SUMMARY
Thiopeptide antibiotics exhibit a profound level of chemical diversity that is installed through cascades of posttranslational modifications on ribosomal peptides. Here we present a technique to rapidly explore the chemical space of the thiopeptide GE37468 through codon randomization, yielding insights into thiopeptide maturation as well as structure and activity relationships. In this incarnation of the methodology, we randomized 7 residues of the prepeptide coding region, enabling the generation of 133 potential thiopeptide variants. Variant libraries were subsequently queried in two ways. First, high through-put MALDI-TOF mass spectrometry was applied to colony-level expressions to sample mutants which permitted full maturation of the antibiotic. Second, the activity of producing mutants was detected in an antibiotic overlay assay. In total, 29 of the 133 variants were found to produce mature compound, 12 of which retained antibiotic activity and one which had improved activity against Methicillin-resistant Staphylococcus aureus (MRSA).
INTRODUCTION
The thiopeptides are a chemically diverse group of natural products known for their potent inhibition of protein synthesis in Gram-positive bacteria and are potential therapeutic agents against dangerous drug-resistant pathogens such as MRSA (Walsh, et al., 2010; Walsh, et al., 2012). Thiopeptides are derived from a cascade of posttranslational modifications (PTMs) that transforms ribosomally-generated linear peptides into highly elaborated macrocyclic architectures rich in thiazole, oxazole, and dehydro amino acids (Arndt, et al., 2009; Bagley, et al., 2005). As opposed to non-ribosomal peptide synthesis (NRPS) – responsible for installing the peptide bonds in many small molecule peptide-based natural products such as penicillins, vancomycin, daptomycin, and cyclosporin – this form of post-ribosomal peptide synthesis (PRPS) has the virtue that DNA sequence is directly coupled to the peptide structure which scaffolds the mature antibiotic (McIntosh, et al., 2009; Nolan and Walsh, 2009). This biosynthetic logic facilitates the use of gene mutagenesis to evaluate the chemical space that is tolerated by the dedicated PTM enzymes to convert nascent peptides into complex molecular structures with potentially improved antibiotic properties (Field, et al., 2010).
Thiopeptide antibiotics represent promising targets for this type of synthetic biology as their highly constrained macrocyclic architectures are the most heavily modified class of PRPS natural products. They are generated from small, 50–60 residue, ribosomal prepeptides that undergo more than a dozen PTMs in the C-terminal 14–16 residues to achieve an extraordinary level of chemical diversity starting from only the 20 proteinogenic amino acids (Walsh, et al., 2010; Walsh, et al., 2012). These transformations represent a convergence of biosynthetic logic from (a) the lantibiotics to create dehydroalanine (dhA) and dehydrobutyrine residues (dhB) residues from the dehydration of Ser and Thr, respectively, (Chatterjee, et al., 2005) and (b) from the cyanobactins, plantazolicin, and microcin B17 pathways to create thiazoles, oxazoles, and methyloxazoles from the cyclodehydration of Cys, Ser, and Thr, respectively (Melby, et al., 2011). The signature element of thiopeptides is the central nitrogenous heterocycle, typically presented as a 2,3,6-trithiazolyl pyridine (illustrated for GE37468 in Figure 1). The pyridine is formed at a late step from cycloaddition of two serine-derived dhA residues at the same time macrocyclization is effected (Bowers, et al., 2010); the spacing of these Ser residues determines the number of atoms in the macrocycle. Net cycloaddition between Ser1 and Ser10 (numbering of the core peptide) gives the 26-membered (atoms) macrocycle found in the thiocillin, thiostrepton, and nosiheptide subclass, whereas coupling between Ser1 and Ser11 yields the 29-membered macrocycle in GE2270, GE37468, and thiomuracin (Morris, et al., 2009) (Figure S1). The 26-membered trithiazolylpyridine scaffolds target the 50S ribosome between the L11 protein and adjacent 23S rRNA helices to inhibit protein translation (Baumann, et al., 2010). In a distinct mode of action, the 29-membered macrocyclic antibiotics target the conditional GTPase, EF-Tu, and block its ability to chaperone aminoacylated tRNAs to the ribosome (Parmeggiani, et al., 2006). While total syntheses of a small number of both 26- and 29-membered thiopeptides have been achieved (Muller, et al., 2007; Nicolaou, et al., 2005), because of the structural complexity of both architectures, these molecules are not readily susceptible to structure activity relationship (SAR) determination via chemical approaches. Thus, in vivo mutational approaches are well suited to access optimized antibiotic analogs which may improve our understanding of how macrocyclic ring size and chemical functionality dictate targeting.
Figure 1.
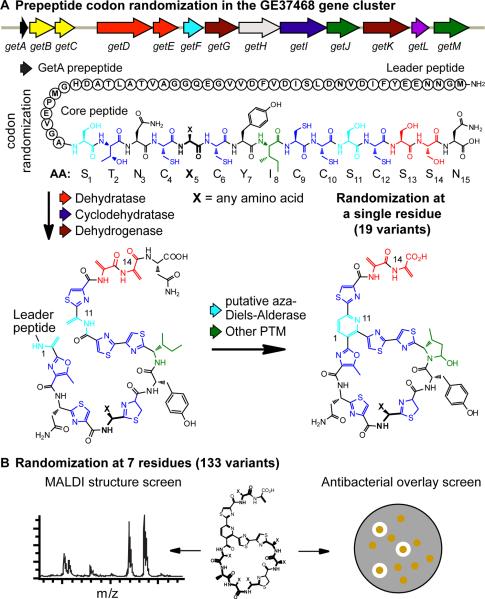
GE37468 biosynthesis and residue randomization. (A) Codon randomization created libraries of variant GetA prepeptides which were subject to a cascade of in vivo PTM provided by enzymes encoded in the GE37468 cluster (top). Randomization at residue 5 (represented by X) is shown as an example. (B) In total, codon randomization was conducted at 7 residues (illustrated by X's). Variants were screened by MALDI-TOF mass spectroscopy to assay for PTM maturation (bottom left) and by overlay with B. subtilis for activity (bottom right).
In previous efforts we have used synthetic genes to carry out alanine and serine scans of the 14 C-terminal core residues of the thiocillin prepeptide gene that ultimately get morphed into the mature thiocillin antibiotic (Acker, et al., 2009; Bowers, et al., 2010; Bowers, et al., 2012; Bowers, et al., 2010; Young and Walsh, 2011). Similar prepeptide gene replacement efforts have been applied to produce thiostrepton antibiotic variants following identification of the tsr biosynthetic gene cluster (Li, et al., 2012). These strategies generate mutants one at a time with bias in the design of the variants. In this study, we designed an approach that allows codon randomization in the 15 C-terminal prepeptide core residues of GE37468 in order to correlate mutants with both PTM maturations and SAR in the 29-membered macrocycle subclass of thiopeptides (Figure 1). GE37468 harbors a diverse array of chemical functionalities and is remarkably potent against Gram-positive bacteria (minimal inhibitory concentration (MIC) of 0.016–0.03 μg/mL against MRSA (Stella, et al., 1995; Stella, et al., 1997)). Our recent demonstration of facile heterologous production makes it ideally suited for diversification studies (Young and Walsh, 2011). In addition, as a representative of the 29-membered subclass, GE37468 targets EF-Tu (Ferrari, et al., 1995; Marinelli, et al., 1996), complementing previous mutasynthetic studies of the 50S ribosome-targeting thiocillins and thiostrepton. Finally, there is significant pharmaceutical interest in developing the 29-membered class of thiopeptides exemplified by the recent advancement of a GE2270 analog to clinical trial for treatment of Clostridium difficile colitis (LaMarche, et al., 2012).
RESULTS
Improved GE37468 expression with engineered Streptomyces “superhosts”
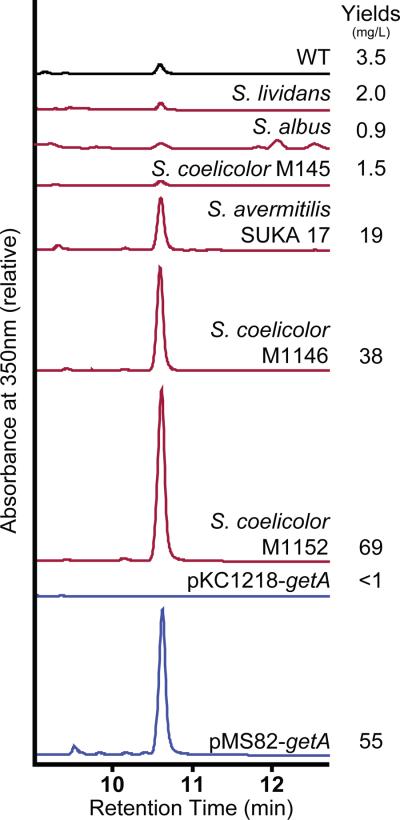
Previously, we reported the heterologous expression of the biosynthetic gene cluster for GE37468 from Streptomyces strain ATCC 55365 in S. lividans and construction of a small number of analogs via prepeptide gene mutagenesis (Young and Walsh, 2011). Although expression in S. lividans was sufficient to produce variants focused on understanding the transformation of Ile8 to the unique β-methyl-δ-hydoxy-leucine (mhP) residue, the proposed screening of prepeptide codon randomization libraries necessitated a more robust expression system. To achieve this, we compared GE37468 expression in the original heterologous host S. lividans with other model hosts, S. coelicolor and S. albus, as well as three engineered Streptomyces strains (Figure 2). S. coelicolor M145 gave yields comparable to S. lividans while S. albus gave approximately half the yields of fully matured GE37468 under our expression conditions. Increased expression was achieved through the use of Streptomyces derivatives referred to as “superhosts” for their engineered ability to increase the biosynthesis of heterologously incorporated clusters through the deletion of endogenous natural product gene clusters (Baltz, 2010). In this vein, engineered S. avermitilis strain SUKA17 gave yields of GE37468 which were 10-fold greater than S. lividans (Komatsua, et al., 2010). A further increase in yield was achieved with the engineered S. coelicolor M145 derivative, M1146, which lacks four endogenous natural product biosynthetic clusters and gave a 19-fold increase over our initial system (Gomez-Escribano and Bibb, 2011). Yields were maximized to 30-fold better than the original S. lividans system with S. coelicolor strain M1152 – a M1146 derivative which harbors a C1298T point mutation in rpoB (RNA polymerase β-subunit) previously reported to increase heterologous expression pleiotropically (Gomez-Escribano and Bibb, 2011). Expression of GE37468 and analogs in this work was thus performed in this engineered strain.
Figure 2.
GE37468 yields. Liquid chromatography traces (UV350) of mycelia methanolic extracts from expressions using the pSETGE1 vector described in (Young and Walsh, 2011). Streptomyces ATCC 55365 is shown at top (WT, black). Red chromatograms (middle) represent GE37468 heterologous expression hosts: S. lividans TK24, S. albus, S. coelicolor M145 and engineered strains S. avermitilis SUKA17, S. coelicolor M1146, and S. coelicolor M1152. Blue chromatograms (bottom) show ectopic complementation of the getA knockout cassette pSETGE-getA∷FRT, with the getA gene on the pKC1218 or pMS82 plasmid. Yields from pKC1218 were only detectable by HR-LCMS (see Figure S2a).
Complementation of the getA prepeptide gene on a mutagenizable plasmid
To optimize construction and assay of GE37468 mutants in M1152, a two plasmid complementation system was tested. The previously reported pSET-getA::FRT plasmid is a non-expressing variant of the GE37468 gene cluster cassette (pSETGE1) in which the getA prepeptide gene was knocked out leaving a flippase recognition target (FRT) scar, while all other get genes required for PTM remained in frame (Young and Walsh, 2011). With this plasmid incorporated into M1152 as a host, we tested ectopic expression of getA from the strong ermE promoter (derived from the erythromycin gene cluster (Bibb, et al., 1985)) in the autonomously replicating pKC1218 plasmid (Bierman, et al., 1992). Complementation in this fashion gave low yields of GE37468 (Figure 2, Figure S2a), indicating additional elements surrounding the getA gene may be necessary for regulation/expression of the prepeptide. Therefore, the getA gene along with ~350 bp of upstream DNA and ~150 bp downstream DNA was placed in the promoterless pMS82 E. coli-Streptomyces shuttle vector (Gregory, et al., 2003) (Figure S2b). Ectopic expression from this construct gave 80% the yield of GE37468 as was found from the single plasmid cassette pSETGE1, thus constituting a useful small plasmid for mutagenesis in E. coli and subsequent conjugative transfer to M1152.
Evaluation of tolerance for mosaic prepeptide genes in the GE37468 maturation pathway
Understanding the specificity of enzymes within thiopeptide gene clusters to act on exogenous, naturally occurring, prepeptide genes is of interest as an early step in testing the viability of assembling combinatorial biosynthetic systems. Thus, pMS82 complementation was employed to assess the potential for the PTM enzymes of the GE37468 biosynthetic cluster to process prepeptide coding sequences for thiomuracin or GE2270 (Morris, et al., 2009). Although the full length prepeptides for thiomuracin and GE2270 are similar to GE37468 (67% and 54% identity with GetA, respectively, Figure S1), we focused on the coding region which scaffolds the mature antibiotic. This sequence for thiomuracin differs from GE37468 by only the T2C mutation. Incorporation of this mutation into the getA prepeptide created a mosaic gene consisting of the GE37468 leader peptide and thiomuracin coding region. Expression of this construct gave robust yields providing the first example of a thiopeptide gene cluster acting on the coding sequence for a naturally occurring relative (see below for further discussion and characterization of this mutant). The coding region for the GE2270 prepeptide contains the T2C mutation and three additional mutations: F5V, Y7G, and I8F. Expression of this mosaic construct containing all four mutations in the getA coding region was not tolerated and produced no matured product. Because intermediates were not detected, it is unknown which step(s) in maturation of the variant core prepeptide was affected. This indicated that amino acid changes significantly altered the maturation of the prepeptide. Therefore, to assess impact of residue changes on peptide maturation as they relate to PTM substrate tolerances, we devised the codon randomization and screening methodology described below.
Prepeptide gene randomization and rapid screening for novel cyclic scaffolds by MALDI-TOF mass spectrometry
To interrogate the GE37468 PTM pathway for permissible mutations we turned to codon randomization (saturation mutagenesis). To assess the viability of this method, 6 residues within the GE37468 macrocycle and one residue outside of the macrocycle were chosen for randomization, one codon at a time. In an effort to sample mutation at each possible type of PTM in GE37468, two azole-generating residues (Thr2 and Cys4), three unmodified residues (Asn3, Phe5, and Tyr7), one dehydrated residue (Ser13), and one oxygenated residue (Ile8) were chosen. Serines 1 and 11 which contribute to the central pyridine ring were not modified in order to maintain a stable scaffold for construction of the pyridine core in the mature antibiotic. Libraries were created using NNK degenerate primers (N = any nucleotide, K = G,T) for getA oligonucleotide-directed randomization (Nov, 2012; Zheng, et al., 2004). Mutation in this way incorporates at least one codon for each amino acid using 32 total codons, while eliminating nonsense mutations from two of the stop codons (TGA and TAA). This allows encodement of all 20 amino acid mutants at each of the 7 sites for a total of 133 getA variants (not including 7 wild type sequences). To confirm diversity, 10 clones from each library (70 total clones) were sequenced. Seventeen of the 20 amino acids were represented in this sample size, in good agreement with the expected fractional completeness of the library approximated by the Poisson distribution (Figure S3a) (Patrick, et al., 2003).
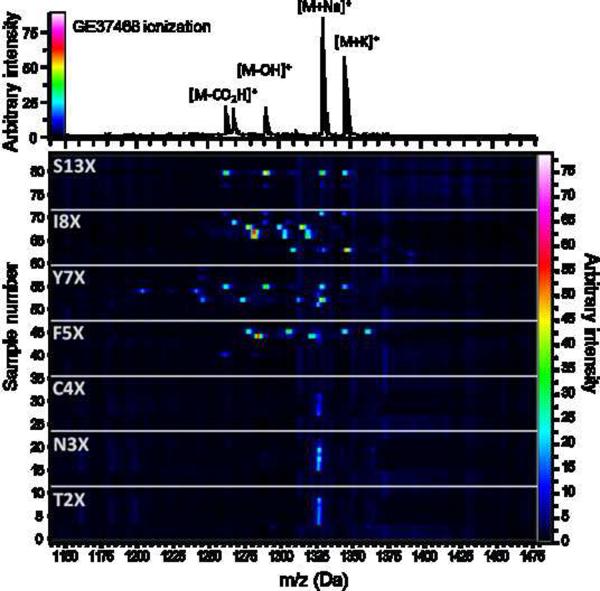
To sample the production of mature GE37468 variants from the genetic libraries, liquid culture and extraction - which can be laborious when screening large numbers of mutants - was circumvented by initial evaluation of expression from colonies on solid media (vide infra). Codon randomized getA libraries in M1152 were grown on agar plates at a density to produce individual colonies. Sixty-eight colonies were randomly chosen from each library (expected to cover approximately 90% of the diversity, see Figure S3a) and picked to a microtiter plate containing methanol. The colonies in methanol were directly mixed with MALDI-TOF matrix, without prior removal of the cell debris, and the entire mixture spotted onto a 384-well MALDITOF plate for high-throughput analysis. Wild type GE37468 produces a distinct pattern of ionization under these conditions (Figure 3 top); this four mass fingerprint could be used to identify producing mutants. To rapidly scan for these masses a heat map was created by stacking acquired spectra in the vertical axis and correlating peak height with color intensity (Figure 3, Figure S3b) (Esquenazi, et al., 2011). Examination of this plot for unique mass sets in the 1200–1400 m/z mass range showed mutation is permissible at six of the seven residues, with Cys4 as the exception. Residues Phe5, Tyr7, and Ile8 had the most tolerance for mutation, while residues Thr2, Asn3, and Ser13 accepted fewer mutations (summarized in Figure 4). Spectra lacking masses in this range indicated that the getA gene variants were either incapable of production or produced macrocyclic compound below the detection limit of this method (expression could be detected at least 250-fold lower than wild type production, or 0.25 mg/L, Figure S3c). Detailed inspection of all spectra revealed 29 unique mass spectra (not including wild type masses) in the library sampling, corresponding to 22% of the 133 possible variants that could be fully processed to the mature macrocycle at reasonable yields (Figure S4a).
Figure 3.
MALDI-TOF heat map. MALDI-TOF mass spectrometry performed on individual library colonies produced a four mass fingerprint for expressing variants (top). Spectra were converted into a “heat map” (bottom) by correlating peak intensity with color. For clarity, 12 randomly chosen samples are shown to illustrate permissibility for mutation at each residue (X = any amino acid). The full data set of 476 samples is presented in Figure S3b. The mass at approximately 1325 Da in the heat map is an unrelated S. coelicolor metabolite.
Figure 4.
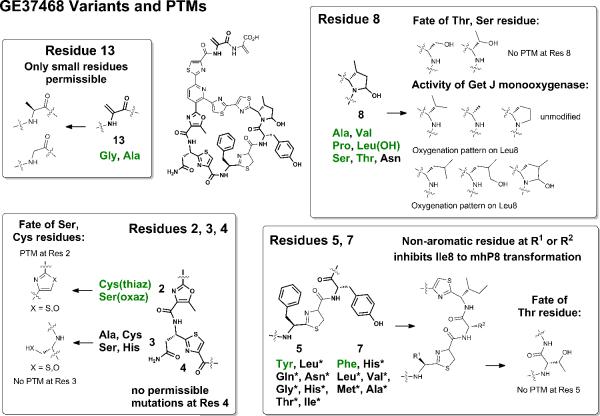
GE37468 mutants and impacts on PTM processing. The 12 active variants indentified in the overlay assay are listed in green. Inactive mutants found in MALDI-TOF assay, but not in the overlay assay, are listed in black. An asterisk indicates downstream processing is affected in the dominant product. No mutants were tolerated at residue 4. See also Figures S3b–gg, 4a.
A portion of the parent colony (that had not been subjected to extraction) which corresponded to these unique spectra was cultured in liquid media and investigated by sequencing of the getA gene and by high-resolution liquid chromatography mass spectrometry (HR-LCMS) analysis of culture extracts. Collectively, this analysis identified 153 matured compounds (including partially or alternatively processed intermediates, see Figure S3c) resulting from the 29 unique prepeptide scaffolds which allowed insights into PTM timing and specificity.
Substitution at residue Thr2 (processed to the methyloxazole in mature GE37468) was only tolerated by substitution with Cys and Ser. Both of these residue substitutions were fully processed to a thiazole or oxazole, respectively (Figure 4). This may reflect a necessity for a heterocycle at this locus to set up late stage pyridine ring-forming macrocyclization. Mutation of Asn3 to Ser or Cys was also permissible. At residue 3, the major fraction of the resultant GE37468 variants lacked PTM; however, small amounts (less than 5% of the major product) of dehydrated (dhA) or cyclodehydrated Ser3 (oxazoline) (indistinguishable by mass) could be detected by HR-LCMS as well as small amounts of full conversion to the oxazole for N3S (less than 1%, Figure S3c,h). Although formally the alternate processing of other heterocycles in the peptide cannot be ruled out, the lack of corresponding masses for other mutants discussed below provides evidence that the GetI cyclodehydratase and GetG/GetK dehydrogenases that work at residues 2, 4, 6, 9, 10, 12 may not be fully regiospecific and may act, albeit inefficiently, for cyclization and aromatization in the N3S mutant.
Residues Phe5 and Tyr7 accepted a wider range of mutations in the library sampling. Non-aromatic residues at either of these positions prevented the double oxygenative processing of Ile8 to mph8 by the monooxygenase GetJ (Figure 4). Additionally, the dominant product for molecules with a non-aromatic substitution at residues 5 or 7 and an unmodified Ile at residue 8 was a dihydro variant (+2 Da by mass spectrometry) which was produced at 2–5 times the yields of the dehydrogenated product (Figure S3c). This dihydro compound is likely the result of incomplete dehydrogenation at one of the 3 thiazoles or methyloxazole and was previously detected as the dominant product of the mhP8Ile variant produced by GetJ knockout (Young and Walsh, 2011). Mutation of Ile8 to small residues was well tolerated, which except for Leu, were not substrates for the monooxygenase GetJ. Oxygenation of Leu8 occurred to the δ-alcohol (δ-OHLeu) as the major product and complete transformation to the hemiaminal γ-methyl-δ-hydoxyproline (γ-mhP) produced at 15% the yield of the major alcohol product as determined by HR-LCMS.
Identification of linear precursors from S13X variants
Substitution at Ser13 gave only Ala13 or Gly13 cyclic variants; however, additional compounds were identified by HR-LCMS of culture extracts that corresponded to uncyclized (linear) prepeptides. These compounds were identified based on their absorption at 254 nm, but not at 350 nm, indicating the presence of thiazoles, but lack of extended conjugation through a trithiazolylpyridine core as previously reported (Bowers, et al., 2010). To confirm these compounds were linear precursors two additional variants, S13N and S13L, for which the signature mass fingerprint for cyclic compounds shown in Figure 4 was not found by MALDI-TOF, were expressed in liquid shake flasks, extracted, and analyzed by HR-LCMS. The major product of both S13N and S13L corresponded to linear products which were confirmed by mass spectrometry fragmentation (MS/MS) (Figure S4b–g). In each variant, the linear precursor was found to lack dehydration at Ser11, indicating the mutation of Ser13, especially to larger amino acids, disrupted dehydration by the GetD/GetE dehydratases. The absence of a dhA11 residue subsequently prohibited macrocyclization by GetF.
Co-culture overlay assay with codon randomized libraries
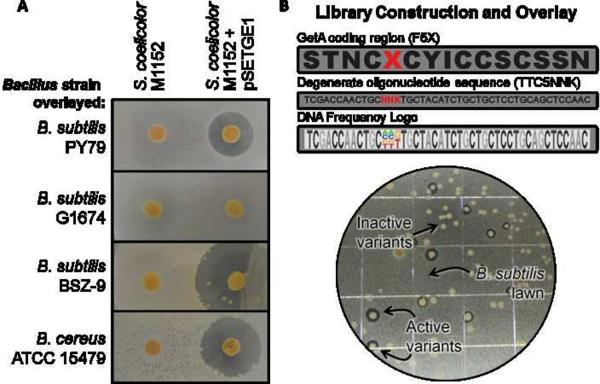
We next developed an assay to screen the codon randomized libraries for antibiotic activity in an unbiased, scalable format using an antibiotic overlay method on agar plates. This method exploited the robust expression profile from S. coelicolor M1152. Additionally, the deletion of endogenous natural product clusters in M1152 minimized host antibiotic production that might interfere with assaying the activity of expressed GE37468 analogs. B. subtilis PY79 was chosen as a model Gram-positive organism for overlays based on its sensitivity to GE37468, non-virulence, and precedent for coculture with S. coelicolor (Yang, et al., 2009).
To test the utility of this method, colony expression from M1152 expressing wild type GE37468 from pSETGE1 was optimized on BTT agar plates by overlay with B. subtilis PY79 in soft agar. After overnight incubation, strain M1152 containing pSETGE1 produced a clear zone of inhibition, while M1152 alone did not antagonize B. subtilis growth (Figure 5a). To confirm the phenotype was a result of EF-Tu inhibition, colonies were overlaid with B. subtilis G1674 which harbors an insensitive G275S (E. coli numbering) mutation in EF-Tu (Sosio, et al., 1996). As expected, growth of G1674 was unaffected. To further confirm the resistance mechanism was specific to EF-Tu, overlay was tested with B. subtilis BSZ-9 or B. cereus ATCC 15479 which exhibit resistance to 26-member thiazolyl peptides through a knockout or mutant L11 protein, respectively (Baumann, et al., 2010; Zhang, et al., 2001). B. cereus ATCC 15479 is responsible for thiocillin production and also encodes export pumps predicted to confer thiocillin resistance. Neither BSZ-9 nor B. cereus ATCC 15479 exhibited resistance to M1152 harboring pSETGE1 confirming the EF-Tu target of GE37468 is distinct from ribosomal resistance.
Figure 5.
Bacillus antibiotic overlay assay. (A) Colonies of S. coelicolor M1152 or S. coelicolor M1152 + pSETGE1 were grown on BTT agar plates followed by overnight incubation with the designated Bacillus strain. B. subtilis PY79 has no engineered resistance, strain G1674 is resistant to 29-membered thiopeptides, and strain BSZ-9 is resistant to 26-membered thiopeptides. B. cereus ATCC 15479 is resistant to 26-membered thiopeptides. The B. subtilis BSZ-9 zone of inhibition exhibits small colonies of S. coelicolor due to the exceptionally slow growth rate of BSZ-9. (B) Randomization and screening at residue Phe5 (red X = any amino acid) is illustrated as an example. The core region of the GetA prepeptide is shown (top) with degenerate oligonucleotide sequence used for codon randomization and DNA frequency logo. Incorporation of this library into S. coelicolor M1152 and overlay with B. subtilis PY79 is shown at bottom.
Six hundred colonies from each codon randomization library were grown on BTT plates for 6 days followed by overlay with B. subtilis PY79. Screening this number of colonies corresponded to an 18.75-fold degeneracy and gave a >99.99% confidence that all 32 sequence variants (resulting from NNK mutagenesis) were represented (Figure S3a). Colonies which produced zones of inhibition (Figure 5b) were restreaked on fresh plates and the getA gene sequenced to identify the library variant (resultant variants colored green in Figure 4). Those which did not produce zones of inhibition indicated mutants that were either incapable of production as noted above, were not secreted, or gave products that lacked sufficient antibiotic activity to produce a zone of inhibition (an MIC less than at least 0.5 μg/mL as determined by the weakest antibiotic MIC found in the screen). Twelve of the 29, or 41%, of the permissible mutations identified by MALDI-TOF were found to retain activity in the overlay screen and were then purified from expressions in liquid shake flasks for quantification of antibiotic activity against B. subtilis PY79 and two strains of MRSA (Table 1).
Table 1.
MIC of GE37468 variants produced in this study.
| MIC (μg/mL)a | ||||
|---|---|---|---|---|
| Variant | PTM | B. subtilis b | MW2c | COLd |
| GE37468 | wild type | 0.016 | 0.016 | 0.016 |
| T2C | thiazole | 0.0078 | 0.0078 | 0.0078 |
| T2S | oxazole | 0.016 | 0.016 | 0.016 |
| F5Y | 0.13 | 0.13 | 0.25 | |
| Y7F | 0.063 | 0.063 | 0.13 | |
| I8P | 0.063 | 0.031 | 0.063 | |
| I8L | γ-hydroxy | 0.063 | 0.031 | 0.031 |
| I8L | γ-mhP | 0.031 | 0.016 | 0.031 |
| I8S | 0.25 | 0.13 | 0.13 | |
| I8T | 0.063 | 0.031 | 0.063 | |
| I8V | 0.25 | 0.063 | 0.13 | |
| I8A | 0.063 | 0.031 | 0.063 | |
| S13A | 0.25 | 0.25 | 0.25 | |
| S13G | 0.5 | 0.5 | 0.5 | |
Minimum inhibitory concentrations determined by overnight serial dilution culture.
Bacillus subtilis PY79.
Methicillin-resistant S. aureus MW2.
Methicillin-resistant S. aureus COL.
See Figure S5 for compound isolation.
Two of the 6 permissible mutants from the left half of the molecule (residues 2, 3, and 4) retained activity. Mutant T2S had equal activity to the wild type compound (0.016 μg/mL) while T2C had 2-fold improved activity (0.0078 μg/mL) indicating that the methyloxazole, thiazole, and oxazole residues have small, but significant, impacts on antibiotic activity. None of the permissible mutations at Asn3, which is conserved in GE2270, thiomuracin, and GE37468, retained antibiotic activity.
On the lower half of the molecule (residues 5 and 7), 2 of the 12 detected mutants from the MALDI-TOF sampling retained activity. Mutant Y7F was 4 to 8-fold less active (0.063 – 0.13 μg/mL depending on strain) while mutant F5Y was 8 to 16-fold less active (0.13 – 0.25 μg/mL) than wild type. It is unknown whether this marked decrease in antibiotic activity is the result of altered macrocyclic ring conformation or a direct modulation of the mutant's interaction with EF-Tu. No mutation to non-aromatic residues retained activity, likely because of the additional defects in processing (lack of oxygenation at Ile8 and dehydrogenation) associated with these mutations noted above.
Mutation at residue 8 was the most promiscuous with six amino acids (Ala, Val, Pro, Leu, Ser, Thr) able to replace the native mhP8 residue with retention of on-target antibiotic activity. These mutations were between 2 and 16-fold less active (0.031 – 0.25 μg/mL) than wild type GE37468. Residue mhP8 was previously hypothesized to contribute to activity via the structural constraint imparted on the macrocycle (Young and Walsh, 2011). Notably, the swap to proline, mhP8Pro, is a 2 to 4-fold (0.031 – 0.063 μg/mL) weaker antibiotic, while replacement with the cyclic γ-mhP8 residue (derived from Leu8) is equal to 2-fold weaker (0.016 – 0.031 μg/mL) than wild type, indicating that the methyl and hydroxyl groups play a significant role in EF-Tu binding. Both the Ala and Gly mutations at residue 13 retained activity, albeit markedly decreased. S13A exhibited a 16-fold decrease in activity (0.25 μg/mL) while S13G was 32-fold less active (0.5 μg/mL) than wild type GE37468.
DISCUSSION
Methodology considerations
Using saturation mutagenesis we have tested the substrate specificity of the GE37468 cluster to process variants of the GetA prepeptide. This methodology has the benefit of being unbiased in its design for exploration of chemical space. A similar approach from researchers at Novacta Biosystems used saturation mutagenesis to generate all possible single point mutation variants at individual residues of the lantibiotics mersacidin and actagardine (Appleyard, et al., 2009; Boakes, et al., 2012); however, the methodology was limited by the necessity to construct 19 separate plasmids to achieve saturation at each residue. A codon randomization approach using NNT mutagenesis was employed by A. James Link's lab to explore the biosynthetic tolerances of the ribosomally synthesized lasso peptide microcin J25 (Pan and Link, 2011); however, the NNT codon only encodes 15 of the 20 amino acids. To encode all 20 amino acid variants at a single codon, Sonomoto and coworkers used NNK mutagenesis to randomize residues in the lantibiotic nukacin (Islam, et al., 2009). The NNK codon allows reduced degeneracy while encoding all 20 amino acids single oligonucleotide-directed mutagenesis experiment and as such was used for saturation mutagenesis in this work.
In codon randomization approaches, the confidence that all encodable members of an NNK mutagenesis library are represented in a screen is proportional to the number of colonies screened. By analyzing 68 colonies from each library in the MALDI-TOF screen we expected to sample approximately 90% of library diversity (17–18 of the 20 possible amino acids) which was supported by the Poisson-like distribution of randomly sequenced clones (Figure S3a). On the other hand, 600 colonies were screened in our overlay assay corresponding to a >99.99% confidence that all library clones were represented (Patrick, et al., 2003). Library screening in both methods was streamlined at the colony level to circumvent the laborious process of separate fermentations, allowing us to cumulatively sample hundreds of library members and ultimately focus on 12 active variants.
Execution of this strategy depended on sufficient peptide expression to increase the probability of detecting weakly expressing analogs. This was accomplished by expression in the modified S. coelicolor M1152 strain, which increased yields more than 30-fold (to 69 mg/L, Figure 2) over the original heterologous producer and allowed detection of variants at ~0.5% the yield of wild type GE37648 (Figure S3c). MALDI-TOF mass spectrometry analysis from single colony extracts allowed efficient visualization of the production of novel scaffolds in the absence of assayable antibiotic activity. Expression of variants from M1152 was also critical to detect the activities of GE37468 variants in overlay assays. This was aided by the quadruple knockout of gene clusters that encode actinorhodin, prodiginine, CPK (cryptic polyketide synthase), and CDA (calcium dependent antibiotic) in the S. coelicolor M1152 strain which limited endogenous antibiotics that could interfere with assaying the activity. This screen was a preamble to larger growth and isolation for MIC quantification of the 12 active variants. Thus, the combination of the two approaches provided valuable insight into the SAR of maturation, production, and activity of the thiopeptide variants.
Structural considerations
Using these screening strategies we were able to witness unanticipated PTM patterns that occur from unbiased mutation. For instance, non-aromatic residues at positions 5 or 7 prevented Ile8 processing to mhp8 suggesting the native aromatic residues are necessary for substrate recognition by the GetJ monooxygenase. This oxygenative transformation is set prior to ring closure, evidenced by its presence in linear precursor peptides found in the S13X mutants (Figure S4b–g). Inhibition of this PTM yielded an unmodified Ile at position 8 and lead to the production of a dihydro analog of the expected product. This suggests that the mhP8 residue is integral in substrate recognition by the GetG/GetK dehydrogenases for thiazole/methyloxazole aromatization; however, further investigation is warranted to determine which thiazole or methyloxazole lacks dehydrogenation.
The conservation of small amino acids at position 13 provided a somewhat surprising result. Placed outside the macrocycle, it may be expected that mutation of this residue would have little impact on ring maturation. Here, we traced the inability of S13X variants to macrocyclize to a lack of dehydration at the cycloaddition-fated residue Ser11 through identification of linear peptide precursors. In general, dehydration of Ser11 and subsequent macrocyclization decreased with increasing size of the amino acid at residue 13 (S13G>S13A>S13N>S13L) (Figure S4b) suggesting a steric interaction with the GetD or GetE dehydratase. The major linear product of the S13L variant contained a Ser11 that had failed to undergo dehydration and Ser14 that had been hydrolyzed off to yield a C-terminal acid (rather than amide). N-terminal proteolytic trimming had released residues upstream of the dhA1 which subsequently hydrolyzed to the methylketone (Bowers, et al., 2010) (Figure S4c). All PTMs besides dehydration of Ser11 were set in this molecule, indicating GE37468 macrocyclization is a late stage modification as found in the thiocillins. Unlike oxygenation of Val6 of thiocillin (Bowers, et al., 2010), we found double oxygenative processing of Ile8 had proceeded prior to ring closure (Figure S4d,g). Although linear variants were not readily detectable in other mutants beyond S13X, these findings suggest that detection of additional linear, partially processed intermediates from other variants (i.e., from ring expansion or contraction experiments (Bowers, et al., 2012) as well as knockout mutants (Bowers, et al., 2010)) will give insights into the timing of the gauntlet of PTM events in the GE37468 pathway and other related pyridine-containing thiopeptide antibiotics.
A major goal of synthetic biology in this context is a comprehensive understanding of the logic controlling thiopeptide biosynthesis in order to engineer molecules with custom tailored chemistries (Benner and Sismour, 2005; Young, et al., 2011). Previously we showed oxygenative transformation of Ile8 to mhP8 is carried out by the GetJ monooxygenase through a proposed regiospecific tandem double hydroxylation/cyclization mechanism (Young and Walsh, 2011). TpdJ, an analog of GetJ in the thiomuracin cluster, produces seven different oxygenation states on thiomuracin's Ile8 and has been shown to modulate activity against different species of bacteria (Morris, et al., 2009). The ability to control this oxygenation may be a useful synthetic biology technique to design more active variants. Here we report substrate mutation as alternate (and complementary) approach to producing oxygenation variants with GetJ. Mutation of Ile8 to Leu8 yielded two new oxygenated residues, γ-methyl-δ-hydroxy-proline and δ-hydroxyl-leucine, not yet found to occur naturally in thiopeptides. This lays the groundwork for future research focused on swapping oxygenases among clusters, which, combined with prepeptide substrate mutation, has the potential to yield an arsenal of oxidation patterns for in vivo synthetic biology.
SAR considerations
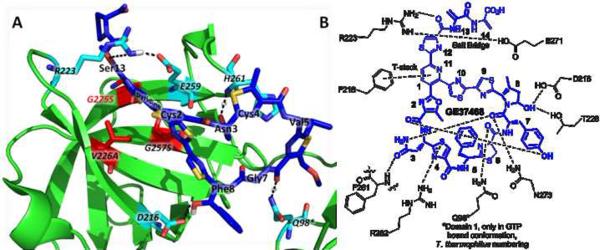
Twelve GE37468 variants resulting from the 29 permissible mutations found in the MALDI-TOF sampling experiments retained antibiotic activity, with one variant (T2C) eliciting slightly improved activity, validating this methodology in the construction of novel antibiotics. The activity, or lack thereof, for variants can be assessed by examining the reported crystal structure of GE2270 complexed with EF-Tu and the GTP analog GDPNP (Figure 6, Table S1) (Parmeggiani, et al., 2006). Important contacts between the GE2270 and EF-Tu are shown in Figure 6a and those relevant to GE37468 charted in Figure 6b (Heffron and Jurnak, 2000; LaMarche, et al., 2012; Lamarche, et al., 2012; Leeds, et al., 2011; Parmeggiani, et al., 2006). GE2270 and GE37468 have very similar antimicrobial profiles and are expected to engage in similar binding mechanisms (Stella, et al., 1995; Stella, et al., 1997). Notably, Asn3 forms two hydrogen bonds, one with the backbone amide of F261 on EF-Tu and one intramolecular transannular hydrogen bond with the carbonyl of Tyr7. It is therefore not surprising that this residue is conserved in GE2270 and thiomuracin and could not be mutated in GE37468 with retention of activity. Another hydrogen bond bifurcated between the β-hydroxylated Phe8 of GE2270 and Asp216 and Thr228 of EF-Tu may explain why oxygenation at residue 8 in thiomuracin, GE2270, and GE37468 is conserved. Although not isosteric with the β-OHPhe in GE2270, the hydroxyl group of mhP8 in GE37468 may also hydrogen bond with these residues or the neighboring Glu215. Interestingly, GE37468 variants with hydroxyl groups which are isosteric with the β-OHPhe hydroxyl group (I8S and I8T) do not retain full activity. The most potent Ile8 mutant was γ-mhP mutant (derived from I8L) supporting evidence that both macrocyclic ring conformation imparted by the mhP ring constraint and hydrogen bonding from the δ-hydroxyl group are important in achieving full activity in GE37468 variants.
Figure 6.
Major contacts between 29-membered thiopeptides and EF-Tu. (A) Crystal structure of GE2270A (dark blue sticks) complexed with T. thermophilus EF-Tu (green ribbon, E. coli numbering) and GDPNP (GTP analog, not visible) adapted from PDB: 2C77 (Parmeggiani, et al., 2006). Residues forming important contacts are illustrated in light blue. Residues involved in resistance mutations are noted in red: G275S occurs in B. subtilis G1674 used in this work. (B) EF-Tu contacts (black, E. coli numbering) identified from GE2270A-EF-Tu structures complexed with GDP or GTP and mapped onto GE37468 (blue). GE37468 residues in bold were subject to randomization. Contacts and references are tabulated in Table S1. Contact with residue Q98 from T. thermophilus EF-Tu is only formed in the GTP bound state.
The structure does not provide clear evidence for why the conservative mutations F5Y and Y7F cause a marked decrease in antibiotic activity. GTP binding has been shown to elicit a large conformational change in EF-Tu which allows domain 1 to contact the southern half of the antibiotic in the region (Figure 6) (Parmeggiani, et al., 2006). These aromatic rings have been shown to be in proximity on the southern half of the molecule by NMR NOE experiments (Morris, et al., 2009). It is possible that the addition or removal of the phenolic oxygen from residue 5 or 7 alters the interaction between the antibiotic and domain 1 and leads to destabilization of this structure. In the Y7F mutant, disruption of a transannular hydrogen bond between the phenolic oxygen of Tyr7 and carbonyl 2, identified in the recently solved thiomuracin-EF-Tu structure (Lamarche, et al., 2012), may destabilize the antibiotic structure.
Mutations S13A and S13G lose 16-fold and 64-fold potency, respectively, compared with wild type dhA13. The C-terminal dhA residues of GE37468 and thiomuracin are not conserved in GE2270 and it is therefore difficult to rationalize what structures in this region of the molecule would be required for optimal binding. However, a protein tunnel formed by a salt-bridge between residues R223–E271 may require a well defined conformation around the bound antibiotic tail. This conformation may be disrupted through the loss of the dhA13 sp2 center by replacement with Ala or Gly. It is worthy to note that significant semisynthetic SAR efforts by a group at Novartis have been applied to the GE2270 C-terminal tail immediately following thiazole 12. This work has provided evidence for the importance of chemical functionalities in this region of 29-membered thiopeptides for EF-Tu binding as well as increasing solubility (LaMarche, et al., 2012; Leeds, et al., 2011).
SIGNIFICANCE
The highly constrained macrocyclic architectures of thiopeptides may be considered privileged structures for bioactivity due to their reduced entropic costs of binding and proteolytic stability, and as such, are ideal molecules for scaffolding peptide evolution. While single amino acid point mutations and gene deletions have been employed to manipulate thiopeptide structures in other contexts, here we use a technique that has the potential to exhaustively explore chemical space at individual residues through codon randomization. Peptide diversification to explore chemical space in natural product peptides is a complementary effort to other in vivo peptide evolution methodologies such as split intein methods or the use of unnatural amino acids in natural product biosynthesis (Tianero, et al., 2012; Young, et al., 2011) and holds the potential to create novel molecules with enhanced or modulated bioactivities. The work presented here goes beyond the existing methods to produce molecules that provide a wealth of information not only on SAR but also on the biosynthetic capacity of the thiopeptide PTM pathway. Application of this methodology to the construction of peptide libraries designed to target resistant EF-Tu proteins may provide a method of counter-evolving thiopeptide antibiotics to evolving resistance. Furthermore, this methodology may be readily adapted to the study of other thiopeptide subclasses such as thiocillins and thiostrepton, as well as to other PRPS natural products such as the lantibiotics and cyanobactins.
EXPERIMENTAL PROCEDURES
GE37468 expression and extraction
General methods for the growth, transformation, and storage of Streptomyces adhered to guidelines found in Practical Streptomyces Genetics (Kieser, et al., 2000). Expression and extraction of GE37468 and analogs was performed as previously described (Young and Walsh, 2011) and is detailed in the Supplemental Experimental Details.
Subcloning procedures
The getA gene was subcloned to pKC1218 (Kieser, et al., 2000) with a spectinomycin resistant marker and exogenous Erm promoter (Bibb, et al., 1985) by PCR amplification of the getA gene using primers `getA PstI RBS F' and `getA XbaI R' (Table S2). PCR product was digested with restriction enzymes PstI and XbaI and ligated into the similarly digested pKC1218 vector to create pKC1218-PErmE-getA. To subclone getA to pMS82 (Gregory, et al., 2003), getA with flanking DNA (Figure S2b) was PCR amplified with primers `GetA-pMS82 F' and `GetA-pMS82 R', digested with HindIII and KpnI, and ligated to the similarly digested pMS82 plasmid. To facilitate cloning of mutants, an XhoI site was added immediately following the structural gene using PCR directed mutagenesis (Zheng, et al., 2004) with primers `A XhoI F' and `A XhoI R' to create pMS82-getA. PCR mutagenesis was used to create the thiomuracin coding sequence (T2C) using primers `A T2C F' and `A T2C R' and the GE2270 coding sequence (T2C, F5V, Y7G, I8F) using primers `A GE2270 F' and `A GE2270 R'. Libraries were created using the `GetA F new primer' forward primer and the `MS82 NNK2 R', `MS82 NNK3 R', `MS82 NNK4 R', `MS82 NNK5 R', `MS82 NNK7 R', `MS82 NNK8 R', or `MS82 NNK13 R', reverse primers to randomize residues 2, 3, 4, 5, 7, 8, and 13, respectively. PCR products were digested with ScaI and XhoI and ligated into the similarly digested pMS82-getA plasmid. To achieve consistent expression from colonies on agar plates it was necessary to transfer libraries from pMS82 into the pSETGE1 plasmid. This was accomplished by incorporating an SpeI site immediately following the getA gene in pSETGE1 by PCR directed mutagenesis with primers `SpeI F' and `SpeI R'. Libraries were then transferred to an ScaI and SpeI digested pSETGE1 plasmid by PCR from pMS82-getA with primers `GetA Gib F' and `GetA Gib R' and ligation using the Gibson/Venter method of isothermal assembly (Gibson, et al., 2008).
MALDI-TOF Assay
Colonies produced using methods described in the Supplemental Experimental Details were mixed with MALDI-TOF Matrix (40 mg of Sigma-Aldrich, 50% 2,5-dihydroxybenzoic acid, 50% 2-cyano-3-(4-hydroxyphenyl)acrylic acid in 1 mL of 78% acetonitrile, 21.9% water, 0.1% trifluoracetic acid) and spotted on a 384-well Bruker stainless steel target plate. Spectra were acquired on a Autoflex Speed MALDI-TOF mass spectrometer (Bruker Daltonics) using parameters outlined in the Supplemental Experimental Details and analyzed using flexAnalysis 3.3 (Bruker Daltonics). Heat maps were created using ClinProTools 2.2 (Bruker Daltonics).
Antibacterial overlay and quantification
Spore stocks of libraries in M1152 were spread on BTT agar at a density of 2 colonies/cm2. After incubation for 6 days at 30 °C, plates were overlaid with 15 mL of B. subtilis in soft agar (1:10,000 dilution of a saturated overnight B. subtilis culture in LB broth containing 0.7% agar at 35–40 °C). Overlaid plates were incubated for an additional 16 hrs at 30 °C. Colonies which produced clear zones of inhibition as illustrated in Figure 4 were picked from beneath the overlaid soft agar and restruck on SFM plates containing apramycin (50 μg/mL) and nalidixic acid (25 μg/mL) to select for growth of the Streptomyces M1152 library variant. Colony sequencing was performed as above. Quantification of antibacterial activity (MIC assays) against B. subtilis PY79, MRSA COL, and MRSA MW2 was performed using the overnight culture/serial dilution assay as previously reported (Acker, et al., 2009; Bowers, et al., 2010; Bowers, et al., 2012; Young and Walsh, 2011). Additional details on MIC assays and sample purification is described in Figure S5.
Supplementary Material
HIGHLIGHTS
Thiopeptide GE37468 expression was increased 30-fold above previous reports
Codon randomization enabled rapid exploration of chemical space
Thirteen new thiopeptide analogs had activity against MRSA
Identification of linear precursor peptides allowed insights into biosynthetic timing
ACKNOWLEDGEMENTS
We thank Jane Yang for MALDI-TOF mass spectroscopy expertise, Haruo Ikedeo for S. avermitilis strains, Mervyn Bibb for engineered S. coelicolor strains, Margaret Smith for the pMS82 plasmid, Margherita Sosio for B. subtilis strain G1674, and William Haldenwang for B. subtilis strain BSZ-9. This work was supported in part by a NIH grant GM20011 (C.T.W.), the New England Regional Center of Excellence grant NIAID 057159 (C.T.W.), and an NIH postdoctoral fellowship F32GM098051 (T.S.Y.). Support for MALDI-TOF mass spectrometry was provided by The Bruker Therapeutic Discovery Mass Spectrometry Center and NIH GM S10RR029121 (P.C.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acker MG, Bowers AA, Walsh CT. Generation of Thiocillin Variants by Prepeptide Gene Replacement and in Vivo Processing by Bacillus cereus. J. Am. Chem. Soc. 2009;131:17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard AN, Choi S, Read DM, Lightfoot A, Boakes S, Hoffmann A, Chopra I, Bierbaum G, Rudd BA, Dawson MJ, et al. Dissecting structural and functional diversity of the lantibiotic mersacidin. Chem Biol. 2009;16:490–498. doi: 10.1016/j.chembiol.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt HD, Schoof S, Lu JY. Thiopeptide antibiotic biosynthesis. Angew Chem Int Ed Engl. 2009;48:6770–6773. doi: 10.1002/anie.200901808. [DOI] [PubMed] [Google Scholar]
- Bagley MC, Dale JW, Merritt EA, Xiong X. Thiopeptide antibiotics. Chem Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- Baltz RH. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol. 2010;37:759–772. doi: 10.1007/s10295-010-0730-9. [DOI] [PubMed] [Google Scholar]
- Baumann S, Schoof S, Bolten M, Haering C, Takagi M, Shin-ya K, Arndt HD. Molecular determinants of microbial resistance to thiopeptide antibiotics. J Am Chem Soc. 2010;132:6973–6981. doi: 10.1021/ja909317n. [DOI] [PubMed] [Google Scholar]
- Benner SA, Sismour AM. Synthetic biology. Nat. Rev. Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb MJ, Janssen GR, Ward JM. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38:215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Boakes S, Ayala T, Herman M, Appleyard AN, Dawson MJ, Cortes J. Generation of an actagardine A variant library through saturation mutagenesis. Appl Microbiol Biotechnol. 2012 doi: 10.1007/s00253-012-4041-0. [DOI] [PubMed] [Google Scholar]
- Bowers AA, Acker MG, Koglin A, Walsh CT. Manipulation of thiocillin variants by prepeptide gene replacement: structure, conformation, and activity of heterocycle substitution mutants. J Am Chem Soc. 2010;132:7519–7527. doi: 10.1021/ja102339q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers AA, Acker MG, Young TS, Walsh CT. Generation of Thiocillin Ring Size Variants by Prepeptide Gene Replacement and In Vivo Processing by Bacillus cereus. J. Am. Chem. Soc. 2012 doi: 10.1021/ja302820x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers AA, Walsh CT, Acker MG. Genetic Interception and Structural Characterization of Thiopeptide Cyclization Precursors from Bacillus cereus. J. Am. Chem. Soc. 2010;132:12182–12184. doi: 10.1021/ja104524q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee C, Paul M, Xie LL, van der Donk WA. Biosynthesis and mode of action of lantibiotics. Chemical Reviews. 2005;105:633–683. doi: 10.1021/cr030105v. [DOI] [PubMed] [Google Scholar]
- Esquenazi E, Jones AC, Byrum T, Dorrestein PC, Gerwick WH. Temporal dynamics of natural product biosynthesis in marine cyanobacteria. Proc Natl Acad Sci U S A. 2011;108:5226–5231. doi: 10.1073/pnas.1012813108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari P, Colombo L, Stella S, Selva E, Zerilli LF. Antibiotic GE37468 A: a novel inhibitor of bacterial protein synthesis. II. Structure elucidation. J Antibiot (Tokyo) 1995;48:1304–1311. doi: 10.7164/antibiotics.48.1304. [DOI] [PubMed] [Google Scholar]
- Field D, Hill C, Cotter PD, Ross RP. The dawning of a 'Golden era' in lantibiotic bioengineering. Mol. Microbiol. 2010;78:1077–1087. doi: 10.1111/j.1365-2958.2010.07406.x. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. Complete Chemical Synthesis, Assembly, and Cloning of a Mycoplasma genitalium Genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano JP, Bibb MJ. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MA, Till R, Smith MC. Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J Bacteriol. 2003;185:5320–5323. doi: 10.1128/JB.185.17.5320-5323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron SE, Jurnak F. Structure of an EF-Tu complex with a thiazolyl peptide antibiotic determined at 2.35 A resolution: atomic basis for GE2270A inhibition of EF-Tu. Biochemistry. 2000;39:37–45. doi: 10.1021/bi9913597. [DOI] [PubMed] [Google Scholar]
- Islam MR, Shioya K, Nagao J, Nishie M, Jikuya H, Zendo T, Nakayama J, Sonomoto K. Evaluation of essential and variable residues of nukacin ISK-1 by NNK scanning. Mol Microbiol. 2009;72:1438–1447. doi: 10.1111/j.1365-2958.2009.06733.x. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Crowes; Norwich, UK: 2000. [Google Scholar]
- Komatsua M, Uchiyama T, Omura S, Cane DE, Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMarche MJ, Leeds JA, Amaral A, Brewer JT, Bushell SM, Deng G, Dewhurst JM, Ding J, Dzink-Fox J, Gamber G, et al. Discovery of LFF571: an investigational agent for Clostridium difficile infection. J Med Chem. 2012;55:2376–2387. doi: 10.1021/jm201685h. [DOI] [PubMed] [Google Scholar]
- Lamarche MJ, Leeds JA, Dzink-Fox J, Gangl E, Krastel P, Neckermann G, Palestrant D, Patane MA, Rann EM, Tiamfook S, et al. Antibiotic optimization and chemical structure stabilization of thiomuracin a. J Med Chem. 2012;55:6934–6941. doi: 10.1021/jm300783c. [DOI] [PubMed] [Google Scholar]
- Leeds JA, LaMarche MJ, Brewer JT, Bushell SM, Deng G, Dewhurst JM, Dzink-Fox J, Gangl E, Jain A, Lee L, et al. In vitro and in vivo activities of novel, semisynthetic thiopeptide inhibitors of bacterial elongation factor Tu. Antimicrob Agents Chemother. 2011;55:5277–5283. doi: 10.1128/AAC.00582-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhang F, Kelly WL. Mutagenesis of the thiostrepton precursor peptide at Thr7 impacts both biosynthesis and function. Chem Commun (Camb) 2012;48:558–560. doi: 10.1039/c1cc14281j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli F, Gastaldo L, Toppo G, Quarta C. Antibiotic GE37468A: a new inhibitor of bacterial protein synthesis. III. Strain and fermentation study. J Antibiot (Tokyo) 1996;49:880–885. doi: 10.7164/antibiotics.49.880. [DOI] [PubMed] [Google Scholar]
- McIntosh JA, Donia MS, Schmidt EW. Ribosomal peptide natural products: bridging the ribosomal and nonribosomal worlds. Natural Product Reports. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby JO, Nard NJ, Mitchell DA. Thiazole/oxazole-modified microcins: complex natural products from ribosomal templates. Curr. Opin. Chem. Biol. 2011;15:369–378. doi: 10.1016/j.cbpa.2011.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, et al. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J Am Chem Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- Muller HM, Delgado O, Bach T. Total synthesis of the thiazolyl peptide GE2270 A. Angew. Chem.-Int. Edit. 2007;46:4771–4774. doi: 10.1002/anie.200700684. [DOI] [PubMed] [Google Scholar]
- Nicolaou KC, Zak M, Safina BS, Estrada AA, Lee SH, Nevalainen M. Total synthesis of thiostrepton. Assembly of key building blocks and completion of the synthesis. J. Am. Chem. Soc. 2005;127:11176–11183. doi: 10.1021/ja052934z. [DOI] [PubMed] [Google Scholar]
- Nolan EM, Walsh CT. How Nature Morphs Peptide Scaffolds into Antibiotics. Chembiochem. 2009;10:34–53. doi: 10.1002/cbic.200800438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nov Y. When Second Best Is Good Enough: Another Probabilistic Look at Saturation Mutagenesis. Applied and Environmental Microbiology. 2012;78:258–262. doi: 10.1128/AEM.06265-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan SJ, Link AJ. Sequence diversity in the lasso peptide framework: discovery of functional microcin J25 variants with multiple amino acid substitutions. J Am Chem Soc. 2011;133:5016–5023. doi: 10.1021/ja1109634. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A, Krab IM, Okamura S, Nielsen RC, Nyborg J, Nissen P. Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu. Biochemistry. 2006;45:6846–6857. doi: 10.1021/bi0525122. [DOI] [PubMed] [Google Scholar]
- Patrick WM, Firth AE, Blackburn JM. User-friendly algorithms for estimating completeness and diversity in randomized protein-encoding libraries. Protein Eng. 2003;16:451–457. doi: 10.1093/protein/gzg057. [DOI] [PubMed] [Google Scholar]
- Sosio M, Amati G, Cappellano C, Sarubbi E, Monti F, Donadio S. An elongation factor Tu (EF-Tu) resistant to the EF-Tu inhibitor GE2270 in the producing organism Planobispora rosea. Mol Microbiol. 1996;22:43–51. doi: 10.1111/j.1365-2958.1996.tb02654.x. [DOI] [PubMed] [Google Scholar]
- Stella S, Montanini N, Le Monnier F, Ferrari P, Colombo L, Marinelli F, Landini P, Ciciliato I, Goldstein BP, Selva E, et al. Antibiotic GE37468 A: a new inhibitor of bacterial protein synthesis. I. Isolation and characterization. J Antibiot (Tokyo) 1995;48:780–786. doi: 10.7164/antibiotics.48.780. [DOI] [PubMed] [Google Scholar]
- Stella S, Montanini N, LeMonnier FJ, Colombo L, Selva E, Denaro M. Antibiotics GE 37468 A, B, and C. Gruppo Lepetit SpA, Gerenzano, ITX; United States: 1997. [Google Scholar]
- Tianero MD, Donia MS, Young TS, Schultz PG, Schmidt EW. Ribosomal route to small-molecule diversity. J Am Chem Soc. 2012;134:418–425. doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT, Acker MG, Bowers AA. Thiazolyl peptide antibiotic biosynthesis: a cascade of post-translational modifications on ribosomal nascent proteins. J Biol Chem. 2010;285:27525–27531. doi: 10.1074/jbc.R110.135970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT, Malcolmson SJ, Young TS. Three ring posttranslational circuses: insertion of oxazoles, thiazoles, and pyridines into protein-derived frameworks. ACS Chem Biol. 2012;7:429–442. doi: 10.1021/cb200518n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YL, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5:885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TS, Walsh CT. Identification of the thiazolyl peptide GE37468 gene cluster from Streptomyces ATCC 55365 and heterologous expression in Streptomyces lividans. Proc Natl Acad Sci U S A. 2011;108:13053–13058. doi: 10.1073/pnas.1110435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TS, Young DD, Ahmad I, Louis JM, Benkovic SJ, Schultz PG. Evolution of cyclic peptide protease inhibitors. Proc Natl Acad Sci U S A. 2011;108:11052–11056. doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Scott JM, Haldenwang WG. Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor sigma(B) J Bacteriol. 2001;183:2316–2321. doi: 10.1128/JB.183.7.2316-2321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.