis
| Brain Region | Clock gene |
Age | Amplitude (dpm/mg) |
Amplitude: Age difference* |
Acrophase (Peak time, h) |
Acrophase: Age Difference # |
|---|---|---|---|---|---|---|
| CA2 | Bmal1 | Young Old |
29.19 arrhythmic |
NA | 4.53 NA |
NA |
| CA3 | Bmal1 | Young Old |
29.13 arrhythmic |
NA | 6.45 | NA |
| Cingulate cortex | Bmal1 | Young Old |
19.77 9.72 |
10.05(50.8%) | 23.32 0.03 |
+0.71 |
| Piriform cortex | Bmal1 | Young Old |
3.69 1.95 |
1.74(47.2%) | 1.10 0.20 |
+0.90 |
| CA1 | Per2 | Young Old |
47.3 21.5 |
25.8(54.5%) | 11.32 13.54 |
−2.22 |
| DG | Per2 | Young Old |
49.6 32.2 |
17.4(35.1%) | 10.56 13.29 |
−2.73 |
Numbers in parentheses represent differences as a percentage of young value
Positive values represent phase advances; negative values represent phase delays
NA, values not available due to arrhythmicity in old animals, as indicated by lack of significance of cosine fit model.
Bmal1 expression in skeletal muscle
Bmal1 mRNA from young and old animals exhibited a clear circadian rhythm of expression in hamster gastrocnemius muscle (P<0.0001) (Figure 2). Peak expression was at ZT1, an approximate 6-hour delay from peak expression in the SCN, and similar to results in vastus muscle of hamsters after short-term exposure to constant darkness (Guo et al., 2006) and in gastrocnemius muscle of Wistar/Kyoto rats (Miyazaki et al., 2011). However, no significant effect of aging (P=0.147), or an interaction of age and time of day on Bmal1 expression (P=0.333) was observed.
Per2 expression in hippocampus
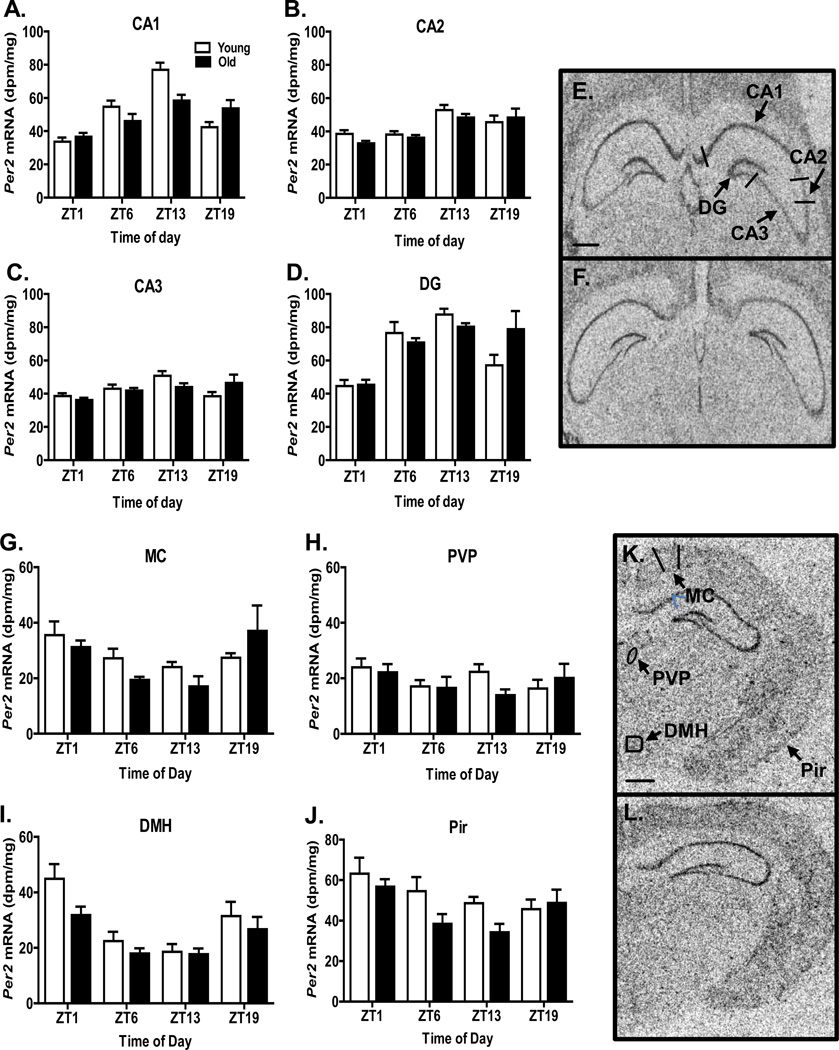
Per2 expression varied with time of day in all hippocampal regions (CA1, P<0.0001; CA2, P<0.0001; CA3, P<0.01; DG, P<0.0001) and in the dorsomedial hypothalamus (P<0.0001), the piriform cortex (P<0.02), and the motor cortex (P<0.005), but not in the posterior paraventricular thalamic nucleus (P=0.465) (Figure 3). Aging significantly affected Per2 expression in the dorsomedial hypothalamus (P<0.05) and the piriform cortex (P<0.05), but not in other regions. An interaction between aging and time of day on Per2 expression was observed only in the CA1 (P<0.01) and DG (P<0.05). As revealed by cosinor analysis, the Per2 expression rhythm in the CA1 exhibited an age-related phase delay of 2.22 h and an age-related amplitude reduction of ~55% (Table 1). In the DG, the Per2 expression rhythm also showed an age-related phase delay (2.73 h) and an amplitude reduction (~35%) (Table 1).
Figure 3.
Per2 expression in selected brain regions. Bars represent the mean + S.E.M. level of expression for each age group (N=8–12/group); open bars indicate young animals and filled bars indicate old animals. In each hippocampal subfield (indicated on the top of each panel), and in the MC, DMH, and Pir, Per2 expression varied significantly across the day. Per2 expression was significantly different in old animals in the DMH and Pir, and there was a significant interaction effect in the CA1 and DG. E & F. Autoradiograms depicting Per2 expression in the hippocampus from a representative young (E) and old (F) hamster euthanized at ZT13. K & L. Autoradiograms depicting Per2 expression in the hippocampus from a representative young (K) and old (L) hamsters euthanized at ZT1. Scale bars at the bottom of E and K represent 1750 microns (F and L are the same magnification as E and K, respectively). DMH, dorsomedial hypothalamus; MC, motor cortex; Pir, piriform cortex; PVP, paraventricular thalamic nucleus, posterior.
3. Discussion
The current study of hamsters shows that natural aging is associated with decreased expression of Bmal1, an essential gene for the expression of circadian oscillations, in the hippocampus and the cingulate cortex, as hypothesized, and in most of the other brain regions examined. Old hamsters exhibited attenuated expression of Bmal1 in all major hippocampal subfields, i.e., the CA1, CA2, CA3 and DG, but statistically significant loss of Bmal1 rhythmicity was observed only in the CA2 and CA3. These findings reveal that the age-related decrease of Bmal1 expression previously reported in the SCN of constant dark-exposed hamsters (Kolker et al., 2003) also occurs in a variety of brain regions, and is not rescued by exposure to an alternating light:dark cycle. Age-related attenuation of Bmal1 expression was not limited to regions essential for cognition and memory, but was also observed in nearly every region investigated, including the motor cortex, piriform cortex, and caudate putamen, indicating that this phenomenon may be a common characteristic of brain aging. Because ablation of diurnal Bmal1 expression rhythms was only observed in the CA2 and CA3 of old animals, the oscillators in these regions appear to be especially vulnerable to impairment during aging. Although the mechanisms underlying selective loss of the Bmal expression rhythm in the CA2 and CA3 have not been elucidated, it is possible that these hippocampal regions are weak oscillators that are more dependent on extrinsic synchronizing signals than the CA1 and DG. In contrast to age-related attenuation of Bmal1 expression in most brain regions, aging did not significantly alter Bmal1 expression in the skeletal muscle tissue.
Our findings on Bmal1 expression in the hamsters brain contrast with recent reports of diurnal rhythms in Bmal1-immunoreactivity (Bmal1-ir) in young (4 months old) and middleaged mice (16 months old) (Wyse and Coogan, 2010). In young adult mice, Bmal1-ir rhythms were observed in several brain regions, including the CA1, CA3, and paraventricular thalamic nucleus (PVT) but surprisingly, not in the SCN. In middle-aged mice, these BMal1-ir rhythms persisted only in the CA3 and PVT. Aging decreased Bmal1-ir in the CA3 and lateral habenula, as well as in other mouse brain areas in which rhythmic expression was not observed. In contrast to these studies in mice, our data indicate that Bmal1 expression in hamsters shows diurnal rhythms in the SCN, all hippocampal regions (CA1, CA2, CA3, and DG), the cingulate, motor, and piriform cortices, the PVA, and the caudate putamen. Aging attenuates Bmal1 expression in all of these hamster brain regions except the PVA, but ablates the diurnal rhythm only in the CA2 and CA3. The factors contributing to the differences between the present study and the previous study in mice may include the species difference as well as the endpoint, i.e., mRNA versus protein expression.
In contrast to the decrease in Bmal1 expression observed in nearly all brain regions examined in the old hamsters, Per2 expression per se was significantly lower only in the dorsomedial hypothalamus and the piriform cortex of these animals. However, the diurnal rhythms of Per2 expression in the CA1 and the dentate gyrus of old hamsters had lower amplitude and were phase delayed, as compared to young animals. Thus, aging appears to differentially affect Per2 expression in various brain regions.
The present observations of diurnal rhythms in Bmal1 and Per2 expression in the hamster hippocampus and cortex support and extend previous reports of clock gene oscillations in the rodent hippocampus. For example, Bmal1 expression exhibits 24-h variations in the mouse hippocampus (Jilg et al., 2010). Also, diurnal rhythms of expression of Per1 and Per2 mRNA or protein have been reported in all hippocampal regions (CA1, CA2, CA3, and DG) in rats and mice (Amir et al., 2006; Jilg et al., 2010; Wang, L. M. et al, ). In mice, these rhythms persist in continuous darkness and are thus endogenously generated, or circadian, in nature (Wang, L. M. et al, ). Furthermore, circadian rhythms in Per2 gene expression are also observed in mouse hippocampal slices maintained in vitro (Wang, L. M. et al, ), indicating that hippocampal circadian rhythms in Per2 expression are intrinsic in origin.
The current study, which investigated the rhythmic expression of Bmal1 and Per2 in two populations of animals using samples collected at discrete points, has several limitations. It was not possible to determine clock gene expression rhythms in individual animals or to obtain high resolution of the timing (phase) of the rhythms in the two age groups sampled at only four time points. In spite of these limitations, age-related decreases in expression of Bmal1 were consistently observed throughout the brain. Also, the high neuroanatomical precision provided by in situ hybridization permitted detection of the age-related decrements in Bmal1 expression within discrete regions within the hippocampus.
Deletion of circadian genes is associated with cognitive impairment. As mentioned above, global, embryonic deletion of Bmal1 is associated with impaired short- and long-term memory in adult mice (Kondratova et al., 2010). The interpretation of this finding is confounded by multiple effects of the global Bmal1 deletion, such as the loss of behavioral circadian rhythms and the early aging phenotype (Kondratov et al., 2006). Mutation of the Per1 and Per2 genes also compromises memory. For example, Per2-mutant mice exhibit decreased ability to recall trace-fear conditioning between 1–7 days after training (Wang, L. M. et al, ). Also, Per2-mutant mice show abnormal long-term potentiation in the hippocampus, in particular, deficits in synaptic potentiation evoked by a strong LTP protocol while basic synaptic transmission was not affected (Wang, L. M. et al, ). Per1 knock-out mice exhibit deficits in spatial memory when tested in a radial arm maze (Jilg et al., 2010). Because the mice used in these studies had global genetic mutations, the specific role of the loss of the Period genes in the hippocampus on the cognitive deficits could not be determined. In order to determine the role of hippocampal expression of the Period genes or Bmal1 in regulating memory, studies inducing hippocampal-selective deletion of these genes are needed. As recently reported in abstract form, a conditional tissue-specific deletion of Bmal1 in the hippocampus of post-natal mice led to impaired performance in hippocampal-dependent tasks such as classical fear conditioning and spatial-object recognition, without alterations in circadian locomotor rhythms (Venkataraman et al., 2012).
Circadian rhythms constitute an important factor regulating learning and memory (Chaudhury et al., 2005; Chaudhury and Colwell, 2002). Furthermore, disruption of circadian rhythms by a variety of treatments leads to deficits in memory. In rats, for example, disruption of circadian rhythms by chronic phase advancement of the daily light:dark cycle leads to memory impairment in the Morris water maze (Devan et al., 2001a). Also, rapid changes in the lighting cycle that mimic jet lag disrupt fear-conditioned memory in mice (Loh, D. H. et al, ). In another paradigm, Siberian hamsters were rendered arrhythmic by light exposure at night followed by phase delay of the lighting cycle the next day; this treatment strongly impaired novel object recognition (Ruby et al., 2008). Although the SCN has been considered the master pacemaker driving circadian rhythms, it is now known that circadian oscillations are exhibited by many tissues and brain regions, including the hippocampus, which is a crucial structure for memory (Jilg et al., 2010; Wang et al., 2009). Spatial memory, which is assessed in the Morris water maze and novel object recognition tests, depends on the hippocampus, as does contextual fear conditioning, which is also influenced by other structures including the anterior cingulate cortex (Steenland et al., 2012). Previous studies have demonstrated an association between attenuation of circadian rhythms and memory impairment during aging. In older community-dwelling women, decreased amplitude and robustness of circadian activity rhythms was associated with increased odds of developing mild cognitive impairment or dementia within five years (Tranah et al., 2011). In hamsters, fragmentation of circadian rhythms was associated with deficits in spatial and contextual memory (Antoniadis et al., 2000).
In the current study, the stability of Bmal1 expression in the gastrocnemius muscle of old hamsters suggests that Bmal1 expression in skeletal muscle may not be affected by aging, but since only one muscle was examined, no definitive conclusion can be made. To the best of our knowledge, investigations of age-related alterations in skeletal muscle expression of Bmal1 or other clock genes have not been previously reported. Bmal1 was examined in the present study because deletion of this gene causes profound alterations in the structure and function of skeletal muscle (Andrews et al., 2010; Kondratov et al., 2006). Furthermore, skeletal muscle-specific rescue of Bmal1 expression in Bmal1 knock-out mice increases activity levels and improves survival over a 10-month period (McDearmon et al., 2006). The limited results of the current study tentatively suggest that attenuation of skeletal muscle expression of Bmal1 expression in old hamsters does not mediate the age-related reduction of running wheel activity previously observed in this species (Duncan et al., 2010). It was interesting to note that aging was associated with decreased expression of Bmal1 in two brain regions, the caudate putamen and piriform cortex, which regulate motor function.
The mechanisms mediating age-related alterations in Bmal1 and Per2 expression in the hippocampus or other brain regions have not been elucidated. It seems unlikely that these changes are caused by cell loss, because previous studies in rats using unbiased, stereological assessments have indicated that normal aging is not associated with loss of cells in the suprachiasmatic nucleus or hippocampus (Madeira et al., 1995; Rasmussen et al., 1999). Instead, aging of the hippocampus is associated with pronounced changes in gene expression, especially genes involved in inflammation/immune function, myelination, cholesterol metabolism, phospholipid trafficking (Blalock et al., 2003; Haberman et al., 2011; Rowe et al., 2007; Verbitsky et al., 2004). During aging, altered expression of genes regulating inflammation and immune function leads to increased levels of cytokines and reactive oxygen species, and a state of chronic, low grade inflammation. A relative increase in pro-inflammatory over anti-inflammatory mediators leads to a pro-inflammatory Th-1 (cellular) type state, associated with increased reactivity to immune stimulation based on priming of brain microglial cells and peripheral macrophages (Oxenkrug, 2011). Interferon-gamma (INF ), which exhibits higher levels of expression during aging, often acts as the priming stimulus (Lyons et al., 2011). In middle-aged non-human primates, alterations in circadian rhythms were associated with circulating levels of INF (Cayetanot et al., 2009). Chronic treatment of rat SCN cultures with INF decreases the amplitude of the Per1-luc expression rhythm, as well as the amplitude, frequency, and regularity of neuronal firing (Kwak et al., 2008). Studies in mice also indicate that the aged SCN exhibits greatly increased responsiveness to INF -induced activation of astrocytes and microglia (Deng et al., 2010). These finding suggest that increased levels of or sensitivity to inflammatory cytokines may play a role in mediating age-related changes in clock gene expression in the SCN, hippocampus, and perhaps other brain regions.
4. Experimental Procedures
4.1 Animals and housing conditions
Male Syrian hamsters (Mesocricetus auratus, Harlan, HsdHan:AURA ), of two ages (young, 3–5 months; old, 17–21 months) were used. The longevity for this species is ~22 months (Kirkman and Yau, 1972). The hamsters were housed individually in polycarbonate cages with rodent chow and water available continuously. All hamsters were exposed to a 14:10 light:dark cycle with average light intensity of ~ 250 lux, for at least 10 days. This photoperiod was used because it maintain reproductive activity in hamsters, in contrast to a 12L:12D photoperiod, which induces gonadal regression to a prepubertal condition. At four specified zeitgeber times (ZT, where ZT 12 by convention is defined as the time of lights off), ZT1, ZT6, ZT13, and ZT19, the hamsters were deeply anesthetized with xylazine/ketamine and decapitated (N=9–14 hamsters/time/age). The animal procedures were approved by the Institutional Animal Care and Use Committees at the University of Kentucky and were consistent with AAALAC guidelines.
4.2 Tissue preparation
After decapitation, brains were removed, frozen on crushed dry ice, and stored at −80°C. Coronal sections (20 µm) were prepared with a cryostat microtome and mounted on negatively-charged slides, which were stored at −80°C. At the time of euthanasia, gastrocnemius muscles were also dissected, frozen on crushed dry ice, and stored at −80°C.
4.3 RNA purification
Total RNA was purified from 80–150 mg of muscle tissue (n=8/group). Samples were ground in mortars chilled in dry ice, transferred to Trizol reagent (1 ml/100 mg tissue; Invitrogen, Carlsbad, CA), and homogenized on ice with a VirTishear homogenizer (VirTis, Gardiner, NY) for 30 seconds. After incubation at room temperature for 5 minutes, samples were mixed with a 20% final volume of chloroform, centrifuged, and aqueous supernatants were removed. RNA was precipitated with a final concentration of 35% isopropanol, centrifuged, and resulting pellets were washed twice with 75% ethanol. Pellets were then dried, RNA was dissolved in RNAsecure (Applied Biosystems, Carlsbad, CA), aliquotted, and stored at −80°C. RNA concentration and purity were determined by spectophotometric absorbance at 260/280 nm. RNA integrity was verified by denaturing formaldehyde agarose gel electrophoresis. DNA was removed from RNA preparations using a DNA-free® Kit (Applied Biosystems).
4.4 In situ hybridization
Expression of Bmal1 mRNA in the SCN, the CA1, CA2, CA3, and DG regions of the hippocampus, the cingulate, motor, and piriform cortices, the anterior paraventricular thalamic nucleus, and the caudate putamen, and of Per2 in the hippocampus, motor and piriform cortices, the posterior paraventricular thalamic nucleus, and the dorsomedial hypothalamus were determined by in situ hybridization (ISH). Two oligonucleotide (oligo) antisense probes to Bmal1 (GenBank ID AF_070917) and to Per2 (GenBank® ID NM_011066.3) mRNA were purchased from IDT® (Coralville, IA). Oligos were labeled by the addition of a 3’ poly-33P-dATP tail using terminal transferase (NEB®, Beverly, MA). The labeled probes were then separated from unincorporated nucleotide through mini Quick-Spin oligo purification columns (Roche®, Indianapolis, IN).
Four brain sections from each animal from each region were used in each of the ISH experiments. Slide-mounted brain sections received a sequence of pre-hybridization treatments, including fixation in 4% paraformaldehyde/0.1 M PB (pH 7.4) solution, several washes in 1X PBS and 1X PBS/27 mM glycine, acetylation using 0.25% acetic anhydride in 0.1 M triethanolamine solution (pH 8.0), and dehydration/delipidation using graded ethanol solutions and chloroform. Slides were then air dried and sections were hybridized to 33P-labeled oligo probes diluted in a hybridization cocktail (final concentration of 37.5% deionized formamide, 7.5% dextran sulfate, 0.75X Denhardt’s solution, 15 mM Tris-HCl pH 7.4, 225 mM NaCl, 0.75 mM EDTA, 75 mM DTT, 250 µg/ml salmon sperm DNA, and 25 µg/ml yeast RNA) and overlayed with coverslips. After an 18 hour incubation at 37°C, slides were rinsed in 2X SSC/10 mM DTT/0.25% Na2S2O3 at room temperature to remove coverslips, and the sections were then subjected to a series of post-hybridization washes, which consisted of one wash in 2X SSC/10 mM DTT/0.25% Na2S2O3 solution at room temperature, four washes in 1X SSC/10 mM Na2S2O3 at 55°C, and two additional washes in the same solution at room temperature. Slides were then quickly dipped in sterile water and then 95% ethanol, air dried, and apposed to autoradiography films (Biomax MR™, Kodak, Rochester, NY) together with a set of 14C radioactive standards (Amersham Life Sciences, Piscataway, NJ) in X-ray cassettes. After exposure times of 14 days, the experimental films were developed.
The hybridization signal in each region of interest was determined using an MCID image analysis system (Imaging Research Inc., St. Catherines, Ontario, Canada). A standard curve was established using optical densities of the 14C standards and curve fitting (3rd-degree-polynomial). The hybridization signal of each sample was determined from the standard curve. The level of background signal was also measured from each brain section, and its average value was subtracted from the hybridization signals of the corresponding regions of interest. Background-subtracted hybridization signals for each region of interest were averaged across all four sections used for each animal. For each region, same group values were subjected to Grubb’s outlier calculations. Outliers were excluded from the final group analyses. Gene expression was analyzed by a 2-way analysis of variance (ANOVA).
4.4 RT-PCR
First-strand cDNA synthesis was achieved by transcription of 1 µg of RNA using a Superscript® Vilo™ cDNA Synthesis Kit (Invitrogen, Carlsbad, CA). Real-time PCR assays were performed on an Applied Biosystems 7500 Fast Real-Time PCR System using Power SYBR Green PCR reagents (Applied Biosystems, Carlsbad, CA). The Bmal1 amplicon was a 184-base pair fragment generated from two primers from nucleotide 1413–1597 of the coding sequence (forward—TGGACGAAGACAATGAGCCAGACA; reverse— AGCTGTTGCCCTCTGGTCTACAAA; IDT, Coralville, IA). GAPDH, which was pre-determined to have stable constituitive expression in both age groups, was amplified as the housekeeping gene (primers: forward—GACCCCTTCATTGACCTCAACTAC; reverse— GCCAGTAGACTCCACAACATACTC; IDT, Coralville, IA). The relative quantification method (Pfaffl, 2001) was used to determine ratio (R) values of gene expression in each sample relative to that of a calibrator cDNA from muscle of a young hamster euthanized at the ZT1 time point. R values were analyzed by a 2-way analysis of variance (ANOVA).
4.5 Data analysis
Data were subjected to 2-way analysis of variance (ANOVA, Graph-Pad Prism, version 5) to investigate the main effects of age and time of day as well as a potential interaction between these main effects. Also, 1-way ANOVA was conducted within each age group to test the effect of time of day. When significant P values (P<0.05), were obtained, post-hoc analysis was conducted using Bonferroni’s post-hoc test. Where the results of the ANOVA indicated age by time of day interactions (P<0.05), cosinor analyses were conducted to estimate the amplitude and phase of the observed rhythms for each age group, as described by (Tong, 1976).
5. Conclusions
The current findings show that aging is associated with decreased expression of Bmal1 in the SCN, cortex, hippocampus, and caudate putamen, as well as attenuated or altered Per2 oscillations in the CA1, DG, dorsomedial hypothalamus, and piriform cortex. In conjunction with previous reports that deletion of circadian clock genes compromises memory, the current findings suggest that age-related changes in circadian clock genes may increase vulnerability to age-related impairments in memory and perhaps other neural functions. Furthermore, these findings add to a growing database indicating that aging alters hippocampal gene expression, by showing that aging also affects genes that regulate circadian rhythms. Bmal1 and Per2 have different roles in the generation of molecular oscillations, acting as components of the positive and negative limbs of the core feedback loop, respectively. Therefore, the current findings show that aging impacts two components of the molecular circadian clock. Finally, aging may differentially influence clock gene expression in the brain and peripheral tissues such as skeletal muscle, in view of the lack of effect of aging on Bmal1 expression in the gastrocnemius muscle.
Acknowledgements
These studies were supported by NIH 2R01AG13418 (MJD). We appreciate the assistance of Dr. Jan Fahrenkrug with cosinor analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amir S, Harbour VL, Robinson B. Pinealectomy does not affect diurnal PER2 expression in the rat limbic forebrain. Neurosci. Lett. 2006;399:147–150. doi: 10.1016/j.neulet.2006.01.041. [DOI] [PubMed] [Google Scholar]
- Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Rusell B, Campbell K, Arbogast S, Reid MB, Walker JR, Hogenesch JB, Takahashi JS, Esser KA. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl. Acad. Sci. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniadis EA, Ko CH, Ralph MR, McDonald RJ. Circadian rhythms, aging and memory. Behav. Brain Research. 2000;111:25–37. doi: 10.1016/s0166-4328(00)00145-5. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Porter NM, Foster TC, Landfield PW. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J. Neurosci. 2003;23:3807–3819. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayetanot F, Nygard M, Perret M, Kristensson K, Aujard F. Plasma levels of interferon-gamma correlate with age-related disturbances of circadian rhythms and survival in a non-human primate. Chronobiol. Internat. 2009;26:1587–1601. doi: 10.3109/07420520903398518. [DOI] [PubMed] [Google Scholar]
- Chaudhury D, Colwell CS. Circadian modulation of learning and memory in fear-conditioned mice. Behav. Brain Research. 2002;133:95–108. doi: 10.1016/s0166-4328(01)00471-5. [DOI] [PubMed] [Google Scholar]
- Chaudhury D, Wang LM, Colwell CS. Circadian regulation of hippocampal long-term potentiation. J. Biol. Rhythms. 2005;20:225–236. doi: 10.1177/0748730405276352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X-H, Bertini G, Palomba M, Xu Y-Z, Bonaconsa M, Nygard M, Bentivoglio M. Glial transcripts and immune-challenged glia in the suprachiasmatic nucleus of young and aged mice. Chronobiol. Internat. 2010;27:742–767. doi: 10.3109/07420521003681498. [DOI] [PubMed] [Google Scholar]
- Devan BD, Goad EH, Petri HL, Antoniadis EA, Hong NS, Ko CH, Leblanc L, Lebovic SS, Lo Q, Ralph MR, McDonald RJ. Circadian phase-shifted rats show normal acquisition but impaired long-term retention of place information in the water task. Neurobiol. Learn. Mem. 2001a;75:51–62. doi: 10.1006/nlme.1999.3957. [DOI] [PubMed] [Google Scholar]
- Devan BD, Goad EH, Petri HL, Antoniadis EA, Hong NS, Ko CH, Leblanc L, Lebovic SS, Lo Q, Ralph MR, McDonald RJ. Circadian phase-shifted rats show normal acquisition but impaired long-term retention of place information in the water task. Neurobiol. Learn. Mem. 2001b;75:51–62. doi: 10.1006/nlme.1999.3957. [DOI] [PubMed] [Google Scholar]
- Duncan MJ. Aging of the mammalian circadian timing system: Changes in the central pacemaker and its regulation by photic and nonphotic signals. Neuroembryol. Aging. 2007;4:85–101. [Google Scholar]
- Duncan MJ, Hester JM, Hopper JA, Franklin KM. The effects of aging and chronic fluoxetine treatment on circadian rhythms and suprachiasmatic nucleus expression of neuropeptide genes and 5-HT1B receptors. Eur. J. Neurosci. 2010;31:1646–1654. doi: 10.1111/j.1460-9568.2010.07186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MJ, Hill SA, Herron JM. Aging selectively suppresses vasoactive intestinal peptide messenger RNA expression in the suprachiasmatic nucleus. Mol. Brain Res. 2001;87:196–203. doi: 10.1016/s0169-328x(01)00015-8. [DOI] [PubMed] [Google Scholar]
- Guo H, Brewer JM, Lehman MN, Bittman EL. Suprachiasmatic regulation of circadian rhythms of gene expression in hamster peripheral organs: Effects of transplanting the pacemaker. J. Neurosci. 2006;26:6406–6412. doi: 10.1523/JNEUROSCI.4676-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Brewer JM, Champhekar A, Harris RBS, Bittman EL. Differential control of peripheral circadian rhythms by suprachiasmatic-dependent neural signals. Proc. Natl. Acad. Sci. 2005;102:3111–3116. doi: 10.1073/pnas.0409734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberman RP, Colantuoni C, Stocker AM, Schmidt AC, Pedersen JT, Gallagher M. Prominent hippocampal CA3 gene expression profile in neurocognitive aging. Neurobiol. Aging. 2011;32:1678–1692. doi: 10.1016/j.neurobiolaging.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilg A, Lesny S, Peruzki N, Schwegler H, Selbach O, Dehghani F, Stehle JH. Temporal dynamics of mouse hippocampal clock gene expression support memory processing. Hippocampus. 2010;20:377–388. doi: 10.1002/hipo.20637. [DOI] [PubMed] [Google Scholar]
- Kirkman H, Yau PKS. Longevity of male and female, intact and gonadectomized, untreated and hormone-treated, neoplastic and non-neoplastic Syrian hamsters. Am. J. Anat. 1972;135:205–220. doi: 10.1002/aja.1001350208. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006;15:R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Kolker DE, Fukuyama H, Huang DS, Takahashi JS, Horton TH, Turek FW. Aging alters circadian and light-induced expression of clock genes in golden hamsters. J. Biol. Rhythms. 2003;18:159–169. doi: 10.1177/0748730403251802. [DOI] [PubMed] [Google Scholar]
- Kondratov R, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1878. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratova AA, Dubrovsky YV, Antoch MP, Kondratov R. Circadian clock proteins control adaptation to novel environment and memory formation. Aging. 2010;2:285–297. doi: 10.18632/aging.100142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak Y, Lundkvist GB, Brask J, Davidson M, Menaker M, Kristensson K, Block GD. Interferon-gamma alters electrical activity and clock gene expression in suprachiasmatic nucleus neurons. J. Biol. Rhythms. 2008;23:150–159. doi: 10.1177/0748730407313355. [DOI] [PubMed] [Google Scholar]
- Loh DH, Navarro J, Hagopian A, Wang LM, Deboer T, Colwell CS. Rapid changes in the light/dark cycle disrupt memory of conditioned fear in mice. PlosONE. 5:e12546. doi: 10.1371/journal.pone.0012546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons A, Murphy K, Clarke R, Lynch M. Atorvastatin prevents age-related and amyloid-beta-induced microglial activation by blocking interferon-gamma release from natural killer cells in the brain. J. Neuroinflam. 2011;8:27. doi: 10.1186/1742-2094-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WP, Cao J, Tian M, Cui MH, Han HL, Yang YX, Xu L. Exposure to chronic constant light impairs spatial memory and influences long-term depression in rats. Neurosci. Res. 2007;59:224–230. doi: 10.1016/j.neures.2007.06.1474. [DOI] [PubMed] [Google Scholar]
- Madeira MD, Sousa N, Santer RM, Paula-Barbosa MM, Gundersen HJG. Age and sex do not affect the volume, cell numbers, or cell size of the suprachiasmatic nucleus of the rat: An unbiased stereological study. J. Comp. Neurol. 1995;361:585–601. doi: 10.1002/cne.903610404. [DOI] [PubMed] [Google Scholar]
- McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, Takahashi JS. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Schroder E, Edelmann SE, Hughes ME, Kornacker K, Balke CW, Esser KA. Age-associated disruption of molecular clock expression in skeletal muscle of the spontaneously hypertensive rat. PLoS ONE. 2011;6:e27168. doi: 10.1371/journal.pone.0027168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk TH. Aging human circadian rhythms: Conventional wisdom may not always be right. J. Biol. Rhythms. 2005;20:366–374. doi: 10.1177/0748730405277378. [DOI] [PubMed] [Google Scholar]
- Oxenkrug G. Interferon-gamma - inducible inflammation: Contribution to aging and aging-associated psychiatric disorders. Aging and Disease. 2011;2:474–486. [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. A new mathematical model for relative quantification in real time RT PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen DD, Boldt BM, Wilkinson CW, Yellon SM, Matsumoto AM. Daily melatonin administration at middle age suppresses male rat visceral fat, plasma leptin, and plasma insulin to youthful levels. Endocrinol. 1999;140:1009–1012. doi: 10.1210/endo.140.2.6674. [DOI] [PubMed] [Google Scholar]
- Rowe WB, Blalock EM, Chen KC, Kadish I, Wang D, Barrett JE, Thibault O, Porter NM, Rose GM, Landfield PW. Hippocampal expression analyses reveal selective association of immediate-early, neuroenergetic, and myelinogenic pathways with cognitive impairment in aged rats. J. Neurosci. 2007;27:3098–3110. doi: 10.1523/JNEUROSCI.4163-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby NF, Hwang CE, Wessells C, Fenrandez F, Zhang P, Sapolsky R, Heller HC. Hippocampal-dependent learning requires a functional circadian system. Proc. Natl. Acad. Sci. 2008;105:15593–15598. doi: 10.1073/pnas.0808259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenland HW, Li X-Y, Zhuo M. Predicting aversive events and terminating fear in the mouse anterior cingulate cortex during trace fear conditioning. J. Neurosci. 2012;32:1082–1095. doi: 10.1523/JNEUROSCI.5566-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y. Parameter estimation in studying circadian rhythms. Biometrics. 1976;32:85–94. [PubMed] [Google Scholar]
- Tranah GJ, Blackwell T, Stone KL, Ancoli-Israel S, paudel MS, Ensrud KE, Cauley JA, Redline S, Hillier TA, Cummings SR, Yaffe K. Circadian activity rhythms and risk of incident dementia and mild cognitive impairment in older women. Ann. Neurol. 2011;70:722–732. doi: 10.1002/ana.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman A, Havekes R, Abel T, Hogenesch J. Circadian regulation of learning and memory. Soc.Research Biol.Rhythms 13th Meeting Prog.Abstr. P173. 2012 [Google Scholar]
- Verbitsky M, Yonan AL, Malleret G, Kandel ER, Gilliam TG, Pavlidis P. Altered hippocampal transcript profile accompanies an age-related spatial memory deficit in mice. Learn. Mem. 2004;11:253–260. doi: 10.1101/lm.68204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LM, Dragich JM, Kudo T, Odom IH, Welsh DK, O'Dell TJ, Colwell CS. Expression of the circadian clock gene Period2 in the hippocampus: possible implications for synaptic plasticity and learned behaviour. ASN NEURO.1, art.e00012. doi: 10.1042/AN20090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyse CA, Coogan AN. Impact of aging on diurnal expression patterns of CLOCK and BMAL1 in the mouse brain. BR. 2010;1337:21–31. doi: 10.1016/j.brainres.2010.03.113. [DOI] [PubMed] [Google Scholar]
- Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong H-K, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. P. N. A. S. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]