Abstract
Although TAM receptor tyrosine kinases play key roles in immune regulation, cancer metastasis, and viral infection, the relative importance of the two TAM ligands – Gas6 and Protein S – has yet to be resolved in any setting in vivo. We have now performed a genetic dissection of ligand function in the retina, where the TAM receptor Mer is required for the circadian phagocytosis of photoreceptor outer segments by retinal pigment epithelial cells. This process is severely attenuated in Mer mutant mice, which leads to photoreceptor death. We find that retinal deletion of either Gas6 or Protein S alone yields retinae with a normal number of photoreceptors. However, concerted deletion of both ligands fully reproduces the photoreceptor death seen in Mer mutants. These results demonstrate that Protein S and Gas6 function as independent, bona fide Mer ligands, and are, to a first approximation, interchangeable with respect to Mer-driven phagocytosis in the retina.
Keywords: Mertk, phagocytosis, retinal pigment epithelium, Gas6, Protein S, retinal degeneration
INTRODUCTION
Genetic studies have demonstrated that the three TAM RTKs – Tyro3, Axl, and Mer (Lai and Lemke, 1991) – play essential regulatory roles in the mature immune, nervous, vascular, and reproductive systems (Burstyn-Cohen et al., 2009; Lemke and Rothlin, 2008; Lu et al., 1999; Scott et al., 2001). In general, these receptors are specialized to control homeostatic responses in cells and tissues that are subject to constant challenge and renewal throughout adult life. In the immune system, for example, Axl functions as a pleiotropic inhibitor of the inflammatory response of dendritic cells and macrophages subsequent to their encounter with bacteria, viruses, and other pathogens (Lemke and Rothlin, 2008; Rothlin et al., 2007). And in these same cells, Mer (protein designation Mer, c-Mer, or Mertk; gene name Mertk) is required for the efficient phagocytosis of apoptotic cells that accumulate following infection (Lemke and Burstyn-Cohen, 2010; Scott et al., 2001). In endothelial cells of the vasculature, Axl is engaged subsequent to both acute and chronic vessel injury and remodeling (Korshunov et al., 2006); and in the testis, all three receptors are required in Sertoli cells for the phagocytosis of the tens of millions of apoptotic germ cells that are generated during every cycle of spermatogenesis (Lemke and Burstyn-Cohen, 2010; Lu et al., 1999). While operation of the TAM system is required in each of these tissues throughout adult life, the TAM family is unique among RTKs in that it is dispensable with respect to embryonic development: mice triply mutant for Tyro3, Axl, and Mertk loss-of-function deletions are fully viable at birth, and are superficially indistinguishable from wild-type for the first two weeks thereafter (Lu et al., 1999).
TAM signaling plays an especially prominent role in the retinal pigment epithelial (RPE) cells of the adult eye. These pigmented cells form a single-layer epithelial sheet at the back of the retina, and are immediately apposed to the opsin-containing outer segments (OS) of photoreceptors (PRs) (Strauss, 2005). The apical microvilli of RPE cells extend deep into the OS layer, where they actively pinch off and phagocytose the distal ends of OS (Kevany and Palczewski, 2010; Strauss, 2005). This phagocytic excision occurs on a regular circadian schedule, around subjective dawn, throughout adult life, and is essential for the removal of toxic oxidative products that are generated during phototransduction (Strauss, 2005). PRs insert fresh, newly-synthesized membrane into the basal aspect of their OS each day, and so the phagocytic pruning of OS distal ends by RPE cells maintains a constant OS length.
The apical microvilli of RPE cells express Mer and Tyro3 (Prasad et al., 2006), and analyses across multiple species have shown that Mer is absolutely required for the phagocytosis of distal OS membrane. The retinae of Mertk−/− mice, for example, develop normally, with a full complement of all retinal cell types and a normal histology by two weeks after birth (Nandrot and Dufour, 2010; Prasad et al., 2006). However, beginning shortly thereafter, and coincident with eye opening, the PRs of these mice undergo apoptotic cell death; by 12 weeks after birth most PRs have been lost from the Mertk−/− retina (Duncan et al., 2003a). This death is cell non-autonomous, in that it reflects the loss of Mer specifically from RPE cells (Duncan et al., 2003b; Vollrath et al., 2001), which fail to phagocytose PR outer segments. Consistent with these findings in Mertk−/−mice, the PR degeneration seen in the RCS rat, a decades-old model of human retinitis pigmentosa (Bourne et al., 1938; Edwards and Szamier, 1977), has been found to be due to a loss-of-function deletion within the rat Mertk gene (D’Cruz et al., 2000). Most dramatically, in humans, more than a dozen distinct pathogenic sequence variants in the Mertk gene have now been shown to result in inherited retinitis pigmentosa and related retinal dystrophies (Gal et al., 2000; Li et al., 2011; Mackay et al., 2010; Ostergaard et al., 2011).
These findings notwithstanding, the ligand or ligands that normally activate Mer and trigger phagocytosis by RPE cells have yet to be defined in vivo. Of the two closely-related proteins known to activate TAM receptors in various cells in culture, Gas6 was originally thought, based on in vitro experiments, to be required for RPE phagocytosis (Hall et al., 2001). However, the retinae of Gas6−/− mouse knockouts were subsequently found to have normal numbers of PRs throughout life (Prasad et al., 2006). More critically for the field in general, the relative contribution of Gas6 and Protein S (gene name Pros1) to TAM activation has never been assessed genetically in any context in vivo, and there remains considerable confusion, debate, and uncertainty as to which ligand may or may not contribute to various TAM actions in vivo (Caberoy et al., 2010; Godowski et al., 1995; Lemke and Rothlin, 2008; Morizono et al., 2011; Stitt et al., 1995).
We have now addressed this issue genetically, in RPE cells of the retina. In these cells the TAM receptor composition is known, and the Mertk−/− mutant phenotype is reproducible with respect to severity and age of onset. We have analyzed both conventional Gas6 and Pros1 mutants, as well as conditional (’floxed’) Pros1fl/fl alleles crossed with either Nestin- or Trp1-Cre drivers in multiple combinations, and have quantitated photoreceptor cell death in all genotypes at 12 weeks of age, a time at which the Mertk−/− degeneration phenotype is fully developed. We find that the number of PRs is equivalent to wild-type in complete retinal knock-outs of either Gas6 or Protein S. However, retinal removal of both ligands fully reproduces the PR death seen in Mertk−/− mice. These results demonstrate unequivocally that both Gas6 and Protein S function as Mer ligands in vivo, and that these ligands are, to a first approximation, independent and interchangeable for Mer-expressing RPE cells of the retina.
RESULTS
Measurement of PR degeneration in TAM receptor and ligand mutants
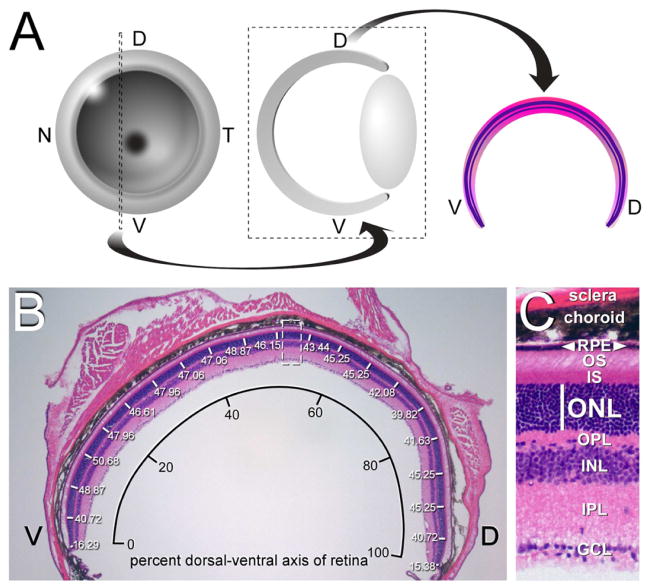
We quantitated PR death by measuring the thickness of the outer nuclear layer (ONL) of the retina, which is composed exclusively of PR nuclei, at 12 weeks after birth. As schematized in Figure 1, we performed all of these measurements on dorsal-ventral (DV) retinal sections taken from the same location – immediately nasal to the optic disk – from the left eye of all mice analyzed (Figure 1A). Sections were stained with hematoxylin and eosin (H&E), photographed, and ONL thickness was measured at 5% intervals across the full DV axis of each section (Figure 1B). Measurements were performed on sections taken from three different mice of a given genotype, and the results at each position averaged. The ONL is easily distinguished from the PR inner segments (IS) above, and the outer plexiform layer (OPL) of fibers below (Figure 1C).
Figure 1. Analysis of retinal degeneration in the retina of Gas6 and Protein S mutants.
(A) Retinal sections. Left eyes were sectioned at 12 weeks along the dorsal-ventral (DV) plane for all genotypes analyzed, at the indicated position just nasal (N) to the optic disk (left, dotted lines), oriented with dorsal to the right, and H&E stained (middle, right). (B) Measurement of the thickness of the outer nuclear layer (white lines, numbers in microns), which is composed of PR nuclei, across 20 segments (5% increments) of the DV axis of a wild-type retinal section. This DV axis is plotted as the x-axis of Figure 2A–C. A section equivalent to the dashed box at center is enlarged in C. (C) The thickness of the outer nuclear layer (ONL; white line) can be measured reliably. This thickness is plotted as the y-axis of Figure 2A–C. Other abbreviations: GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; IS, inner segment; OS, outer segment; RPE, retinal pigment epithelium.
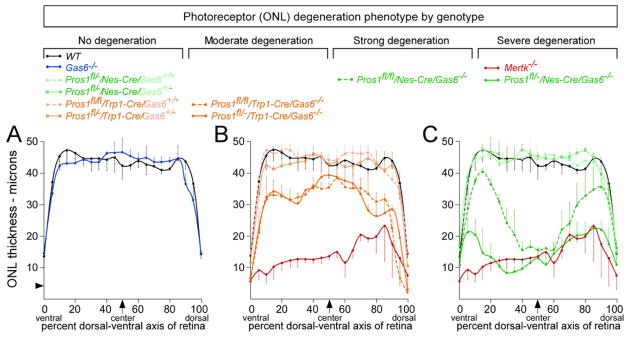
We plotted the data from these measurements as displayed in Figure 2, where the x-axis of the plot is relative position of the ONL expressed as percent of the retinal DV axis (ventral = 0, dorsal = 100%) and the y-axis is ONL thickness in microns (μm), both measured as in Figure 1B. These plots provide a complete description of the PR degeneration phenotype across the entire retina. This proved to be an important consideration, since for some of the genotypes described below there is significant phenotypic variation across the DV axis. In wild-type mice, we found that the thickness of the ONL is essentially constant from 10–90% of the DV axis, ranging from 42 to 47 μm. The number of PR nuclei normally decreases, to an ONL thickness of ~14 μm, at both the dorsal and ventral extremes of the retina (Figure 2A, black curve). There is minimal variation between wild-type individuals within these ranges (Figure 2A).
Figure 2. Relative roles of Protein S and Gas6 with respect to photoreceptor (PR) viability at 12 weeks.
(A) Measurement of ONL thickness was performed on stained sections as illustrated in Figure 1B, and average values in microns plotted across the DV axis of the retina. ONL thickness across the Gas6−/− retina (blue curve) is equivalent to wild-type (black curve). Arrowhead on y-axis represents the approximate thickness of a single PR nucleus. Error bars in this and all subsequent plots represent 1 S.D. from the mean, for independent measurements at the equivalent position in retinal sections from 3 separate mice. (B) Mertk−/− mice (red curve) display a severe loss of PRs, with only 2–4 PR nuclei present in the ONL across the DV axis of the retina. Retinal sections from mice that have Protein S eliminated from cells of the RPE (Pros1fl/fl/Trp1-Cre or Pros1fl/−/Trp1-Cre) and are also either Gas6+/+ or Gas6+/− (light orange curves) have a normal distribution of ONL thickness across the retina. However, sections from mice in which Gas6 is completely knocked out and Protein S is also eliminated from the RPE display an intermediate phenotype (dark orange solid and dashed curves), with partial PR loss relative to wild-type (black curve). As highlighted in Supplemental Figure S1C, D, the reduction in ONL thickness is especially severe in these Pros1fl/fl/Trp1-Cre/Gas6−/− and Pros1fl/−/Trp1-Cre/Gas6−/− mice at the extreme ventral and dorsal ends of the retina. (C) Pros1fl/−/Nes-Cre/Gas6−/− mice (dark green solid curve) display the same severe reduction in ONL thickness seen in Mertk−/− mice. Pros1fl/fl/Nes-Cre/Gas6−/− mice (dark green dashed curve) show an equivalent PR loss only in the central-most 30% of the retina, but this restriction in phenotype is due to reduced recombination activity of the Nestin-Cre driver in peripheral retinal locations (see Supplemental Figure S2). Adding a single wild-type Gas6 allele back to the Pros1fl/−/Nes-Cre genotype restores the distribution of ONL thickness across the retina to wild-type (light green curves). Genotypes are ordered with respect to phenotype severity above the plots.
Removal of retinal Gas6 or Protein S alone does not result in photoreceptor death at 12 weeks
The distribution of ONL thickness across the DV axis of the Gas6−/− retina (Angelillo-Scherrer et al., 2001) is statistically indistinguishable from wild-type at 12 weeks (Figure 2A, blue curve). Consistent with our earlier observations (Prasad et al., 2006), these results indicate that, in a normal Pros1 background, Gas6 is dispensable with respect to the maintenance of a normal number of PR nuclei. (See below for an outer segment length phenotype in Gas6−/− mice.) In marked contrast, loss of the Mer receptor leads to massive PR degeneration in the retina at this same time (D’Cruz et al., 2000; Duncan et al., 2003a; Prasad et al., 2006), such that the Mertk−/− ONL is only 2–3 nuclei thick (10–12μm) across most of its DV extent (Figure 2B, red curve). In general, we observed that the Mertk−/− PR degeneration phenotype is, for unknown reasons, less severe and more variable in the mid-dorsal aspect of the retina (70–90% of the DV axis, Figure 2B, C).
Given that the absence of Mer leads to profound PR death whereas the absence of its ligand Gas6 has no effect on PR number, we examined the effects of retinal deletion of the other identified TAM ligand – Protein S. Complete Pros1 mouse knock-outs cannot be studied, since they yield a lethal phenotype during late embryogenesis due to fulminant blood clotting and concomitant hemorrhage (Burstyn-Cohen et al., 2009). We therefore analyzed both Pros1+/− heterozygotes and conditional Pros1 ‘floxed’ alleles (Burstyn-Cohen et al., 2009) crossed with two different Cre driver lines. We first assessed conditional Pros1 mice crossed with the Trp1-Cre line, which drives recombination between floxed sites in cells of the RPE and the contiguous pigmented epithelia of the ciliary body (CB) (Mori et al., 2002). These studies were motivated by earlier observations that RPE cells express Protein S (Hall et al., 2005; Prasad et al., 2006). Mice that are Pros1fl/fl/Trp1-Cre (both Pros1 alleles floxed) or Pros1fl/−/Trp1-Cre (one Pros1 allele floxed, the other allele completely inactivated) – but that are either wild-type or heterozygous for Gas6 knock-out – have a normal ONL that is indistinguishable from wild-type (Figure 2B, light orange curves). Removing Protein S alone from the RPE and CB has no effect on the number of PRs.
Concerted removal of both Gas6 and Protein S results in severe photoreceptor degeneration
However, when these alleles are crossed with a Gas6 knock-out to generate Pros1fl/fl/Trp1-Cre/Gas6−/− and Pros1fl/−/Trp1-Cre/Gas6−/− mice, a significant (30–35%) reduction in the thickness of the ONL is seen across most of the retina in both compound mutants (Figure 2B, dark orange curves). [Trp1-Cre is an especially effective Cre driver in the RPE (Kim et al., 2008; Mori et al., 2002).] This reduction is notably more severe at the extreme edges of the retina, where essentially all PR nuclei are eliminated (Figure 2B, dark orange curves; see also Figs. S1C, D). At these extremes, the RPE abuts the contiguous ciliary body, which is also pigmented and also Trp1-positive. Thus, Protein S does indeed function as a ligand for the Mer receptor expressed by RPE cells, and a fraction of this Protein S is produced by the RPE and CB.
These effects notwithstanding, the PR loss seen in the Pros1fl/−/Trp1-Cre/Gas6−/− and Pros1fl/fl/Trp1-Cre/Gas6−/− mice is still less severe than that of the Mertk−/− mice (Figure 2B). We therefore used a second, nervous-system-restricted Cre driver, Nestin-Cre (Tronche et al., 1999), which should recombine floxed Pros1 alleles in all cells of the retina, including the RPE and CB. We again crossed this driver with both Pros1fl/fl and Pros1fl/− mice, which were simultaneously either Gas6+/+, Gas6+/−, or Gas6−/−. Most dramatically, retinae from Pros1fl/−/Nes-Cre/Gas6−/− mice, in which retinal expression of both ligands is eliminated, display a severe loss of ONL nuclei that is statistically identical to the PR death seen in the Mertk−/− mutants (Figure 2C, solid dark green curve). Adding a single wild-type Gas6 allele back to this genotype - to generate Pros1fl/−/Nes-Cre/Gas6+/− mice - completely restores the ONL to a wild-type thickness (Figure 2C, solid light green curve, outlined data points). Thus, a retina with no neural Protein S and no Gas6 displays the same severe PR loss and retinal degeneration seen in a retina with no Mer; but a retina with no neural Protein S and only half the normal level of Gas6 has a normal number of PRs (Figure 2C). This is also the case for a retina of the reciprocal genotype - Pros1fl/−/Gas6−/− - which has no Gas6 and only half the normal level of Protein S: this retina also has an ONL of normal thickness that extends all the way to its ends (Figure S1G, H). In summary, only half the normal retinal level of either ligand – in the complete absence of the other – is sufficient to maintain a normal number of PRs in the 12-week mouse retina.
There is no difference in PR number across the retina between Pros1fl/−/Nes-Cre/Gas6+/− mice and Pros1fl/−/Nes-Cre/Gas6+/+ mice, both of which display a wild-type profile (Figure 2C, light green curves). In contrast, Pros1fl/fl/Nes-Cre/Gas6−/− mice display PR degeneration that is comparable to the Mertk−/− and Pros1fl/−/Nes-Cre/Gas6−/− mice, but only in the center of the retina – from ~35–65% of the retinal DV axis (Figure 2C, dark green dashed curve). At more peripheral positions – both ventral and dorsal from the center – PR degeneration becomes progressively less pronounced in these Pros1fl/fl/Nes-Cre/Gas6−/− mice. This effect is due to incomplete recombination of the floxed Pros1 allele under the influence of the Nestin-Cre driver at peripheral retinal locations. When the 1Kln/J Nes-Cre driver was crossed with Rosa26 nuclear lacZ or tdTomato red reporter lines, we observed strong reporter expression only in the central retina, with diminished expression at more peripheral locations and almost no expression at the extreme ends of the retina adjacent to the ciliary body (Figure S2).
Retinal histology across genotypes
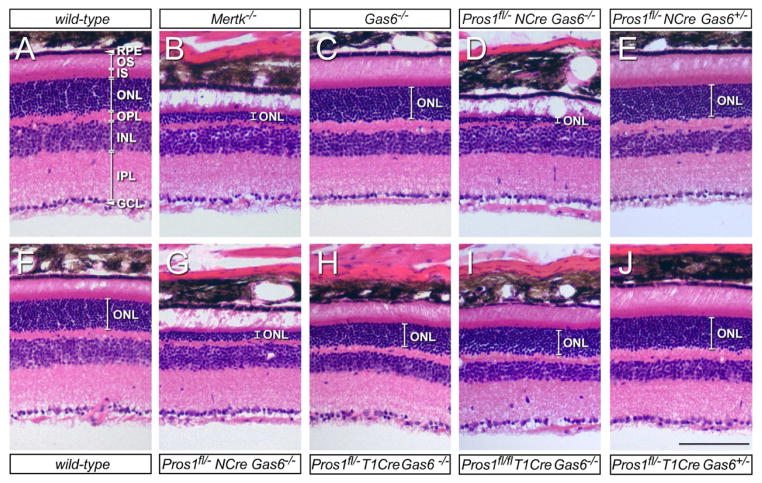
Illustrative H&E-stained retinal sections from informative genotypes are presented in Figure 3. Each of these representative images is taken from 30% of the DV axis of the retina. While the wild-type ONL has an average thickness of ~45μm, consisting of 12–15 compact, darkly-staining PR nuclei (Figure 3A, F are examples from different mice), the ONL of Mertk−/− retinae are only 2–4 nuclei thick (Figure 3B), and outer segments (OS) are almost entirely eliminated (white expanse above ONL in Figure 3B). In comparison, the Gas6−/− retina has a normal ONL thickness, and dense, well-elaborated outer segments (Figure 3C; see below). Pros1fl/−/Nes-Cre/Gas6−/− mice (Figs. 3D and G are representative examples from two different animals) display the same severe ONL depletion as that seen in the Mertk−/− mice (Figure 3B), with few surviving PR nuclei and an almost complete obliteration of the OS layer. Very dramatically, adding back just one allele of Gas6 to these badly damaged retinae restores the ONL to a normal configuration at 12 weeks (Figure 3E). Removing Protein S from RPE cells with the Trp1-Cre driver, combined with complete elimination of Gas6, yields an intermediate ONL depletion phenotype, with partial PR loss and a thinning of the OS layer (Figs. 3H, I). This phenotype is again restored to normal by the provision of just a single wild-type Gas6 allele (Figure 3J).
Figure 3. Retinal histology of mutant phenotypes.
All images are H&E stained sections from 30% of the retinal DV axis, as defined in Figs. 1 and 2. (A, F) Wild-type. Abbreviations are as for Figure 1C. (B) Mertk−/−. ONL is reduced to a thickness of only 2–4 PR nuclei, and PR OS are almost entirely absent. RPE and other retinal laminae are intact. (C) Gas6−/−. ONL is of normal thickness, and OS are longer and more densely packed than wild-type. See also Figure 4. (D, G) Pros1fl/−/Nes-Cre/Gas6−/−, in which both Protein S and Gas6 are eliminated from the retina. ONL is nearly obliterated, as in Mertk−/− mice (compare with B). (E) Pros1fl/−/Nes-Cre/Gas6+/−. Adding a single wild-type Gas6 allele to the genotype in (D, G) completely restores the ONL. OS are longer than wild-type; see also Figure 4. (H, I) Pros1fl/−/Trp1-Cre/Gas6−/− and Pros1fl/fl/Trp1-Cre/Gas6−/−, respectively, in which Gas6 is eliminated from the entire retina and Protein S from the RPE. ONL thickness is reduced by ~30% relative to wild-type. (See also Figure 2B.) (J) Adding a single wild-type Gas6 allele to the genotype in (H) completely restores the ONL. OS are again longer than wild-type; see also Figure 4. Measurements across the complete DV axis of multiple mice indicate that a statistically significant diminution in INL thickness in Gas6−/− mutants relative to wild-type is not a consistently observed phenotype. Scale bar is 100 μm.
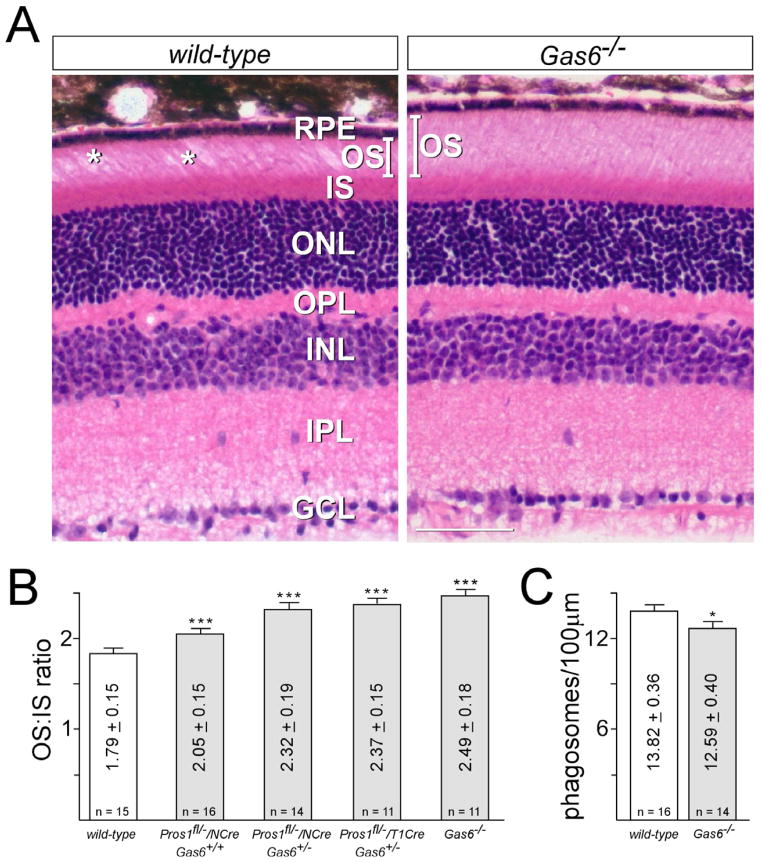
Although retinae in which only one TAM ligand gene is inactivated display a wild-type ONL phenotype (Figure 2A–C), careful comparison of outer segment histology revealed subtle but significant differences between these mutants and wild-type mice. The OS layer of the Gas6−/− mice, for example, is actually fuller (denser) and longer than wild-type (a representative comparison is shown in Figure 4A). We measured the average outer-to-inner segment length (OS:IS) ratio at the center of the wild-type retina at 1.79 ± 0.15, whereas the same ratio in the Gas6 knock-outs was 2.49 ± 0.18 (Figure 4B). [This measurement stands in contrast to an earlier anecdotal report (Hall et al., 2005).] This increase is due entirely to an increase in OS length in Gas6−/− individuals (Figure 4A; compare also Figure 3C to Figs. 3A/F). Similarly, while removing all of the Protein S from the retina in a Gas6+/+ background has no effect on ONL thickness in the central retina at 12 weeks (Figure 2), Pros1fl/−/Nes-Cre/Gas6+/+ mice also display an increase in their OS:IS length ratio, albeit a more modest one, to 2.05 ± 0.15 (Figure 4B). Inactivating one Gas6 allele in these mice (in Pros1fl/−/Nes-Cre/Gas6+/− individuals) increases this ratio to 2.32 ± 0.19 (Figure 4B; compare also OS length in Figs. 3E versus 3A/F). Finally, a Pros1fl/−/Trp1-Cre/Gas6+/− retina also displays an obviously greater OS:IS ratio (Figure 4B; compare also OS in Figure 3J to 3A/F).
Figure 4. Outer segments (OS) in TAM ligand single mutants are longer than those of wild-type.
(a) A representative side-by-side comparison of central retinal sections, stained with H&E, taken from wild-type (left) versus Gas6−/− (right) mice at 12 weeks. The sections are entirely comparable, except for (i) a longer average OS length in the Gas6−/− mice (bars in left and right panels, IS length is unchanged); and (ii) a greater frequency of gaps in the OS layer (asterisks in left panel) in wild-type section. Scale bar is 50 μm. (B) Quantitation of the central retinal ratio of the length of the outer segment to the length of the inner segment (OS:IS ratio) performed on two retinal sections from two separate mice of each indicated genotype. (n is the total number of measurements performed.) Variation is 1 standard deviation from the mean. *** indicates highly significant difference (p value <0.0001) between the indicated genotypes and wild-type upon analysis by two-tailed t test. (C) Quantitation of opsin-positive phagosomes, as described in Experimental procedures, per 100μm length of RPE in sections of WT (white bar) versus Gas6−/− (gray bar) retinae, at 30 min after subjective dawn. (n is the total number of sections counted, from two eyes per genotype). Variation is 1 standard error of the mean. * indicates significant difference (p value =0.03) between Gas6−/− and wild-type upon analysis by two-tailed t test.
These increases in steady-state OS length are consistent with a modestly diminished rate of OS phagocytosis by RPE cells, which is seen in any genotype in which at least all of one ligand is eliminated, and are most pronounced in Gas6−/− mice. These effects are too small to result in any PR death or change in ONL thickness in any genotype (Figs. 2A, 3C, 3E, 3J, and 4A), but are clearly apparent when OS histology is examined. A slightly diminished rate of phagocytosis by RPE cells, coupled with an unchanged rate of new basal membrane insertion by PRs, would establish a new set point for the balance between synthesis and phagocytosis, and would result in the observed increases in OS length in Pros1fl/−/Nes-Cre/Gas6+/+, Pros1fl/−/Nes-Cre/Gas6+/−, Pros1fl/−/Trp1-Cre/Gas6+/−, and Gas6−/− mutants (Figure 4B). To examine this possibility directly, we stained wild-type and Gas6−/− retinal sections, obtained at 30 minutes after subjective dawn (when the rate of RPE phagocytosis of OS is high in the mouse), with anti-opsin antibodies, and counted phagosome vesicles within RPE cells, as described previously (Nandrot et al., 2007). We measured 13.82 ± 0.36 phagosomes/100μm of RPE length in wild-type mice, and 12.59 ± 0.40/100μm in Gas6−/− mutants (Figure 4C). Although the OS of Gas6−/− PRs are longer than wild-type, the morphology of these mutant OS, as examined by transmission electron microscopy at the RPE-OS interface, is indistinguishable from wild-type (Figure S3).
TAM receptor and ligand expression in the eye
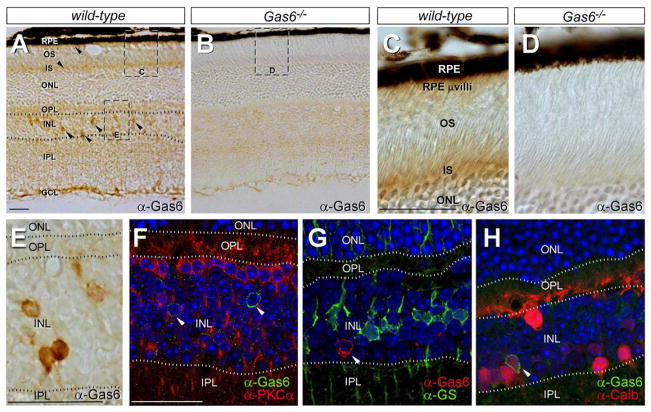
Protein S and Gas6 protein and/or mRNA have been detected previously - by northern blot, western blot, RT-PCR, or in situ hybridization - in RPE cells, and also in the neural retina proper (Hall et al., 2005; Kociok and Joussen, 2007; Prasad et al., 2006). We used immunohistochemistry (IHC) with Gas6 antibodies to localize Gas6 expression more precisely on retinal sections. Gas6 was detected in the inner segments of PRs (Figure 5A–D), and in a region occupied by the apical microvilli of RPE cells (Figure 5C, D; see also Gas6 mRNA expression in isolated RPE cells in Figure 6 below). Given (a) the intimate association of PR OS and RPE cells, and (b) the fact that TAM ligands bridge a TAM-receptor-positive phagocyte to the membrane of its engulfment target (Lemke and Rothlin, 2008), PRs and RPE cells may be major sources of the Gas6 that is delivered to the Mer receptor expressed on the RPE apical microvilli (Prasad et al., 2006). In addition to these cell types, we detected Gas6 in a subset of cells located in the inner nuclear layer (Figure 5A, E). We co-stained sections with antibodies to Gas6 and PKCα (Figure 5F), glutamine synthetase (Figure 5G), parvalbumin (not shown), and calbindin (Figure 5H), which serve as markers for rod bipolar cells, Müller glia (MG), and amacrine cell subsets and horizontal cells (parvalbumin/calbindin), respectively (Haverkamp and Wassle, 2000). We detected co-expression of Gas6 only in a subset of calbindin-positive cells (Figure 5H). Although available Protein S antibodies function well on western blots, none of a panel of four different antibodies we tested, both polyclonal and monoclonal (see Experimental procedures), yielded IHC signals that were not also present when we analyzed Protein S-deficient retinal sections.
Figure 5. Gas6 expression in the retina.
(A, B) Anti-Gas6 IHC with HRP-conjugated secondary antibodies across all retinal layers (labels as for Figure 1C) in wild-type (A) and Gas6−/− (negative control) mice (B). Arrowheads in A denote Gas6-positive cells (brown signal) in the inner nuclear layer (INL), the cytoplasm-rich inner segments (IS) of PRs, and the apical microvilli of RPE cells, which penetrate into the PR OS layer. [These same apical microvilli are positive for both Mer and Tyro 3 (Prasad et al., 2006).] Images equivalent to (but distinct from) the indicated boxed areas are enlarged in panels C, D, and E. (C, D) Anti-Gas6 IHC in PR IS layer and region populated by RPE microvilli in wild-type (C) and Gas6−/− (D) mice. (E) High power view of anti-Gas6 IHC in the INL, equivalent to the box indicated in A. The positions of retinal laminae are indicated by the dotted lines. (F) Co-staining with anti-Gas6 (green) and anti-PKCα (red); nuclei are counter-stained with a Hoechst dye (blue). (G) Co-staining with anti-Gas6 (red) and anti-glutamine synthetase (GS; green); nuclei are blue. (H) Co-staining with anti-Gas6 (green) and anti-calbindin (Calb; red); nuclei are blue. Scale bars are 25 μm.
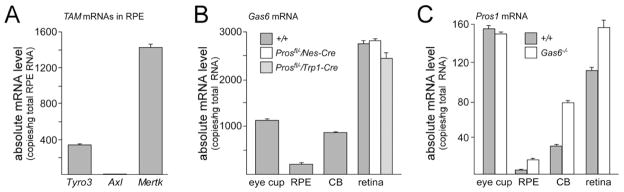
Figure 6. TAM receptor and ligand mRNA expression in the eye.
(A) TAM receptor mRNAs in the RPE. The RPE cell layer was dissected from the retina, and the absolute level of Tyro3, Axl, and Mertk mRNAs determined by qRT-PCR (see Experimental Procedures). Mertk mRNA is 4.2-fold more abundant than Tyro3 mRNA; Axl mRNA is not detectable. (B, C) The eye cup (primarily choroid and sclera remaining after removal of neural retina and RPE), the single-cell RPE layer, the pigmented ciliary body at the retinal periphery that is contiguous with the RPE (CB), and the neural retina were each dissected from the retina, and the absolute level of Gas6 (B) and Pros1 (C) mRNAs determined by qRT-PCR. Gas6 mRNAs were also measured in retina obtained from Pros1fl/−/Nes-Cre and Pros1fl/−/Trp1-Cre mice (B, white and gray-striped bars, respectively); and Pros1 mRNAs were measured in all eye tissues obtained from Gas6−/− mice (C, white bars). Both TAM ligand mRNAs are expressed broadly in ocular tissues.
We also measured TAM receptor and ligand mRNAs in the retina and eye using quantitative RT-PCR with mRNA prepared from multiple isolated tissues - RPE, choroid, eye cup (tissue remaining after removal of neural retina and RPE), ciliary body (CB), and neural retina (minus RPE; see Experimental Procedures) (Figure 6). Although the apical microvilli of RPE cells express both Mer and Tyro3 (Prasad et al., 2006), Mertk−/− single gene mutants yield a strong PR degeneration phenotype (Figs. 2 and 3), whereas Tyro3−/− mutants have a normally configured ONL (Prasad et al., 2006). To assess the relative expression level of the two TAM receptor genes in these cells, we measured the relative expression of Mertk and Tyro3 mRNAs in isolated RPE. As before (Prasad et al., 2006), we used a collagenase/hyaluronidase dissociation protocol to cleanly peel off the RPE layer from adult 129/C57Bl/6 retinae, and prepared mRNA from this purified RPE layer. Quantitative RT-PCR revealed that RPE cells express slightly more than four times as much Mertk mRNA as Tyro3 mRNA, and no detectable Axl mRNA (Figure 6A). In these same qRT-PCR experiments, we measured Gas6 and Pros1 mRNA levels. Gas6 (Figure 6B) and Pros1 (Figure 6C) mRNAs were detected in all ocular tissues, with measured Gas6 mRNA levels being approximately an order of magnitude higher than those for Pros1 mRNA throughout the eye. We observed a modest (~2.5-fold) up-regulation of Pros1 mRNA expression in the CB of Gas6−/− mutants, with no substantial change in this mRNA in other Gas6−/− ocular tissues (Figure 6C). Similarly, there was no substantial change in Gas6 mRNA levels measured in the retina of either Pros1fl/−/Nes-Cre or Pros1fl/−/Trp1-Cre mice (Figure 6B). The complete degeneration phenotype evident in the central retina of the Pros1fl/fl/Nes-Cre/Gas6−/− mice suggests that blood-derived Protein S is unlikely to be a major reservoir of ligand for the Mer receptor of the RPE.
DISCUSSION
The TAM RTKs play key roles in immune regulation, the phagocytosis of apoptotic cells (ACs) and membranes, the facilitation of viral infection, and the progression of cancer (Lemke and Burstyn-Cohen, 2010; Lemke and Rothlin, 2008; Meertens, 2012; Morizono et al., 2011; Verma et al., 2011). However, the relative importance and contribution of the ligands that activate these receptors has yet to be assessed genetically in any setting in vivo. Although there have been previous biochemical studies that document Protein S and/or Gas6 binding to or activation of Tyro3, Axl, or Mer (Lemke and Rothlin, 2008), most of these in vitro studies were performed with cultured cells or membranes in which endogenous TAM receptor and ligand expression were unknown. There is therefore considerable confusion and uncertainty in the literature as to whether Gas6 or Protein S - or additional proteins, such as those of the Tubby family (Caberoy et al., 2010) - might function differentially as genuine TAM ligands in vivo. The possibility that TAM receptors might act independently of; that is, without a requirement for, their proposed ligands has also been advanced (Ruan and Kazlauskas, 2012). Our study is the first genetic analysis to address these fundamental questions.
We draw five conclusions from our findings. First, Protein S and Gas6 both function as bona fide TAM receptor ligands in vivo in the mouse. Second, either ligand is sufficient to activate Mer and trigger Mer-dependent RPE cell phagocytosis of PR outer segments in the mouse retina. Third, for this process, the two ligands are to a first approximation interchangeable. Fourth, although it may exist, there is no absolute signaling requirement for the formation of a Gas6-Protein S heterodimer. Finally, loss of both ligands from the retina phenocopies the loss of Mer, which (a) obviates an absolute requirement for a TAM ligand in addition to Gas6 and Protein S, and (b) also argues against the possibility that Mer function in the eye is TAM-ligand independent.
We emphasize that our conclusions on TAM receptor-ligand pairing apply to RPE cells in the retina, and that other cells display different TAM receptor and ligand expression profiles. Macrophages and dendritic cells (DCs) of the immune system, for example, express Axl and Mer, but little or no Tyro3 (Rothlin et al., 2007), while pyramidal neurons of the brain express high levels of Tyro3 but essentially no Axl or Mer (Prieto et al., 2007). Similarly, the diverse populations of TAM-positive cells in the body may be selectively exposed to either Gas6 or Protein S in vivo. In addition to TAM receptor composition, the expression of TAM interacting receptors may also vary between cells, and this may in turn affect the ability of Gas6 or Protein S to activate TAM signaling. RPE cells, for example, express the αvβ5 integrin, which cooperates with Mer and also plays a role in the circadian RPE phagocytosis of OS membranes (Finnemann and Nandrot, 2006; Nandrot et al., 2008).
These points notwithstanding, our results provide a definitive demonstration that Protein S functions as a Mer ligand in the mouse, where it stimulates RPE phagocytosis of OS membranes. Protein S also potently stimulates the phagocytosis of ACs by cultured human macrophages (Anderson et al., 2003; McColl et al., 2009), and Mer is again a key receptor for this process (McColl et al., 2009; Scott et al., 2001). If Mer-expressing macrophages and DCs transit through the blood, they are exposed to the high levels of Protein S (~300 nM) that are present in the circulation (Burstyn-Cohen et al., 2009; Dahlback, 2000). [In contrast, Gas6 is expressed at very low levels (<0.2 nM) in the blood (Ekman et al., 2010).] In tissues, these same cells may be exposed to Protein S produced by activated T cells (Smiley et al., 1997). Defects in the phagocytic clearance of ACs from lymphoid tissues are strongly linked to the development of human autoimmune disorders (Gaipl et al., 2007; Nagata et al., 2010); and Protein S deficiency is associated with the development of both systemic lupus and inflammatory bowel disease (Alkim et al., 2011; Suh et al., 2010). Together, these observations suggest that, in select settings, Protein S may be the TAM ligand of greatest biological significance.
EXPERIMENTAL PROCEDURES
Mice
The Tyro3−/−, Axl−/−, and Mertk−/− mutants (Lu et al., 1999), the Pros1fl/fl conditional and Pros1−/− mutants (Burstyn-Cohen et al., 2009), the Gas6−/− mutants (Angelillo-Scherrer et al., 2001), the Trp1-Cre driver line (Mori et al., 2002), and the Nestin-Cre driver line (Tronche et al., 1999) have all been described previously.
Antibodies
We used the following primary antibodies for the analyses of Figure 5: anti-Gas6 (AF986; R&D Systems); anti-PKCalpha (1608-1; Epitomics); anti-Calbindin D28K (CB-38a; Swant); anti-Glutamine Synthetase (G-2781; Sigma). We also tested the following Protein S antibodies for IHC: anti-protein S (AB15928; Millipore); anti-protein S (sc-25836; Santa Cruz); anti-protein S (AF4036; R&D Systems) and anti-protein S (P5180; Sigma). We used anti-opsin (MAB5356; Millipore) for the phagosome counts of Figure 4C.
Quantitative RT-PCR
For quantitative PCR studies, RPE cells were isolated as previously described (Prasad et al. 2006). RNA was prepared using Qiagen RNeasy kits (Qiagen). Reverse transcription was carried out using Superscript III Reverse transcriptase (Invitrogen), and PCR reactions were carried out on an ABI Prism 7000 Sequence Detection System using Sybr Green Assay (Applied Biosystems). Data was analyzed using SDS 2.0 software. Calibration curves were generated using plasmid DNA ranging from 10–108 copies and used for absolute quantitation of respective mRNAs in eye samples with primers listed in Table S1.
Tissue dissections and histology
12-week-old mice were anesthetized with 2.5% avertin/saline solution delivered intraperitoneally. For immunohistochemistry and light microscopy studies, mice were subsequently perfused with a 20U/mL heparin/PBS solution and followed by a 4% paraformaldehyde/PBS solution. The eyes were marked nasally and removed. The cornea, lens, and iris epithelium were removed, and the eyes were then immersion fixed overnight in 4% paraformaldehyde/PBS at 4°C. After fixation, the eyes were infiltrated with 30% sucrose/PBS at 4°C overnight, then frozen in tissue freezing medium. Eyes were cut into 10μm thick sections in a nasal to temporal orientation. Sections were air dried overnight at room temperature before freezing at −70°C, or were used immediately.
Before immunostaining, heat-induced epitope retrieval was applied and sections were immersed in 0.1M citrate buffer (pH 6.00) prewarmed (95–100°C) and then boiled in a microwave for 3 min. Sections were allowed to cool in solution. Slides were rinsed in distilled water twice, washed twice in PBS, and then incubated for 30 min in 0.5% H2O2 solution in PBS. After washing twice in PBS-Tween 0.1%, sections were incubated (O/N; 4°C) with primary antibodies diluted in a fish gelatin blocking solution (PBS1x, pH 7.4, 0.5% Tween, 10% glycerol (v/v), 18% D(+)-Glucose (w/v), 4.5% fish skin gelatin (G-7765; Sigma)). DAB staining was performed using a Vectastain ABC kit (Vector Labs) and Peroxidase substrate DAB kit (Vector Labs), following the supplier’s instructions. Sections were mounted using VectaMount (Vector Labs). Immunofluorescence was performed using the appropriate conjugated secondary antibodies (Jackson ImmunoResearch). Sections were mounted using Fluoromont-G (SouthernBiotech). Colocalization analyses were performed using a LSM 780 confocal microscope (Zeiss) with Zen 2011 software. Electron microscopy of retinal sections was performed as described previously (Prasad et al., 2006)
Phagosome counts were performed as described previously (Nandrot et al., 2007), using 8μm fixed retinal sections stained with an anti-opsin antibody (see above). Sections were prepared from mice sacrificed and perfused at 6:30 a.m., 30 min after lights-on in our animal facility. Opsin-positive vesicles contained within the RPE layer (visualized at 80×) were scored for entire retinal sections, and the observer was blind to the genotype of the section. The length of the single-cell RPE layer in each section was measured using ImageJ, and the results expressed as phagosomes per 100μm RPE length.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 AI077058 to G.L.), the European Union (Marie Curie grant IRG-256319 to T. B.-C.), and the Israel Science Foundation (grant 984/12 to T. B.-C.), by the Salk Institute, and by postdoctoral fellowships from the Leukemia and Lymphoma Society (to E.D.L.), and the Fundación Ramón Areces (to P.G.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alkim H, Ayaz S, Alkim C, Ulker A, Sahin B. Continuous active state of coagulation system in patients with nonthrombotic inflammatory bowel disease. Clin Appl Thromb Hemost. 2011;17:600–604. doi: 10.1177/1076029611405034. [DOI] [PubMed] [Google Scholar]
- Anderson HA, Maylock CA, Williams JA, Paweletz CP, Shu H, Shacter E. Serum-derived protein S binds to phosphatidylserine and stimulates the phagocytosis of apoptotic cells. Nat Immunol. 2003;4:87–91. doi: 10.1038/ni871. [DOI] [PubMed] [Google Scholar]
- Angelillo-Scherrer A, de Frutos P, Aparicio C, Melis E, Savi P, Lupu F, Arnout J, Dewerchin M, Hoylaerts M, Herbert J, et al. Deficiency or inhibition of Gas6 causes platelet dysfunction and protects mice against thrombosis. Nat Med. 2001;7:215–221. doi: 10.1038/84667. [DOI] [PubMed] [Google Scholar]
- Bourne MC, Campbell DA, Tansley K. Hereditary Degeneration of the Rat Retina. Br J Ophthalmol. 1938;22:613–623. doi: 10.1136/bjo.22.10.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstyn-Cohen T, Heeb MJ, Lemke G. Lack of protein S in mice causes embryonic lethal coagulopathy and vascular dysgenesis. J Clin Invest. 2009;119:2942–2953. doi: 10.1172/JCI39325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberoy NB, Zhou Y, Li W. Tubby and tubby-like protein 1 are new MerTK ligands for phagocytosis. EMBO J. 2010;29:3898–3910. doi: 10.1038/emboj.2010.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Cruz PM, Yasumura D, Weir J, Matthes MT, Abderrahim H, LaVail MM, Vollrath D. Mutation of the receptor tyrosine kinase gene Mertk in the retinal dystrophic RCS rat. Hum Mol Genet. 2000;9:645–651. doi: 10.1093/hmg/9.4.645. [DOI] [PubMed] [Google Scholar]
- Dahlback B. Blood coagulation. Lancet. 2000;355:1627–1632. doi: 10.1016/S0140-6736(00)02225-X. [DOI] [PubMed] [Google Scholar]
- Duncan HJ, LaVail MM, Yasumura D, Matthes MT, Yang H, Trautmann N, Chappelow AV, Feng W, Earp HS, Matsushima GK, Vollrath D. An RCS-like retinal dystrophy phenotype in mer knockout mice. Invest Ophthalmol Visual Sci. 2003a;44:826–838. doi: 10.1167/iovs.02-0438. [DOI] [PubMed] [Google Scholar]
- Duncan JL, LaVail MM, Yasumura D, Matthes MT, Yang H, Trautmann N, Chappelow AV, Feng W, Earp HS, Matsushima GK, Vollrath D. An RCS-like retinal dystrophy phenotype in mer knockout mice. Invest Ophthalmol Vis Sci. 2003b;44:826–838. doi: 10.1167/iovs.02-0438. [DOI] [PubMed] [Google Scholar]
- Edwards RB, Szamier RB. Defective phagocytosis of isolated rod outer segments by RCS rat retinal pigment epithelium in culture. Science. 1977;197:1001–1003. doi: 10.1126/science.560718. [DOI] [PubMed] [Google Scholar]
- Ekman C, Stenhoff J, Dahlback B. Gas6 is complexed to the soluble tyrosine kinase receptor Axl in human blood. J Thromb Haemost. 2010;8:838–844. doi: 10.1111/j.1538-7836.2010.03752.x. [DOI] [PubMed] [Google Scholar]
- Finnemann SC, Nandrot EF. MerTK activation during RPE phagocytosis in vivo requires alphaVbeta5 integrin. Adv Exp Med Biol. 2006;572:499–503. doi: 10.1007/0-387-32442-9_69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaipl US, Munoz LE, Grossmayer G, Lauber K, Franz S, Sarter K, Voll RE, Winkler T, Kuhn A, Kalden J, et al. Clearance deficiency and systemic lupus erythematosus (SLE) J Autoimmun. 2007;28:114–121. doi: 10.1016/j.jaut.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Gal A, Li Y, Thompson DA, Weir J, Orth U, Jacobson SG, Apfelstedt-Sylla E, Vollrath D. Mutations in MERTK, the human orthologue of the RCS rat retinal dystrophy gene, cause retinitis pigmentosa. Nat Genet. 2000;26:270–271. doi: 10.1038/81555. [DOI] [PubMed] [Google Scholar]
- Godowski PJ, Mark MR, Chen J, Sadick MD, Raab H, Hammonds RG. Reevaluation of the roles of protein S and Gas6 as ligands for the receptor tyrosine kinase Rse/Tyro 3. Cell. 1995;82:355–358. doi: 10.1016/0092-8674(95)90424-7. [DOI] [PubMed] [Google Scholar]
- Hall MO, Obin MS, Heeb MJ, Burgess BL, Abrams TA. Both protein S and Gas6 stimulate outer segment phagocytosis by cultured rat retinal pigment epithelial cells. Exp Eye Res. 2005;81:581–591. doi: 10.1016/j.exer.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Hall MO, Prieto AL, Obin MS, Abrams TA, Burgess BL, Heeb MJ, Agnew BJ. Outer segment phagocytosis by cultured retinal pigment epithelial cells requires Gas6. Exp Eye Res. 2001;73:509–520. doi: 10.1006/exer.2001.1062. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, Wassle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology (Bethesda) 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Kang KH, Burrola P, Mak TW, Lemke G. Retinal degeneration triggered by inactivation of PTEN in the retinal pigment epithelium. Genes Dev. 2008;22:3147–3157. doi: 10.1101/gad.1700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kociok N, Joussen AM. Varied expression of functionally important genes of RPE and choroid in the macula and in the periphery of normal human eyes. Graefes Arch Clin Exp Ophthalmol. 2007;245:101–113. doi: 10.1007/s00417-006-0266-x. [DOI] [PubMed] [Google Scholar]
- Korshunov VA, Mohan AM, Georger MA, Berk BC. Axl, a receptor tyrosine kinase, mediates flow-induced vascular remodeling. Circ Res. 2006;98:1446–1452. doi: 10.1161/01.RES.0000223322.16149.9a. [DOI] [PubMed] [Google Scholar]
- Lai C, Lemke G. An extended family of protein-tyrosine kinase genes differentially expressed in the vertebrate nervous system. Neuron. 1991;6:691–704. doi: 10.1016/0896-6273(91)90167-x. [DOI] [PubMed] [Google Scholar]
- Lemke G, Burstyn-Cohen T. TAM receptors and the clearance of apoptotic cells. Ann N Y Acad Sci. 2010;1209:23–29. doi: 10.1111/j.1749-6632.2010.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke G, Rothlin CV. Immunobiology of the TAM receptors. Nat Rev Immunol. 2008;8:327–336. doi: 10.1038/nri2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xiao X, Li S, Jia X, Wang P, Guo X, Jiao X, Zhang Q, Hejtmancik JF. Detection of variants in 15 genes in 87 unrelated Chinese patients with Leber congenital amaurosis. PLoS One. 2011;6:e19458. doi: 10.1371/journal.pone.0019458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Gore M, Zhang Q, Camenisch T, Boast S, Casagranda F, Lai C, Skinner MK, Klein R, Matsushima GK, et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature. 1999;398:723–728. doi: 10.1038/19554. [DOI] [PubMed] [Google Scholar]
- Mackay DS, Henderson RH, Sergouniotis PI, Li Z, Moradi P, Holder GE, Waseem N, Bhattacharya SS, Aldahmesh MA, Alkuraya FS, et al. Novel mutations in MERTK associated with childhood onset rod-cone dystrophy. Mol Vis. 2010;16:369–377. [PMC free article] [PubMed] [Google Scholar]
- McColl A, Bournazos S, Franz S, Perretti M, Morgan BP, Haslett C, Dransfield I. Glucocorticoids induce protein S-dependent phagocytosis of apoptotic neutrophils by human macrophages. J Immunol. 2009;183:2167–2175. doi: 10.4049/jimmunol.0803503. [DOI] [PubMed] [Google Scholar]
- Meertens L, Carnec X, Perera Lecoin M, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. TIM and TAM receptors mediate dengue virus infection. Cell Host & Microbe. 2012 doi: 10.1016/j.chom.2012.08.009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Metzger D, Garnier JM, Chambon P, Mark M. Site-specific somatic mutagenesis in the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2002;43:1384–1388. [PubMed] [Google Scholar]
- Morizono K, Xie Y, Olafsen T, Lee B, Dasgupta A, Wu AM, Chen IS. The soluble serum protein Gas6 bridges virion envelope phosphatidylserine to the TAM receptor tyrosine kinase Axl to mediate viral entry. Cell Host Microbe. 2011;9:286–298. doi: 10.1016/j.chom.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Hanayama R, Kawane K. Autoimmunity and the clearance of dead cells. Cell. 2010;140:619–630. doi: 10.1016/j.cell.2010.02.014. [DOI] [PubMed] [Google Scholar]
- Nandrot EF, Anand M, Almeida D, Atabai K, Sheppard D, Finnemann SC. Essential role for MFG-E8 as ligand for alphavbeta5 integrin in diurnal retinal phagocytosis. Proc Natl Acad Sci U S A. 2007;104:12005–12010. doi: 10.1073/pnas.0704756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot EF, Chang Y, Finnemann SC. Alphavbeta5 integrin receptors at the apical surface of the RPE: one receptor, two functions. Adv Exp Med Biol. 2008;613:369–375. doi: 10.1007/978-0-387-74904-4_43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandrot EF, Dufour EM. Mertk in daily retinal phagocytosis: a history in the making. Adv Exp Med Biol. 2010;664:133–140. doi: 10.1007/978-1-4419-1399-9_16. [DOI] [PubMed] [Google Scholar]
- Ostergaard E, Duno M, Batbayli M, Vilhelmsen K, Rosenberg T. A novel MERTK deletion is a common founder mutation in the Faroe Islands and is responsible for a high proportion of retinitis pigmentosa cases. Mol Vis. 2011;17:1485–1492. [PMC free article] [PubMed] [Google Scholar]
- Prasad D, Rothlin CV, Burrola P, Burstyn-Cohen T, Lu Q, Garcia de Frutos P, Lemke G. TAM receptor function in the retinal pigment epithelium. Mol Cell Neurosci. 2006;33:96–108. doi: 10.1016/j.mcn.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Prieto AL, O’Dell S, Varnum B, Lai C. Localization and signaling of the receptor protein tyrosine kinase Tyro3 in cortical and hippocampal neurons. Neuroscience. 2007;150:319–334. doi: 10.1016/j.neuroscience.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G. TAM receptors are pleiotropic inhibitors of the innate immune response. Cell. 2007;131:1124–1136. doi: 10.1016/j.cell.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Ruan GX, Kazlauskas A. Axl is essential for VEGF-A-dependent activation of PI3K/Akt. EMBO J. 2012;31:1692–1703. doi: 10.1038/emboj.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RS, McMahon EJ, Pop SM, Reap EA, Caricchio R, Cohen PL, Earp HS, Matsushima GK. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 2001;411:207–211. doi: 10.1038/35075603. [DOI] [PubMed] [Google Scholar]
- Smiley ST, Boyer SN, Heeb MJ, Griffin JH, Grusby MJ. Protein S is inducible by interleukin 4 in T cells and inhibits lymphoid cell procoagulant activity. Proc Natl Acad Sci U S A. 1997;94:11484–11489. doi: 10.1073/pnas.94.21.11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stitt TN, Conn G, Gore M, Lai C, Bruno J, Radziejewski C, Mattsson K, Fisher J, Gies DR, Jones PF, et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell. 1995;80:661–670. doi: 10.1016/0092-8674(95)90520-0. [DOI] [PubMed] [Google Scholar]
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- Suh CH, Hilliard B, Li S, Merrill JT, Cohen PL. TAM receptor ligands in lupus: protein S but not Gas6 levels reflect disease activity in systemic lupus erythematosus. Arthritis Res Ther. 2010;12:R146. doi: 10.1186/ar3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Verma A, Warner SL, Vankayalapati H, Bearss DJ, Sharma S. Targeting axl and mer kinases in cancer. Mol Cancer Ther. 2011;10:1763–1773. doi: 10.1158/1535-7163.MCT-11-0116. [DOI] [PubMed] [Google Scholar]
- Vollrath D, Feng W, Duncan JL, Yasumura D, D’Cruz PM, Chappelow A, Matthes MT, Kay MA, LaVail MM. Correction of the retinal dystrophy phenotype of the RCS rat by viral gene transfer of Mertk. Proc Natl Acad Sci U S A. 2001;98:12584–12589. doi: 10.1073/pnas.221364198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.