Summary
Wnt/β-catenin signaling plays a key role in the pathogenesis of colon and other cancers; emerging evidence indicates that oncogenic β-catenin regulates several biological processes essential for cancer initiation and progression. To decipher the role of β-catenin in transformation, we classified β-catenin activity in 85 cancer cell lines in which we performed genome scale loss-of-function screens and found that β-catenin active cancers are dependent on a signaling pathway involving the transcriptional regulator YAP1. Specifically, we found that YAP1 and the transcription factor TBX5 form a complex with β-catenin. Phosphorylation of YAP1 by the tyrosine kinase YES1 leads to localization of this complex to the promoters of anti-apoptotic genes including BCL2L1 and BIRC5. A small molecule inhibitor of YES1 impeded the proliferation of β-catenin-dependent cancers in both cell lines and animal models. These observations define a β-catenin-YAP1-TBX5 complex essential to the transformation and survival of β-catenin-driven cancers.
Introduction
β-catenin signaling plays a key role in colon development and cancer (Clevers, 2006). The destruction complex composed of AXIN1, GSK3β and adenomatous polyposis coli (APC) phosphorylates serine residues in β-catenin, which leads to its proteasomal degradation (Clevers, 2006). Binding of Wnts to the LPR6-Frizzled receptor inactivates this complex, leading to accumulation and nuclear translocation of β-catenin. In the nucleus, β-catenin forms a complex with TCF4 that drives the transcription of genes that contribute to cell proliferation (Klaus and Birchmeier, 2008). Individuals carrying APC germline mutations (Familial adenomatous polyposis) develop colonic polyps that progress to colon cancer (Kinzler and Vogelstein, 1996), and mutations in the tumor suppressor APC or the oncogene β-catenin have been found in the majority of spontaneously arising colon cancers (TCGA, 2012).
β-catenin is a component of the adherent junctions (Baum and Georgiou, 2011) and in the nucleus binds to TCF4 and several transcriptional regulators. For example, when cancer cell lines are cultured under hypoxic conditions, β-catenin forms a complex with HIF-1 leading to hypoxia adaptation (Kaidi et al., 2007), and in prostate cancer cells, a β-catenin-androgen receptor (AR) complex increases the transcription of AR (Mulholland et al., 2002). β-catenin and YAP1 also co-regulate genes that are essential for cardiac development (Heallen et al., 2011). These observations suggest that through interactions with different partners, β-catenin regulates many biological processes.
Yes-associated protein 1 (YAP1) is a transcriptional modulator that has been implicated in stem cell differentiation and the control of organ size (Pan, 2010). YAP1 regulates several context-specific transcriptional programs (Badouel et al., 2009) and promotes proliferation and tumor growth (Overholtzer et al., 2006; Zhao et al., 2008). Indeed, YAP1 is recurrently amplified in hepatocellular cancer, where YAP1 is essential for survival of tumors that harbor YAP1 amplifications (Zender et al., 2006). Furthermore, inducible transgenic expression of a stabilized YAP1 mutant (S127A) in mice induced liver hyperplasia and colonic adenomas (Camargo et al., 2007).
YAP1 transcriptional activity is regulated by several mechanisms. In quiescent cells, Hippo pathway-mediated serine phosphorylation of YAP1 inhibits nuclear import and promotes its degradation (Zhao et al., 2012). In contrast, YES1-mediated phosphorylation of YAP1 activates YAP1 in embryonic stem cell self-renewal (Tamm et al., 2011), and ABL-mediated phosphorylation of YAP1 in response to DNA damage results in transcription of pro-apoptotic genes (Levy et al., 2008). Recent work suggests that YAP1 also plays a role in mechanotransduction in a Hippo-independent manner (Dupont et al., 2011).
Although stabilization and localization of β-catenin contributes to adenoma formation, our understanding of β-catenin regulation and function in cancer remains incomplete. For example, Rac1-mediated phosphorylation of β-catenin has been shown to affect β-catenin activation and localization (Wu et al., 2008). Moreover, in zebrafish and some human cell lines, APC loss alone resulted in impaired differentiation but failed to induce nuclear localization of β-catenin and transformation (Phelps et al., 2009). To gain insights into β-catenin activity in malignant transformation, we classified β-catenin activity in a panel of human cancer cell lines in which we have systematically characterized genetic alterations, gene expression and gene essentiality. Here we report the identification of an alternative transcriptional regulatory complex required for the β-catenin-driven transformation and tumor maintenance.
Results
Identification of essential genes in β-catenin active cancer cell lines
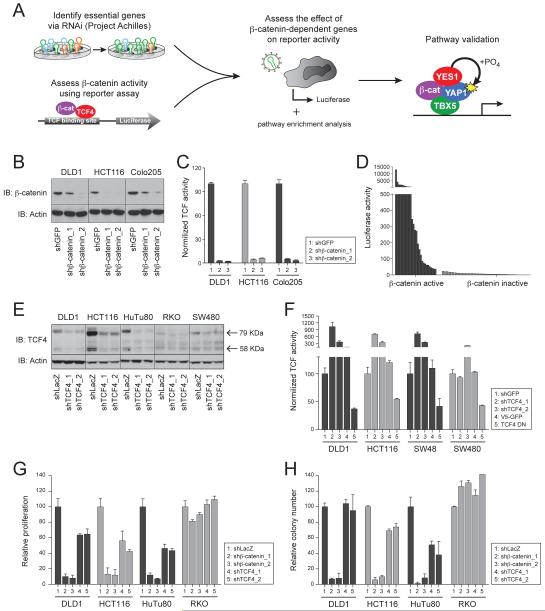
To identify genes whose expression is essential in cell lines that exhibit β-catenin activity, we used a β-catenin/TCF4 reporter (Fuerer and Nusse, 2010) to classify β-catenin activity in 85 cancer cell lines in which we had previously performed genome scale loss of function screens (Cheung et al., 2011), transcriptional profiling and global copy number analyses (Barretina et al., 2012) (Figure 1A). To evaluate the specificity of this reporter, we used colon cancer cell lines (DLD1, Colo205 and HCT116) that harbor mutations in components of the Wnt/β-catenin pathway. Expression of two distinct β-catenin-specific shRNAs suppressed β-catenin expression (Figure 1B) and inhibited β-catenin/TCF4 reporter activity (Figure 1C) in these cell lines. Of the 85 cell lines, 19 showed reporter activity that was at least 10-fold above background (Figure 1D, Table 1, S1). We note that two colon cancer cell lines that harbor APC mutations (HT29 and LS411N) exhibited little β-catenin activity and were classified as reporter inactive.
Figure 1. Identification of genes essential for β-catenin active cell lines.
(A) Strategy to identify genes required in β-catenin active cell lines. (B) β-catenin expression in cells expressing β-catenin-specific shRNAs. IB indicates immunoblotting. (C) Activity of the β-catenin/TCF4 reporter following suppression of β-catenin. LacZ was used for normalization. (D) β-catenin/TCF4 reporter activity in 85 cell lines. (E) TCF4 expression following introduction of TCF4-specific shRNAs. (F) β-catenin/TCF4 activity following suppression of TCF4 or expression of DN-TCF4 (G) Proliferation or (H) AI growth following suppression of TCF4. Data presented as mean ± SD for 3 independent experiments. See also Figure S1.
Table 1.
Cancer cell lines profiled for β-catenin activity.
| Origin | β-catenin inactive | β-catenin active |
|---|---|---|
| Colon | 6 | 13 |
| Esophageal | 8 | 1 |
| Liver | 1 | |
| Bone | 1 | |
| Gastric | 0 | 2 |
| Pancreas | 9 | |
| Ovarian | 20 | 1 |
| GBM | 6 | |
| Lung | 7 | 1 |
| Meningioma | 2 | 1 |
| Endometrium | 1 | |
| Multiple myeloma | 1 | |
| Melanoma | 1 | |
| Osteosarcoma | 1 | |
| Breast | 2 |
We applied a two class permutation analysis (Extended Experimental Procedures) to cell lines classified as either β-catenin active or inactive and identified genes whose expression was essential for the survival/proliferation of β-catenin active cells (File S1). We identified β-catenin as the top candidate and found 49 other genes that scored as significantly essential for the proliferation/survival of β-catenin active cells (q-value<0.25).
We tested whether suppressing each of the top 250 genes that scored in the above analysis affected β-catenin/TCF4 reporter activity in an APC mutated cell line (Figure S1A). Suppression of APC or CSNK1A1 induced increased reporter activity, and suppression of β-catenin or BCL9L inhibited reporter activity (Figure S1B). Although suppression of 44 genes inhibited β-catenin/TCF4 reporter activity in the DLD1 cell line, more than 80% of the genes that scored as selectively essential in β-catenin active cell lines did not regulate the this reporter, suggesting that the majority of these essential genes represent co-dependencies not directly related to β-catenin/TCF4-regulated gene regulation.
β-catenin binds TCF/LEF family transcription factors to regulate gene expression (Klaus and Birchmeier, 2008). Since we failed to identify TCF4 as required for the survival of β-catenin active cell lines, we manipulated the expression of TCF4 using TCF4-specific shRNAs (Figure 1E) or perturbed TCF4 function by expressing a dominantly interfering allele of TCF4 [TCF4 DN, (Korinek et al., 1997)]. Expression of the TCF4 DN allele inhibited β-catenin/TCF4 reporter activity (Figure 1F). In contrast, suppression of TCF4 increased the activity of the β-catenin/TCF4 reporter (Figure 1F), as recently reported (Tang et al., 2008). We then assessed the consequences of suppressing β-catenin or TCF4 on the proliferation and anchorage independent (AI) growth of β-catenin active (HuTu80, DLD1 and HCT116) or inactive (RKO) cells. Suppression of β-catenin or TCF4 failed to affect the proliferation (Figure 1G) or AI growth (Figure 1H) of RKO cells (β-catenin reporter inactive) but inhibited these phenotypes in β-catenin active cell lines. In contrast, depletion of TCF4 only partially inhibited (30-40%) the proliferation and AI growth of β-catenin active cell lines (Figure 1G, H). Although we cannot exclude the possibility that residual TCF4 remains in these cells or that other TCF family members compensate for TCF4 suppression, these observations suggest that β-catenin acts in part in a TCF4-independent manner.
Based on these observations, we examined the list of the top 50 scoring genes (q<0.25) that were essential for the proliferation of β-catenin active cell lines and identified a striking enrichment for proteins related to the transcriptional regulator YAP1 (Table 2). When we classified the cell lines based on the mutational status of oncogenes commonly found in colon cancers, such as KRAS, BRAF or PIK3CA (Wood et al., 2007), we failed to find any enrichment for genes related to YAP1 (Table 2). These observations suggested that these essential genes are specific for β-catenin active cells.
Table 2.
Significance scores for genes related to YAP1 following classification of cell lines by the mutational status of PIK3CA, BRAF, KRAS or based on β-catenin activity.
| β-catenin active vs. rest | PIK3CA mut vs. Rest | BRAF mut vs. rest | KRAS mut vs. rest | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rank | Gene | p-value | q-value | Rank | Gene | p-value | q-value | Rank | Gene | p-value | q-value | Rank | Gene | p-value | q-value |
| 1 | CTNNB1 | 2*10−5 | 0.04 | 1 | PIK3CA | 2*10−5 | 0.04 | 1 | BRAF | 2*10−5 | 0.14 | 1 | KRAS | 2*10−5 | 0.02 |
| 1 | BCL2L1 | 2*10−5 | 0.04 | 144 | BCL2L1 | 0.006 | 0.3 | 1442 | BCL2L1 | 0.23 | 0.97 | 218 | BCL2L1 | 0.02 | 0.74 |
| 32 | YAP1 | 0.001 | 0.24 | 269 | YAP1 | 0.01 | 0.38 | 283 | YAP1 | 0.047 | 0.97 | 249 | YAP1 | 0.02 | 0.77 |
| 30 | YES1 | 0.001 | 0.24 | 2573 | YES1 | 0.32 | 0.91 | 868 | YES1 | 0.14 | 0.97 | 3098 | YES1 | 0.42 | 0.98 |
| 27 | BIRC5 | 0.0008 | 0.22 | 329 | BIRC5 | 0.01 | 0.4 | 508 | BIRC5 | 0.09 | 0.97 | 2336 | BIRC5 | 0.31 | 0.97 |
| 8 | TBX5 | 0.0001 | 0.1 | 2728 | TBX5 | 0.34 | 0.92 | 2290 | TBX5 | 0.35 | 0.97 | 1273 | TBX5 | 0.16 | 0.95 |
| 16 | FAT1 | 0.0003 | 0.16 | 2723 | FAT1 | 0.34 | 0.92 | 2208 | FAT1 | 0.34 | 0.97 | 246 | FAT1 | 0.02 | 0.77 |
YAP1 is essential for the transforming properties of β-catenin active cell lines
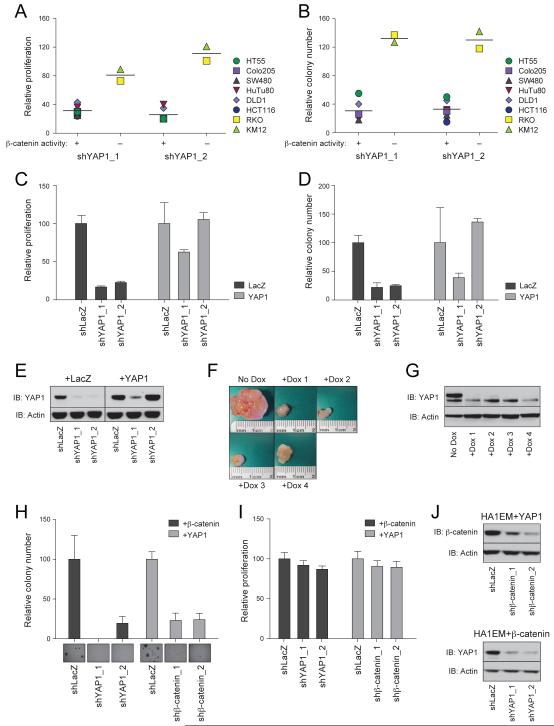
YAP1, is a transcriptional co-activator that scored as essential for the proliferation/survival of β-catenin active cells (Rank 32, q-value=0.24, Table 2). To confirm the observed dependency on YAP1 in β-catenin active cell lines, we introduced two independent YAP1-specific shRNAs into a panel of colon cancer cell lines with high or undetectable β-catenin activity (Table 1). We found that YAP1 expression was selectively required for the proliferation and AI growth of β-catenin active cells (Figure 2A, B, S2A). When we examined the two cell lines that harbored APC inactivating mutations but did not exhibit β-catenin/TCF4 reporter activity (Table S1), we found that these cell lines (HT29 and LS411N) were dependent on YAP1 and β-catenin (Figure S2B, C). To eliminate the possibility that the observed effects were due to off-target effects, we expressed LacZ or YAP1 in parallel cultures of HuTu80 cells expressing control shRNAs or the two YAP1-specific shRNAs, one of which (shYAP1 2) targets the YAP1 3′ untranslated region (UTR). We found that forced expression of YAP1 rescued the proliferation and AI growth of HuTu80 cells in which YAP1 was suppressed (Figure 2C-E).
Figure 2. YAP1 is essential for tumorigenicity of β-catenin dependent cancer cell lines.
(A) Proliferation and (B) AI growth of the indicated cancer cell lines following suppression of YAP1. Classification of β-catenin activity in each cell line is noted. (C) Proliferation, (D) AI growth and (E) expression of YAP1 in HuTu80 cell lines overexpressing wild type (WT) YAP1 and the indicated YAP1-specific or control (shLacZ) shRNAs. (F) Effects of suppressing YAP1 on orthotopic colon tumors. (G) YAP1 expression in tumors shown in (F). (H-J) HA1EM cells expressing β-catenin or YAP1 and YAP1- or β-catenin-specific shRNAs. (H) AI growth, (I) proliferation or (J) protein levels. In (H) inset shows representative images from AI growth assay. Data presented as mean ± SD for 3 independent experiments. See also Figure S2 and S3.
The YAP1 related protein TAZ has been reported to bind to YAP1 and also to regulate Wnt signaling by inhibiting DVL1 (Varelas et al., 2010). TAZ did not score as essential for β-catenin active cell lines, and when we suppressed the expression of TAZ in β-catenin active cells (Figure S2D), we failed to observe an effect on proliferation (Figure S2E). These observations suggest that TAZ is not required in cells that exhibit β-catenin activity.
We found that suppression of YAP1 failed to affect the activity of the β-catenin/TCF4 reporter (Figure S1A). Since YAP1 was reported to affect reporter activity in SW480 cells (Zhou et al., 2011), we suppressed YAP1 in 4 additional colon cancer lines that harbor mutations that activate the Wnt/β-catenin pathway and failed to detect decreased reporter activity (Figure S2F) or alterations in the transcription of known β-catenin/TCF4 target genes such as c-Myc, AXIN2 and SOX4 (Figure S2G) (He et al., 1998; Yan et al., 2001). Moreover, suppression of YAP1 failed to affect the stability of β-catenin (Figure S2H).
To determine whether YAP1 is required for tumorigenicity, we developed an orthotopic colon cancer model in which subcutaneous tumor xenografts derived from an established colon cancer cell line are implanted into the cecum of a second host. Orthotopic implantation of these tumors resulted infiltration of the colon and liver metastases (Figure S3A). We used this model to determine whether β-catenin or YAP1 were required for tumor growth. Specifically, we developed vectors that harbored doxycycline-inducible shRNAs targeting either β-catenin or YAP1 and introduced these vectors into HCT116 cells. ShRNA expression was induced post cecal implantation with doxycycline. We found that tumors expressing the inducible β-catenin-specific shRNAs showed diminished expression of β-catenin and were substantially smaller (Figure S3B, C). When we analyzed tumors expressing an inducible YAP1-specific shRNA, we found that suppression of YAP1 also inhibited tumor growth by 80-90% (Figure 2F, G), indicating that YAP1 was essential for tumorigenic growth.
These observations confirmed that YAP1 expression is required for the tumorigenicity of β-catenin active cells. To determine whether YAP1 contributes to cell transformation, we expressed a stabilized form of β-catenin (S33A, S37A, T41A, S45A) that cannot be phosphorylated (Morin et al., 1997) or YAP1 (Zhao et al., 2010) in HA1EM cells, a non tumorigenic immortalized kidney epithelial cell line that is rendered tumorigenic by the forced expression of myristoylated AKT1 (Boehm et al., 2007). Expression of stabilized β-catenin or YAP1 sufficed to promote AI growth (Figure 2H), indicating that expression of either YAP1 or activated β-catenin transforms these cells. These immortalized cells were not dependent on stabilized β-catenin or YAP1 for proliferation (Figure 2I). However, suppression of β-catenin inhibited the AI growth of cells expressing stabilized YAP1, and suppression of YAP1 reduced the AI growth of cells expressing stabilized β-catenin (Figure 2H, J). Together these observations implicate YAP1 as an essential gene in β-catenin-mediated transformation and suggest that YAP1 and β-catenin cooperate to induce transformation.
YAP1, β-catenin and TBX5 form a complex
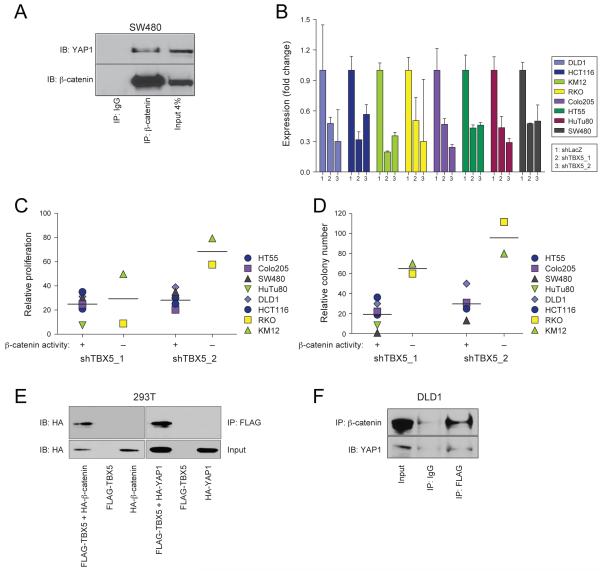
YAP1 and β-catenin have recently been shown to co-regulate genes critical for cardiac development (Heallen et al., 2011). Using SW480 and HuTu80 cells, we found that endogenous YAP1 and β-catenin interact. Specifically, we found that β-catenin-specific but not control immunoglobulin immune complexes contained endogenous YAP1 (Figure 3A). Moreover, when we isolated YAP1 immune complexes, we detected endogenous β-catenin (Figure 6A).
Figure 3. YAP1, β-catenin and TBX5 interact.
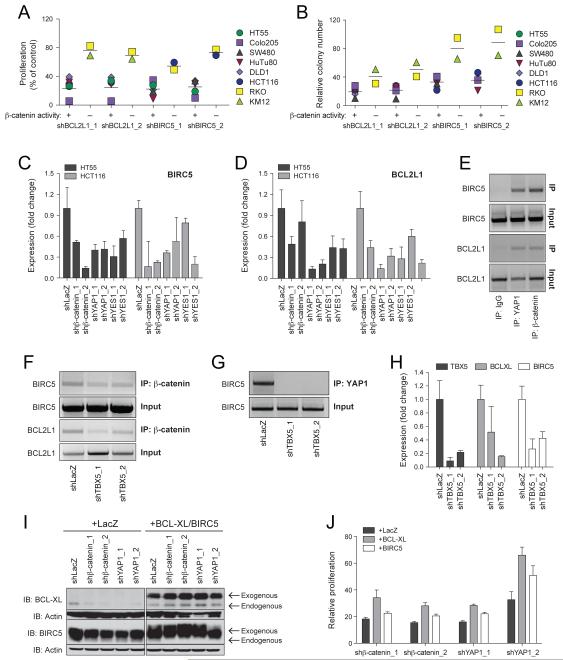
(A) β-catenin or control IgG immune complexes were isolated from SW480 lysates and the indicated proteins were analyzed by immunoblotting. (B) TBX5 mRNA levels measured by quantitative PCR in cells expressing TBX5-specific or control (shLacZ) shRNAs. Data presented as mean ± SD for 3 independent experiments. (C) Proliferation or (D) AI growth of the indicated cells following suppression of TBX5. (E) The indicated expression vectors were introduced into 293T cells, FLAG immune complexes were isolated and analyzed by immunoblotting with an anti-HA antibody. (F) FLAG-immune complexes were isolated from DLD1 cells stably expressing a FLAG-epitope tagged TBX5 protein and analyzed by immunoblotting with the indicated antibodies.
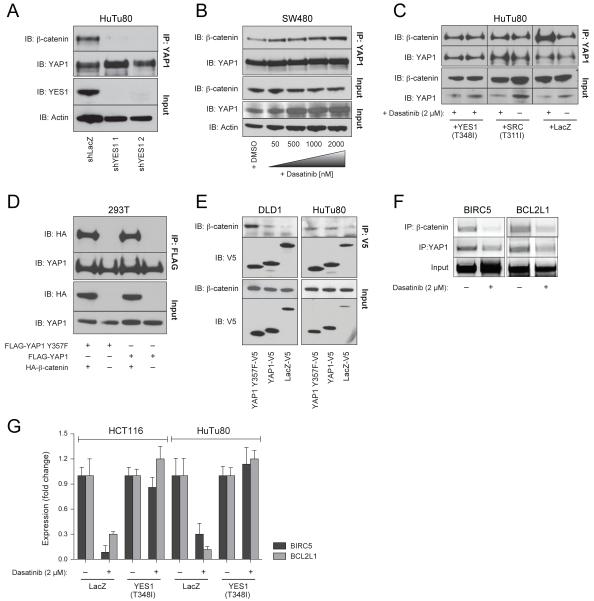
Figure 6. Expression of YES1 is essential for formation of the YAP1-β-catenin-TBX5 complex.
(A) YAP1 immune complexes were isolated from HuTu80 cells expressing YES1-specific or control shRNAs and β-catenin abundance was analyzed by immunoblotting. (B) SW480 cells were treated for 6 h with increasing concentrations of dasatinib and β-catenin-YAP1 complexes were assessed as in (A). (C) HuTu80 cells expressing dasatinib resistant YES1 or SRC mutants were treated with 2μM of dasatinib for 6 h and the β-catenin-YAP1 interaction was assessed as in (A). (D) 293T cells were transfected with FLAG-epitope tagged WT or Y357F YAP1 (5 μg) with or without HA-epitope tagged β-catenin. FLAG immune complexes were assessed for the presence of HA tagged proteins. (E) V5 immune complexes were isolated from DLD1 or HuTu80 colon cancer cell lines stably expressing V5-epitope tagged WT or Y357F YAP1 or control LacZ and the presence of β-catenin was assessed by immunoblotting. (F) β-catenin or YAP1 immune complexes from HCT116 cells treated with 2 μM of dasatinib or vehicle (DMSO) were subjected to ChIP analysis. (G) mRNA levels of BCL2L1 and BIRC5 in HCT116 or HuTu80 cells treated for 1 h with 2 μM of dasatinib.
Both YAP1 and β-catenin are transcriptional co-regulators (Kaidi et al., 2007; Zhao et al., 2008). When we re-examined the list of genes that were required for proliferation/viability of β-catenin active cells, we failed to find the known β-catenin partner TCF4 or the TEAD transcription factor family, a known YAP1 partner (Zhao et al., 2008) but noted that the transcription factor TBX5 was highly ranked in this analysis (Rank 8, q-value=0.1, Table 2). To confirm that TBX5 was required in β-catenin active cells, we expressed TBX5-specific shRNAs in β-catenin active cells (Figure 3B) and found that TBX5 suppression induced a 60-80% decrease in proliferation and AI growth (Figure 3C, D). Although suppression of TBX5 expression also inhibited the proliferation of β-catenin inactive cell lines to some degree (Figure 3C), these observations indicate that β-catenin active cell lines are also dependent on TBX5 expression.
To determine whether TBX5 interacts with YAP1 and β-catenin, we isolated TBX5-specific immune complexes from 293T or DLD1 cells and found that TBX5 binds to both β-catenin and YAP1 (Figure 3E, F). These findings corroborate a prior report showing that TBX5 and YAP1 interact in 293T cells when overexpressed (Murakami et al., 2005) and identify a complex composed of YAP1, β-catenin and TBX5.
BCL2L1 and BIRC5 are transcriptional targets of the β-catenin-YAP1-TBX5 complex
Among the genes that we identified as required in β-catenin active cell lines were BIRC5 (survivin) and BCL2L1 (Table 2). Using BIRC5- or BCL2L1-specific shRNAs (Figure 4SA, B), we found that depletion of BIRC5 or BCL2L1 impaired the proliferation and AI growth of β-catenin-dependent cell lines (Figure 4A, B). These observations corroborate recent observations that cancer cell lines that harbor activating mutations in β-catenin are particularly sensitive to navitoclax, an inhibitor of BCL2 family members including BCL-XL (Personal communication, Stuart Schreiber).
Figure 4. BCL2L1 and BIRC5 are transcriptional targets of the β-catenin-YAP1-TBX5 complex.
(A) Proliferation or (B) AI growth following suppression of BCL2L1 or BIRC5 in the indicated cell lines. (C, D) mRNA levels of BCL2L1 and BIRC5 in HT55 or HCT116 cells expressing β-catenin-specific, YAP1-specific or control shRNAs. (E) β-catenin, YAP1 or control immune complexes were isolated from HuTu80 cells and subjected to ChIP analysis with primers derived from the promoter regions of BCL2L1 (1-1000 bp) and BIRC5 (−952-0 bp). (F) β-catenin or (G) YAP1 immune complexes were derived from HuTu80 cells expressing TBX5-specific shRNAs were subjected to ChIP analysis using primers for BIRC5. (H) mRNA levels of BCL2L1 and BIRC5 in HCT116 cells expressing TBX⍰-specific shRNAs. (I) Immunoblot analysis of BCL-XL or BIRC5 in HuTu80 cells overexpressing BCL-XL, BIRC5 or LacZ and YAP1-specific, β-catenin-specific or control shRNAs. (J) Proliferation of the cell lines described in (I). Data presented as mean ± SD for 3 independent experiments. See also Figure S4.
To test whether BIRC5 and BCL2L1 are transcriptional targets of the β-catenin-YAP1-TBX5 complex, we examined the mRNA levels of BIRC5 and BCL2L1 in cell lines (HT55 and HCT116) in which we had suppressed either YAP1 or β-catenin. We found that the expression of both BIRC5 and BCL2L1 were dependent on the presence of β-catenin and YAP1 (Figure 4C, D) suggesting that the β-catenin-YAP1-TBX5 complex is involved in the transcriptional regulation of these genes.
To determine whether β-catenin and YAP1 directly regulate BCL2L1 and BIRC5 expression, we performed a chromatin immunoprecipitation assay (ChIP) focused on sites in the BCL2L1 and BIRC5 promoters identified by β-catenin-specific ChIP seq and found that both β-catenin and YAP1 were bound to these promoters (Figure 4E). Furthermore, suppression of TBX5 expression in HuTu80 cells abrogated the binding of β-catenin (Figure 4F) or YAP1 (Figure 4G) to these promoters. Similar to what we found when we suppressed YAP1, YES1 or β-catenin (Figure 4C, D), suppression of TBX5 expression resulted decreased expression of BIRC5 and BCL2L1 (Figure 4H).
To investigate whether BCL2L1 and BIRC5 contribute to the proliferation arrest observed following suppression of either β-catenin or YAP1 in β-catenin active cancer cell lines, we stably expressed the anti-apoptotic isoform of BCL2L1 (BCL-XL) or BIRC5 in HuTu80 (β-catenin active). Following overexpression of these genes, we expressed YAP1- or β-catenin-specific shRNAs. Ectopic expression of BCL-XL or BIRC5 rendered the levels of these proteins independent of β-catenin or YAP1 (Figure 4I) and partially restored the proliferation of cell lines in which we suppressed either β-catenin or YAP1 (Figure 4J), suggesting that these genes are targets of the β-catenin-YAP-TBX5 complex.
YES1 kinase activity is essential for the transforming properties of β-catenin dependent cancers
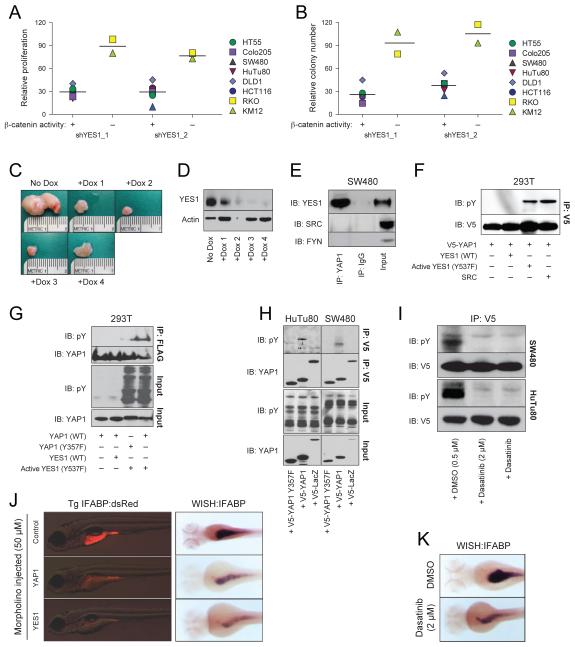
YAP1 was originally identified as a YES1-associated protein (Sudol et al., 1995). We found that the SRC family tyrosine kinase YES1 was essential for the growth of β-catenin active cell lines (rank 30, q-value=0.24, Table 2). As we observed for YAP1, suppression of YES1 inhibited the proliferation, AI growth and tumor formation of β-catenin active cell lines (Figure 5A-D, S5A). Furthermore, when we suppressed YES1 expression, we also found reduced levels of BIRC5 and BCL2L1 (Figure 4C, D). We confirmed that YES1-specific shRNAs did not alter the expression of the closely related kinase SRC (Figure S5A, B). These observations confirmed that YES1 expression was required in β-catenin active cell lines.
Figure 5. YES1 is essential for tumorigenicity of β-catenin active cells.
(A) AI growth or (B) proliferation following suppression of YES1 expression. (C) Effects of suppressing YES1 on orthotopic HCT116 colon tumors. (D) YES1 expression in tumors shown in (C). (E) YAP1 immune complexes were isolated from SW480 cells, and the indicated proteins were analyzed by immunoblotting. (F) V5-epitope tagged YAP1 and WT or activated (Y537F) YES1 or SRC expression constructs were introduced into 293T cells. V5 immune complexes were analyzed by immunoblotting with the PY99 phosphotyrosine-specific antibody. (G) FLAG-epitope tagged WT or mutated (Y357F) YAP1 were transfected into 293T cells together with activated (Y537F) YES1. FLAG-epitope immune complexes were isolated and analyzed by immunoblotting with a phosphotyrosine antibody. (H) V5 immune complexes were isolated from HuTu80 or SW480 cells stably expressing WT or Y357F V5-epitope tagged YAP1 or control V5-LacZ. Immune complexes were analyzed by immunoblotting with a phosphotyrosine antibody. (I) HuTu80 or SW480 cells stably expressing V5-epitope tagged WT YAP1 were treated for 6 h with 0.5 or 2 μM of dasatinib. V5 immune complexes were analyzed by immunoblotting with a phosphotyrosine antibody. (J) Transgenic IFABP:RFP zebrafish were injected with 50 μM of YAP1- or YES1-specific morpholinos, and red fluorescence was assessed 4 d post fertilization (dpf) or IFABP expression was assessed 3 dpf using whole mount in situ hybridization with an IFABP-specific probe. (K) Embryos were exposed to 2 μM of dasatinib at 2 dpf, and IFABP expression was assessed after 24 h using whole mount in situ hybridization. See also Figure S5-S7.
YAP1 binds to YES1 and is phosphorylated by SRC family kinases in embryonic stem cells (Tamm et al., 2011). We confirmed that YES1 and YAP1 interact in the β-catenin active colon cancer cell line SW480 (Figure 5E). Previous studies have shown that YAP1 is able to bind to other SRC family members such as SRC in HeLa cells (Zaidi et al., 2004). However, in colon cancer cell lines, we failed to detect an interaction between YAP1 and SRC or FYN (Figure 5E).
To determine whether YES1 or SRC phosphorylates YAP1, we expressed YAP1 in 293T cells and assessed YAP1 tyrosine phosphorylation when co-expressed with YES1 or SRC (Figure 5F). We detected phosphorylated YAP1 only when YAP1 was co-expressed with SRC or with activated mutant version of YES1 (Y537F). We failed to detect phosphorylation of YAP1 when co-expressed with WT YES1 indicating that YAP1 phosphorylation requires the active form of YES1 (Figure 5F). Although both YES1 and SRC phosphorylated YAP1, suppression of SRC failed to inhibit the proliferation and AI growth of β-catenin active cell lines (Figure S5C-E). Thus, we concluded that both YES1 and SRC are able to phosphorylate YAP1 but only YES1 is essential for the survival of β-catenin active cell lines.
In 293T cells, we did not detect phosphorylated YAP1 when expressed alone (Figure 5F). In contrast, we readily detected YAP1 tyrosine phosphorylation in HuTu80 or SW480 cells expressing WT YAP1 (Figure 5H). Furthermore, treatment of colon cancer cells expressing WT YAP1 with the tyrosine kinase inhibitor dasatinib (Lombardo et al., 2004) inhibited the tyrosine phosphorylation of YAP1 (Figure 5I). These results confirm reported observations that demonstrated that YES1 is activated in colon cancer cell lines and tumors (Pena et al., 1995).
Prior work has demonstrated that SRC family members phosphorylate tyrosine residues contained with the sequence motif YXXP (Levy et al., 2008). YAP1 harbors one tyrosine residue with this motif (tyrosine 357). Under conditions where active YES1 phosphorylated WT YAP1 in 293T cells, we failed to detect tyrosine phosphorylation of the YAP1 Y357F mutant in either 293T or colon cancer cell lines (Figure 5G, 5H). These observations confirm that YES1 phosphorylates YAP1 at tyrosine 357.
We then tested whether phosphorylation of YAP tyrosine 357 was essential for YAP1 function. Specifically, when we expressed WT or mutant Y357F YAP1 in HuTu80 cells expressing YAP1-specific shRNAs, we found that WT but not Y357F YAP1 was able to rescue the anti-proliferative and AI growth effects of the YAP1-specific shRNA (compare Figure 2C-D to S5F-G), when expressed at equivalent levels (Figures 2E, S5H). Together, these observations confirm that YES1 is essential for the tumorigenicity of β-catenin dependent cell lines and suggests that YES1 mediated phosphorylation of tyrosine 357 regulates YAP1 activity.
To assess the relationship between YES1 and YAP1 in vivo, we examined the effect of suppressing these genes on zebrafish development. Microinjection of zebrafish embryos with a high concentration (200 μM) of YAP1- or YES1-specific morpholinos resulted in severe developmental phenotypes (Figure S5I). Specifically, the YAP1 morphants developed craniofacial abnormalities and cardiac edema, whereas the YES1 morphants exhibited craniofacial abnormalities associated with pharyngeal defects (Figure S5I). These phenotypes resemble defects observed when high concentrations of β-catenin-specific morpholinos were injected (Zhang et al., 2012), and confirm previous reports showing that YAP1 and YES1 are essential for early embryonic development in zebrafish (Jiang et al., 2009; Tsai et al., 2005).
Microinjection of YAP1 or YES1 morpholinos at lower doses (50 μM) avoided global toxicity but impaired gut development (Figure S5I). Intestinal fatty acid-binding protein (IFABP, FAPB2) is expressed in intestinal epithelial cells, where it plays a key role in gut metabolism, and is used as a marker of gut development (Goessling et al., 2008). Morpholino-mediated suppression of YAP1 or YES1 expression dramatically inhibited gut formation as determined by both fluorescence microscopy of Tg(fabp2:RFP)as200 gut reporter embryos and by examination of IFABP expression by in situ hybridization (Figure 5J). Furthermore, treatment of zebrafish embryos post fertilization (dpf) with 2 μM dasatinib inhibited gut formation to a similar extent as the YAP1- or YES1-specific morpholinos (Figure 5K), indicating that YES1 kinase activity is essential for zebrafish gut development. Since the Wnt/β-catenin pathway has been shown to be crucial for gut development in zebrafish (Cheesman et al., 2011), we concluded that phosphorylation of YAP1 by YES1 is essential for developmental and malignant processes that are dependent on the function of β-catenin.
Previous studies have shown that in response to cell contact inhibition, activation of the Hippo pathway induces serine 127 phosphorylation and cytosolic accumulation of YAP1 (Zhao et al., 2012). Using immunofluorescence, we found that both YAP1 and β-catenin were constitutively localized in the nucleus in colon cancer cell lines regardless of cell density or β-catenin activity (Figure S6A) and that suppression of β-catenin failed to alter YAP1 localization (Figure S6B). Collectively these observations suggest that in contrast to non-transformed cell lines (Zhao et al., 2007), culture density does not regulate YAP localization in colon cancer cell lines.
YES1 kinase activity regulates the activity of the YAP1-β-catenin-TBX5 complex
To determine whether the interaction between β-catenin and YAP1 was regulated by YES1, we expressed two distinct YES1-specific shRNAs in HuTu80 cells and found that suppression of YES1 expression abrogated the formation of the β-catenin-YAP1 complex (Figure 6A).
Treatment of zebrafish embryos with dasatinib, which inhibits YES1, resulted in a similar phenotype to that of suppressing YES1 expression (Figure 5J, K). Thus, we used dasatinib to test whether YES1 kinase activity was essential for the β-catenin-YAP1 interaction. In contrast to what we observed when we suppressed YES1 expression, treatment of the SW480 colon cancer cell line with dasatinib increased the interaction between β-catenin and YAP1, indicating that YES1 kinase activity is not required for formation of the β-catenin-YAP1 complex (Figure 6B). The dasatinib-induced increase in β-catenin-YAP1 interaction was reversed by expression of a dasatinib-resistant form of YES1 or SRC (Figure 6C). Furthermore, we found that the YAP1 mutant (YAP1 Y357F), which cannot be tyrosine phosphorylated, interacted with β-catenin when expressed in 293T cells or in colon cancer cell lines (Figure 6D,E). Thus, the interaction of YES1 with YAP1 and β-catenin is essential for formation of the β-catenin-YAP1 complex in a manner independent of YES1 kinase activity.
Since YES1 suppression disrupted the activity of the β-catenin-YAP1-TBX5 complex, we tested whether YES1 kinase activity was required for binding of the β-catenin-YAP1-TBX5 complex to specific target promoters. Treatment of HCT116 cells with dasatinib inhibited the binding of β-catenin and YAP1 to the BCL2L1 and BIRC5 promoters (Figure 6F). Moreover, treatment of HCT116 or HuTu80 with dasatinib resulted in down regulation of BCL2L1 and BIRC5 expression, which was reversed by expression of a dasatinib-resistant form of YES1 (Figure 6G). These observations suggest that phosphorylation of YAP1 by YES1 is required for the activity of the β-catenin-YAP1-TBX5 complex.
β-catenin active cancers are sensitive to SRC family inhibitors
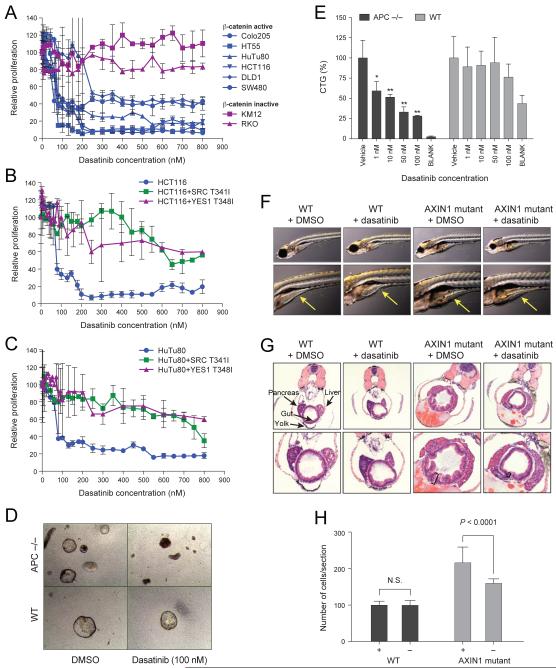
The observation that the β-catenin-YAP1-TBX5 complex is required for the survival of β-catenin active cells suggests that disrupting the activity of this complex may selectively affect β-catenin active cancers. To test this hypothesis, we exposed β-catenin active and inactive cell lines a wide range of dasatinib concentrations. Indeed, we found that β-catenin active cell lines were 6.4-16 times more sensitive to dasatinib than cells that lack β-catenin activity (Figure 7A, Table 3).
Figure 7. Dasatinib impairs the proliferation of β-catenin active cell lines.
(A) Proliferation (7 d) following dasatinib treatment. Proliferation of (B) HCT116 or (C) HuTu80 cells following dasatinib treatment of cells stably expressing dasatinib resistant YES1 (T348I) or SRC (T341I) mutants. Data presented as mean ± SD for 4 independent experiments. (D) Representative images of colon organoids derived from WT or APC null mice treated for 6 d with 100 nM of dasatinib. (E) Quantification of results in D. Error bars represent SD from 4 replicates. Blank mark wells where no organoids were added. (F) WT or AXIN1 mutant (Masterblind) zebrafish were treated with 2 μM of dasatinib from 6-8 dpf. Arrow indicates developing gut. (G) H&E staining of zebrafish in (F). Width of epithelium noted by bars. (H) The number of epithelial cells/section was measured in WT or AXIN1 mutant zebrafish treated with 2 μM of dasatinib or DMSO. Error bars represent the SD from 20 different sections from 5 treated fish. P value was calculated using student’s t test.
Table 3.
IC50 of dasatinib in colon cancer cell lines.
| Cell line | Status of β-catenin pathway | IC50 Dasatinib (nM) |
|---|---|---|
| Colo205 | active | 80 |
| HCT116 | active | 68 |
| HT55 | active | 70 |
| HuTu80 | active | 70 |
| DLD1 | active | 125 |
| SW480 | active | 50 |
| KM12 | Non-active | >800 |
| RKO | Non-active | >800 |
Since dasatinib inhibits a broad range of tyrosine kinases (Lombardo et al., 2004), we tested whether the observed effects on cell proliferation were due to its effects on SRC family members. Specifically, we expressed dasatinib resistant YES1 or SRC mutants (Du et al., 2009) in HuTu80 or HCT116 cells and then tested the sensitivity of these cells to dasatinib. We found that expression of either of these mutants rescued the proliferation arrest induced by dasatinib (Figure 7B, C). These observations confirm that the tyrosine kinase activity of YES1 is required for the proliferation of β-catenin active cell lines.
To corroborate these findings, we investigated the effects of inhibiting YES1 in colonic organoids and zebrafish. Primary colon organoids can be propagated in vitro as explants in air-liquid interface cultures (Ootani et al., 2009). Under these conditions, colon organoids recapitulate multilineage differentiation and Lgr5+ intestinal stem cells. We cultured colon organoids from Apcflox/flox; villin-CreER mice, which were exposed to tamoxifen in vitro to delete Apc deletion. These WT or Apc-null organoids were then treated with dasatinib (1-100 nM). We found that Apc-null organoids were (p<0.005) more sensitive to dasatinib than WT organoids (Figure 7D, E). Specifically, we observed a 70% decrease in growth of APC-null organoids treated with 50 nM of dasatinib compared to a 5% growth inhibition of WT organoids treated with dasatinib (Figure 7E). These observations demonstrate that inhibition of YES1 kinase activity in APC null epithelium reverses the hyperproliferation induced by APC loss.
Stabilization of β-catenin in AXIN1 temperature sensitive mutant zebrafish (Masterblind) induces a β-catenin dependent hyperproliferation of intestinal epithelial cells (Cheesman et al., 2011). Using this model, we treated WT or AXIN1 mutant zebrafish at 6 dpf with 2 μM dasatinib for 48 h, which suppressed intestinal hyperplasia in the AXIN1 mutants as assessed by morphology (Figure 7F) or H&E staining (Figure 7G). Furthermore, the number of epithelial cells was significantly (p<0.0001) decreased in AXIN1 mutant zebrafish treated with dasatinib (Figure 7H). In contrast, we failed to observe changes in the intestinal structure or cell number in WT zebrafish treated with dasatinib, indicating that the effect of dasatinib was specific to AXIN1 mutant animals (Figure 7G, H). We concluded that inhibition of YES1 kinase activity inhibits the β-catenin dependent proliferation in cultured human cancer cells, in colon epithelial organoids, and in a zebrafish model of intestinal hyperplasia.
Discussion
Inactivating mutations in the tumor suppressor APC occur in the majority of colon cancers and lead to stabilization of β-catenin. As a consequence, β-catenin accumulates in the nucleus and contributes to adenoma formation. Here, we demonstrate that β-catenin forms a ternary complex with the transcriptional regulator YAP1 and the transcription factor TBX5, which promotes colon cancer cell survival and contributes to malignant transformation. These observations reveal hitherto unidentified components of the β-catenin pathway that contribute to colon cancer and that may provide new targets for therapeutic intervention.
Identification of co-dependent genes in cells that exhibit constitutively active β-catenin signaling
Using a loss of function approach to identify essential genes in a large panel of cancer cell lines (Cheung et al., 2011), we identified 50 genes whose expression was preferentially required for the proliferation of cell lines that exhibit β-catenin activity (FDR q<0.25). The use of a large number of cell lines allowed us to perform permutation analyses to define the false discovery rate for each candidate and ensure that the genes identified by this approach robustly distinguished the two groups. Indeed, when we arbitrarily assigned cell lines to two classes, we failed to identify genes that distinguished these groups.
Among these top candidates, we found that only two genes, β-catenin and BCL9L, a known β-catenin binding protein (Adachi et al., 2004), affected the activity of this reporter in multiple β-catenin active cell lines. These observations suggested that other genes beyond the known components of the Wnt/β-catenin pathway play key roles in survival of β-catenin-active cells.
Cancer cell lines that exhibit β-catenin activity require YAP1
YAP1 has been implicated as an effector of the Hippo pathway (Zhao et al., 2011), an oncogene in hepatocellular cancers (Zender et al., 2006) and as a protein involved in mechanotransduction (Dupont et al., 2011). Here we found that YAP1 is essential for both proliferation and tumor maintenance of β-catenin active cancer cell lines. We also found that YAP1, β-catenin and TBX5 form a complex that regulates the expression of genes that promote survival, including BIRC5 and BCL2L1. Using immortalized cell lines, we found that YAP1 expression is required for β-catenin driven transformation suggesting that the β-catenin-YAP1 complex plays a key role in tumor progression.
In hepatocellular carcinomas that harbor amplifications of 11q22, YAP1 cooperates with a co-amplified gene CIAP1 to accelerate tumor formation (Zender et al., 2006). Genetically engineered mice that express a doxycycline inducible stabilized YAP1 mutant (S127A) develop colonic adenomas and liver hyperplasia (Camargo et al., 2007). These observations implicate YAP1 as an oncogene in hepatocellular cancers. YAP1 has also been found to be overexpressed in many epithelial cancers (Steinhardt et al., 2008). However, when we analyzed YAP1 expression in 807 cancer cell lines (Barretina et al., 2012), we found that YAP1 was expressed ubiquitously in epithelial cell lines regardless of tissue origin (Figure S7), and we failed to identify a correlation between YAP1 dependence and YAP1 copy number or expression. Based on the strong correlation between YAP1 and β-catenin dependency, we conclude that YAP1 induces transformation in β-catenin active cancers primarily through its interactions with β-catenin.
The Hippo pathway controls organ size by regulation of YAP1. Specifically, upon cell contact inhibition the Hippo pathway is triggered leading to a cascade of phosphorylation events resulting in activation of the MST1/2 kinases. Activated Mst1/2 phosphorylates LATS1/2, which, in turn, phosphorylate YAP1 on serine 127 leading to inactivation of YAP1 by proteasomal degradation and cytosolic retention of YAP1 (Zhao et al., 2011). In genetically engineered mice, inactivation of the Hippo pathway by tissue specific germline deletion of Mst1/2 induced hepatocellular cancers (Zhou et al., 2009) or colonic hyperplasia and adenomas (Zhou et al., 2011). However, recurrent deletions or loss of heterozygosity involving these genes have not been identified in human cancers. Prior work failed to identify a correlation between Hippo pathway activity and dependence on YAP1 in 11 colon cancer cell lines (Zhou et al., 2011). We also failed to find a correlation between YAP1 nuclear localization or expression and YAP1 dependency. Since YAP1 is regulated by Hippo-dependent and independent mechanisms (Dupont et al., 2011), further studies will be necessary to determine the role of Hippo signaling in colon cancer pathogenesis.
β-catenin has been shown to regulate YAP1 expression by binding to the YAP1 promoter (Konsavage et al., 2012). Specifically, both β-catenin and TCF4 bind to sequences upstream of YAP1, and suppression of β-catenin expression decreased the levels of YAP1 mRNA. However, we found comparable levels of YAP1 across a large number of cell lines including β-catenin inactive colon cancer cell lines (Figure S2A, S7), suggesting that the regulation of YAP1 by β-catenin is not the primary driver of sensitivity in these cancer cell lines. However, it remains possible that the β-catenin-TCF4 complex plays a role as a feedback loop enhancing the expression of YAP1.
The β-catenin-YAP1-TBX5 complex
TBX5, a member of the T-box family of transcription factors, plays key roles in cardiac muscle development and limb identity (Rodriguez-Esteban et al., 1999). Germline mutations in TBX5 have been associated with the Holt-Oram syndrome (Mori and Bruneau, 2004). TBX5 has also been shown to form a complex with TAZ and YAP1 that induces transcription of atrial natriuretic factor (Murakami et al., 2005). Here we found that TBX5 forms a complex with β-catenin-YAP1 that is found at the BCL2L1 and BIRC5 promoters. These observations extend prior work that showed that TBX5 binds the BIRC5 and BCL2L1 promoters when overexpressed in 293T cells (He et al., 2011).
YAP1 interacts with the TEAD family of transcriptional factors to regulate both developmental and cancer-associated phenotypes (Zhao et al., 2008). We did not find that TEAD family members were differentially required for proliferation of β-catenin active cells in the Project Achilles dataset (Cheung et al., 2011). Although the YAP1-TEAD complex regulates other cancer phenotypes (Lamar et al., 2012), our observations implicate TBX5 as a key transcription factor target of the β-catenin-YAP1 complex. Moreover, since β-catenin interacts with different transcription factors such as the AR (Mulholland et al., 2002) and HIF-1 (Kaidi et al., 2007), these observations suggest that both YAP1 and β-catenin regulate several transcriptional programs through interactions with distinct transcription factors.
TCF4 dependency in β-catenin dependent cancers
Although the β-catenin-TCF4 complex plays an important role in colon adenoma initiation, several lines of evidence suggest that β-catenin may also contribute to cancer in a TCF4 independent manner. Specifically, although expression of a dominantly interfering allele of TCF4 inhibits the β-catenin/TCF4 reporter activity in β-catenin-dependent colon cancer cell lines [(Korinek et al., 1997), Fig. 1H], suppression of TCF4 induces a substantial increase in β-catenin/TCF4 reporter activity and only partially affected the proliferation and AI growth of β-catenin dependent cell lines (Figure 1I, J)(Tang et al., 2008). Moreover, two colon cancer cell lines (HT29 and LS411N) that harbor APC mutations and depend on β-catenin expression for survival failed to exhibit detectable β-catenin/TCF4 reporter activity yet were dependent on YAP1 for survival.
Furthermore, compound genetically engineered mice that express both the APCmin allele and deletion of one TCF4 allele, developed aggressive, metastatic colon cancers (Angus-Hill et al., 2011). Whole genome sequencing of colon cancer genomes identified a recurrent TCF4-VTI1A translocation that creates a dominantly interfering allele of TCF4 (Bass et al., 2011) as well as inactivating mutations and copy number loss involving TCF4 (TCGA, 2012). In aggregate, these observations suggest that TCF4 may initially contribute directly to adenoma formation but then is mutated or lost to foster malignant transformation. We cannot exclude the possibility that a residual amount of TCF4 remains in the experiments presented herein, and these observations do not exclude the possibility that other TCF or LEF proteins may be essential for β-catenin mediated transformation.
YES1 regulates the formation and activity of the β-catenin-YAP1-TBX5 complex
We found that the SRC family member YES1 is also specifically essential for the proliferation and transformed phenotype of β-catenin active cancer cell lines, both because YES1 is necessary for the formation of the β-catenin-YAP1-TBX5 complex and because phosphorylation of YAP1 on Y357 by YES1 is required for the localization and activity of this complex.
YAP1 is phosphorylated by several tyrosine kinases including YES1 (Tamm et al., 2011), SRC (Zaidi et al., 2004) and ABL (Levy et al., 2008). We found that suppression of SRC or ABL failed to affect the proliferation/survival of β-catenin active cells, demonstrating that in this context, YES1 plays a dominant role in regulating the formation of the β-catenin-YAP1-TBX5 complex. Moreover, treatment of β-catenin active cells with dasatinib inhibited the activity of the β-catenin-YAP1-TBX5 complex and the survival of β-catenin active cancer cell lines in a manner that is rescued by expression of dasatinib resistant YES1 or SRC mutants. Extending these findings, we found that dasatinib induces an anti-proliferative effect in both murine and fish experimental models of APC loss/WNT pathway activation. These findings corroborate a recent report that showed that treatment of APCmin mice with dasatinib decreases intestinal adenomas (Nautiyal et al., 2011). Together, these observations support a role for YES1 phosphorylation in β-catenin-driven cancers.
Conclusions
We have identified a novel β-catenin-YAP1-TBX5 complex required for the survival and transformation of β-catenin active cancer cell lines. The tyrosine kinase YES1 regulates the formation of this complex and localization to specific promoters, which is dispensable for β-catenin/TCF4 activity, but which regulates transcription of pro-survival genes. These observations demonstrate that deregulated β-catenin stability and function drives malignant transformation through interactions with at least two distinct transcriptional complexes (β-catenin-YAP1-TBX5 and β-catenin-TCF4). Although further work is necessary to decipher the specific roles of each of these complexes in tumorigenesis, these observations provide a framework to explain recent observations that loss of TCF4 activity is associated with tumor progression (Angus-Hill et al., 2011).
Although no specific inhibitors of YES1 exist, the sensitivity of β-catenin active cancer cell lines and animal models to dasatinib suggests that YES1 is an attractive target in β-catenin-driven cancers. Moreover, we found that suppression of BCL2L1 and BIRC5 also inhibited the proliferation/survival of β-catenin active cell lines. Since small molecule inhibitors of BCL2L1 and BIRC5 are currently under investigation, these molecules may also prove useful for targeting of β-catenin active cancers.
Experimental Procedures
In vivo orthotopic tumor model
4×106 HCT116 cells were injected into the flanks of immunodeficient mice (NCr Nude, Taconic). Tumors were extracted, cut into 1 mm3 cubes, and implanted into a pouch created in the cecum of a second mouse. For experiments with inducible shRNAs, the mice were fed a doxycycline diet 2 d post-cecal implantation. Tumors were examined 3 wk post implantation.
Three-dimensional primary intestinal organoid culture
Colons from Apcflox/flox; villin-CreER mice were dissected lengthwise and washed in cold PBS. 0.5-1 cm segment per dish was minced extensively on ice and embedded in a 3D collagen gel using a double-dish culture system (Ootani et al., 2009). Tamoxifen (Sigma, 2 μM in ethanol) or vehicle (ethanol) was applied on the day of initial plating for 7 d to generate APC-null or WT organoids. Organoids were recovered from collagen gel by collagenase IV incubation followed by 0.05% trypsin/EDTA incubation to dissociate organoids into single cells. 5000 cells per well were seeded into 96 well transwell plates (Fisher Scientific). Organoids were treated with the indicated concentration of dasatinib (in DMSO) for 7 d and quantified using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega).
Zebrafish experiments
Zebrafish were maintained according to institutional animal care and use committee (IACUC-BIDMC) protocols. Validated morpholinos (MO) (GeneTools, PhiloMath, OR) designed against the ATG site of YES1 (5′-CCTCTTTACTCTTGACACAGCCCAT-3′)(Jopling and Hertog, 2007) or YAP1 (5′-AGCAACATTAACAACTCACTTTAGG-3′) (Skouloudaki et al., 2009) were injected into WT or gut reporter (Tg(fabp2:RFP)as200) zebrafish at the one-cell-stage. At 4 dpf, the gut morphology of intestinal reporter embryos was imaged by fluorescent microscopy (Discovery, Carl Zeiss). Whole-mount in situ hybridization experiments were conducted using standard zebrafish protocols (www.zfin.org) and the gut tissue was visualized using the established marker IFABP (Mudumana et al., 2004). The axin1tm213 mutant line Masterblind, was reared at 30°C (temperature required for homozygote phenotype to be fully penetrant). Larvae were fixed overnight in 4% PFA, processed and embedded in JB-4 resin, cut into 7 μM sections and stained with Hematoxylin and Eosin (Sullivan-Brown et al., 2011). The total number of intestinal epithelial cells and intestinal wall thickness were quantified in sections at a location of the intestinal bulb that had a comparable amount of pancreatic and liver tissue (20 sections quantified represent 4 sections of 5 animals).
Supplementary Material
Highlights.
-
-
β-catenin-dependent cancers require YAP1 expression for survival.
-
-
β-catenin, YAP1 and TBX5 form a complex that drives expression of BIRC5 and BCL2L1.
-
-
YES1 regulates the activity of the β-catenin-YAP1-TBX5 complex.
-
-
The YES1 inhibitor dasatinib inhibits the proliferation of β-catenin-active cells.
Acknowledgements
We thank the members of the Hahn lab for helpful discussions and L. Gafney and L. Solomon for assistance with graphic arts. This work was supported in part by NIH/NCI grants R01 CA140545 (W.C.H.), RC2 CA148268 (W.C.H.), U54 CA143798 (R.B.), U01 DK085527 (C.J.K), U01 CA151920 (C.J.K), R01 DK085720 (C.J.K), U54 CA112962 (A.T., J.P.M., W.C.H.), R01 DK090311 (W.G.), the Pew Charitable Trust (W.G.), a Fidelity Foundation grant (C.J.K), a Stanford University Dean’s Fellowship (J.T.N), a DoD breast cancer postdoctoral fellowship W81XWH-10-1-0062 (X.W.), Conquer Cancer Foundation Young Investigator Award and Sass Foundation Fellowship (D.N.), a SPORE career development award P50 CA127003, (J.R.) and a NIH F32 postdoctoral fellowship F32 GM090437 (J.R.). J.R. is a John Svenson postdoctoral fellow. W.C.H. and R.B. are consultants for Novartis Pharmaceuticals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi S, Jigami T, Yasui T, Nakano T, Ohwada S, Omori Y, Sugano S, Ohkawara B, Shibuya H, Nakamura T, et al. Role of a BCL9-related beta-catenin-binding protein, B9L, in tumorigenesis induced by aberrant activation of Wnt signaling. Cancer Res. 2004;64:8496–8501. doi: 10.1158/0008-5472.CAN-04-2254. [DOI] [PubMed] [Google Scholar]
- Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi M. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:4914–4919. doi: 10.1073/pnas.1102300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouel C, Garg A, McNeill H. Herding Hippos: regulating growth in flies and man. Curr Opin Cell Biol. 2009;21:837–843. doi: 10.1016/j.ceb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nature Genetics. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B, Georgiou M. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J Cell Biol. 2011;192:907–917. doi: 10.1083/jcb.201009141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Current Biology. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4570–4577. doi: 10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HW, Cowley GS, Weir BA, Boehm JS, Rusin S, Scott JA, East A, Ali LD, Lizotte PH, Wong TC, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci U S A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Du J, Bernasconi P, Clauser KR, Mani DR, Finn SP, Beroukhim R, Burns M, Julian B, Peng XP, Hieronymus H, et al. Bead-based profiling of tyrosine kinase phosphorylation identifies SRC as a potential target for glioblastoma therapy. Nature Biotech. 2009;27:77–83. doi: 10.1038/nbt.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessling W, North TE, Lord AM, Ceol C, Lee S, Weidinger G, Bourque C, Strijbosch R, Haramis AP, Puder M, et al. APC mutant zebrafish uncover a changing temporal requirement for wnt signaling in liver development. Dev Biol. 2008;320:161–174. doi: 10.1016/j.ydbio.2008.05.526. [DOI] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Liu D, Gong Y, Wang Y, Sun S, Gui Y, Song H. yap is required for the development of brain, eyes, and neural crest in zebrafish. Biochem Biophys Res Commun. 2009;384:114–119. doi: 10.1016/j.bbrc.2009.04.070. [DOI] [PubMed] [Google Scholar]
- Jopling C, Hertog J. Essential role for Csk upstream of Fyn and Yes in zebrafish gastrulation. Mech Dev. 2007;124:129–136. doi: 10.1016/j.mod.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Kaidi A, Williams AC, Paraskeva C. Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nature Cell Biol. 2007;9:210–217. doi: 10.1038/ncb1534. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nature Review Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- Konsavage WM, Kyler SL, Rennoll SA, Jin G, Yochum GS. Wnt/beta-catenin signaling regulates Yes-associated protein (YAP) gene expression in colorectal carcinoma cells. J Biol Chem. 2012;287:11730–9. doi: 10.1074/jbc.M111.327767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A. 2012;109:E2441–2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)-piperazin-1-yl)-2-methylpyrimidin-4-ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- Mori AD, Bruneau BG. TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Curr Opin Cardiol. 2004;19:211–215. doi: 10.1097/00001573-200405000-00004. [DOI] [PubMed] [Google Scholar]
- Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Mudumana SP, Wan H, Singh M, Korzh V, Gong Z. Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: liver, intestine, and exocrine pancreas. Dev Dyn. 2004;230:165–173. doi: 10.1002/dvdy.20032. [DOI] [PubMed] [Google Scholar]
- Mulholland DJ, Cheng H, Reid K, Rennie PS, Nelson CC. The androgen receptor can promote beta-catenin nuclear translocation independently of adenomatous polyposis coli. J Biol Chem. 2002;277:17933–17943. doi: 10.1074/jbc.M200135200. [DOI] [PubMed] [Google Scholar]
- Murakami M, Nakagawa M, Olson EN, Nakagawa O. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc Natl Acad Sci U S A. 2005;102:18034–18039. doi: 10.1073/pnas.0509109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal J, Banerjee S, Kanwar SS, Yu Y, Patel BB, Sarkar FH, Majumdar AP. Curcumin enhances dasatinib-induced inhibition of growth and transformation of colon cancer cells. Int J Cancer. 2011;128:951–961. doi: 10.1002/ijc.25410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena SV, Melhem MF, Meisler AI, Cartwright CA. Elevated c-yes tyrosine kinase activity in premalignant lesions of the colon. Gastroenterology. 1995;108:117–124. doi: 10.1016/0016-5085(95)90015-2. [DOI] [PubMed] [Google Scholar]
- Phelps RA, Chidester S, Dehghanizadeh S, Phelps J, Sandoval IT, Rai K, Broadbent T, Sarkar S, Burt RW, Jones DA. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137:623–634. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Esteban C, Tsukui T, Yonei S, Magallon J, Tamura K, Izpisua Belmonte JC. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature. 1999;398:814–818. doi: 10.1038/19769. [DOI] [PubMed] [Google Scholar]
- Skouloudaki K, Puetz M, Simons M, Courbard JR, Boehlke C, Hartleben B, Engel C, Moeller MJ, Englert C, Bollig F, et al. Scribble participates in Hippo signaling and is required for normal zebrafish pronephros development. Proc Natl Acad Sci U S A. 2009;106:8579–8584. doi: 10.1073/pnas.0811691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Path. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module--the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- Sullivan-Brown J, Bisher ME, Burdine RD. Embedding, serial sectioning and staining of zebrafish embryos using JB-4 resin. Nat Protoc. 2011;6:46–55. doi: 10.1038/nprot.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm C, Bower N, Anneren C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J Cell Sci. 2011;124:1136–1144. doi: 10.1242/jcs.075796. [DOI] [PubMed] [Google Scholar]
- Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L. A genome-wide RNAi screen for Wnt/beta-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci U S A. 2008;105:9697–9702. doi: 10.1073/pnas.0804709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WB, Zhang X, Sharma D, Wu W, Kinsey WH. Role of Yes kinase during early zebrafish development. Dev Biol. 2005;277:129–141. doi: 10.1016/j.ydbio.2004.08.052. [DOI] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 2008;133:340–353. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wiesmann M, Rohan M, Chan V, Jefferson AB, Guo L, Sakamoto D, Caothien RH, Fuller JH, Reinhard C, et al. Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta -catenin signaling is activated in human colon tumors. Proc Natl Acad Sci U S A. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, Medina R, Ito Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 2004;23:790–799. doi: 10.1038/sj.emboj.7600073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhang J, Lin SC, Meng A. beta-Catenin 1 and beta-catenin 2 play similar and distinct roles in left-right asymmetric development of zebrafish embryos. Development. 2012;139:2009–2019. doi: 10.1242/dev.074435. [DOI] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, Dawson D, Willis JE, Markowitz SD, Camargo FD, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A. 2011;108:E1312–1320. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.