Abstract
The globin gene disorders including the thalassemias are among the most common human genetic diseases with more than 300,000 severely affected individuals born throughout the world every year. Because of the easy accessibility of purified, highly specialized, mature erythroid cells from peripheral blood, the hemoglobinopathies were among the first tractable human molecular diseases. From the 1970s onward, the analysis of the large repertoire of mutations underlying these conditions has elucidated many of the principles by which mutations occur and cause human genetic diseases. This work will summarize our current knowledge of the α-thalassemias, illustrating how detailed analysis of this group of diseases has contributed to our understanding of the general molecular mechanisms underlying many orphan and common diseases.
Mutations in the α-globin cluster cause α-thalassemias. These mutations and their effects on gene expression may provide insight into general mechanisms underlying human genetic diseases.
The molecular basis of α-thalassemia has been extensively reviewed many times in the past (Higgs et al. 1989; Harteveld and Higgs 2010), so why should we take the opportunity to revisit this topic; what can this contribute to our further understanding of how globin synthesis may be perturbed and how this relates more generally to human genetic disease? In my view, despite the extensive new knowledge of human genetic disease that is being obtained from the application of new generation sequencing of material from patients with a wide range of inherited and acquired human genetic diseases, many of the principles by which mutations cause human disease have been established for many years since the application of modern molecular and cellular biology to the globin gene disorders. It is underrecognized that what we have learned from the hemoglobinopathies applies to most human genetic diseases and further analysis of these uniquely well-characterized gene clusters will continue to elucidate important new mechanisms by which mammalian genes are normally switched on and off during cell fate decisions, and how this is perturbed in both common and orphan genetic diseases.
An argument that is commonly put forward is that the globin genes are very unusual, atypical genes and what we learn from them may be exceptions to the rule. On the contrary, it is hard to think of a single example in which what has been established from analyzing the globin genes (involving enhancers, locus control regions, boundary elements, promoters, RNA processing, and numerous aspects of protein translation, structure, and function) has been limited to these genes rather than establishing general principles of mammalian genome organization and how this relates to gene expression. Furthermore, the molecular mechanisms by which globin gene mutations arise (chromosomal rearrangements, telomere truncations, homologous and illegitimate recombination, gene conversion, copy number variation, nucleotide polymorphism, changes in tandem repeats, abnormal methylation, the involvement of antisense RNAs, and many more) are now known to be common to many human diseases and yet were first recognized in the globin gene disorders. Their analysis has been at the forefront of identifying the mechanisms by which mutations at every level of gene expression may cause human disease. Rather than being unusual outliers, the globin genes are in most ways, typical mammalian genes, which when mutated cause typical human genetic diseases by common mechanisms and have pioneered our understanding of how natural mutations cause human genetic disease.
In the future, it will be important to continue using the knowledge gained from the well-characterized globin gene disorders to further develop our understanding of the mechanisms by which both common and orphan human genetic diseases may arise. In this article, I summarize some of what we have learned from studying mutations of the α-globin cluster and how this may provide insight into how we may understand the mechanisms underlying human genetic disease in general.
IDENTIFYING INDIVIDUALS WITH α-THALASSEMIA
Down-regulation of α-globin synthesis causes α-thalassemia, which is characterized by underproduction of fetal (HbF, α2γ2) and adult (HbA, α2β2) hemoglobin (Higgs et al. 1989). The considerable diversity of mutations that have been identified is explained by the fact that carriers for α-thalassemia are (to some extent) protected from falciparum malaria in a manner that we still do not fully understand (reviewed in Higgs 2009a; Williams and Weatherall 2012). Consequently, in tropical and subtropical regions of the world (where malaria is endemic), α-thalassemia reaches very high frequencies, making it one of the most common of all human genetic disease traits (Higgs 2009a; Williams and Weatherall 2012). Among people originating from these areas, compound heterozygotes and homozygotes for some mutations may have severe hematologic phenotypes. When α-globin synthesis is reduced to ∼25% or less, patients may suffer from a moderately severe hemolytic anemia associated with readily detectable excess β-globin chains in the form of β4 tetramers (referred to as HbH); hence, the condition is called HbH disease (Chui et al. 2003; Vichinsky 2012). When α-globin synthesis is even further reduced (or even abolished), this gives rise to a severe intrauterine anemia associated with excess γ-globin chains in the form of γ4 tetramers (referred to as Hb Bart’s); hence, this condition is referred to as the Hb Bart’s hydrops fetalis syndrome (BHFS) (Lorey et al. 2001; Vichinsky 2012). The high frequency of α-thalassemia trait, HbH disease, and BHFS means that tens of thousands of patient samples have been analyzed and that probably most of the natural mutations affecting these genes are now known. Consequently, in most instances, accurate genetic counseling and (when appropriate) prenatal testing can be made available in countries where the health care infrastructure is able to support this. However, even when all known mutations have been excluded, new, often rare but informative genetic variants are still being found.
THE NORMAL STRUCTURE AND EXPRESSION OF THE α-GLOBIN CLUSTER
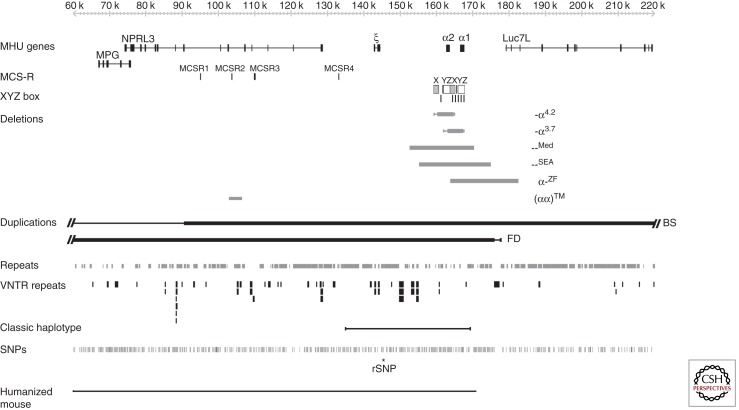
Expression of the α-like and β-like globin chains is regulated by clusters of genes on chromosomes 16 and 11, respectively (Higgs and Wood 2008; Noordermeer and de Laat 2008). The α-like gene cluster is located close to the telomere of chromosome 16 (16p13.3) including an embryonic gene (ζ) and two fetal/adult genes arranged along the chromosome in the order telomere-ζ-α2-α1-centromere, surrounded by widely expressed genes (Fig. 1). The normal cluster is denoted αα. Approximately 25–65 kb upstream of the α-globin genes there are four highly conserved noncoding sequences, or multispecies conserved sequences (MCS), called MCS-R1 to -R4, which are thought to be involved in the regulation of the α-like globin genes (see Fig. 1). It is of interest that three of these elements (MCSR1–3) lie within the introns of the adjacent widely expressed gene NPRL3 (Kowalczyk et al. 2012b). Of these elements to date, only MCS-R2 (also known as HS-40) has been shown to be essential for α-globin expression (Higgs and Wood 2008).
Figure 1.
The human α-globin cluster (5′-ζ-α2-α1-3′) surrounded by widely expressed genes (MPG, NPRL3, and Luc7L) on chromosome 16 (16p13.3). Below this, the multispecies conserved elements (MCS-Rs) are shown. The X, Y, and Z boxes are the regions of duplication that play a part in the generation of the common α-thalassemias, as discussed in the text. The most common deletions removing one (-α3.7 and -α4.2) or both (--Med and --SEA) α genes and causing thalassemia are shown as horizontal bars, as are the unusual deletions α-ZF and (αα)TM. Duplications of the cluster BS and FD are also indicated. For a full catalog of all deletions that cause α-thalassemia, see Higgs (2009a). At the bottom of the figure the positions of common repeats, variable number random repeats (VNTRs) and single-nucleotide polymorphisms (SNPs) are shown. The region containing all SNPs and VNTRs corresponding to the classic haplotype used in population studies (as discussed in the text) is illustrated as a thin horizontal line. The regulatory SNP (rSNP) that creates new functional GATA1-binding sites seen in the Melanesian nondeletional α-thalassemia is shown with an asterisk. The extent of the α-globin locus present in the humanized mouse is shown as a thin line.
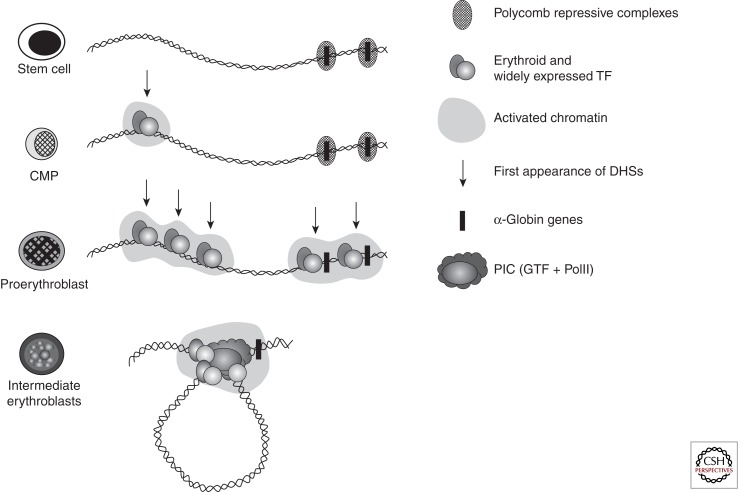
As multipotent hemopoietic progenitors commit to the erythroid lineage and begin to differentiate to form mature red blood cells, key erythroid transcription factors (e.g., GATA-2, GATA-1, SCL, Sp/XKLF, NF-E2) and various cofactors (e.g., FOG, pCAF, p300) progressively bind to the upstream MCS-R elements and the promoters of the α-like globin genes (Fig. 2). Binding of these factors is associated with widespread modifications of the associated chromatin reflecting activation (e.g., histone acetylation). Finally, RNA polymerase II (PolII) is recruited to both the upstream regions and the globin promoters as transcription starts in early erythroid progenitors (Anguita et al. 2004; Vernimmen et al. 2007). At the same time, the upstream elements and promoters of the globin genes interact with each other via the formation of chromatin loops. It has recently been shown that HS-40 is critically required for looping to occur and for the stable recruitment of PolII to the α-globin promoters (Vernimmen et al. 2007, 2009).
Figure 2.
In stem cells and early progenitors, the cluster is silenced by the Polycomb repressive complex. In multipotent cells (CMP), the cluster is primed in the upstream region (MCSR-2) by multiprotein complexes containing SCL and NF-E2 nucleated by GATA-2. In committed erythroid progenitors (U-MEL, proerythroblast stage), additional remote regulatory sequences are bound by multiprotein complexes containing various combinations of SCL, NF-E2, and GATA1 replacing GATA2. At this stage, the α-globin promoter is also occupied by a combination of factors including NF-Y and is poised for expression. In differentiating erythroid cells, the preinitiation complex (PIC), including PolII, is recruited to the enhancers in a cooperative manner but independently of the promoter. Krüppel-like transcription factors are also recruited, independently of the upstream elements and to the promoter. At this final stage, the α-globin promoter is now occupied by a multiprotein complex that represents a docking site for the recruitment of the PIC, which is entirely dependent on the presence of the upstream elements that interact with the promoter, forming a loop.
This series of up-regulatory events taking place in erythroid cells is only a part of the complete story of α-globin regulation. Although the α-globin genes are expressed in a strictly tissue-specific manner, they are contained in a large chromosomal region, which is broadly transcriptionally active and bears the hallmarks of constitutively active chromatin (Flint et al. 1997; Daniels et al. 2001). Therefore, it was predicted that within such a region, mechanisms should exist to maintain the α-globin genes in a silent state in nonerythroid cells. An extensive survey of chromatin modifications in erythroid and nonerythroid cells showed that the α-globin cluster is marked by a specific signature (H3K27me3) in nonerythroid cells but less so in erythroid cells (Garrick et al. 2008). The H3K27me3 mark is imposed by a histone methyltransferase (EZH2), which forms part of a repressive Polycomb complex called PRC2 (Morey and Helin 2010). Consistent with this, components of this complex (EZH2 and SUZ12) have also been found at the α-globin cluster in nonerythroid cells. The PRC2 complex recruits histone deacetylases (HDACs) and another repressive Polycomb complex (called PRC1) (Lynch et al. 2012). Experimental data suggest that together, these complexes (PRC1, PRC2, and HDACs) maintain the silence of the α genes in multipotent cells and differentiated nonerythroid cells (Garrick et al. 2008; Lynch et al. 2012) but that they are cleared from the α-globin promoter before the activation steps described (see Fig. 2).
Activation of the α-globin genes can thus be thought of as a multistep process in which transcription of α globin is at first somewhat leaky, albeit expressed at very low levels, in hematopoietic stem cells when the α genes are partially repressed by the Polycomb system. As cells differentiate down the nonerythroid pathway, the genes become increasingly silenced by Polycomb so that in mature nonerythroid cells no transcription can be detected. In contrast, as cells differentiate along the erythroid pathway, Polycomb silencing is removed as the activating events are played out on the cluster (see Fig. 2) (De Gobbi et al. 2011). All results to date have been obtained by analyzing populations of cells so that we only see the average effects. Seeing the true order of events in single cells is a challenge for the future.
NORMAL VARIATION IN THE α-GLOBIN CLUSTER
Over the past few years, whole genome analysis has revealed the scale of variation across the human genome in apparently normal individuals (Lupski and Stankiewicz 2005; Day 2010). Such variation results from single-nucleotide polymorphisms (SNPs), variations in the numbers of tandem repeats (VNTRs), variations in microsatellites, copy number variants (CNVs, caused by homologous or illegitimate recombination), segmental duplications and deletions, and chromosomal translocations (Lupski and Stankiewicz 2005; Day 2010). Although heralded as a surprise, in fact the extent of variation and mechanisms underlying such polymorphic changes were anticipated from detailed characterization of the regions containing the globin loci in large numbers (thousands) of nonthalassemic individuals. In addition to SNPs forming ancestral haplotypes (Higgs et al. 1986), it had been shown that extensive polymorphic variations caused by genomic rearrangements were found in hematologically normal individuals. These variants are summarized in detail by Higgs (2009a) but the most notable small-scale variations in the α cluster occur in the many G-rich VNTRs in this region (Flint et al. 1997) together with the small- and large-scale variations commonly observed in the subtelomeric region of chromosome 16 (Wilkie et al. 1991). Studies of the different arrangements of these polymorphisms and VNTRs in various population samples have made it possible to define a series of α-globin gene haplotypes that have been of considerable value both in the analysis of evolutionary aspects of the gene clusters and in defining the origins of many of the α-thalassemia mutations (Higgs et al. 1986; Higgs 2009b). In addition to their value as genetic markers, these variants have been useful in distinguishing functionally important areas of the α-globin cluster from regions that appear to be of little functional significance (Rugless et al. 2008).
α-THALASSEMIA CAUSED BY SEQUENCE VARIATIONS IN THE STRUCTURAL GENES
Although most sequence variants in the α-globin cluster (including variation within HS-40) are caused by neutral SNPs, we currently know of 69 point mutations or oligonucleotide variants that alter gene expression, referred to as nondeletional forms of α-thalassemia (denoted αTα or ααT depending on whether the α2 or α1 gene is affected). As for many other human genetic diseases, these mutations may affect the canonical sequences that control gene expression, including the CCAAT and TATA box sequences associated with the promoter, the initiation codon (ATG), splicing signals (GT/AG), the termination codon (TAA), and the polyadenylation (polyA) signal (AATAAA). In addition to these mutations, α-thalassemia may also be caused by in-frame deletions, frame-shift mutations, and nonsense mutations (often leading to nonsense-mediated decay of the RNA) and/or to the production of abnormal protein. Some variants alter the structure of the hemoglobin molecule making the dimer (αβ) or tetramer (α2β2) unstable. Such molecules may precipitate in the red cell, forming insoluble inclusions that damage the red cell membrane. Over the past few years, it has become apparent that some α-globin structural variants are so unstable that they undergo very rapid postsynthetic degradation and thereby cause the phenotype of α-thalassemia. A full list of sequence variations in the structural genes that cause α-thalassemia is available (Higgs 2009a). An even greater number of similar point mutations have been described for the β-like globin genes (summarized in Giardine et al. 2011). Of all such variants, a major class of mutations that is not found in the globin genes is trinucleotide expansions (Lopez Castel et al. 2010). Furthermore, to date, no mutations affecting mRNAs or long noncoding RNAs have been found.
CHROMOSOMAL TRANSLOCATIONS AND DUPLICATIONS AFFECTING THE α-GLOBIN CLUSTER
Rarely, chromosomal translocations involving 16p13.3 place the α-globin locus at the tip of another chromosome, as seen, for example, in some relatives of patients with the ATR-16 syndrome (Gibbons 2012) (see below). To date, we know of 16 individuals with such balanced translocations and none of them has α-thalassemia (Steinberg et al. 2009). Because the closest centromeric breakpoint of these chromosomal translocations lies only 1.2 Mb from the α-globin genes, these findings show that the cis-acting sequences required for full α-globin regulation are contained within this region and that expression is not perturbed by rearrangements on this scale. In two individuals with unbalanced translocations and three copies of 16p13.3, the α/β-globin chain synthesis ratios were 1.5 and 1.6 (Wainscoat et al. 1981; Buckle et al. 1988), again indicating that the additional, mislocalized copy of the α complex is expressed even though its genomic position has changed.
Two large, internal duplications of the terminal region of chromosome 16 have also been described in patients with β-thalassemia intermedia. Because these patients are simply heterozygotes for the β-thalassemia mutations, the implication is that their relatively severe phenotype results from the production of excess α-globin chains from the α genes in the duplicated regions. In one pedigree, three α clusters (αα:αα:αα) are present on one copy of chromosome 16 (Fichera et al. 1994). Provisional data suggest that at least two and possibly all three clusters in the duplicated region are fully active. A carrier for this abnormal chromosome (αα:αα:αα/αα), with a total of eight α genes, has an α/β-globin chain synthesis ratio of 2.7 (Fichera et al. 1994). A recent study has more fully characterized what appears to be a very similar rearrangement in another Italian family with β-thalassemia intermedia (Harteveld et al. 2008) revealing a duplication of ∼260 kb (see BS in Fig. 1). These investigators also characterized another duplication of ∼175 kb of chromosome 16 lying between the end of the α cluster and the telomere (see FD in Fig. 1). Again the phenotype of a compound heterozygote for this rearrangement (αα:αα/αα) and β-thalassemia trait suggested that the additional α clusters in the duplicated segment are fully active, indicating that all sequences required for fully regulated α-globin expression lie in this duplicated segment of chromosome 16. These findings, delimiting the region required to direct fully regulated expression of the human α-globin cluster, are consistent with experimental data from a mouse model in which the mouse α-globin cluster was replaced with ∼135 kb of the human α-globin cluster (Wallace et al. 2007). This region contains all sequences within a region of conserved synteny between the human and mouse, including the globin genes and their regulatory elements. The pattern and levels of expression of the human transgenes in this segment of DNA suggest that this region of ∼135 kb contains all of the sequences required to express the α-globin genes correctly from an appropriate chromosomal environment, although, as discussed previously (Wallace et al. 2007), the level of expression in a mouse environment is less than in the human.
α-THALASSEMIA CAUSED BY DELETIONS REMOVING ONE OR MORE OF THE DUPLICATED STRUCTURAL GENES
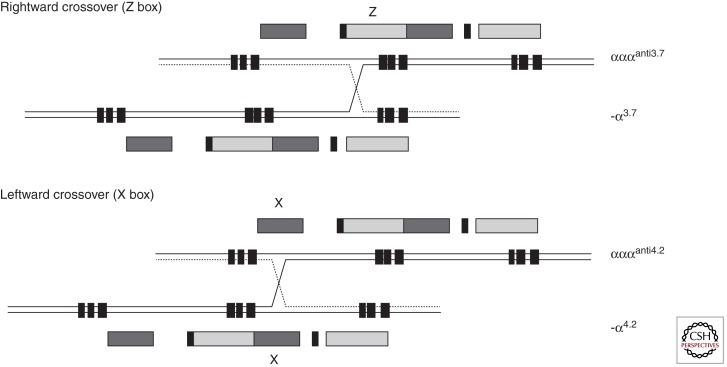
Heteroduplex and DNA sequence analysis has shown that the duplicated α-globin genes (αα) are embedded within two highly homologous, 4-kb duplication units whose sequence identity appears to have been maintained throughout evolution by gene conversion and unequal crossover events (Lauer et al. 1980; Zimmer et al. 1980; Michelson and Orkin 1983; Hess et al. 1984). These regions are divided into homologous subsegments (X, Y, and Z) by nonhomologous elements (I, II, and III) (see Figs. 1 and 3). Reciprocal homologous recombination between Z segments, which are 3.7 kb apart, produces chromosomes with only one α gene (-α3.7, rightward deletion) (see Figs. 1 and 3) (Embury et al. 1980) that cause α-thalassemia and others with three α genes (αααanti3.7) (Goossens et al. 1980). Recombination between homologous X boxes, which are 4.2 kb apart, also gives rise to an α-thalassemia determinant (-α4.2, leftward deletion) (see Figs. 1 and 3) (Embury et al. 1980) and an αααanti4.2 chromosome (Trent et al. 1981). Further recombination events between the resulting chromosomes (α, αα, and ααα) may give rise to quadruplicated α genes (αααα) (De Angioletti et al. 1992), or quintuplicated (ααααα) (Cook et al. 2006) or other unusual patchwork rearrangements. Although these long-standing observations have pointed to the mechanism by which -α and ααα chromosomes arise, it has now been shown (using single DNA molecule polymerase chain reaction) how they may occur in vivo (Lam and Jeffreys 2007). From this work, the overall picture is one of reciprocal recombination (and unequal exchange) occurring in mitosis (premeiotic) in the germ line. The estimated frequencies of -α and ααα arrangements in sperm are in the order of 1–5 × 105. Deletions and duplications may also occur in somatic tissues by related mechanisms, although deletions detected in blood occur by intrachromosomal rather than interchromosomal recombination.
Figure 3.
The mechanism by which the common deletions underlying α-thalassemia occur. Crossovers between misaligned Z boxes give rise to the -α3.7 and αααanti3.7 chromosomes. Crossovers between misaligned X boxes give rise to -α4.2 and αααanti4.2 chromosomes.
In addition to the common -α chromosomes, several rare deletions that remove either the α1 or α2 gene (leaving one gene intact, -α) have been described. In general, these remove either the α1 or α2 gene by nonhomologous recombination. All of these deletions leave one α gene intact, and it is therefore possible to assess the influence of these deletions on expression of the remaining gene. When all of the data from these deletions that cause α-thalassemia are considered alongside the observations from nonthalassemic variants (see above), it appears that large segments of the α-globin cluster are not essential for α-globin expression (Rugless et al. 2008). Figures showing the full list of currently known deletions removing a single α gene are presented by Rugless and colleagues (2008).
It is now known that many human diseases (referred to as copy number variants) ranging from color blindness to inherited neuropathies (Carvalho et al. 2010) result from unequal exchange of low copy number duplicated sequences in exactly the same way as described in the globin clusters. In these diseases, too, the phenotypes associated with gain or loss of gene product may be quite different.
α-THALASSEMIA CAUSED BY DELETIONS REMOVING BOTH OF THE DUPLICATED STRUCTURAL GENES
There are currently approximately 50 deletions from the α-globin cluster that either completely or partially delete both α-globin genes, and consequently no α-chain synthesis is directed by these chromosomes in vivo. Examples of two common deletions --MED and --SEA are shown in Fig. 1. Homozygotes for such chromosomes (--/--) have the Hb BHFS. Compound heterozygotes for these deletions and deletions removing a single α gene (see above) have HbH disease (--/-α). With completion of the DNA sequence of 16p13.3 and beyond (International Human Genome Sequencing Consortium 2004), it has been possible to define the full extent of many of the deletions that remove both α genes (--). These deletions can be grouped into those (like --MED and --SEA) (see Fig. 1) that lie entirely within the α-globin cluster and deletions that extend up to ∼800 kb beyond the α cluster to include the flanking genes. Although these deletions remove other genes, affected heterozygotes appear phenotypically normal apart from their α-thalassemia: in some patients, α-thalassemia trait (--/αα) and, in others, HbH disease (--/-α). In patients with more extensive deletions, with monosomy for a large segment of 16p13.3, α-thalassemia is associated with developmental abnormalities and mental retardation (so-called ATR-16 syndrome; see below) (Gibbons 2012).
It is interesting to note that all of the α-thalassemia deletions that occur at polymorphic frequencies in human populations are limited to the α cluster and do not extend into the surrounding genes, suggesting that deletion of these genes (even in heterozygotes) may result in a selective disadvantage. Detailed analysis of several of these determinants of α-thalassemia indicates that they often result from illegitimate or nonhomologous recombination events (e.g., Nicholls et al. 1987; Rugless et al. 2008). Such events may involve short regions of partial sequence homology at the breakpoints of the molecules that are rejoined, but they do not involve the extensive sequence matching required for homologous recombination as described in the previous section. Sequence analysis has shown that members of the dispersed family of Alu repeats are frequently found at or near the breakpoints of these deletions. Alu-family repeats occur frequently in the genome (3 × 105 copies) and seem to be particularly common in and around the α-globin cluster, where they make up ∼25% of the entire sequence. These repeats may simply provide partially homologous sequences that promote DNA-strand exchanges during replication, or possibly a subset of Alu sequences may be more actively involved in the process. Detailed sequence analysis of the junctions of the α-globin deletions has revealed several interesting features including palindromes, direct repeats, and regions of weak homology. Some deletions involve more complex rearrangements that introduce new pieces of DNA bridging the two breakpoints of the deletion. In two deletions, this inserted DNA originates from upstream of the α cluster, and appears to have been incorporated into the junction in a manner suggesting that the upstream segment lies close to the breakpoint regions during replication (Nicholls et al. 1987; Rugless et al. 2008). Orphan sequences from unknown regions of the genome are frequently found bridging the sequence breakpoints of other α-thalassemia deletions. Such complex deletion events are now being recognized with increasing frequency by new generation sequencing (Zhang et al. 2009). At least two of the deletions result from chromosomal breaks in the 16p telomeric region that have been “healed” by the direct addition of telomeric repeats (TTAGGG)n (Flint et al. 1994). This mechanism is described below in further detail. All currently known deletions that remove both α genes are summarized in Higgs (2009a).
LARGE DELETIONS EXTENDING BEYOND THE α-GLOBIN CLUSTER
Nearly all patients with large deletions (up to 900 kb) from the end of chromosome 16p (removing one copy of up to 52 genes) appear phenotypically normal apart from the presence of α-thalassemia. However, in 40 patients analyzed to date, deletions of >900 kb have been associated with a variety of developmental abnormalities; such patients are said to have the ATR-16 syndrome (Wilkie et al. 1990; Gibbons 2012). Relating phenotype to genotype is difficult in these cases because of the rather heterogeneous nature of the underlying abnormalities. Using a combination of conventional cytogenetics, fluorescent in situ hybridization, and molecular analysis, at least three types of chromosomal rearrangements (translocation, inversion/deletion, and truncation) have now been found in ATR-16 patients. To date, few breakpoints have been fully characterized. However, in some cases, telomeric truncations have been documented; in these cases, it appears that the affected chromosomes have been broken, truncated, and “healed” by the direct addition of telomeric repeats (TTAGGG)n, as described above for some less extensive 16p deletions in patients with α-thalassemia.
Altogether, 11 cases of ATR-16 have been shown to have pure monosomy for 16p13.3 and deletions of 900–1700 kb with various developmental abnormalities (e.g., facial dysmorphism, speech delay, and skeletal abnormalities). How might monosomy for 16p13.3 cause such developmental abnormalities? One possibility is that deletion of a large number of genes from one copy of chromosome 16 may unmask mutations in its homolog; the more genes that are deleted the greater the probability of this occurring. A further possibility is that some genes in 16p are imprinted (Reik and Walter 2001) so that deletions could remove the only active copy of the gene. At present there is no evidence for imprinting of the 16p region (reviewed in Schneider et al. 1996), and in the relatively few ATR-16 cases analyzed there appear to be no major clinical differences between patients with deletions of the maternally or paternally derived chromosomes (Gibbons 2012). It therefore seems more likely that there are some genes in the 16p region that encode proteins whose effect is critically determined by the amount produced—so-called dosage-sensitive genes. Examples of such genes include those encoding proteins that form heterodimers, those required at a critical level for a rate-determining step of a regulatory pathway, and tumor suppressor genes (e.g., TSC2). Removal of genes from one copy of 16p13.3 consistently reduces their levels of expression to ∼50% of normal (V Buckle and colleagues, unpubl.). If the deletion includes one or more dosage-sensitive genes, this could account for the clinical effects seen in ATR-16 patients. The region lying between 900 and 1700 kb from the 16p telomere, deleted in all patients with the characteristic features of ATR-16 syndrome, contains genes and gene families of known function that have been implicated in a wide range of disorders with few or no features in common with ATR-16. One of these (SOX8) was considered as a strong candidate because it is involved in the regulation of embryonic development and is strongly expressed in the brain (Pfeifer et al. 2000). However, a recently described Brazilian patient with a deletion that removes both the α-globin locus and SOX8 was not associated with mental retardation (MR) or any dysmorphism (Bezerra et al. 2008). It is clear that further examples of ATR-16 owing to monosomy for 16p13.3 would have to be characterized to identify the gene(s) responsible for the MR and other developmental abnormalities associated with this condition. However, we cannot rule out the fact that similar deletions may cause different phenotypes owing to variations in the genetic background. The ATR-16 syndrome has served as an important model for improving our general understanding of the molecular basis for mental retardation (Gibbons 2012). The ATR-16 syndrome provided the first examples of mental retardation caused by a cryptic chromosomal translocation and truncation. Further work showed that such telomeric rearrangements may underlie a significant proportion of unexplained mental retardation (Flint et al. 1995). The current challenge is to understand in detail the mechanisms by which monosomy causes developmental abnormalities; the ATR-16 syndrome provides an excellent model for addressing this issue. All deletions currently associated with ATR-16 syndrome are shown in Higgs (2009a).
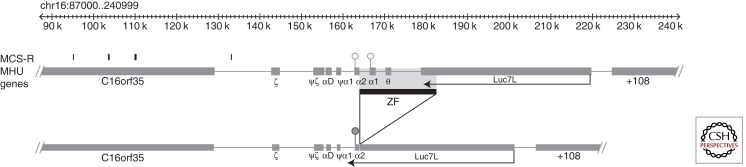
A RARE MUTATION CAUSING α-THALASSEMIA VIA AN ANTISENSE RNA
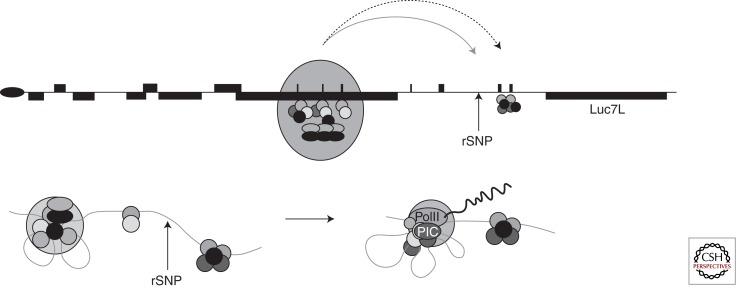
During a study to identify thalassemia in families from the Czech Republic, Indrak and colleagues (1993) reported a novel deletion (>18 kb) involving the α1 and θ gene (denoted α-ZF) (see Figs. 1 and 4). Heterozygotes for this deletion have a mild hypochromic microcytic anemia with a reduced α/β-globin chain biosynthesis ratio and Hb H inclusions. These findings suggested that although the α2 gene appeared to be intact, it had been inactivated by the deletion. Barbour and colleagues (2000) and Tufarelli and colleagues (2003) further characterized this mutation and showed that the deletion juxtaposes a downstream gene (Luc7L) next to the structurally normal α2-globin gene. Although this α2 gene retains all of its local (e.g., promoter) and remote (e.g., MCS-R2/HS-40) cis-acting elements, its expression is silenced and its associated CpG island (see Fig. 4) becomes completely methylated during early development, and the chromatin associated with the promoter remains inactive and inaccessible, even in erythroid cells. From the analysis of experimental models recapitulating this deletion and from further characterization of the affected individual (Tufarelli et al. 2003), it was shown that transcription of antisense mRNA from Luc7L through the α2-globin gene was responsible for methylation of the associated CpG island and silencing of α-globin expression. Since this original report, the authors have identified two additional individuals (also from Poland) with the same mutation. The mutation is not only important for understanding the molecular basis for this rare form of thalassemia, but also illustrates a new mechanism underlying human genetic disease. Since this description, it has become increasingly clear that antisense RNAs may play important roles in regulating mammalian gene expression (Kowalczyk et al. 2012a). In terms of causing human genetic disease, there has been a report of a similar mechanism silencing the mismatch repair gene MSH2 in patients with susceptibility to colorectal cancer (Lynch syndrome) (Ligtenberg et al. 2009).
Figure 4.
The key features of the α-ZF mutation. In the normal cluster, the promoters of the α-globin genes lie in unmethylated CpG-rich islands. Partial deletion of Luc7L juxtaposes this truncated gene next to the remaining α2-globin gene and RNA transcripts from Luc7L extend through the α2-globin gene. This process is thought to attract de novo DNA methylases early in development, methylating the α2 CpG island and silencing it.
DELETIONS REMOVING THE UPSTREAM REGULATORY ELEMENTS OF THE α-GLOBIN CLUSTER
As discussed earlier, expression of the α genes is critically dependent on a multispecies conserved, noncoding regulatory sequence (MCS-R) that lies 40 kb upstream of the ζ2-globin gene. This region (called MCS-R2) is associated with an erythroid-specific DNase I hypersensitive site, referred to as HS-40. Detailed analysis of MCS-R2 has shown that it contains multiple binding sites for the erythroid-restricted trans-acting factors GATA-1 and NF-E2, and binding sites for the ubiquitously expressed Sp/XKLF family of transcription factors (Higgs et al. 2008).
The first indication that remote regulatory sequences controlling α-globin expression might exist came from observations on a patient with α-thalassemia (Hatton et al. 1990). Analysis of the abnormal chromosome (αα)RA from this patient showed a 62-kb deletion from upstream of the α complex that includes HS-40. Although both α genes on this chromosome are intact and entirely normal, they appear to be nonfunctional. Since this original observation, many more patients with α-thalassemia caused by deletions of HS-40 and a variable amount of the flanking DNA have been described, summarized in Higgs and Wood (2008). Until recently, the smallest of these deletions, which completely abolishes α-globin expression, removed both MCS-R1 and MCS-R2. Recently, an even smaller deletion, which removes ∼3.3 kb of DNA including MCS-R2 but no other MCS-R element, was described (see Fig. 1) (Phylipsen et al. 2010). The phenotype (HbH disease) of the proband, which carries the same small deletion on both copies of chromosome 16, is consistent with expression of both cis-linked α genes being down-regulated but not completely abolished (Coelho et al. 2010). This suggests that other cis-acting elements on this chromosome (possibly MCS-R1) can activate α-globin expression. Clearly, these new observations demand further analysis of the other upstream MCS elements (MCS-R1, -R3, and -R4), which contain similar combinations of binding sites to MCS-R2 but whose functions are not yet clear. Together with similar deletions removing the regulatory elements controlling the β-globin cluster (Kioussis et al. 1983; Grosveld et al. 1987), these observations were among the first to illustrate in detail how mutation of distal regulatory elements can cause human genetic disease.
The mechanisms by which these natural mutations have arisen are quite diverse. In one, the deletion resulted from a recombination event between partially homologous Alu repeats that are normally 62 kb apart (Hatton et al. 1990). In another, the deletion arose via a subtelomeric rearrangement (Flint et al. 1996). The chromosomal breakpoint was found in an Alu element located ∼105 kb from the 16p subtelomeric region. The broken chromosome was stabilized with a new telomere acquired by recombination between this Alu element and a subtelomeric Alu repeat associated with the newly acquired chromosome end. In at least five cases, the chromosomes appear to have been broken and then stabilized by the direct addition of telomeric repeats to nontelomeric DNA (Flint et al. 1994). Sequence analysis suggests that these chromosomes are “healed” via the action of telomerase, an enzyme that is normally involved in maintaining the integrity of telomeres. In the remaining cases, the mechanism has not yet been established. However, it is interesting that some (e.g., [αα]IJ, [αα]Sco) (Liebhaber et al. 1990; Viprakasit et al. 2003), but not all, of these mutations appear to have arisen de novo, because neither parent has the abnormal chromosome.
α-THALASSEMIA RESULTING FROM COMPETITION FOR THE UPSTREAM REGULATORY ELEMENTS
α-Thalassemia is common throughout Melanesia and is frequently caused by the known -α3.7 and -α4.2 mutations. However, it is also documented that in some Melanesian patients (from Papua New Guinea and Vanuatu) with α-thalassemia (α-thalassemia trait and HbH disease), the α-globin genes are intact, suggesting a nondeletional form of α-thalassemia (αTα/αα or αTα/αTα) (Fig. 5). In these patients, detailed mapping and DNA sequence analysis of the α genes and all of the upstream MCS elements was normal, and yet further studies showed that this form of α-thalassemia is linked to the α-globin cluster at 16p13.3. To identify the mutation responsible for this unusual form of α-thalassemia, De Gobbi and colleagues (2006) cloned this region from an affected homozygote and resequenced ∼213 kb of DNA containing and flanking the α-globin cluster, identifying 283 SNPs. The SNP responsible for the mutation was identified when the SNPs were aligned with a tiled microarray analyzed using labeled RNA from the patient’s erythroid cells. This configuration revealed a new peak of mRNA expression (located between the ζ and ψζ genes), which coincides with a SNP that creates a GATA-1 binding site. In association studies, this SNP is always linked to the phenotype of α-thalassemia. Like the MCS-R elements, this new GATA site binds erythroid transcription factors in vivo and becomes activated in erythroid cells. Why should creating a new promoter-like sequence between the α genes and their regulatory elements cause α-thalassemia? Perhaps the most likely explanation is that, because it lies closer to the MCS elements, this new promoter competes with the α-globin promoters and “steals” the activity of the upstream regulatory elements, thus resulting in α-thalassemia. Several lines of evidence suggest that the α-globin promoters may indeed compete for the activity of the upstream regulatory elements, in particular, MCS-R2(HS-40). First, it has been noted that although the sequences of the duplicated α genes and their promoters are identical, the gene (α2) nearest MCS-R2 is expressed at a higher level than the more distal gene (α1). Furthermore, additional duplications of the α genes (ααα, αααα, and ααααα) do not lead to a linear increase in α-globin expression1; genes located further from MCS-R2 are expressed at progressively lower levels. When one α gene is deleted from the chromosome (-α), the remaining α gene appears to recruit more PolII and is expressed at increased levels (Rugless et al. 2008). It was recently shown that MCS-R2 also regulates expression of another gene (NME4) located 300 kb away from this element on chromosome 16 (Lower et al. 2009). The α-globin genes lie between MCS-R2 and NME4. When both α genes are deleted, expression of NME4 increases by a factor of eightfold. Together these findings suggest that promoters may compete for the activity associated with enhancers, and this may provide the explanation for this form of α-thalassemia seen in patients from the south Pacific. It seems likely that other similar “decoy promoters” will be found to cause changes in gene expression during the course of genome-wide studies.
Figure 5.
The key features of the Melanesian form of α-thalassemia. A regulatory SNP (rSNP) located between the ζ- and α2-globin genes creates a new GATA1-binding site, which in turn also creates a new promoter. This promoter preferentially interacts with the upstream enhancer elements (gray curved arrow) and steals activity from the α genes (dashed curved arrow). In the hypothetical looping model (below), the upstream element interacts with the new promoter but not the α genes.
SUMMARY
The α-globin cluster provides one of the best characterized models to develop our understanding of how the integration of a transcriptional program and an epigenetic program via a key set of cis-acting elements dispersed across a large chromosomal region, switch genes on and off at the correct time and place during development and differentiation. In particular, by analyzing the normal cluster, the natural mutants, and experimental models we should gain significant insight into how long-range regulatory elements control gene expression. There is now sufficient knowledge of the mutations underlying α-thalassemia to develop comprehensive genetic counseling and prenatal diagnosis wherever there is sufficient expertise and resources to support such a program. Further identification of natural mutations of the globin genes should be pursued because these observations continue to elucidate the general principles underlying human molecular genetics.
Footnotes
Editors: David Weatherall, Alan N. Schechter, and David G. Nathan
Additional Perspectives on Hemoglobin and Its Diseases available at www.perspectivesinmedicine.org
REFERENCES
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J 23: 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour VM, Tufarelli C, Sharpe JA, Smith ZE, Ayyub H, Heinlein CA, Sloane-Stanley J, Indrak K, Wood WG, Higgs DR 2000. α-Thalassemia resulting from a negative chromosomal position effect. Blood 96: 800–807 [PubMed] [Google Scholar]
- Bezerra MA, Araujo AS, Phylipsen M, Balak D, Kimura EM, Oliveira DM, Costa FF, Sonati MF, Harteveld CL 2008. The deletion of SOX8 is not associated with ATR-16 in an HbH family from Brazil. Br J Haematol 142: 324–326 [DOI] [PubMed] [Google Scholar]
- Buckle VJ, Higgs DR, Wilkie AO, Super M, Weatherall DJ 1988. Localisation of human α globin to 16p13.3—pter. J Med Genet 25: 847–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Zhang F, Lupski JR 2010. Evolution in health and medicine Sackler colloquium: Genomic disorders: A window into human gene and genome evolution. Proc Natl Acad Sci 107: 1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chui DH, Fucharoen S, Chan V 2003. Hemoglobin H disease: Not necessarily a benign disorder. Blood 101: 791–800 [DOI] [PubMed] [Google Scholar]
- Coelho A, Picanco I, Seuanes F, Seixas MT, Faustino P 2010. Novel large deletions in the human α-globin gene cluster: Clarifying the HS-40 long-range regulatory role in the native chromosome environment. Blood Cells Mol Dis 45: 147–153 [DOI] [PubMed] [Google Scholar]
- Cook RJ, Hoyer JD, Highsmith WE 2006. Quintuple α-globin gene: A novel allele in a Sudanese man. Hemoglobin 30: 51–55 [DOI] [PubMed] [Google Scholar]
- Daniels RJ, Peden JF, Lloyd C, Horsley SW, Clark K, Tufarelli C, Kearney L, Buckle VJ, Doggett NA, Flint J, et al. 2001. Sequence, structure and pathology of the fully annotated terminal 2 Mb of the short arm of human chromosome 16. Hum Mol Genet 10: 339–352 [DOI] [PubMed] [Google Scholar]
- Day IN 2010. dbSNP in the detail and copy number complexities. Hum Mutat 31: 2–4 [DOI] [PubMed] [Google Scholar]
- De Angioletti M, Lacerra G, Castaldo C, Cutolo R, de Bonis C, Buonanno G, Carestia C 1992. ααααanti-3.7 type II: A new α-globin gene rearrangement suggesting that the α-globin gene duplication could be caused by intrachromosomal recombination. Hum Genet 89: 37–41 [DOI] [PubMed] [Google Scholar]
- De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de Jong P, et al. 2006. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science 312: 1215–1217 [DOI] [PubMed] [Google Scholar]
- De Gobbi M, Garrick D, Lynch M, Vernimmen D, Hughes JR, Goardon N, Luc S, Lower KM, Sloane-Stanley JA, Pina C, et al. 2011. Generation of bivalent chromatin domains during cell fate decisions. Epigenetics Chromatin 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embury SH, Miller JA, Dozy AM, Kan YW, Chan V, Todd D 1980. Two different molecular organizations account for the single α-globin gene of the α-thalassemia-2 genotype. J Clin Invest 66: 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichera M, Rappazzo G, Spalletta A, et al. 1994. Triplicated α-globin gene locus with translocation of the whole telomeric end in association with β-thalassemia trait, results in a severe syndrome. Blood 84: 260a [Google Scholar]
- Flint J, Craddock CF, Villegas A, Bentley DP, Williams HJ, Galanello R, Cao A, Wood WG, Ayyub H, Higgs DR 1994. Healing of broken human chromosomes by the addition of telomeric repeats. Am J Hum Genet 55: 505–512 [PMC free article] [PubMed] [Google Scholar]
- Flint J, Wilkie AO, Buckle VJ, Winter RM, Holland AJ, McDermid HE 1995. The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet 9: 132–140 [DOI] [PubMed] [Google Scholar]
- Flint J, Rochette J, Craddock CF, Dode C, Vignes B, Horsley SW, Kearney L, Buckle VJ, Ayyub H, Higgs DR 1996. Chromosomal stabilisation by a subtelomeric rearrangement involving two closely related Alu elements. Hum Mol Genet 5: 1163–1169 [DOI] [PubMed] [Google Scholar]
- Flint J, Thomas K, Micklem G, Raynham H, Clark K, Doggett NA, King A, Higgs DR 1997. The relationship between chromosome structure and function at a human telomeric region. Nat Genet 15: 252–257 [DOI] [PubMed] [Google Scholar]
- Garrick D, De Gobbi M, Samara V, Rugless M, Holland M, Ayyub H, Lower K, Sloane-Stanley J, Gray N, Koch C, et al. 2008. The role of the polycomb complex in silencing α-globin gene expression in nonerythroid cells. Blood 112: 3889–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Borg J, Higgs DR, Peterson KR, Philipsen S, Maglott D, Singleton BK, Anstee DJ, Basak AN, Clark B, et al. 2011. Systemic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat Genet 43: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens M, Dozy AM, Embury SH, Zachariades Z, Hadjiminas MG, Stamatoyannopoulos G, Kan YW 1980. Triplicated α-globin loci in humans. Proc Natl Acad Sci 77: 518–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G 1987. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell 51: 975–985 [DOI] [PubMed] [Google Scholar]
- Harteveld CL, Higgs DR 2010. α-Thalassaemia. Orphanet J Rare Dis 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harteveld CL, Refaldi C, Cassinerio E, Cappellini MD, Giordano PC 2008. Segmental duplications involving the α-globin gene cluster are causing β-thalassemia intermedia phenotypes in β-thalassemia heterozygous patients. Blood Cells Mol Dis 40: 312–316 [DOI] [PubMed] [Google Scholar]
- Hatton CS, Wilkie AO, Drysdale HC, Wood WG, Vickers MA, Sharpe J, Ayyub H, Pretorius IM, Buckle VJ, Higgs DR 1990. α-Thalassemia caused by a large (62 kb) deletion upstream of the human α globin gene cluster. Blood 76: 221–227 [PubMed] [Google Scholar]
- Hess JF, Schmid CW, Shen CK 1984. A gradient of sequence divergence in the human adult α-globin duplication units. Science 226: 67–70 [DOI] [PubMed] [Google Scholar]
- Higgs DR 2009a. The molecular basis of α thalassemia. In Disorders of hemoglobin (ed. Steinberg MH, et al. ), pp. 241–265 Cambridge University Press, Cambridge [Google Scholar]
- Higgs DR 2009b. The pathophysiology and clinical features of α thalassemia. In Disorders of hemoglobin (ed. Steinberg MH, et al. ), pp. 266–295 Cambridge University Press, Cambridge [Google Scholar]
- Higgs DR, Wood WG 2008. Long-range regulation of α globin gene expression during erythropoiesis. Curr Opin Hematol 15: 176–183 [DOI] [PubMed] [Google Scholar]
- Higgs DR, Wainscoat JS, Flint J, Hill AV, Thein SL, Nicholls RD, Teal H, Ayyub H, Peto TE, Falusi AG, et al. 1986. Analysis of the human α-globin gene cluster reveals a highly informative genetic locus. Proc Natl Acad Sci 83: 5165–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR, Vickers MA, Wilkie AO, Pretorius IM, Jarman AP, Weatherall DJ 1989. A review of the molecular genetics of the human α-globin gene cluster. Blood 73: 1081–1104 [PubMed] [Google Scholar]
- Higgs DR, Vernimmen D, Wood B 2008. Long-range regulation of α-globin gene expression. Adv Genet 61: 143–173 [DOI] [PubMed] [Google Scholar]
- Indrak K, Gu YC, Novotny J, Huisman TH 1993. A new α-thalassemia-2 deletion resulting in microcytosis and hypochromia and in vitro chain imbalance in the heterozygote. Am J Hematol 43: 144–145 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Kioussis D, Vanin E, deLange T, Flavell RA, Grosveld FG 1983. β-Globin gene inactivation by DNA translocation in γ β-thalassaemia. Nature 306: 662–666 [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Higgs DR, Gingeras TR 2012a. Molecular biology: RNA discrimination. Nature 482: 310–311 [DOI] [PubMed] [Google Scholar]
- Kowalczyk MS, Hughes JR, Babbs C, Sanchez-Pulido L, Szumska D, Sharpe JA, Sloane-Stanley JA, Morriss-Kay GM, Smoot LB, Roberts AE, et al. 2012b. Nprl3 is required for normal development of the cardiovascular system. Mamm Genome 23: 404–415 [DOI] [PubMed] [Google Scholar]
- Lam KW, Jeffreys AJ 2007. Processes of de novo duplication of human α-globin genes. Proc Natl Acad Sci 104: 10950–10955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer J, Shen CK, Maniatis T 1980. The chromosomal arrangement of human α-like globin genes: Sequence homology and α-globin gene deletions. Cell 20: 119–130 [DOI] [PubMed] [Google Scholar]
- Liebhaber SA, Griese EU, Weiss I, Cash FE, Ayyub H, Higgs DR, Horst J 1990. Inactivation of human α-globin gene expression by a de novo deletion located upstream of the α-globin gene cluster. Proc Natl Acad Sci 87: 9431–9435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, et al. 2009. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet 41: 112–117 [DOI] [PubMed] [Google Scholar]
- Lopez Castel A, Cleary JD, Pearson CE 2010. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol 11: 165–170 [DOI] [PubMed] [Google Scholar]
- Lorey F, Charoenkwan P, Witkowska HE, Lafferty J, Patterson M, Eng B, Waye JS, Finklestein JZ, Chui DH 2001. Hb H hydrops foetalis syndrome: A case report and review of literature. Br J Haematol 115: 72–78 [DOI] [PubMed] [Google Scholar]
- Lower KM, Hughes JR, De Gobbi M, Henderson S, Viprakasit V, Fisher C, Goriely A, Ayyub H, Sloane-Stanley J, Vernimmen D, et al. 2009. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci 106: 21771–21776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P 2005. Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet 1: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MD, Smith AJ, De Gobbi M, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, et al. 2012. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J 31: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson AM, Orkin SH 1983. Boundaries of gene conversion within the duplicated human α-globin genes. Concerted evolution by segmental recombination. J Biol Chem 258: 15245–15254 [PubMed] [Google Scholar]
- Morey L, Helin K 2010. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 35: 323–332 [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Fischel-Ghodsian N, Higgs DR 1987. Recombination at the human α-globin gene cluster: Sequence features and topological constraints. Cell 49: 369–378 [DOI] [PubMed] [Google Scholar]
- Noordermeer D, de Laat W 2008. Joining the loops: β-Globin gene regulation. IUBMB Life 60: 824–833 [DOI] [PubMed] [Google Scholar]
- Pfeifer D, Poulat F, Holinski-Feder E, Kooy F, Scherer G 2000. The SOX8 gene is located within 700 kb of the tip of chromosome 16p and is deleted in a patient with ATR-16 syndrome. Genomics 63: 108–116 [DOI] [PubMed] [Google Scholar]
- Phylipsen M, Prior JF, Lim E, Lingam N, Vogelaar IP, Giordano PC, Finlayson J, Harteveld CL 2010. Thalassemia in Western Australia: 11 novel deletions characterized by multiplex ligation-dependent probe amplification. Blood Cells Mol Dis 44: 146–151 [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J 2001. Genomic imprinting: Parental influence on the genome. Nat Rev Genet 2: 21–32 [DOI] [PubMed] [Google Scholar]
- Rugless MJ, Fisher CA, Old JM, Sloane-Stanley J, Ayyub H, Higgs DR, Garrick D 2008. A large deletion in the human α-globin cluster caused by a replication error is associated with an unexpectedly mild phenotype. Hum Mol Genet 17: 3084–3093 [DOI] [PubMed] [Google Scholar]
- Schneider AS, Bischoff FZ, McCaskill C, Coady ML, Stopfer JE, Shaffer LG 1996. Comprehensive 4-year follow-up on a case of maternal heterodisomy for chromosome 16. Am J Med Genet 66: 204–208 [DOI] [PubMed] [Google Scholar]
- Steinberg MH, Forget BG, Higgs DR, Weatherall DJ, ed. 2009. Disorders of hemoglobin. Cambridge University Press, New York [Google Scholar]
- Trent RJ, Higgs DR, Clegg JB, Weatherall DJ 1981. A new triplicated α-globin gene arrangement in man. Br J Haematol 49: 149–152 [DOI] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34: 157–165 [DOI] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J 26: 2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernimmen D, Marques-Kranc F, Sharpe JA, Sloane-Stanley JA, Wood WG, Wallace HA, Smith AJ, Higgs DR 2009. Chromosome looping at the human α-globin locus is mediated via the major upstream regulatory element (HS -40). Blood 114: 4253–4260 [DOI] [PubMed] [Google Scholar]
- Viprakasit V, Kidd AM, Ayyub H, Horsley S, Hughes J, Higgs DR 2003. De novo deletion within the telomeric region flanking the human α globin locus as a cause of α thalassaemia. Br J Haematol 120: 867–875 [DOI] [PubMed] [Google Scholar]
- Wainscoat JS, Kanavakis E, Weatherall DJ, Walker J, Holmes-Seidle M, Bobrow M, Donnison AB 1981. Regional localisation of the human α-globin genes. Lancet 2: 301–302 [DOI] [PubMed] [Google Scholar]
- Wallace HA, Marques-Kranc F, Richardson M, Luna-Crespo F, Sharpe JA, Hughes J, Wood WG, Higgs DR, Smith AJ 2007. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell 128: 197–209 [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Buckle VJ, Harris PC, Lamb J, Barton NJ, Reeders ST, Lindenbaum RH, Nicholls RD, Barrow M, Bethlenfalvay NC, et al. 1990. Clinical features and molecular analysis of the α thalassemia/mental retardation syndromes. I: Cases due to deletions involving chromosome band 16p13.3. Am J Hum Genet 46: 1112–1126 [PMC free article] [PubMed] [Google Scholar]
- Wilkie AO, Higgs DR, Rack KA, Buckle VJ, Spurr NK, Fischel-Ghodsian N, Ceccherini I, Brown WR, Harris PC 1991. Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell 64: 595–606 [DOI] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR 2009. Complex human chromosomal and genomic rearrangements. Trends Genet 25: 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer EA, Martin SL, Beverley SM, Kan YW, Wilson AC 1980. Rapid duplication and loss of genes coding for the α chains of hemoglobin. Proc Natl Acad Sci 77: 2158–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]