SUMMARY
In the ubiquitin proteasome system, the E3 ligase SCF-Skp2 and its accessory protein Cks1 promote proliferation largely by inducing the degradation of the CDK inhibitor p27. Overexpression of Skp2 in human cancers correlates with poor prognosis, and deregulation of SCF-Skp2-Cks1 promotes tumorigenesis in animal models. We identified small molecule inhibitors specific to SCF-Skp2 activity using in silico screens targeted to the binding interface for p27. These compounds selectively inhibited Skp2-mediated p27 degradation by reducing p27 binding through key compound-receptor contacts. In cancer cells, the compounds induced p27 accumulation in a Skp2-dependent manner and promoted cell-type specific blocks in the G1 or G2/M phases. Designing SCF-Skp2 specific inhibitors may be a novel strategy to treat cancers dependent on the Skp2-p27 axis.
INTRODUCTION
The ubiquitin proteasome system (UPS) is essential for the turnover of almost all cellular proteins, maintaining homeostatic levels in normal cells while controlling levels of oncogenes and tumor suppressors in transformed cells. In an ATP-dependent process, ubiquitin is transferred from the ubiquitin-activating enzyme (E1) to the ubiquitin-conjugating enzyme (E2), and covalently attached via an isopeptide linkage to a target protein bound to an ubiquitin ligase (E3) (Ciechanover, 2005). Chains of 4 or more ubiquitin domains trigger degradation by the 26S proteasome.
FDA approval of the proteasome inhibitor Bortezomib (Velcade™, Millennium Pharmaceuticals Inc.) established the UPS as a validated target for treatment of multiple myeloma and mantle cell lymphoma (Bross, et al., 2004; Kane, et al., 2007). Yet advances in the clinical use of Bortezomib for solid tumors are lacking, resistance is developing, and peripheral neuropathy is a major side effect (Argyriou, et al., 2008; Orlowski and Kuhn, 2008). Recent investigations are now focused on inhibiting UPS targets upstream of the proteasome (Ceccarelli, et al., 2012; Orlicky, et al., 2010; Soucy, et al., 2009). Of particular interest are inhibitors specific to E3 ligases in the hope of reducing off-target effects (Sun, 2006).
The Skp1-Cullin1-F-box (SCF) family is a multi-protein RING-finger E3 ligase that drives each stage of the cell cycle by controlling the protein levels of cyclins and cyclin-dependent kinase inhibitors (CKIs) (Cardozo and Pagano, 2004). Through a coordinated repertoire of protein-protein interactions, the scaffold protein Cullin-1 (Cul1) binds both the Ring-box protein 1 (Rbx1), recruiting the E2-ubiquitin complex, and the adaptor protein Skp1, recruiting the F-Box E3 ligase (Petroski and Deshaies, 2005). The F-box family members dictate the substrate by binding a degron that is usually, but not always, post-translationally modified (Skowyra, et al., 1997).
The F-box protein Skp2 (S-phase kinase-associated protein 2) is overexpressed in human cancers and implicated in multiple murine cancer models (Frescas and Pagano, 2008; Lin, et al., 2010; Nakayama and Nakayama, 2006). SCF-Skp2 degrades known tumor suppressors CKIs p27, p21, and p57 (Carrano, et al., 1999; Kamura, et al., 2003; Yu, et al., 1998). Recognition of the p27 degron is unique, being bound by a complex consisting of Skp2 and an accessory protein, Cdc kinase subunit 1 (Cks1), after phosphorylation on Thr-187 by CyclinE-CDK2 (Ganoth, et al., 2001; Montagnoli, et al., 1999; Tsvetkov, et al., 1999). Additional non-phosphorylated residues of the p27 degron reinforce this trimeric complex for a high rate of p27 ubiquitylation (Hao, et al., 2005; Sitry, et al., 2002; Wang, et al., 2004; Wang, et al., 2003).
Small molecule inhibitors have been successfully developed against other E3 ligase-substrate interfaces, including Mdm2-p53 and IAPs-caspases (Vassilev, et al., 2004; Wang, et al., 2004). High-throughput screens designed to detect small molecules that stabilize p27 identified compounds that either inhibited 26S proteasome activity, prevented Skp2 from incorporating into the SCF complex, or downregulated Skp2 mRNA (Chen, et al., 2008; Nickeleit, et al., 2008; Rico-Bautista, et al., 2010). No inhibitors specifically and directly targeted to the E3 ligase activity of Skp2 have been identified however.
We hypothesized that such inhibitors could be identified using structure-based drug discovery to target specific three-dimensional (3D) molecular surfaces, or pockets, at the substrate’s binding site (Cardozo and Abagyan, 2005; Cardozo and Pagano, 2007). Our laboratory previously identified the first reported selective inhibitors against PERK catalytic activity using a pocket-targeted approach (Wang, et al., 2010). In the present study, we adapted this approach to target a protein-protein interface with an in silico structure-based discovery tool, virtual ligand screening (VLS), against a pocket identified at the p27-binding interface formed by Skp2-Cks1. The combination of VLS, chemical similarity searches, in vitro functional screens, and counterscreens identified four selective inhibitors of Skp2 ligase activity. The inhibitors increased both p27 protein level and half-life in metastatic melanoma cell lines, with this activity dependent on Skp2. Inhibitor treatments in various cancer cells also shifted the population of cells into G1, or G2/M phase, and this phenotype was both p27 and cell type dependent.
RESULTS
Identification of small molecule inhibitors
The published Skp2-Cks1-p27 crystal structure (Figure 1A, top) was interrogated with ICM-PocketFinder (Molsoft LLC, La Jolla, San Diego) to identify a pocket (blue geometric object) formed jointly by Skp2 and Cks1 (Figure 1A, bottom) and flanked by residues Skp2-R294, Skp2-Y346, Cks1-R44, and Cks1-Q52, which are essential for p27 binding and/or ubiquitylation (Hao, et al., 2005; Sitry, et al., 2002; Ungermannova, et al., 2005; Wang, et al., 2004). The pocket’s area and volume fall within a range calculated to be permissive for drug-like small molecule binding (An, et al., 2004; Cardozo and Abagyan, 2005). This pocket was targeted in a virtual screen (ICM-VLS) of 315,000 diverse compounds (ChemBridge Corp., La Jolla, CA) and from the 202 VLS hits 96 compounds were selected based on calculated binding score and Lipinski properties (Lipinski, et al., 2001).
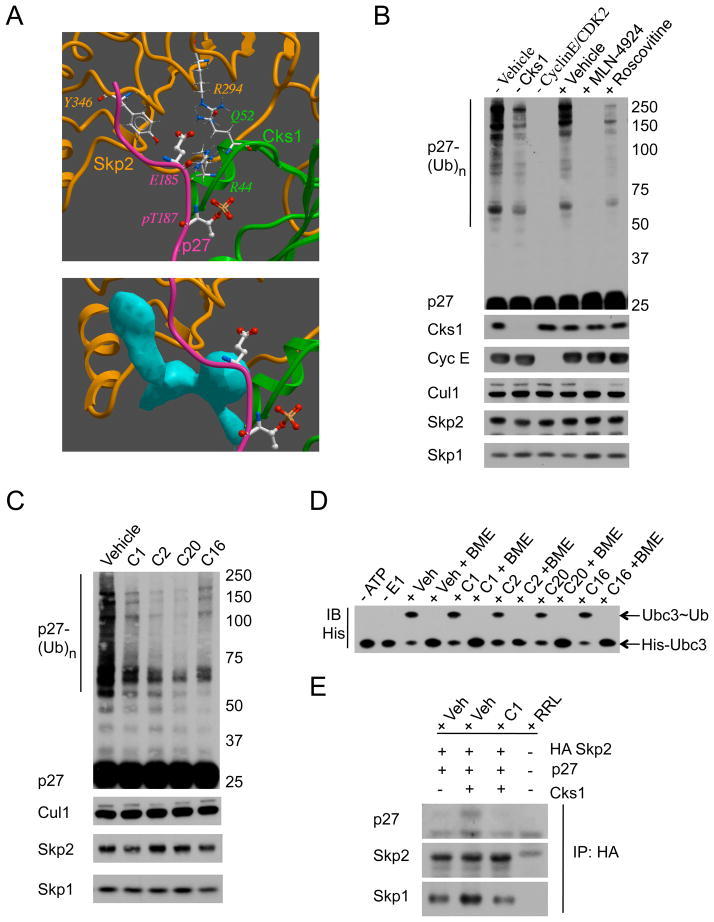
Figure 1. In silico and in vitro screens for Skp2 ligase inhibitors.
(A) Top: ICM 3D receptor model of Skp2 (gold ribbon), Cks1 (green ribbon), and p27 phospho-peptide (purple ribbon, phospho-T187 and E185 in ball and stick). Key residues important for D185 interactions are highlighted in wire. Bottom: Blue pocket exposed with removal of p27 peptide and antagonizes p27-D185 interactions.
(B) Calibration of the in vitro ubiquitylation assay. Immunoblotting for p27 poly-ubiquitylation (ladder, p27-(Ub)n), Cks1, Cyclin E, Cul1, Skp2, and Skp1 levels in the absence of Vehicle, Cks1, or CyclinE/CDK2; or in the presence of Vehicle (0.1% DMSO), 1 μM NEDD8-activating enzyme (NAE) inhibitor MLN-4924, or 50 μM CDK2 inhibitor Roscovitine.
(C) Representative primary screen for activity. Immunoblotting for p27-(Ub)n, Cul1, Skp2, and Skp1 in ubiquitylation assays treated with Vehicle (0.1% DMSO) or 50 μM inhibitor (C1, C2, C20, or C16).
(D) E2 charging unaffected by active compound. Immunoblotting for His-tagged Ubc3-ubiquitin conjugates (Ubc3~Ub) in the presence of Vehicle (0.1% DMSO, Veh) or 50 μM inhibitor (C1, C2, C20, or C16) under non-reducing or reducing (Beta-mercaptoethanol, BME) conditions.
(E) Compound treatment blocks the physical p27-Skp2 interaction. Immunoblotting for p27, Skp2, and Skp1 from HA-immunoprecipitates of ubiquitylation assays treated with Vehicle (0.1% DMSO, Veh) or 50 μM C1. No background using rabbit reticulocyte lysate (RRL) only.
See also Figure S1.
The 96 hits were tested in an in vitro ubiquitylation assay driven by in vitro transcribed/translated Skp2, Cks1, and p27 (Sitry, et al., 2002). Assay sensitivity was established for various criteria (Figure 1B): enhanced ubiquitylation with Cks1 (lane 2), kinase activity requirement (lane 3), and responsiveness to known inhibitors of Cullin (lane 5) or Cdk2 (lane 6) activity (Meijer, et al., 1997; Soucy, et al., 2009). Compounds showing at least 50% inhibition of Skp2-mediated p27 ubiquitylation (Figure 1C, data not shown) were counter-screened for inactivity against two anti-target E3 ligases: MDM2 and SCF-βTrCP (Figure S1A, S1B). Compounds C1 and C2 were identified from the primary VLS and C16 and C20 were identified from a second VLS (Figure 2).
Figure 2. Structure-function approach to identify key contacts.
(A) Chemical structure of C1 and C2. Red box= active group used as query in chemical similarity search.
(B) Predicted conformations with lowest energy for C1 and C2 (ball and stick) interacting with residues (wire) in Skp2 (gold ribbon) and Cks1 (green ribbon). Hydrogen bonds depicted as small balls.
(C) Chemical structure of C16 and C20 with active groups (red box) from 2A.
(D) Removal of active groups reverses activity. Immunoblotting for p27, Skp2, and Skp1 from ubiquitylation assays treated with Vehicle (0.1% DMSO), 50 μM inhibitors (C2, C20, C1, or C16), or 50 μM Unmatched compound (UM, see Figure S3A) lacking active groups.
(E) Cks1 mutation reverses inhibitor activity, as predicted by binding model. Immunoblotting for p27, Cks1, Skp2, and Skp1 from ubiquitylation assays using Wild Type-Cks1 (Left) or Q52L-Cks1 (Right) treated with Vehicle (0.1% DMSO) or 50 μM inhibitor (C1 or C16).
See also Figure S2 and S3.
Secondary bioassays confirmed the selectivity for Skp2-Cks1 interface by testing compound effects on ubiquitin transfer to either E2-Ubc3 or E2-Ubc5, as observed when ATP or E1 is omitted (Figure 1D, data not shown), level of CyclinE/CDK2 phosphorylation of p27 (Figure S1C), and dose dependent inhibition of Skp2 ligase activity (Figure S1D). In addition, C1 reduced the amount of p27, but not Skp1, interacting with Skp2 (Figure 1E) to a level similar of p27 bound in the absence of Cks1 (lane 1 versus 3), suggesting that inhibitor activity is dependent on Cks1. Taken together, our approach identified a set of inhibitors that fit into a molecular surface pocket at the Skp2-Cks1 interface and block p27 ubiquitylation in vitro, but do not block the non-Skp2-p27 interfaces of the active SCF.
Identification of contacts responsible for inhibitor activity
In order to identify the chemical groups responsible for inhibitor activity, divergent compounds sharing a common R-group with C1 or C2 (Figure 2A, highlighted in box) were selected from the PubChem database and docked in silico to the pocket at the Skp2-Cks1 interface. Compounds were tested in vitro if they preferentially docked in positions similar to C1 or C2 (Figure 2B): either forming predicted electrostatic interactions with Cks1-Q52 and/or hydrogen bonding to Cks1-R44 or Skp2-R344 (as in C1) or predicted cation-π interaction with Skp2-R294 and/or a hydrogen bond to Cks1-R44 (as in C2). C16 and C20 were identified using this method (Figure 2C) and showed similar inhibition as C1 or C2 (Figure 2D, lane 2 versus 5, lane 7 versus 9) and parallel docking poses (Figure S2A, data not shown).
To test the necessity of the predicted chemical contact group, compounds sharing the same scaffold but differing in the key R-group listed above (unmatched compounds; UM) were identified for each of the 4 active compounds (Figure S3A). With loss of the specific contact R-group, levels of p27 ubiquitylation were restored to vehicle control levels (Figure 2D, lane 1 versus 4, 6, 8, 10). Notably, UM-C2a (unmatched compound for C2) partially reduced p27 ubiquitylation (Figure 2D, lane 1 versus 3), suggesting that both the distance and charge of the contact group is important for C2 activity. Compound C1 contains a potentially promiscuous rhodanine core, but this core chemical group was not sufficient to inhibit ubiquitylation (Figure S3B, data not shown). Thus, chemical fingerprints guided the identification of additional Skp2 ligase inhibitors, with key 3D structural chemical groups confirmed necessary for inhibiting p27 ubiquitylation.
To interrogate the compound-contacting residues on the Skp2-Cks1 interface, mutants of Skp2 and Cks1 were designed based on the in silico dockings. The predicted electrostatic interactions between the positive charge on the NH2 side group in Q52-Cks1 with the COOH-group in C1 and C16 should be lost when mutated to a neutral-charge leucine (Figure S2B). In vitro ubiquitylation assays confirm that C1 and C16 are active against Wild-Type Cks1 but are inactive against the Q52L-Cks1 mutant (Figure 2E). In contrast, C2 and C20 strongly inhibited the activity of both the wild-type Cks1 and the Q52L-Cks1 mutant (data not shown). This confirms that the Q52-Cks1 residue is important for only C1 and C16 activity, as predicted by the in silico dockings. Notably, the basal level of ubiquitylation for the Q52L-Cks1 mutant is reduced (Figure 2E, lane 1 versus 5) being the first report that Q52 contributes partially to p27 degradation. The Skp2-R294A and the Cks1-R44A/H mutants lacked ubiquitylation activity (Figure S2C) preventing the interrogation of these residues’ contribution to the activity of C2 and C20. R294A-Skp2 was previously shown to reduce p27 binding, suggesting that the predicted site for C2 and C20 binding correlates with ligase activity (Ungermannova, et al., 2005). Thus, the predicted three-dimensional mode of interaction of the compounds with Skp2-Cks1 was validated at both the chemical and protein level.
Reduction of p27 binding to Skp2 by the inhibitors
The docked poses predict that the active compounds sterically clash with p27 (Figure S2A). To test this model, immunoprecipitations for HA-Skp2 were performed from the in vitro ubiquitylation assay mix. The compounds (10 μM) exhibited various degrees of Skp2 ligase inhibition (Figure 3A, left), which corresponded with a reduction of p27 binding to HA-Skp2 (Figure 3A, right). The amount of Skp1 and Cul1 bound was not affected, confirming that the compounds are not disrupting SCF formation.
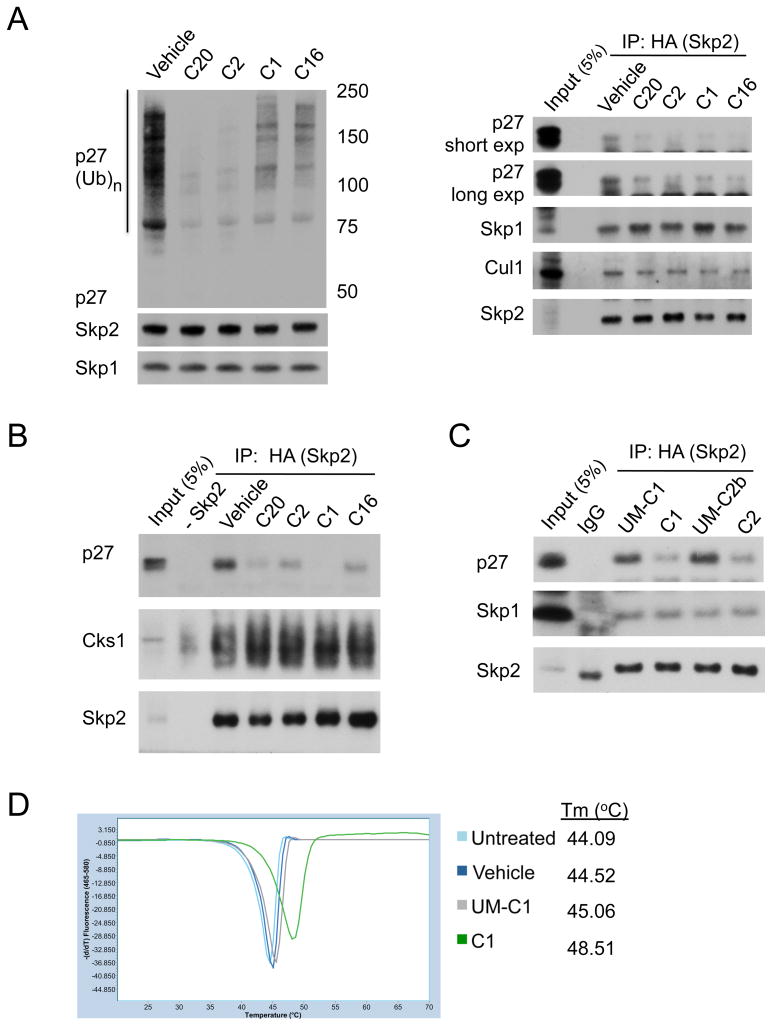
Figure 3. Binding of inhibitors disrupts the Skp2-p27 interaction.
(A) Reduced ubiquitylation correlates with reduced interaction. (Left) Immunoblotting for p27, Skp2, and Skp1 from ubiquitylation assays treated with Vehicle (0.1% DMSO) or 10 μM inhibitor (C20, C2, C1, or C16). (Right) Immunoblotting for p27, Skp1, Cul1, and HA-Skp2 in HA immunoprecipitates from ubiquitylation reactions.
(B) Inhibitors reduce p27 binding to Skp2. HA-Skp2 and Cks1 were pretreated with Vehicle (0.1%DMSO) or 10 μM inhibitor (C20, C2, C1, or C16) before added to p27-pT187. HA-immunoprecipitates were immunoblotted for p27, Cks1, and Skp2.
(C) Loss of chemically active groups restores p27 binding. HA-Skp2/Cks1 was pretreated with 10 μM inhibitor (C1 or C2) or corresponding unmatched compound (UM-C1 or UM-C2b, see Figure S3A) before p27-pT187 addition. HA-immunoprecipitates were immunoblotted for p27, Skp1, and Skp2.
(D) Loss of active groups removes compound binding. Melting temperature (Tm) of recombinant His-6Skp1-Skp2-Cks1 (1.5 μM) preincubated with C1 (75 μM), UM-C1 (75 μM), or Vehicle (0.5% DMSO) determined from melting peaks using differential scanning fluorimetry.
See also Figure S3.
To test if the compounds are specifically targeting the Skp2-Cks1 interface, immunoprecipitations were performed in the absence of ubiquitylation. The amount of p27 bound to HA-Skp2 was reduced in the presence of the inhibitors, whereas Cks1 binding was unaffected (Figure 3B). Knowing which chemical groups in C1 and C2 are necessary for inhibiting ubiquitylation (Figure 2D, S3A), we tested if the same groups are responsible for blocking p27 binding. Unmatched compounds for C1 (UM-C1) and C2 (UM-C2b) lost the ability to inhibit p27 from binding to Skp2 (Figure 3C, lane 3 versus 4, lane 5 versus 6). The same chemical contact was also important in mediating direct interaction with purified Skp2-Cks1 complex. Using differential scanning fluorimetry, C1 but not UM-C1 shifted the melting temperature of purified protein complex (Figure 3D). Surface plasmon resonance assays also confirmed the ability of the compounds to bind Skp2-Cks1 complex (Figure S3C, data not shown). These data validate that the specific chemical groups identified from the VLS docking poses are mediating the compounds interaction at the Skp2-Cks1 interface and blocking p27 binding.
Induction of p27 by inhibitors in cancer cells
To test if these compounds prevent p27 degradation in cells, the metastatic melanoma cell lines 501 Mel, SK-MEL-147, and SK-MEL-173 were treated with each inhibitor (10 μM) or 0.1% DMSO (vehicle). Steady state protein levels of p27 were induced with inhibitors in a dose dependent manner (Figure 4A, 4B, S4A). p21 protein levels also increased, supporting p21 ubiquitylation by SCF-Skp2 is enhanced with Cks1 (Bornstein, et al., 2003). Inhibitor treatment did not alter protein levels of Skp2, Skp1, Cul1, and other Skp2 (Cks1-independent) targets p130, Cdt1, Tob1, and Cyclin E (Figure 4A left, data not shown). 501 Mel cells were also treated with MG-132 (a proteasome inhibitor) or MLN-4924 (the NEDD8-activating enzyme inhibitor) to confirm that p27 and p21 induction were responsive to known UPS inhibitors (Figure 4A, right). MLN-4924 inhibited neddylation of Cul1, an effect not mediated by the ligase inhibitors. Thus, treatment of cells with inhibitors targeting the Skp2-Cks1 interface induces protein levels of specific Skp2-Cks1 substrates.
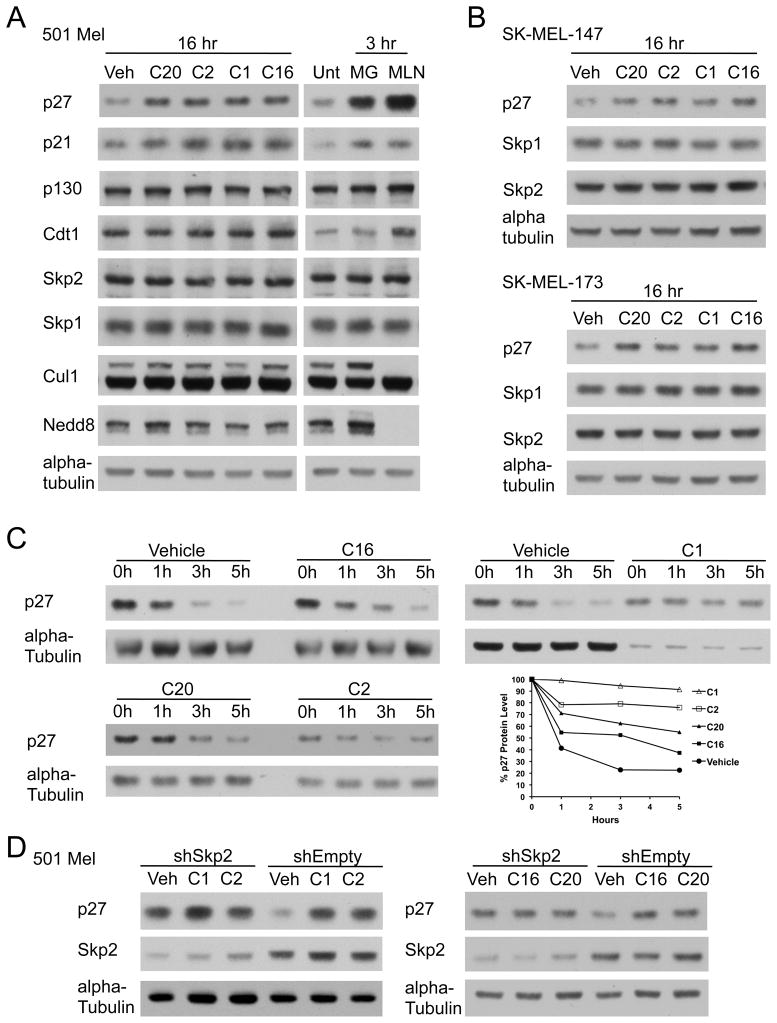
Figure 4. Inhibitors induces p27 protein in melanoma cells.
(A) Protein induction following inhibitor treatment. Immunoblotting for steady state levels of Skp2 substrates (p27, p21, p130, and Cdt1), Skp2, Skp1, Cul1, and Nedd8 in 501 Mel cells (Left) treated with vehicle (0.1% DMSO, Veh) or 10 μM inhibitor (C20, C2, C1, or C16) and (Right) untreated (Unt) or treated with 10 μM proteasome inhibitor MG-132 (MG) or 400 nM NAE inhibitor MLN-4924 (MLN). Loading normalized to alpha-tubulin.
(B) Immunoblotting for p27, Skp1, and Skp2 steady state levels in melanoma cell lines SK-MEL-147 (Top) or SK-MEL-173 (Bottom) treated with vehicle (0.1% DMSO) or 10 μM inhibitor (C20, C2, C1, or C16). Loading normalized to alpha-tubulin.
(C) p27 half-life analysis. Cycloheximide half-life from 501 Mel cells pretreated 16h with Vehicle (0.1% DMSO) or 10 μM inhibitor (C16, C1, C20, or C2). Representative immunoblots at times 0, 1, 3, and 5 hours post-cycloheximide (n=3). Total protein normalized to alpha-tubulin. (Graph): Densitometry levels for % p27 protein remaining.
(D) p27 induction by compounds is dependent on Skp2. Immunoblotting for p27 and Skp2 in 501 Mel cells expressing shRNA against Skp2 (shSkp2) or empty vector (shEmpty) treated with Vehicle (0.1% DMSO, Veh) or 10 μM inhibitor (C1 or C2, Left; C16 or C20, Right). Loading normalized to alpha-tubulin.
See also Figure S4.
To assess the mechanism of p27 induction, cycloheximide half-lives were analyzed in 501 Mel cell lines pretreated with each inhibitor (10 μM) or 0.1% DMSO (vehicle). p27 degrades rapidly with a half-life of 1 hour with vehicle, with this rate slowed by the inhibitors (Figure 4C). C1 and C2 exhibit the strongest stabilization, with a p27 half-life greater than 5 hours, whereas C20 extends it to 5 hours and C16 extends it to 3 hours. Protein levels of Skp2 and Skp1 remained constant (data not shown).
Since p27 has additional reported E3 ligases (Cao, et al., 2011; Kamura, et al., 2004), we tested the dependency of inhibitors for Skp2 using lentiviral tet-inducible short hairpins to Skp2 (shSkp2). Immunoblotting revealed that Skp2 knockdown eliminated inhibitor-mediated p27 induction (Figure 4D, lane 1 versus 2, 3), where as the 501 Mel cells expressing control shEmpty shRNA retained marked induction of p27 (Figure 4D, lane 4 versus 5, 6). Compound induced p27 levels were similar to the basal level of p27 in the Skp2 knockdown cells (Figure 4D, lane 1 versus 5, 6), suggesting that Skp2 is responsible for maintaining low levels of p27 in the 501 Mel cells. Taken together, the small molecules targeted to the Skp2-Cks1 interface inhibited Skp2-dependent p27 degradation in metastatic melanoma cells.
Cell cycle effects of inhibitor treatment
Skp2 siRNA treatment in melanoma cells results in p27 upregulation and reduced cell growth (Katagiri, et al., 2006; Sumimoto, et al., 2006). 501 Mel cells responded to Skp2 ligase inhibitors by inducing p27 (Figure 4, S4A), with a dose dependent effect on cell viability (Figure S4B, left). To address the specific role of p27 in 501 Mel cell cycle progression, p27 was knockdown using tet-inducible shRNAs (shp27-1 or shp27-5) and compared to control empty vector shRNA (shEmpty). FACS analysis confirmed that the reduction of p27 protein levels (Figure 5A, left) correlated with a significant decrease in G1 phase cells (p < 0.001) and an increase in S phase cells (p < 0.01)(Figure 5A, middle). In the cells lacking p27, treatment with the inhibitors (10 μM for 16 hours) did not alter the cell cycle profile (Figure 5A, right and data not shown), suggesting that the inhibitors require p27 to elicit changes in cell cycle. To demonstrate this, shEmpty control cells were treated with the inhibitors (10 μM for 16 hours) and showed p27 protein induction (Figure 5B, left) with a corresponding increase in G1 phase cells (p < 0.001) and a decrease in S phase cells (p < 0.01 for C1, p < 0.05 for C16, p < 0.0001 for C2 and C20) (Figure 5B, right). Since inhibitor-mediated G1/S arrest is dependent on p27 and the inhibitors stabilize p27 protein in a Skp2-dependant manner (Figure 4), the changes observed in the cell cycle most likely results from the inhibitors targeting Skp2 ligase activity.
Figure 5. Skp2 ligase inhibitors induce cell cycle changes.
(A) p27-dependent G1 arrest. 501 Mel cells expressing empty vector (shEmpty) or two different shRNAs (shp27-1 or shp27-5) against p27 were (Left) immunoblotted for p27, Skp2, Skp1, and alpha-tubulin or (Center) stained with Propidium Iodide for FACS analysis. Graph represents mean ± STDEV from n=3. *, p < 0.01; **, p < 0.001 compared with shEmpty (one-way ANOVA followed by Dunnett test). (Right) shp27-1 501 Mel cells treated with Vehicle (0.1% DMSO, Veh) or 10 μM inhibitor (C1, C16, C2, or C20), and stained with Propidium Iodide for FACS analysis. Graph represents mean ± STDEV from n=4.
(B) G1 induction by inhibitors. shEmpty 501 Mel cells treated with Vehicle (0.1% DMSO, Veh) or 10 μM inhibitor (C1, C16, C2, or C20) were (Left) immunoblotted for p27, Skp2, Skp1, and alpha-tubulin or (Right) stained with Propidium Iodide for FACS analysis. Graph represents mean ± STDEV from n=4. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with shEmpty Vehicle (one-way ANOVA followed by Dunnett test).
(C) G1 or G2/M induction by C1 in breast cancer cells. (Left) FACS analysis of T47D and MCF-7 treated with Vehicle (0.1% DMSO, Veh) or 5 μM C1. Graph represents mean ± STDEV from n=4 for T47D and n=3 for MCF-7. *, p < 0.0001 compared with corresponding cell cycle phase in Vehicle (unpaired two-tailed Student’s t test). (Right) FACS analysis of MCF-7 treated with increasing C1 dose. Graph represents mean ± STDEV from n=3. *, p < 0.01; **, p < 0.001 compared with Vehicle (one-way ANOVA followed by Dunnett test).
(D) G1 induction by C2 in prostate cancer cells. FACS analysis of LNCaP cells treated with Vehicle (0.1% DMSO, Veh) or 10 μM C2. Graph represents mean ± STDEV from n=6. *, p < 0.0001 compared with Vehicle (unpaired two-tailed Student’s t test).
(E) p27 protein induction in cancer cells. Immunoblotting for p27, Skp2, Skp1, and alpha-tubulin in MCF-7, T47D, or LNCaP cells treated with Vehicle (0.1% DMSO, Veh), 5 μM C1, or 10 μM C2.
See also Figure S4.
In addition to melanomas, the Skp2-p27 axis regulates growth of breast and prostate cancer cells (Lu, et al., 2002; Shibahara, et al., 2005; Sun, et al., 2007). Breast cancer cell lines MCF-7 and T47D displayed no significant cellular phenotype after treatment with C2, C16, and C20 (data not shown). In contrast, treatment with C1 resulted in a lower rate of active cells (Figure S4B, right), thus C1 treatment was reduced from 10 μM to 5 μM in order to monitor early cell cycle effects. T47D cells treated with C1 (5 μM for 16 hours) displayed an increase in G1 phase (p < 0.0001) and a decrease in S phase (p < 0.0001), correlating with p27 protein induction (Figure 5C left, Figure 5E center). In contrast, MCF-7 cells responded to C1 with a significant reduction in G1 phase (35%, p < 0.0001) and an increase in G2-M phase (43%, p < 0.0001) (Figure 5C left). This G1 reduction and G2/M arrest is dose dependent on C1 (Figure 5C right; p < 0.001 and p < 0.01, respectively) and correlates with increased p27 protein levels (Figure 5E left, S4A top right). LNCaP prostate cancer cells treated with C2 (10 μM for 16 hours) displayed a mild, but significant, increase in G1 cells (p < 0.0001) and a reduction in S phase cells (p < 0.0001) (Figure 5D), correlating with p27 protein induction (Figure 5E, right). Therefore, multiple Skp2 ligase inhibitors induce p27 protein expression in a number of cancer cells, and this induction correlated with changes in cell cycle distribution. However, the cell cycle effects are dependent on cell type, with blocks at either the G1/S or G2/M transition.
DISCUSSION
We combined an in silico high-throughput VLS with low-throughput in vitro bioassays to identify the first set of small molecule inhibitors targeted to block Skp2-mediated p27 ubiquitylation and alter cell cycle progression. Multiple human cancers, including prostate, breast, ovarian, lung, and metastatic melanoma, express low levels of p27 and high Skp2 protein levels, with either protein status independently associated with poor prognosis and low patient survival (Chu, et al., 2008; Florenes, et al., 1998; Hershko, 2008; Li, et al., 2004; Woenckhaus, et al., 2005). Thus, the Skp2-p27 axis is an attractive target for drug discovery and cancers that rely on Skp2 ligase activity for their oncogenic properties should be highly susceptible to specific and direct inhibitors of Skp2-mediated p27 ubiquitylation.
One major challenge we faced was that the target site is a protein-protein interface. We previously proposed that certain protein-protein interfaces may be targeted for drug discovery if they exhibited favorable configurations, including the presence of a drug-binding pocket of sufficient size located at a functionally sensitive location on the interface (Cardozo and Abagyan, 2005). In order to target such surfaces, pocket specific screening approaches, as opposed to high-throughput screening (HTS), must be utilized, of which VLS is perhaps the most suitable. In this study, we successfully put this theory into practice. Furthermore, this study extends the theory by suggesting that pockets formed by the association of two proteins, in this case Skp2 and Cks1, are no different from mono-domain pockets and can be equally targeted with success.
A second major challenge was to confirm the primary target of the inhibitors, given that previous attempts identified compounds that either blocked Skp2 incorporation into an active SCF or reduced Skp2 mRNA levels (Chen, et al., 2008; Rico-Bautista, et al., 2010). Since our in silico screen selects small molecules that are highly structurally compatible with the Skp2-Cks1 protein interface, there is a high likelihood that validated compounds would not non-specifically inhibit any of the multiple proteins/enzymes present in the ubiquitylation process. This included sites other small molecules inhibit: (1) the F-box domain interface with Skp1, (2) CyclinE/Cdk2 kinase activity, (3) E2 Ubc3 (Cdc34) activity, and (4) a known druggable protein interface MDM2-p53 (Aghajan, et al., 2010; Ceccarelli, et al., 2012; Meijer, et al., 1997; Millard, et al., 2011). Our compounds do not exhibit activity against any of these off-targets, showing high selectivity for SCF-Skp2 ligase.
One major advantage of our approach is that the predicted VLS docking poses highlight key chemical groups that can be used as a chemical fingerprint to identify additional Skp2 ligase inhibitors. Interrogating receptor-ligand atomic contacts and using chemical similarity searches identifies more diverse and/or potent inhibitors (Cheltsov, et al., 2010; Wang, et al., 2009). Thus, a blueprint or pharmacophore of the entire Skp2 ligase inhibitor class is preliminary visualized, which may guide the rational synthesis of derivatives of C1, C2, C16, and C20 during future preclinical lead optimization stages. Using thermal shifts and SPR technologies, the compounds display specific binding to the Skp2-Cks1 protein complex, but we have not been able to obtain the co-crystal structures of Skp1-Skp2-Cks1 in complex with the inhibitors (data not shown), possibly due to stringent crystallization conditions. Nevertheless, the compendium of data on these inhibitors leaves little doubt that they act at the targeted pocket formed by Skp2-Cks1. The clear visualization of their pharmacophore also leaves little doubt that chemically diverse, more potent and soluble compounds derived from this pharmacophore will be resolved crystallographically.
By targeting the Skp2-Cks1-p27 interface, we focused on inhibiting cell cycle progression (Bloom and Pagano, 2003). Inhibitor-treated metastatic melanoma cells displayed a significant shift of cells from S phase into G1 phase that was dependent on both p27 and Skp2, validating the inhibitors are regulating p27 levels to elicit cellular changes. Interestingly C1 treatment of MCF-7 breast cancer cells lines did not increase, but rather decreased the percentage of cells in G1 while increasing the G2/M population. This same G2/M phenotype has been observed in Skp2−/− MEFs and was reversed by knocking out p27, suggesting that p27 also plays a pivotal role in G2/M phase (Nakayama, et al., 2004; Pagano, 2004). Elucidation of the specific mechanism may now be interrogated in MCF-7 cells that are especially sensitive to C1 mediated G2/M accumulation. This type of chemical genetic approach would not be possible without the availability of inhibitors specifically blocking Skp2 mediated p27 degradation.
In this study, we proved the principle that Skp2 ligase activity is susceptible to direct inhibition by small molecules. UPS inhibitor research has accelerated since the FDA approval of Bortezomib, and the focus has been on the additional enzymes present in this pathway. Our approach demonstrates that a non-enzymatic protein-substrate interface is druggable with a high degree of selectivity. This added level of specificity may prove indispensable when evaluating combination therapies between inhibitors of the UPS and conventional chemotherapies (Ning, et al., 2007; Wright, 2010). These pharmacological inhibitors may also aid in identifying and treating cancers that rely on the Skp2 ligase activity for their oncogenic properties.
SIGNIFICANCE
Clinical efficacy of the proteasome inhibitor Bortezomib (Velcade™) established the ubiquitin proteasome system (UPS) as a key target in cancer. In a number of human cancers, low levels of the cell-cycle regulatory tumor suppressor, p27, correlate with the overexpression of its UPS E3 ligase SCF-Skp2, suggesting that deregulation of p27 degradation by the UPS promotes carcinogenesis. Thus, inhibitors designed to stabilize p27 by selectively targeting SCF-Skp2 may provide a new avenue of cancer treatment with enhanced specificity over Bortezomib. From a chemistry point of view, however, identifying a specific small molecule inhibitor of the SCF-Skp2/p27 interface is challenging, since this is a protein-protein interface. Using a structure-based approach encompassing both in silico virtual ligand screening (VLS) and in vitro assays, we identified a diverse set of inhibitors of Skp2-mediated p27 degradation that reduce p27 ubiquitylation by specifically and competitively inhibiting the Skp2-p27 interaction. In various cancer cells, these compounds induce p27 and elicit cell cycle arrest. This work establishes a proof-of-principle both for drug discovery targeting Skp2, a potential advance in biology, and for drug discovery targeted this form of protein-protein interface, a potential advance in chemistry.
EXPERIMENTAL PROCEDURE
In silico assays
The in silico protein receptor and ligand preparations, the docking simulations during VLS, and the docking score calculations as well as chemical similarity and substructure searches were carried out with ICM-Pro, ICM-VLS and ICM-Chemistry software (Molsoft LLC, La Jolla, CA) (Abagyan, et al., 1994). Pockets on the crystallographic structure PDB 2ast were identified with the ICM PocketFinder module in ICM-Pro and screened by ICM-VLS against the ChemBridge Library (ChemBridge Corp, San Diego, CA, USA) using default ICM-VLS docking parameters. Dockings used global optimization with a biased probability Monte Carlo (BPMC) conformational search of fully flexible, full-atom models of the ligands within a set of grid potential maps calculated from the coordinates of the atoms in the protein receptor (Abagyan and Totrov, 1994; Abagyan and Totrov, 2001). These grid energy maps account for the hydrophobic, heavy atom and hydrogen van der Waals interactions, hydrogen-bonding interactions, and electrostatic potential. Hits with VLS docking score better than -30U were further filtered by Lipinski’s rules to eliminate non-drug-like compounds. Substructure search were performed using the ChemBridge and ICM MolCart databases, with Tanimoto distance as a measure of chemical similarity.
Plasmids
HA-Skp2, Cks1, Q52L-Cks1, p27, Flag-β-TrCP, and HA-PDCD4 cloned into pcDNA3.1 plasmids were provided by Dr. Michele Pagano (NYUMC). Short hairpins for p27 (cat. no. RHS4740-NM_004064) and Skp2 (cat. no. RHS4740-NM_005983) in doxycycline-regulated TRIPZ lentiviral vector were obtained from Open Biosystems and prepared following manufacturer instructions.
Antibodies and Immunoblotting
Antibodies include mouse monoclonal antisera against Skp2/p45 (Invitrogen), p27 (BD Biosciences), Cks1 (Invitrogen), Cyclin E (Invitrogen), alpha-tubulin (Sigma), and HA (Covance); and rabbit polyclonal antisera against phosphoThr-187 p27 (Invitrogen), total p27 (Cell Signaling), p21 (Cell Signaling), Skp1 (Invitrogen), NEDD8 (Cell Signaling), Cul1 (Invitrogen), p130 (Santa Cruz), Cdt1 (Cell Signaling), His (Santa Cruz), HA (Invitrogen), and β-TrCP (Invitrogen).
Ubiquitylation Assays
In vitro reconstituted assays were performed as previously described (Dorrello, et al., 2006) with the exceptions of the use of (1) unlabeled proteins (wild-type and mutant Skp2 and Cks1, p27, Flag-βTrCP, and HA-PDCD4) generated by in vitro transcription-translation (TNT T7 Coupled Reticulocyte Lysate System, Promega), (2) chain elongation with non-methylated ubiquitin, and (3) immunoblotting to detect poly-ubiquitylation.. The following reagents were obtained from Boston Biochem: UBE1, His-UbcH3, His-UbcH5c, Ubiquitin, and Ubiquitin Aldehyde. Additional reagents include CDK2/CyclinE1 (Sigma-Aldrich), S6K (Invitrogen), Okadaic acid (Sigma), and DMSO (Sigma-Aldrich). For ubiquitin charging, UBE1 (100 nM) and His-UbcH3 (10 ng/μl) were incubated with ubiquitin (2.5 μg/μl), ATP (2 mM) and 10 μM of inhibitors or Vehicle control for 60 min at 30°C. Reactions were stopped in nondenaturing 2X Sample Buffer, with the addition of 10% β-mercaptoethanol as indicated, and analyzed by western blotting.
Inhibitors
Control ubiquitylation inhibitors were Roscovitine (Cayman Chemicals), MG132 (Peptide Institute), and MLN-4924 (Active Biochem). Compounds obtained from ChemBridge Corp (San Diego, CA, USA) include C1 (6719837), C2 (6544607), and C16 (6744881). C20 (A067/0031209) was obtained from Ryan Scientific, Inc (Mount Pleasant, SC, USA).
Binding and Kinase assays
In vitro ubiquitylation assays were incubated with anti-HA affinity matrix (Roche) in IP buffer (50 mM Tris pH 7.6, 150 mM NaCl, 0.1% NP40, and Roche Complete EDTA-free protease tablet) for 16h at 4°C. Beads were washed 3X with IP buffer and eluted with 2X SDS Sample Buffer (Boston Bioproducts) and 10% β-mercaptoethanol. For binding without ubiquitylation, HA-Skp2 IVT (10 μl) and Cks1 IVT (10 μl) were preincubated with anti-HA affinity matrix in IP buffer for 2h, with the addition of p27 kinase reaction (10 μl) for another 16h incubation at 4°C. All reactions were analyzed after SDS/PAGE by western blotting. Where indicated, Skp2 and Cks1 were pretreated with 10 μM inhibitors for 30m. Kinase reactions were performed with p27 IVT (10 μl) and 0.5 μg/μl CyclinE/CDK2 in kinase buffer (50 mM Tris HCL pH 7.5, 10 mM MgCl2, 2 mM ATP, 2.5 μM Okadaic Acid, and 0.6 mM DTT) at 30°C for 1h.
Differential scanning fluorimetry
Protein melt assays were carried out following manufacturer’s protocol on the LightCycler 480 System II (Roche) using Sypro Orange dye (Sigma). Fluorescence (465 nm excitation/580nm detection) was acquired (10 data points/sec) at a ramp rate of 0.06°C/sec between 20°C and 85°C. Melting temperatures were calculated using the Lightcycler 480 Protein Melt analysis tool (Roche).
Cell Lines and knockdown
MCF-7 and T47D were purchased from ATCC and cultured as specified. 501 Mel, SK-MEL-147, and SK-MEL-173 were kindly provided by Dr. Eva Hernando (NYUMC). LNCaP were a kind gift from Dr. Garabedian (NYUMC). 501 Mel were cultured in OPTI-MEM reduced serum supplemented with 5% FBS and 1% pen-strep. SK-MEL lines and LNCaP were cultured in DMEM supplemental with 10% FBS and 1% pen-strep. All cell culture reagents were obtained from GIBCO, except Tet System approved Fetal Bovine Serum (#631101) and doxycycline (#631311) supplied by Clontech. For knockdown, 501 Mel cells were infected with TRIPZ p27, Skp2, or empty vector and treated for 3 days with 2 ng/ml doxycycline. Cells expressing more than 70% RFP were used for further analysis. From the p27 shRNA set, knockdown was achieved with V3THS_410220 (shp27-1) and V3THS_372105 (shp27-5). From the Skp2 shRNA set, knockdown was achieved with V2THS_254607 (shSkp2).
Cell based analysis
Whole cells extracts were prepared and immunoblotted as previously described (Dorrello, et al., 2006). For knockdown of Skp2 or p27, shRNAs were induced with 2 ng/ml doxycycline for 48 hours, with the last 16 hours co-treated with 0.1% Vehicle or 10 μM inhibitors as indicated. For cycloheximide chase, cells were pretreated 16h with 0.1% Vehicle or 10 μM inhibitors, followed by 20 ng/ml cycloheximide (Sigma) and whole cell extracts collected for each time point. Scanning densitometry was performed on immunoblots using Image J software. For cell cycle analysis, cells were treated with 0.1% Vehicle or 10 μM inhibitors for 16h. 1 × 106 cells were fixed in 70% ethanol for 2h, treated with 200μg/ml RNase A (Invitrogen) for 30m, and stained with 20μg/ml Propidium Iodine (Sigma). All samples were assayed in triplicates. Signal was detected using the Becton Dickson LSRII flow cytometer and cell cycle profiles were analyzed with FloJo.
Statistical Methodologies
Statistical significance between and among groups was determined by Student’s paired two-tailed t test or One-way ANOVA, using Dunnett’s multiple comparison tests as the post hoc analysis. p < 0.05 was considered significant. (GraphPad Prism Software).
Supplementary Material
HIGHLIGHTS.
In silico screening against a pocket formed by the Skp2-Cks1 protein interface
Structure-based discovery of Skp2 ligase inhibitors that block p27 ubiquitylation
Inhibition of Skp2 ligase activity stabilizes p27 protein in melanoma cells
Inhibitor induction of p27 results in cell cycle arrest in various cancer cells
Acknowledgments
We thank Dr. Eva Hernando and Dr. Douglas Hanniford for technical assistance on generating the TRIPZ knockdown 501 Mel cell lines, and James Swetnam for the preliminary VLS. This work was supported by National Institutes of Health grants DPOD004631 from the Office of the Director (to TC) and supported in part by grants 1UL1RR029893 from the National Center for Research Resources, 5P30CA016087 from the National Cancer Institute 1R01GM099948 (to BH) from the National Institute of General Medical Sciences, and a Ruth L. Kirschstein National Research Service award 5T32HL007151 (to LW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- Abagyan R, Totrov M. High-throughput docking for lead generation. Curr Opin Chem Biol. 2001;5:375–382. doi: 10.1016/s1367-5931(00)00217-9. [DOI] [PubMed] [Google Scholar]
- Abagyan R, Totrov M, Kuznetsov D. ICM—A new method for protein modeling and design: Applications to docking and structure prediction from the distorted native conformation. Journal of Computational Chemistry. 1994;15:488–506. [Google Scholar]
- Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, et al. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Totrov M, Abagyan R. Comprehensive identification of “druggable” protein ligand binding sites. Genome Inform. 2004;15:31–41. [PubMed] [Google Scholar]
- Argyriou AA, Iconomou G, Kalofonos HP. Bortezomib-induced peripheral neuropathy in multiple myeloma: a comprehensive review of the literature. Blood. 2008;112:1593–1599. doi: 10.1182/blood-2008-04-149385. [DOI] [PubMed] [Google Scholar]
- Bloom J, Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin Cancer Biol. 2003;13:41–47. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- Bross PF, Kane R, Farrell AT, Abraham S, Benson K, Brower ME, Bradley S, Gobburu JV, Goheer A, Lee SL, et al. Approval summary for bortezomib for injection in the treatment of multiple myeloma. Clin Cancer Res. 2004;10:3954–3964. doi: 10.1158/1078-0432.CCR-03-0781. [DOI] [PubMed] [Google Scholar]
- Cao X, Xue L, Han L, Ma L, Chen T, Tong T. WW domain-containing E3 ubiquitin protein ligase 1 (WWP1) delays cellular senescence by promoting p27(Kip1) degradation in human diploid fibroblasts. J Biol Chem. 2011;286:33447–33456. doi: 10.1074/jbc.M111.225565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Abagyan R. Druggability of SCF ubiquitin ligase-protein interfaces. Methods Enzymol. 2005;399:634–653. doi: 10.1016/S0076-6879(05)99042-3. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. Wrenches in the works: drug discovery targeting the SCF ubiquitin ligase and APC/C complexes. BMC Biochem. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2091-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2012;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- Cheltsov AV, Aoyagi M, Aleshin A, Yu EC, Gilliland T, Zhai D, Bobkov AA, Reed JC, Liddington RC, Abagyan R. Vaccinia virus virulence factor N1L is a novel promising target for antiviral therapeutic intervention. J Med Chem. 2010;53:3899–3906. doi: 10.1021/jm901446n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- Florenes VA, Maelandsmo GM, Kerbel RS, Slingerland JM, Nesland JM, Holm R. Protein expression of the cell-cycle inhibitor p27Kip1 in malignant melanoma: inverse correlation with disease-free survival. Am J Pathol. 1998;153:305–312. doi: 10.1016/S0002-9440(10)65572-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. The cell-cycle regulatory protein Cks1 is required for SCF(Skp2)-mediated ubiquitinylation of p27. Nat Cell Biol. 2001;3:321–324. doi: 10.1038/35060126. [DOI] [PubMed] [Google Scholar]
- Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Hershko DD. Oncogenic properties and prognostic implications of the ubiquitin ligase Skp2 in cancer. Cancer. 2008;112:1415–1424. doi: 10.1002/cncr.23317. [DOI] [PubMed] [Google Scholar]
- Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, Hatakeyama S, Nakayama K, Nakayama KI. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc Natl Acad Sci U S A. 2003;100:10231–10236. doi: 10.1073/pnas.1831009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol. 2004;6:1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- Kane RC, Dagher R, Farrell A, Ko CW, Sridhara R, Justice R, Pazdur R. Bortezomib for the treatment of mantle cell lymphoma. Clin Cancer Res. 2007;13:5291–5294. doi: 10.1158/1078-0432.CCR-07-0871. [DOI] [PubMed] [Google Scholar]
- Katagiri Y, Hozumi Y, Kondo S. Knockdown of Skp2 by siRNA inhibits melanoma cell growth in vitro and in vivo. J Dermatol Sci. 2006;42:215–224. doi: 10.1016/j.jdermsci.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Li Q, Murphy M, Ross J, Sheehan C, Carlson JA. Skp2 and p27kip1 expression in melanocytic nevi and melanoma: an inverse relationship. J Cutan Pathol. 2004;31:633–642. doi: 10.1111/j.0303-6987.2004.00243.x. [DOI] [PubMed] [Google Scholar]
- Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, et al. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Lu L, Schulz H, Wolf DA. The F-box protein SKP2 mediates androgen control of p27 stability in LNCaP human prostate cancer cells. BMC Cell Biol. 2002;3:22. doi: 10.1186/1471-2121-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer L, Borgne A, Mulner O, Chong JP, Blow JJ, Inagaki N, Inagaki M, Delcros JG, Moulinoux JP. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- Millard M, Pathania D, Grande F, Xu S, Neamati N. Small-molecule inhibitors of p53-MDM2 interaction: the 2006–2010 update. Curr Pharm Des. 2011;17:536–559. doi: 10.2174/138161211795222649. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Fiore F, Eytan E, Carrano AC, Draetta GF, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, Kitagawa M, Iemura S, Natsume T, Nakayama KI. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev Cell. 2004;6:661–672. doi: 10.1016/s1534-5807(04)00131-5. [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- Nickeleit I, Zender S, Sasse F, Geffers R, Brandes G, Sorensen I, Steinmetz H, Kubicka S, Carlomagno T, Menche D, et al. Argyrin a reveals a critical role for the tumor suppressor protein p27(kip1) in mediating antitumor activities in response to proteasome inhibition. Cancer Cell. 2008;14:23–35. doi: 10.1016/j.ccr.2008.05.016. [DOI] [PubMed] [Google Scholar]
- Ning YM, He K, Dagher R, Sridhara R, Farrell AT, Justice R, Pazdur R. Liposomal doxorubicin in combination with bortezomib for relapsed or refractory multiple myeloma. Oncology (Williston Park) 2007;21:1503–1508. discussion 1511, 1513, 1516 passim. [PubMed] [Google Scholar]
- Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649–1657. doi: 10.1158/1078-0432.CCR-07-2218. [DOI] [PubMed] [Google Scholar]
- Pagano M. Control of DNA synthesis and mitosis by the Skp2-p27-Cdk1/2 axis. Mol Cell. 2004;14:414–416. doi: 10.1016/s1097-2765(04)00268-0. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Rico-Bautista E, Yang CC, Lu L, Roth GP, Wolf DA. Chemical genetics approach to restoring p27Kip1 reveals novel compounds with antiproliferative activity in prostate cancer cells. BMC Biol. 2010;8:153. doi: 10.1186/1741-7007-8-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara T, Onishi T, Franco OE, Arima K, Sugimura Y. Down-regulation of Skp2 is correlated with p27-associated cell cycle arrest induced by phenylacetate in human prostate cancer cells. Anticancer Res. 2005;25:1881–1888. [PubMed] [Google Scholar]
- Sitry D, Seeliger MA, Ko TK, Ganoth D, Breward SE, Itzhaki LS, Pagano M, Hershko A. Three different binding sites of Cks1 are required for p27-ubiquitin ligation. J Biol Chem. 2002;277:42233–42240. doi: 10.1074/jbc.M205254200. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Hirata K, Yamagata S, Miyoshi H, Miyagishi M, Taira K, Kawakami Y. Effective inhibition of cell growth and invasion of melanoma by combined suppression of BRAF (V599E) and Skp2 with lentiviral RNAi. Int J Cancer. 2006;118:472–476. doi: 10.1002/ijc.21286. [DOI] [PubMed] [Google Scholar]
- Sun L, Cai L, Yu Y, Meng Q, Cheng X, Zhao Y, Sui G, Zhang F. Knockdown of S-phase kinase-associated protein-2 expression in MCF-7 inhibits cell growth and enhances the cytotoxic effects of epirubicin. Acta Biochim Biophys Sin (Shanghai) 2007;39:999–1007. doi: 10.1111/j.1745-7270.2007.00361.x. [DOI] [PubMed] [Google Scholar]
- Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- Ungermannova D, Gao Y, Liu X. Ubiquitination of p27Kip1 requires physical interaction with cyclin E and probable phosphate recognition by SKP2. J Biol Chem. 2005;280:30301–30309. doi: 10.1074/jbc.M411103200. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Wang H, Bauzon F, Ji P, Xu X, Sun D, Locker J, Sellers RS, Nakayama K, Nakayama KI, Cobrinik D, et al. Skp2 is required for survival of aberrantly proliferating Rb1-deficient cells and for tumorigenesis in Rb1+/− mice. Nat Genet. 2009;42:83–88. doi: 10.1038/ng.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Blais J, Ron D, Cardozo T. Structural determinants of PERK inhibitor potency and selectivity. Chem Biol Drug Des. 2010;76:480–495. doi: 10.1111/j.1747-0285.2010.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ungermannova D, Chen L, Liu X. Molecular and biochemical characterization of the Skp2-Cks1 binding interface. J Biol Chem. 2004;279:51362–51369. doi: 10.1074/jbc.M405944200. [DOI] [PubMed] [Google Scholar]
- Wang W, Ungermannova D, Chen L, Liu X. A negatively charged amino acid in Skp2 is required for Skp2-Cks1 interaction and ubiquitination of p27Kip1. J Biol Chem. 2003;278:32390–32396. doi: 10.1074/jbc.M305241200. [DOI] [PubMed] [Google Scholar]
- Wang Z, Cuddy M, Samuel T, Welsh K, Schimmer A, Hanaii F, Houghten R, Pinilla C, Reed JC. Cellular, biochemical, and genetic analysis of mechanism of small molecule IAP inhibitors. J Biol Chem. 2004;279:48168–48176. doi: 10.1074/jbc.M405022200. [DOI] [PubMed] [Google Scholar]
- Woenckhaus C, Maile S, Uffmann S, Bansemir M, Dittberner T, Poetsch M, Giebel J. Expression of Skp2 and p27KIP1 in naevi and malignant melanoma of the skin and its relation to clinical outcome. Histol Histopathol. 2005;20:501–508. doi: 10.14670/HH-20.501. [DOI] [PubMed] [Google Scholar]
- Wright JJ. Combination therapy of bortezomib with novel targeted agents: an emerging treatment strategy. Clin Cancer Res. 2010;16:4094–4104. doi: 10.1158/1078-0432.CCR-09-2882. [DOI] [PubMed] [Google Scholar]
- Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci U S A. 1998;95:11324–11329. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.