Abstract
In the absence of treatment, HIV-1 infection, usually starting with a single virion, leads inexorably to a catastrophic decline in the numbers of CD4+ T cells and to AIDS, characterized by numerous opportunistic infections as well as other symptoms, including dementia and wasting. In the 30 years since the AIDS pandemic came to our attention, we have learned a remarkable amount about HIV-1, the responsible virus—the molecular details about how it functions and interacts with the host cell, its evolution within the host, and the countermeasures it has evolved to overcome host defenses against viral infection. Despite these advances, we remain remarkably ignorant about how HIV-1 infection leads to disease and the death of the human host. In this brief article, we introduce and discuss important lessons that we have learned by examining the dynamics of viral populations and infected cells. These studies have revealed important features of the virus–host interaction that now form the basis of our understanding of the importance and consequence of ongoing viral replication during HIV-1 infection.
How does HIV-1 infection lead to AIDS in the human host? Insights have been gained by quantifying viral RNA and DNA in plasma and cells, perturbing viral replication with antiviral therapies, and following genome sequence changes.
TIME COURSE OF INFECTION
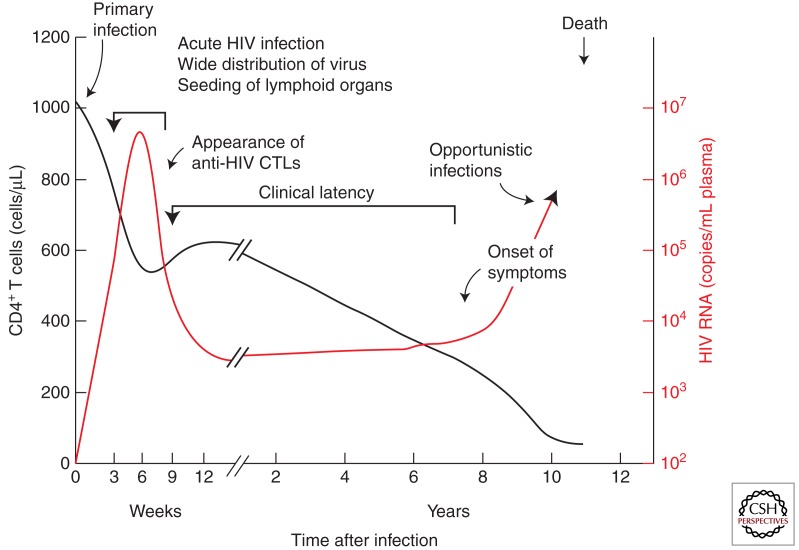
As discussed elsewhere (Shaw and Hunter 2011), HIV-1 infection is usually initiated with a single virion infecting a single target cell at the portal of entry. The subsequent course of infection can be monitored in several ways: overt symptoms, such as fever, wasting, opportunistic infections, neurological symptoms, and so on; blood levels of the CD4+ T-cell target and antiviral antibodies; and viremia (virus in blood), measured by infectivity, immunoassay for viral proteins, and, most accurately, by PCR for viral RNA. A typical time course of infection relating these properties to one another is shown in Figure 1. Although their timing can vary considerably from individual to individual, as can the levels of viremia, the general outline is essentially the same in virtually every infected person who does not receive effective antiviral therapy.
(∼1–2 wk) An eclipse phase, during which the virus is freely replicating and spreading from the initial site of infection to the many tissues and organs that provide the sites for replication. Viremia is undetectable, and neither immune response nor symptoms of infection are yet visible.
(∼2–4 wk) The acute (or primary) infection phase, characterized by relatively high levels of viremia (up to 107 or more copies of viral RNA per milliliter of blood), and large fractions of infected CD4+ T cells in blood and lymph nodes. This phase is often, but not always, accompanied by “flu-like” symptoms—fever, enlarged lymph nodes, and the like. Around the time of peak viremia, the immune response begins to appear, both in the form of antibodies against all viral proteins, and a CD8+ T-cell response against HIV-1 antigens expressed on infected cells. The high levels of viremia that characterize this phase most likely result from the absence of the early immune response and the generation, as part of the host response, of large numbers of activated CD4+ T cells, providing a wealth of targets for viral replication. At the end of the acute phase, the level of viremia declines sharply, 100-fold or more, a result of both partial control by the immune system and exhaustion of activated target cells. This phase is also characterized by a transient decline in the numbers of CD4+ T cells in the blood.
(∼1–20 yr) Chronic infection, or “clinical latency,” is characterized by a constant or slowly increasing level of viremia, usually on the order of 1–100,000 copies/mL, sometimes referred to as the “set point,” and steady, near normal (∼1000 cells/μL), or gradually falling levels of CD4+ T cells. As a rule, patients in this phase are asymptomatic and usually unaware that they have been infected. Despite the term “latency,” the viral infection is far from latent, with large numbers of CD4+ T cells becoming infected and dying every day.
Finally, the number of CD4+ T cells declines to the point (∼200 cells/μL) at which immune control of adventitious infectious agents can no longer be maintained, and opportunistic infections (discussed in Lackner et al. 2011) begin to appear. Control of the HIV-1 infection itself is also lost, and the level of viremia rises during the AIDS phase, culminating in death of the infected patient. Indeed, untreated HIV-1 infection is one of the most uniformly lethal infectious diseases known, with a mortality rate well over 95%.
Figure 1.
Time course of typical HIV infection. Patterns of CD4+ T cell decline and viremia vary greatly from one patient to another. (From Fauci and Desrosiers 1997; reprinted, with permission, from Cold Spring Harbor Laboratory Press © 1997.)
SIGNIFICANCE OF VIREMIA
As clinical researchers were developing tools for diagnosis and prognosis of HIV-1 infection, it became apparent that the most powerful of them is the measurement of viremia using quantitative nucleic acid hybridization or PCR assays for HIV-1 genomic RNA in blood. RNA measurements have the virtue of an extraordinarily wide dynamic range. Routine commercial assays can now accurately measure levels of viremia as low as 50 copies/mL and as high as 107 or more copies/mL. More sensitive research assays have reduced the lower end to much less than 1 copy/mL with good quantitative accuracy (Palmer et al. 2003).
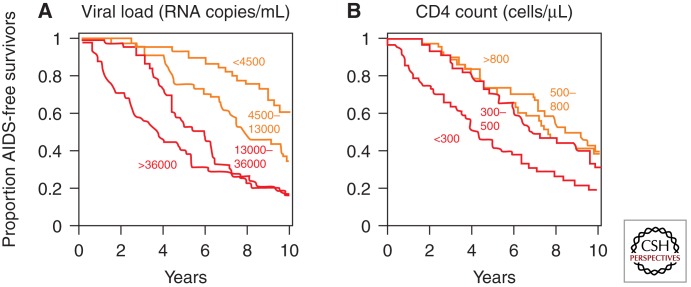
The first recognition of the prognostic importance of viremia came from analysis of a large prospective study of gay men at risk for AIDS, the Multicenter AIDS Cohort Study (MACS), in which volunteers provided blood samples and health information at frequent intervals over long periods of time, during which many men who were initially infected, but still healthy, progressed to AIDS and death (Mellors et al. 1996). When the levels of viremia at study entry were compared with outcome, a strong inverse correlation between the level of viremia and the time of progression to AIDS was observed (Fig. 2). The data showed that the group with the highest level of viremia progressed to AIDS on average about fourfold faster than the lowest group. Thus, the level of virus present in the blood provides a measure of the rate at which viral infection damages the immune system of the infected host.
Figure 2.
Relationship between viral load (viremia) and clinical progression. Shown are Kaplan–Meier plots of AIDS-free survival divided into quartiles according to virus load (A) or CD4 count (B) at the time of diagnosis. (From Fauci and Desrosiers 1997; reprinted, with permission, from Cold Spring Harbor Laboratory Press © 1997.)
The second important insight made possible by the ability to accurately measure levels of viremia was in analyzing the consequences of antiviral therapy, where it was found that the ability of an antiretroviral drug to halt or even reverse progression of an infected patient to AIDS was well correlated with its ability to suppress viremia. Effective therapies, such as combinations of antiviral drugs discussed by Arts and Hazuda (2011), which can lead to sustained reductions in viremia to undetectable levels of <50 copies/mL by standard clinical assays, can forestall progression to AIDS indefinitely, although they do not cure the infection. Indeed, if therapy is suspended, even after many years, the level of viremia will rapidly return to the prior set point.
The correlation between viremia and disease has a straightforward underlying explanation (Coffin 1995). Infected cells produce virions, some fraction of which are released into the blood, from which they are rapidly removed or degraded. Although different cells may produce and release different amounts of virus into the blood, their numbers are very large, and, on average, the amount of virus per infected cell will be about the same from time to time in the same patient and similar from one patient to another. Also, the rate of removal of virions from the blood will be similar (and, as has been measured, quite rapid) from time to time and patient to patient. Because the level of viremia is determined by the balance between virion release into and removal from the bloodstream, assuming a constant rate of release per cell and a constant rate of removal, the level of viremia will be an instantaneous relative measure of the number of productively infected cells at any one time.
INSIGHTS FROM THERAPEUTIC DRUG STUDIES
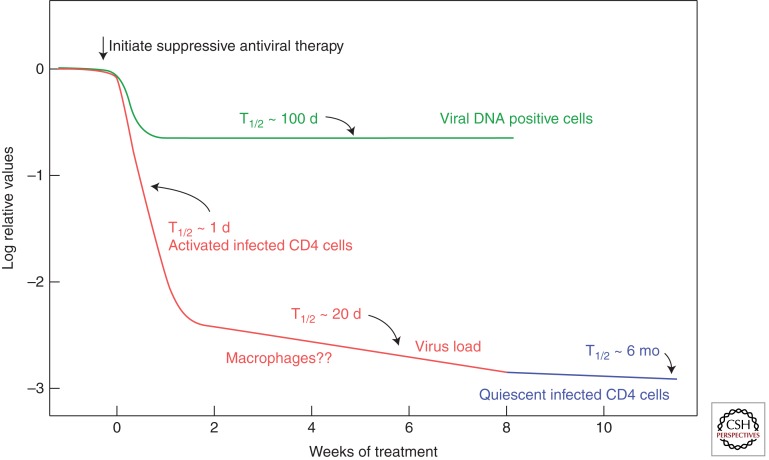
This understanding of the significance of viremia in HIV-1 infection, combined with the new availability of potent antiretroviral drugs (Arts and Hazuda 2011), led to an experiment that provided considerable insight into the process of HIV-1 infection and pathogenesis in vivo (Fig. 3) (Coffin 1995; Ho et al. 1995; Wei et al. 1995). The study was based on the principle that all antiviral drugs affect the ability of the virus to infect cells and do not affect the lifetime of infected cells or their ability to produce viral particles (albeit noninfectious ones in the case of protease inhibitors). Therefore, any decrease in viremia following initiation of suppressive therapy must reflect a combination of the length of time that productively infected cells live and produce virus after infection and the lifetime of the virus in the blood. Because the latter factor is very small, it can be ignored with little effect on the overall result. Thus, viremia was assayed by quantitative PCR at closely spaced time points after initiation of suppressive therapy, with striking results: After a brief lag, viremia drops rapidly, to ∼1%–10% of the set point level with a half-time of ∼1–2 d. Following this first phase of decay, there is a second phase in which most of the remaining viremia decays with a half-time of ∼2 wk or so, down to the detection limit of standard assays. Two more phases, which have been defined with more sensitive assays, are discussed below.
Figure 3.
Decay of circulating virus and infected cells after initiation of suppressive antiviral therapy. These results imply the presence of at least four classes of HIV-infected cells: productively infected, a second class, perhaps macrophages, with a half-life of ∼2 wk; latently infected resting CD4 cells, with a half-life of ∼6 mo; and long-lived DNA positive cells, most of which are nonproductive. A fourth class of infected cells, inferred to have an approximately infinite half-life, is not shown. (Figure modified from Fauci and Desrosiers 1997.)
The first phase of decay almost certainly reflects the lifetime of productively infected CD4+ T cells, although the exact cells and their location have not been identified. The second phase has been speculatively attributed to macrophages (Perelson et al. 1997), but this point is far from established. Given that this phase is much less prominent when the treatment includes an integrase inhibitor, it may reflect at least in part a population of infected cells in which integration is delayed for some reason (Murray et al. 2007).
THE HIV-1 STEADY STATE
The results discussed above, combined with the fact that the characteristic level of viremia (set point) in any infected individual is essentially constant, led to the conclusion that HIV-1 exists in the infected host as a quasi-steady state, in which the level of viremia is largely determined by the number of virus-producing cells, which therefore must also be constant day in and day out throughout most of the course of infection. In any given patient, therefore, there are about the same number of infected cells today as there were yesterday and last year, but they must be different cells arising from infection of fresh target cells, which then rapidly die after producing sufficient virus, on average, to infect the same number of cells again. This concept is key to our current understanding of the HIV–host interaction. It has many important implications.
The replication cycle time of the virus is, on average, ∼1–2 d. Thus, virus in an individual at a year after infection is 200–300 generations removed from the initial infecting virion.
The population size is determined by the steady-state number of productively infected cells, not by the number of virions, except that the latter must be at least large enough to ensure that the same number of cells is infected every day.
The steady state is remarkably robust, changing little throughout the clinically latent phase, and returning to the same value after even prolonged periods (7 yr or more) when it is undetectable on suppressive therapy, or after transient increases due to immune stimulation by vaccination or by infection with some other agent.
The virus exists as a replicating population that is carried forward by repeated infection of large numbers of cells with no evidence of bottlenecks or other major alterations in population size from time to time.
The level of viremia is determined by a combination of the number of available target cells, the infectivity of the virus, the number of virions produced per cell, the productive lifetime of the infected cell, and the clearance rate of the virus.
We know little about the infected cells that are responsible for producing the virus observed in the blood and can only infer their properties indirectly.
The extraordinary extent of genetic diversity that accumulates during HIV-1 infection, relative to other RNA viruses, is not due to its unusually high error rate, but rather to replication of a large population of virus with an average error rate repeated over and over again, accumulating thousands of generations over the course of infection of a single individual.
With these properties in mind, we can look at some specific issues.
Nature and Lifetimes of Infected Cells
The initial therapeutic studies that led to the steady-state concept implied two populations of target cells: one with a half-life of ∼1–2 d and another of ∼2 wk. These conclusions generated considerable optimism because the proportionality of the level of viremia to the number of productively infected cells suggested that sufficiently prolonged treatment could eventually reduce the number of infected cells to <1, effectively curing the patient of the HIV-1 infection. Unfortunately, these hopes were dashed when further studies, using more sensitive techniques, revealed the presence of at least two more phases of decay of HIV-1 viremia on fully suppressive therapy, apparently due to small populations of infected cells capable of surviving and releasing virus many years after infection (Siliciano and Green 2011). Although such cells constitute only a tiny fraction of the total number of infected cells present before therapy, they decay very slowly (with a half-life of ∼6 mo by one estimate) (Zhang et al. 1999) after treatment. Also, much more sensitive PCR assays, with a limit of detection well below 1 copy/mL of viral RNA in plasma (Palmer et al. 2003), revealed that the majority patients on therapy with “undetectable” viremia for more that 1 yr in fact had an average of about 3 copies of HIV-1 RNA per milliliter, or about 1/10,000 of the level before therapy, and that this level declined further with a curve consistent with two more phases of decay: one with a half-life of ∼39 wk, not significantly different from that of inducible cells, perhaps from the same source, and another with a half-life not different from infinity, suggesting the possibility of a dividing reservoir. Viremia was still detectable in all patients as long as 7 yr after the start of therapy (Palmer et al. 2008). The existence of these populations of cells, apparently infected before therapy and capable of producing virus many years later, rules out eradicating infection by antiviral therapy alone.
One more population of infected cells bears discussion. In a typical chronically infected individual, about one CD4+ T cell per 100 to one per 1000 contains HIV-1 DNA, largely in the form of an integrated provirus. Very few cells contain more than one provirus (Josefsson et al. 2011). After the initiation of therapy, there is an initial decline in the numbers of these cells, reflecting the decay in viremia and implying that at least some of the virus in the blood comes from populations of cells also found in the blood. However, as can be seen in Figure 3, the level of viral DNA positive cells soon levels off, typically somewhere around 1%–10% of the starting value, implying that a significant fraction of these cells is not involved in phase 1 or 2 viral production. Furthermore, the numbers of DNA positive cells after prolonged therapy is typically 100-fold greater than the number of cells inducible to produce virus ex vivo (Chun et al. 1997), implying that most of them are incapable of ever producing infectious virus, perhaps because of mutations in the provirus, unfavorable sites of integration, or some undefined cellular factor that prevents expression of the silent provirus. Simple modeling implies that over time, because of their much longer lifetime, cells containing inactive or latent proviruses should accumulate to a considerable extent relative to productively infected cells, most of which die within a few days. Thus, only a small fraction of the population of cells containing detectable proviruses represent cells that are latently infected, that is, that contain a provirus that is not expressed but is capable of being induced to express infectious virus by some means, such as stimulation to reenter the cell cycle. For this reason, it cannot be assumed (although it often is) that the nature of cells in the so-called latent reservoir presumed responsible for the persistence of HIV-1 during years of therapy can be inferred from the properties of all provirus-containing cells (such as sites of integration, expression of transcription factors, etc.), the large majority of which must contain dead proviruses incapable of reincarnation. The physical identification and manipulation of the tiny fraction of latently infected cells in suppressed infection is a major unsolved problem in HIV-1 pathogenesis (Maldarelli 2011; Siliciano and Green 2011).
Persistence of HIV-1 Infection on Therapy
Although it is well accepted that the large majority of patients on suppressive therapy have low levels of viremia as well as latently infected cells capable of being reactivated to produce HIV-1 ex vivo, the link between the two phenomena remains somewhat controversial, with some investigators arguing that the persistent viremia observed in patients on long-term therapy could also be due to a small amount of “escape” replication of virus in anatomic sites or cells that are poorly accessible to antiviral therapy (referred to as “sanctuaries”) (Sharkey and Stevenson 2001). In support of this claim, they point to the ongoing presence of two-LTR circles (Craigie and Bushman 2011) in PBMCs in a fraction of patients (Buzon et al. 2011), an observation interpreted as meaning that productive infection of new cells is continuing to occur. However, the underlying assumption that the circular forms of viral DNA are inherently unstable in cells in vivo remains unsupported, and there are several lines of evidence in favor of the idea that the low-level viremia observed in the majority of patients on long-term suppressive therapy is derived from cells infected before the start of treatment.
The distribution of the levels of viremia among treated patients is independent of the drugs used for treatment (Maldarelli et al. 2007). This result is not what would be expected for replication in a sanctuary because it is improbable that such sites or cells would be equally unavailable to different drugs.
The level of viremia is not affected by the inclusion of another drug. Several studies have been performed in which a suppressive three-drug regimen was intensified by the addition of a fourth inhibitor of a class previously unseen by the patient (Dinoso et al. 2009; Gandhi et al. 2010; McMahon et al. 2010). Despite some differences in design and inhibitors used, all studies gave the same result: There was no difference in the amount of virus during and after the intensification period as compared with before. Again, it is highly improbable that sanctuaries of replication would be equally inaccessible to all drugs, and these results are inconsistent with low-level ongoing replication.
As is discussed in more detail in the next section, accumulation of genetic variation due to the rapid turnover of infected cells is a hallmark of HIV-1 infection. In the chronic infection phase, there is a slow but perceptible turnover of the viral population such that genetically distinct populations arise every few years (F Maldarelli, M Kearney, S Palmer, et al., unpubl.). In contrast, in patients on maximally suppressive therapy, evolution of viral genomes is frozen at the point at which therapy began (Ruff et al. 2002; Tobin et al. 2005), and no evidence for continuing evolution can be seen even after many years. In patients for which therapy is partially suppressive, ongoing evolution can still be observed (Tobin et al. 2005), as is also true in patients known as “elite controllers,” who naturally control HIV-1 infection at levels similar to those seen in patients on suppressive antiviral therapy (Mens et al. 2010). The apparent lack of evolution on therapy is consistent with the absence of new cycles of infection.
As noted, the appearance of 2-LTR circles in a minority of patients undergoing intensification with an integrase inhibitor raises the possibility of a low level of infection occurring in the face of antiviral drugs. However, this result remains to be fully understood or reproduced.
It has recently been suggested, on the basis of mathematical and cell culture modeling, that the high multiplicity of infection that would result from cell–cell spread could greatly reduce the effectiveness of antiviral therapy and allow persistent viral replication (Sigal et al. 2011). However, this model predicts that cells infected in vivo should have multiple proviruses and that genetic variation should continue to accrue on suppressive therapy, neither of which is observed.
Regarding point 2, in the small amount of virus in the blood of some patients whose viremia has been suppressed for more than 3 yr or so, significant subpopulations of virus with genomes identical in sequence can be observed and, in some cases, can become a large fraction of the total population (Bailey et al. 2006). These subpopulations have been called predominant plasma clones (PPCs). Their origin is unknown, but it can be speculated that they may arise from a very small fraction of infected cells that is capable of dividing and releasing virus but that do not die as a result of the expansion. Such clonal populations of virus can also be seen in the blood of patients who have stopped suppressive therapy after long-term suppression (Joos et al. 2008), although the two phenomena have not yet been firmly connected. It seems likely that the last, apparently stable, phase of viremia on therapy reflects clonal viral populations, but this point remains to be established.
It is important to keep in mind that the infected cell populations that give rise to long-lived, persistent viremia on therapy are not created by the therapy; rather, they must exist throughout the course of infection, only to be exposed as the shorter-lived cells of phases 1 and 2 die off and are not replenished and the virus produced by them disappears. The cells responsible for the production of the persistent virus must represent an extremely small fraction of the total population of productively infected cells in chronic infection—somewhere between 10−4 and 10−5 is a reasonable estimate. Given that HIV-1 infection is carried forward, day in and day out, by repeated cycles of infection, viral production, and death of target cells, these long-lived cell populations must be irrelevant to the natural history of HIV-1 infection (as well as SIV infection of the natural hosts). They are of critical importance to attempts to treat HIV-1 infection, however, because the virus they produce is capable of rekindling full-blown infection from the faintest embers remaining after prolonged completely suppressive therapy.
GENETIC VARIATION AND EVOLUTION
One of the first unusual features of HIV-1 infection to become apparent was the remarkable accumulation of genetic diversity during the course of infection of a single individual. Indeed, when first observed, it represented the fastest rate of evolution in any natural eukaryotic system; for example, the extent of HIV-1 diversity that accumulates in one infected individual exceeds that of all influenza virus isolates worldwide in any given year (Korber et al. 2001). (Actually, the record diversity no longer belongs to HIV, having since been eclipsed by hepatitis C virus.) Although it was first thought that the remarkable diversification reflected an extraordinarily high underlying error rate associated with HIV-1 replication, we now know that this rate is about average for an RNA virus and that it is the pace of replication, the duration of infection, and the size of the replicating population, allowing it to evolve rapidly in response to selective influences, that set HIV-1 (and HCV) apart from all other known viral infections. These features, in addition to the ease of generating large numbers of sequences either directly from infected individuals and populations of individuals with well-timed infections or from massive databases of such sequences (Kuiken et al. 2003), have made HIV-1 a favorite topic of study for evolutionary biologists, and they have used it effectively to develop and test new mathematical models for evolution in general, not just for HIV-1 or just for viruses. We do not present any specific models here, but the next section discusses the role of specific factors of evolution that all models must include and how they apply to HIV-1 in an infected human host.
Factors of HIV-1 Evolution
By “evolution,” we mean the process by which genetic change accumulates during generations of replication. For any replicating entity, organisms or viruses, the general principles of evolution are similar, although their application can differ greatly. As a rule, models are based on consideration of four principal factors. We discuss them here in the context of HIV.
Mutation
In the case of HIV-1 and other retroviruses, mutations are primarily attributed to error-prone reverse transcription, but there are actually two other possible sources of error: transcription by host RNA polymerase II, and G-to-A hypermutation mediated by ABOBEC3G (or F). As discussed in other articles in this collection (Hu and Hughes 2011; Karn and Stoltzfus 2011; Malim and Bieniasz 2011), the relative contribution of these mechanisms is a matter of some debate. Whatever the source, the overall single-step point mutation rate for HIV-1 is ∼3 × 10−5 mutations per base per replication cycle, and about 10-fold less for transversions than transitions (Mansky and Temin 1995). Thus, about one genome in three contains a mutation after a single round of replication. Other types of mutation, including frameshifts (insertion or deletion of small numbers of bases), as well as large-scale deletions, insertions, duplications, and inversions, are also common (Svarovskaia et al. 2003). The error rate of retrovirus replication, while much greater than that of cellular and viral DNA polymerases, is about average for RNA viruses.
Selection results from the ability of one variant to replicate more effectively than another, a property referred to as “fitness.” In the case of viruses, fitness differences lead to differences in the number of infectious progeny at each replication cycle. Fitness differences can result from differences at any stage of the series of events leading from one infected cell to the next, including efficiency of entry, rate of infectious progeny production by an infected cell, and its productive lifetime, stability of the infectious virion, and so on. Note that there is no such thing as absolute fitness: The concept is meaningful only in relative terms and only in a specific context (e.g., in vitro vs. in vivo, in the presence or absence of neutralizing antibodies or antiviral drugs). It is often expressed as a selection coefficient, the difference in the relative number of progeny produced by one variant compared with another per generation. Thus, for a variant that yields 1% more infectious progeny than another, the selection coefficient is 0.01. For large populations undergoing multiple repeated cycles of infection, even small selection coefficients can have major effects on the frequency of slightly advantageous or disadvantageous mutations, with the former leading to positive selection and the latter to purifying selection. Because synonymous mutations (those that do not change the corresponding amino acid) generally have much smaller selection coefficients than nonsynonymous ones, the ratio between the two in a population (dN/dS) is a useful indicator of the nature of selective forces acting at any given site or set of sites.
Drift
“Drift” is a term for the change in frequency of a variant independent of selection or mutation, resulting from sampling error that arises when replicating population sizes are small. In the case of viral populations, it is important to bear in mind that the population size is determined by the number of cells infected at each round of replication, not by the number of virions produced by them. In the case of HIV-1 infection, perhaps 1011 virions are produced daily; the number of cells infected in the same time span is still a matter of some debate, but is unlikely to exceed 109. A further important consideration is the effective population size: in simple terms, the number of infected cells multiplied by the probability that any one cell gives rise to progeny that go on to infect other target cells. If there is an infinite supply of target cells and the population of infected and target cells is well mixed, then the effective population size can be similar to the census population size (defined as the total number of infected cells). These conditions obviously do not apply in infected individuals, and the effective population size is probably much smaller. At very large effective population sizes, evolution can be considered to be deterministic, in that the frequency of a mutation at any given time (in generations) in the future can be exactly determined from its present frequency, mutation rate, and selection coefficient, as long as these factors remain constant. At smaller sizes, evolution becomes more and more stochastic, that is, subject to random factors, so that the frequency can only be predicted as an average, with the variance increasing with decreasing size. A useful relationship to keep in mind is that, after many generations of deterministic evolution, slightly deleterious mutations will reach an equilibrium frequency equal to the mutation rate divided by their selection coefficient. In the stochastic case, this calculation will give the average value over many observations but will vary considerably from one individual to the next. This consideration is of more than academic interest because it defines the probability that drug-resistance mutations, which are slightly deleterious in the absence of the drug, will already be present in the replicating viral population before initiation of antiviral therapy (the so-called genetic barrier) (see Arts and Hazuda 2011). Although transition from deterministic to stochastic evolution is continuous with decreasing population size, a useful approximate crossover point to keep in mind is a size equal to the inverse of the mutation rate, at which it becomes probable that all possible single mutations will arise at each round of replication (Rouzine and Coffin 2005).
Linkage
To complicate matters further, selection does not act on individual mutations, but rather on whole genomes, which may contain other mutations as well. In very large deterministic populations, where any mutation is always present in many genomes, this effect is unimportant; in smaller populations, it can prevent selection from operating on individual mutations, and even, in extreme cases (and at least in theory) lead to declining fitness of a population due to gradual accumulation of deleterious mutations (Muller’s ratchet). Linkage can thus be visualized as increasing the role of stochastic effects on evolution, even at large population sizes. The effect of linkage on evolution can be reduced by recombination to allow mutations to be redistributed among genomes. It is important to keep in mind that recombination does not generate diversity, but rather redistributes it among replicating genomes, allowing effects of selection to be felt more strongly. As discussed elsewhere (Hu and Hughes 2011), recombination between genetically distinct genomes in retrovirus replication occurs at very high rates, but only during reverse transcription of genomes from heterozygous progeny of cells containing two or more proviruses. Although it was originally reported that such multiply infected cells are very frequent in HIV-infected individuals (Jung et al. 2002), more recent results suggest that they are much rarer, constituting perhaps 10% of infected CD4+ T cells in blood (Josefsson et al. 2011). Nevertheless, there is ample evidence for the occurrence and importance of recombination in HIV-1 infection, particularly in rare individuals coinfected with two or more genetically distinct viruses (Keele et al. 2008; Kearney et al. 2009).
Course of Evolution following Infection
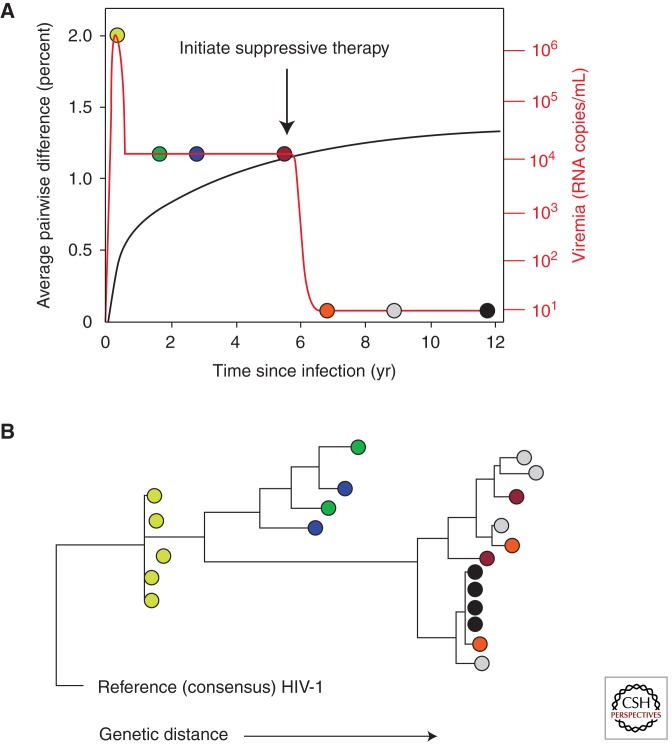
Monitoring of genetic diversity of the virus following HIV-1 infection has provided a very important tool for understanding many important aspects of the virus–host interaction. For reasons discussed above, the sequences of viral genomes in the blood provide the most accurate instantaneous indicator for the genetics of the replicating viral population, particularly when single-genome sequencing (SGS) technology, which avoids many PCR artifacts (Palmer et al. 2005), is used. Figure 4 shows, in cartoon form, the important events in virus evolution during the course of a “typical” infection, starting with a clonal population and proceeding toward the highly diverse populations characteristic of chronic infection. The initial accumulation of diversity is at a rate very close to the mutation rate, with little evidence for selection, except for highly deleterious mutations; as time goes on, purifying selection takes hold, and the increase in diversity begins to level off, approaching an asymptotic value of an average pairwise distance ∼1%–2% for gag, pro, and pol, which are largely subject to purifying selection, as indicated by low dN/dS ratios. Rates are much higher for env, where the greater, and constantly increasing, diversity and higher dN/dS ratios point to constant positive selection resulting from an ever-changing antibody response (Richman et al. 2003). In the case of gag, pro, and pol (as well as accessory genes, particularly nef), even though dN/dS ratios are low, positive selection can be discerned at sites corresponding to T-cell epitopes. Indeed, virtually all of the nonsynonymous diversity accumulating in these genes in untreated infection can be attributed to escape from the cytotoxic T-cell (CTL) response (Kearney et al. 2009; Walker and McMichael 2011). In env, escape from the antibody response is characterized not only by point mutation, particularly affecting the display of glycosylation sites, but also by gross structural changes—insertions and duplications—in the hypervariable loops V1, V2, and V4, which tend to increase in length and numbers of glycosylation sites (the “glycan shield”) during the course of infection. It is important to bear in mind that, given the conditions under which HIV populations are evolving, even weak selective forces (on the order of a few percent) can have dramatic effects on the genetics of the viral population. Thus, it cannot be inferred from these results alone that the immune response plays a significant role in controlling HIV infection. The partial control of viral replication by the host is likely the sum of effects of the immune response and the fitness cost incurred with the initial escape by the virus, a fitness cost that is in part alleviated over time through compensatory mutations.
Figure 4.
Typical evolution of HIV after infection. (A) The level of viremia (orange) and the genetic diversity, measured as the average pairwise difference (black) of the HIV population as a function of time after infection. (B) Phylogenetic relationship of HIV-1 sequences at various stages of infection. The colored circles correspond to sequences of plasma virus RNA sampled at the corresponding time points in A: At peak viremia (yellow), the viral population (in most patients) is highly uniform, diversifying gradually over the first few years of infection. Samples taken from chronic infection (green and blue) are considerably more diverse, but take 3 yr or more to diverge perceptibly (red). Following initiation of suppressive therapy, divergence ceases, even after long times (orange, gray). After years of therapy, clonal populations of virus arise in some patients (black).
A remarkable aspect of HIV genetic diversity during chronic infection is its stability. Except in the case of escape from antiretroviral therapy, genetic turnover of the viral population in the blood is very slow, with no evidence of a genetic or physical bottleneck (Shankarappa et al. 1999; F Maldarelli, M Kearney, and S Palmer, unpubl.). This observation is consistent with a large replicating population that can be steered by immune selection, but without ever having to recover from significant depletion. Because mutations leading to immune escape will often reduce fitness, when that immune response is not present, such as after transmission to another person, these mutations will tend to revert to the consensus sequence, although at highly variable rates. For example, mutations increasing the length of the variable loops tend to reset to shorter length following transmission (Shaw and Hunter 2011).
In addition to immune escape, resistance to antiretroviral therapy is an important consequence of genetic diversity accumulating during chronic HIV-1 infection. Both theory (discussed above) and observation are consistent with the idea that drug-resistance mutations that accumulate to low levels before therapy are an important cause of failure. Indeed, in virtually all cases, treatment of HIV-1 infection with a single agent (monotherapy) leads to rapid reappearance of viremia, rebounding to levels near the pretherapy set point. A striking example is in women treated with a single dose of the nonnucleoside RT inhibitor (NNRTI) nevirapine to prevent mother-to-child transmission (Chersich and Gray 2005). A single pill taken during labor prevents a significant fraction of infections of the newborn but leads to a very high frequency of drug-resistant virus in the mother a few weeks later. The reproducibility of this dramatic effect implies a low (and so far undetectable) frequency of NNRTI-resistant mutants in the replicating viral population combined with a relatively long lifetime of the drug, allowing their selection during the ensuing cycles of HIV-1 replication.
Unlike the immune response, antiviral therapy can impose a significant physical and genetic bottleneck on the HIV-1 population, with drug-resistant virus that rebounds after monotherapy often showing substantially less diversity than the starting population (Kitrinos et al. 2005).
Another important consequence of HIV genetic variation is the appearance of populations of virus infecting specific anatomical compartments and cell types. As discussed by Swanstrom and Coffin (2011), HIV can be found in a large number of organs and tissues. In some sites, such as the central nervous system (CNS), there is a clear genetic separation of independently evolving populations, often leading to macrophage-tropic viruses that likely replicate in either macrophages or microglial cells in the brain. Macrophage-tropic viruses can also be found late in infection at other anatomical sites, but their pathophysiological significance is much less clear. The final important consequence of HIV evolution in vivo is the appearance of virus capable of using alternative coreceptors, such as CXCR4, distinct from the CCR5 chemokine receptor used by the founder virus.
HOW DOES HIV-1 CAUSE AIDS?
As is apparent from this article and the rest of the collection, in the 25+ years since its discovery, we have learned an enormous amount about HIV, but we still cannot answer the one big question: How does HIV-1 cause AIDS? At first thought, the answer is obvious. HIV-1 infects and kills CD4+ T cells. AIDS is a result of loss of CD4+ T cells. QED. Unfortunately, it is not that simple.
For starters, although HIV-1 infection kills infected cells ex vivo, by a mechanism that is still controversial, there is still considerable debate about how infected cells die in vivo. There are three alternatives: virus-mediated killing of infected cells, as in culture systems; immune-mediated effects due to HIV-specific CD8+ T cells or antibodies; or one or more of a variety of indirect effects. In support of the role of such direct killing is the short half-life of infected cells early in infection, before the appearance of antibodies or CTL, ruling out the adaptive immune response as essential for cell killing in vivo. The innate (i.e., interferon) response could still play some role at this point, however. Although the antibody response appears to have little effect on the life of the infected cell, there is evidence for an important role of the CTL response in cell killing because treatment of SIV-infected macaques with antibody directed against CD8 leads to a significant (100-fold) but transient increase in the level of viremia (Jin et al. 1999; Schmitz et al. 1999). The most straightforward explanation for this result is that CTLs shorten the productive lifetime of the infected cell, although more indirect effects have also been proposed (Davenport and Petravic 2010; Klatt et al. 2010; Wong et al. 2010). Two features of this hypothesis are worth noting. First, the productive lifetime of an infected cell is the time from the onset of virus production to cell death by CTL and is likely to be not much shorter than the lifetime defined by virus-mediated killing, perhaps only an hour or so. Second, it is likely that part of the advantage conferred on HIV-1 (and other complex viruses) by the two-step replication strategy mediated by tat and rev is the delay of the synthesis of virion proteins as long as possible in the life cycle to minimize exposure to the CTL response and maximize virion output per cell. Although cells can die from the effects of infection alone, their productive lifetime is somewhat shortened by the CTL response, that is, specifically during the time when virus is being produced. Thus, CTLs can modulate infection, but they do not eliminate it. Effects due to immune activation could include not only degradation of immune function late in infection, but also stimulation of production of activated memory CD4+ T cells, increasing the number of targets available to the virus.
Even if we knew the mechanism of HIV-mediated cell killing, we would not know how HIV-1 causes CD4+ T-cell decline and AIDS in humans. The observation that virus and cell turnover rates in various SIVs in their natural hosts (such as SIVsm in sooty mangabeys), which do not progress to AIDS, are essentially identical to those in humans, who do progress, implies that cell killing alone cannot account for AIDS pathogenesis. Indeed, this result is consistent with the high natural turnover rate of activated effector memory helper T cells, the primary target for HIV-1 infection, on the order of 1010 cells per day, of which only a small fraction are infected after the initial primary infection phase. Clearly, in the natural host, these viruses have evolved to replicate in a population of cells that is replenished constantly and in excess over what is needed for good health. In unnatural hosts, such as HIV-1 infection of humans, or SIVsm infection of macaques, something else must be happening to lead to AIDS. Two features that distinguish the pathogenic from nonpathogenic infection are chronic immune activation, discussed above, and greater levels of infection of other CD4+ T-cell subsets, particularly central memory helper T cells. The former may lead eventually to immune exhaustion and damaged lymphoid architecture, the latter (which might, itself, be a result of immune activation) to loss of critical cells necessary for maintenance of a diverse response to common environmental pathogens. Sorting out the roles of these and other side effects of HIV-1 infection in the causation of AIDS will provide subject matter for HIV-1 researchers for a long time to come.
SUMMARY AND PERSPECTIVE
Quantitation of viral RNA in plasma, of viral DNA in cells, and of cells capable of being stimulated to express quiescent proviruses has led to a conceptual framework of the dynamics of HIV-1 replication and persistence in the host. The availability of suppressive antiviral therapy has brought disease progression under control, and it has also provided a means to perturb the dynamics of viral replication that has revealed additional features of viral persistence, features that show the necessity for lifelong therapy given the current treatment strategies. The dynamic changes in the sequence of the viral genome provide another critical source of information about viral replication and the selective pressures at work.
Given the ongoing generation of HIV-1 sequence information and the dynamic nature of viral populations, this virus will continue to provide the basis for studies of viral evolution and evolution in general. Deep sequencing strategies will reveal features of the viral population that have previously been hidden. There will be continuing efforts to define selective pressures in the context of sequence changes, thus providing a link between the selective pressure and the affected viral protein. Viral dynamics and evolution will continue to be central tools as our understanding of HIV-1 deepens. The lessons learned to date provide the foundation for continuing studies of virus–host interactions and for efforts to develop strategies to eradicate the virus completely.
Footnotes
Editors: Frederic D. Bushman, Gary J. Nabel, and Ronald Swanstrom
Additional Perspectives on HIV available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- *.Arts EJ, Hazuda DJ 2011. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, Haggerty CM, Kamireddi AR, Liu Y, Lee J, et al. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol 80: 6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon MJ, Massanella M, Llibre JM, Esteve A, Dahl V, Puertas MC, Gatell JM, Domingo P, Paredes R, Sharkey M, et al. 2011. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat Med 16: 460–465 [DOI] [PubMed] [Google Scholar]
- Chersich MF, Gray GE 2005. Progress and emerging challenges in preventing mother-to-child transmission. Curr Infect Dis Rep 7: 393–400 [DOI] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, et al. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387: 183–188 [DOI] [PubMed] [Google Scholar]
- Coffin JM 1995. HIV population dynamics in vivo: Implications for genetic variation, pathogenesis, and therapy. Science 267: 483–488 [DOI] [PubMed] [Google Scholar]
- *.Craigie R, Bushman FD 2011. HIV DNA integration. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport MP, Petravic J 2010. CD8+ T cell control of HIV—A known unknown. PLoS Pathog 6: e1000728 10.1371/journal.ppat.1000728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange SJ, Cranmer L, O’Shea A, Callender M, Spivak A, Brennan T, et al. 2009. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci 106: 9403–9408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauci AS, Desrosiers RC 1997. Pathogenesis of HIV and SIV. In Retroviruses (ed. Coffin JM, et al. ), pp. 587–635 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- Gandhi RT, Bosch RJ, Aga E, Albrecht M, Demeter LM, Dykes C, Bastow B, Para M, Lai J, Siliciano RF, et al. 2010. No evidence for decay of the latent reservoir in HIV-1-infected patients receiving intensive enfuvirtide-containing antiretroviral therapy. J Infect Dis 201: 293–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho DD, Neumann AU, Perelson AS, Chen W, Leonard JM, Markowitz M 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV infection. Nature 373: 123–126 [DOI] [PubMed] [Google Scholar]
- *.Hu WS, Hughes SH 2011. HIV-1 reverse transcription. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, et al. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med 189: 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Boni J, Hirschel B, Weber R, Trkola A, Gunthard HF 2008. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci 105: 16725–16730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefsson L, King MS, Makitalo B, Brannstrom J, Shao W, Maldarelli F, Kearney MF, Hu WS, Chen J, Gaines H, et al. 2011. Majority of CD4+ T cells from peripheral blood of HIV-1-infected individuals contain only one HIV DNA molecule. Proc Natl Acad Sci 108: 11199–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung A, Maier R, Vartanian JP, Bocharov G, Jung V, Fischer U, Meese E, Wain-Hobson S, Meyerhans A 2002. Recombination: Multiply infected spleen cells in HIV patients. Nature 418: 144. [DOI] [PubMed] [Google Scholar]
- *.Karn J, Stoltzfus CM 2011. Transcriptional and post-transcriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney M, Maldarelli F, Shao W, Margolick JB, Daar ES, Mellors JW, Rao V, Coffin JM, Palmer S 2009. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol 83: 2715–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci 105: 7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitrinos KM, Nelson JA, Resch W, Swanstrom R 2005. Effect of a protease inhibitor-induced genetic bottleneck on human immunodeficiency virus type 1 env gene populations. J Virol 79: 10627–10637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea U, Estes JD, et al. 2010. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog 6: e1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br Med Bull 58: 19–42 [DOI] [PubMed] [Google Scholar]
- Kuiken C, Korber B, Shafer RW 2003. HIV sequence databases. AIDS Rev 5: 52–61 [PMC free article] [PubMed] [Google Scholar]
- *.Lackner AA, Lederman MM, Rodriguez B 2011. HIV pathogenesis—The host. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F 2011. Targeting viral reservoirs: Ability of antiretroviral therapy to stop viral replication. Curr Opin HIV AIDS 6: 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, Kovacs JA, Davey RT, Rock-Kress D, Dewar R, et al. 2007. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog 3: e46 10.1371/journal.ppat.0030046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Malim MH, Bieniasz PD 2011. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansky LM, Temin HM 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol 69: 5087–5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon D, Jones J, Wiegand A, Gange SJ, Kearney M, Palmer S, McNulty S, Metcalf JA, Acosta E, Rehm C, et al. 2010. Short-course raltegravir intensification does not reduce persistent low-level viremia in patients with HIV-1 suppression during receipt of combination antiretroviral therapy. Clin Infect Dis 50: 912–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellors JW, Rinaldo CRJ, Gupta P, White RM, Todd JA, Kingsley LA 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272: 1167–1170 [DOI] [PubMed] [Google Scholar]
- Mens H, Kearney M, Wiegand A, Shao W, Schonning K, Gerstoft J, Obel N, Maldarelli F, Mellors JW, Benfield T, et al. 2010. HIV-1 continues to replicate and evolve in patients with natural control of HIV infection. J Virol 84: 12971–12981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JM, Emery S, Kelleher AD, Law M, Chen J, Hazuda DJ, Nguyen BY, Teppler H, Cooper DA 2007. Antiretroviral therapy with the integrase inhibitor raltegravir alters decay kinetics of HIV, significantly reducing the second phase. AIDS 21: 2315–2321 [DOI] [PubMed] [Google Scholar]
- Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, et al. 2003. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 41: 4531–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT Jr, Dewar RL, et al. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol 43: 406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, Kempf DJ, Mellors JW, Coffin JM, King MS 2008. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci 105: 3879–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho DD 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387: 188–191 [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci 100: 4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzine IM, Coffin JM 2005. Evolution of human immunodeficiency virus under selection and weak recombination. Genetics 170: 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff CT, Ray SC, Kwon P, Zinn R, Pendleton A, Hutton N, Ashworth R, Gange S, Quinn TC, Siliciano RF, et al. 2002. Persistence of wild-type virus and lack of temporal structure in the latent reservoir for human immunodeficiency virus type 1 in pediatric patients with extensive antiretroviral exposure. J Virol 76: 9481–9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283: 857–860 [DOI] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, Gupta P, Rinaldo CR, Learn GH, He X, et al. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol 7312: 10489–10502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey ME, Stevenson M 2001. Two long terminal repeat circles and persistent HIV-1 replication. Curr Opin Infect Dis 14: 5–11 [DOI] [PubMed] [Google Scholar]
- *.Shaw GM, Hunter E 2011. HIV transmission. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal A, Kim JT, Balazs AB, Dekel E, Mayo A, Milo R, Baltimore D 2011. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 477: 95–98 [DOI] [PubMed] [Google Scholar]
- *.Siliciano RF, Greene WC 2011. HIV latency. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia ES, Cheslock SR, Zhang WH, Hu WS, Pathak VK 2003. Retroviral mutation rates and reverse transcriptase fidelity. Front Biosci 8: D117–D134 [DOI] [PubMed] [Google Scholar]
- *.Swanstrom R, Coffin J 2011. HIV-1 pathogenesis: The virus. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, Pawluk DM, Mohan KM, Lewis PF, Mullins JI, et al. 2005. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: Expression of archival virus and replication of virus. J Virol 79: 9625–9634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Walker B, McMichael A 2011. The T-cell response to HIV. Cold Spring Harb Perspect Med 10.1101/cshperspect.a007054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Ghosh S, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373: 117–122 [DOI] [PubMed] [Google Scholar]
- Wong JK, Strain MC, Porrata R, Reay E, Sankaran-Walters S, Ignacio CC, Russell T, Pillai SK, Looney DJ, Dandekar S 2010. In vivo CD8+ T-cell suppression of siv viremia is not mediated by CTL clearance of productively infected cells. PLoS Pathog 61: e1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson AS, Korber BT, et al. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N Engl J Med 340: 1605–1613 [DOI] [PubMed] [Google Scholar]