Summary
The activity-dependent transcription factor, Myocyte Enhancer Factor-2 (MEF2), induces excitatory synapse elimination in mouse neurons which requires Fragile X Mental Retardation Protein (FMRP), an RNA binding protein implicated in human cognitive dysfunction and autism. We report here that protocadherin-10 (Pcdh10), an autism-spectrum disorders gene, is necessary for this process. MEF2 and FMRP cooperatively regulate the expression of Pcdh10. Upon MEF2 activation, PSD-95 is ubiquitinated by the ubiquitin E3 ligase, murine double minute-2 (Mdm2) and then binds to Pcdh10, which links it to the proteasome for degradation. Blockade of the Pcdh10 -proteasome interaction inhibits MEF2-induced PSD-95 degradation and synapse elimination. In FMRP-lacking neurons, elevated protein levels of eukaryotic translation elongation factor 1-alpha (EF1α), an Mdm2 interacting protein and FMRP target mRNA, sequester Mdm2 and prevent MEF2-induced PSD-95 ubiquitination and synapse elimination. Together our findings reveal novel roles for multiple autism-linked genes in activity-dependent synapse elimination.
Keywords: MEF2, Pcdh10, PSD-95, FMRP, Mdm2, ubiquitination, EF1α, synapse elimination
Introduction
Sensory experience, learning and neuronal activity stabilize or eliminate select excitatory synapses and sculpt the mature neuronal circuits that mediate sensory processing and memory (Lai et al., 2012; Xu et al., 2009; Yang et al., 2009). The cellular mechanisms that underlie activity or experience-dependent synapse elimination in the central nervous system are largely unknown. The discovery that activation of the Myocyte Enhancer Factor 2 (MEF2) family of transcription factors suppresses synapse number provides an important molecular link to understand how neuronal activity leads to synapse elimination (Flavell et al., 2006; Pfeiffer et al., 2010; Tian et al., 2010). MEF2 is activated upon neuronal depolarization and calcium influx which induces the expression of MEF2 target genes that are thought to lead to synapse elimination (Flavell et al., 2006; Flavell et al., 2008; McKinsey et al., 2002; Pulipparacharuvil et al., 2008). The identity of the MEF2 target genes that promote synapse elimination or their mechanisms of action in synapse elimination are unknown. We recently demonstrated that MEF2-induced synapse elimination requires Fragile X Mental Retardation Protein (FMRP) (Pfeiffer et al., 2010), an RNA binding protein that regulates translation of its target mRNAs (Bassell and Warren, 2008). FMRP is encoded by the Fmr1 gene, which is transcriptionally silenced in patients with Fragile X syndrome (FXS), the most common inherited form of intellectual disability and autism (Abrahams and Geschwind, 2008; Kelleher and Bear, 2008). In neurons of the mouse model of FXS, Fmr1 knockout (KO), MEF2-triggered synapse elimination is absent (Pfeiffer et al., 2010) which may contribute to the observed excess of dendritic spines on cortical neurons of FXS patients and the Fmr1 KO mouse (Bagni and Greenough, 2005). The mechanisms that underlie the deficiencies in synapse elimination associated with FXS are unknown. Our previous results indicated that FMRP functions downstream of MEF2-induced transcription to eliminate synapses (Pfeiffer et al., 2010). Based on the known function of FMRP, we hypothesized that FMRP regulated translation of MEF2-generated transcripts.
To identify candidate transcripts involved in synapse elimination, we compared the known MEF2 target genes (Flavell et al., 2008) with FMRP-interacting mRNAs (Darnell et al., 2011). Of the many common MEF2 and FMRP interacting transcripts, we focused on Protocadherin-10 (Pcdh10), also known as OL-protocadherin, a member of the cadherin superfamily of calcium-dependent cell adhesion molecules (Morishita and Yagi, 2007). Importantly, homologous deletion of Pcdh10 in humans is associated with autism (Morrow et al., 2008). Pcdh10 and other family members of the δ2 non-clustered protocadherin family (Pcdh8 and 19) demonstrate weak homophilic binding activity relative to the classical cadherins (Hirano et al., 1999). Furthermore, the diverse structure of protocadherins’ cytoplasmic C-termini is consistent with the multifunctional roles of protocadherins in cell signaling and function, in addition to cell adhesion (Kim et al., 2011; Yasuda et al., 2007). Pcdh10 is primarily expressed in the brain (Hirano et al., 1999) where it is required for the growth of striatal axons and patterning of thalamocortical and corticothalamic projections during embryonic development (Uemura et al., 2007). Interestingly, Pcdh10 is highly expressed in mature cortical neurons where nothing is known of its function (Kim et al., 2011).
Here we demonstrate that Pcdh10 is required for MEF2-induced synapse elimination and functions to deliver ubiquitinated post-synaptic density protein 95 (PSD-95), a critical synaptic scaffolding molecule to the proteasome. Translation of Pcdh10 is altered in Fmr1 KO neurons, but, surprisingly, this does not underlie the deficit in MEF2-induced synapse elimination. Instead, Fmr1 KO neurons have deficits in MEF2-induced ubiquitination of PSD-95 as a result of reduced synaptic localization of the ubiquitin E3 ligase for PSD-95, murine double minute-2 (Mdm2) (Bianchetta et al., 2011; Colledge et al., 2003). Elevated protein levels of elongation factor 1-alpha (EF1α), an FMRP target mRNA and Mdm2-interacting partner (Frum et al., 2007; Sung et al., 2003), sequester Mdm2 in Fmr1 KO neurons and prevent Mdm2 interactions and ubiquitination of PSD-95 upon MEF2 activation. Our results reveal novel mechanisms by which the activity-dependent transcription factor MEF2 refines synaptic connections in wild type neurons as well as identify the molecular basis for the defect in synapse elimination in Fragile X syndrome. This work demonstrates the necessity and a distinct cellular function of two genes linked to autism, Pcdh10 and Fmr1, in the same synapse elimination process and provides evidence for a common synaptic deficit in different genetic causes of autism.
Results
MEF2 and FMRP regulate Pcdh10 expression in dendrites
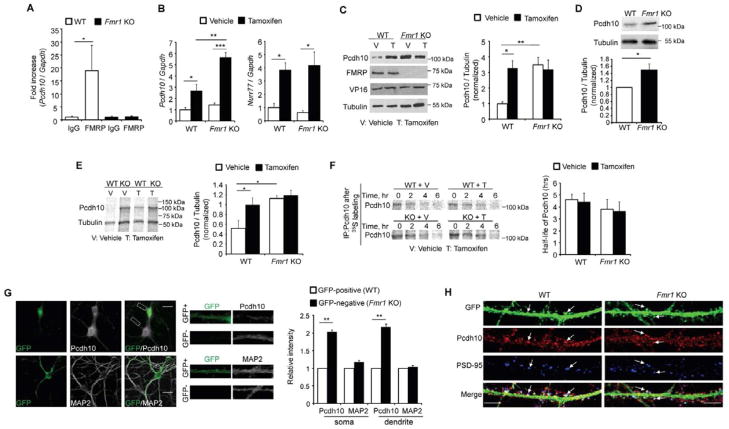
To study the role of Pcdh10 in MEF2 and FMRP regulation of synapse number, we first validated the association of FMRP and Pcdh10 mRNA (Darnell et al., 2011). We performed an RNA-immunoprecipitation with an anti-FMRP antibody and pulled down Pcdh10 mRNA from hippocampal lysates of wild type (WT) but not Fmr1 KO mice (Figure 1A) providing evidence for Pcdh10 as an mRNA target of FMRP. We next examined if Pcdh10 expression is regulated by MEF2 and FMRP, using lentivirus to transfect tamoxifen-inducible, constitutively active MEF2 (MEF2-VP16ERtm) (Flavell et al., 2006; Pfeiffer et al., 2010) into dissociated cortical neuron cultures which are amenable to biochemical assays. Basal levels of Pcdh10 mRNA and another MEF2 target gene Nurr77 were not different between WT and Fmr1 KO neurons (vehicle; white bars, Figure 1B). After 6 hours of 4-Hydroxytamoxifen (Tamoxifen, 1 μm) treatment, which induces translocation of MEF2-VP16ERtm to the nucleus to activate target genes (Flavell et al., 2006), both Pcdh10 and Nurr77 mRNAs were elevated in WT and Fmr1 KO neurons (black bars, Figure 1B). Surprisingly, the level of Pcdh10 mRNA induced with MEF2 activation in Fmr1 KO neurons was greater than in WT neurons, perhaps reflecting a role of FMRP in RNA stability (Zhang et al., 2007). Therefore, FMRP is not required for MEF2 induced transcription of target genes, including Pcdh10, as previously indicated (Pfeiffer et al., 2010).
Figure 1. MEF2 and FMRP co-regulate Pcdh10 expression.
(A) Quantitative real-time RT-PCR of Pcdh10 precipitated with anti-FMRP antibody or IgG from wild type (WT) or Fmr1 KO mouse brains. Plotted is average of Pcdh10/Gapdh mRNA. (n = 5 mice per genotype) (B) Quantitative real-time RT-PCR of Pcdh10/Gapdh and Nurr77/Gapdh mRNA and (C) Western blots of Pcdh10 from vehicle- or 4-Hydroxy-Tamoxifen (Tamoxifen or “T“) treated WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm. (n = 5 independent cultures for real-time RT-PCR and n = 3 for Western blots) (D) Western blots of Pcdh10 from WT or Fmr1 KO hippocampi. (n = 4 mice per genotype) (E) 35S-Met/Cys metabolic labeling from vehicle- or Tamoxifen -treated (T) WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm followed by immunoprecipitation of Pcdh10 and Tubulin. Plotted is average of Pcdh10/Tubulin. (n = 3 cultures) (F) 35S-Met/Cys pulse-chase assay of Pcdh10 from vehicle- or T-treated WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm. Plotted is average half-life of Pcdh10 (n = 3 cultures). (G) Immunohistochemistry of GFP with Pcdh10 or MAP2 from hippocampal neurons of GFP/Fmr1 mosaic mouse. Selected GFP positive (WT) and GFP-negative (Fmr1 KO) areas are enlarged in the middle. Quantification of fluorescence intensity from soma and dendrites is shown on the right. Scale bar: 20 μm. (n = 21 cells for both WT and Fmr1 KO) (H) Immunohistochemistry of Pcdh10 and PSD-95 from dissociated hippocampal neurons of WT or Fmr1 KO mice. Arrows indicate spines. In all figures, error bars represent SEM. Scale bar: 5 μm. (*p < 0.05; **p < 0.01; ***p < 0.001) See also Figure S1.
In contrast to Pcdh10 mRNA, Pcdh10 protein is basally higher in Fmr1 KO neurons and is unresponsive to MEF2 activation (Fig. 1C). Similar to cultured neurons, Pcdh10 protein is elevated in hippocampal lysates of Fmr1 KO mice (Figure 1D and S1A). One function of FMRP is to suppress translation of its mRNA targets (Darnell et al., 2011; Napoli et al., 2008) suggesting that elevated Pcdh10 in Fmr1 KO neurons is due to increased translation rate. To test this possibility, we performed metabolic labeling in WT and Fmr1 KO cultures under basal and MEF2 activated conditions. 35S-Met/Cys was added with vehicle or Tamoxifen for 6 hours to MEF2-VP16ERtm-transfected neurons followed by immunoprecipitation of Pcdh10 or Tubulin (as a normalization control). We observed enhanced newly synthesized Pcdh10 in WT neurons upon Tamoxifen treatment (Figure 1E). In Fmr1 KO neurons, basal translation rates of Pcdh10 were elevated and MEF2 activation failed to further increase Pcdh10 synthesis. To determine if MEF2 regulates the half-life of Pcdh10, we applied a 2-hour pulse of 35S-Met/Cys label to neuronal cultures, and then the medium was replaced with fresh medium containing vehicle or Tamoxifen for the time points indicated in Figure 1F. The half-life of Pcdh10 was not affected by genotype or MEF2 activation (Figure 1F). This data indicates that the elevated basal levels of Pcdh10 in Fmr1 KO neurons are due to elevated protein synthesis rates and not reduced turnover. Furthermore, MEF2-activation of WT neurons causes an increase in transcription and translation of Pcdh10, whereas in Fmr1 KO neurons, MEF2 induces Pcdh10 mRNA, but not protein. This may be because FMRP is required for translational activation of Pcdh10 mRNA (Bassell and Warren, 2008) or basal Pcdh10 synthesis rates are saturated in Fmr1 KO neurons.
To determine if dendritic Pcdh10 protein levels are elevated in Fmr1 KO neurons and to assess the cell autonomous role of FMRP in regulation of Pcdh10, we performed immunohistochemistry for Pcdh10 and Microtubule-associated protein 2 (MAP2), a dendritic marker, in dissociated neuron cultures prepared from GFP/Fmr1 mosaic mice. GFP/Fmr1 mosaic mice were generated by crossing Fmr1 KO mice with mice that harbor a GFP vector on the X chromosome (Hadjantonakis et al., 1998; Hanson and Madison, 2007). Due to random X-inactivation, cells of heterozygous females are mosaic; that is, GFP (+) cells are “wild type” and express FMRP and are intermingled with GFP (−) cells that are Fmr1 KO. Immunohistochemistry confirmed that FMRP and GFP are coexpressed in hippocampal and cortical neurons of GFP/Fmr1 mosaic mice (Niere et al., 2012) (Figure S1B). Pcdh10, but not MAP2, were elevated in both the soma and dendrites of Fmr1 KO neurons (Figure 1G) in comparison to neighboring GFP+, WT neurons. These results support a role for FMRP in translational suppression of Pcdh10, which leads to elevated Pcdh10 in Fmr1 KO neurons.
We also determined whether Pcdh10 is present in dendritic spines and localized to excitatory synapses by performing immunohistochemistry for Pcdh10 and the postsynaptic excitatory synapse marker, PSD-95 in dissociated hippocampal neurons that were transfected with GFP to visualize dendritic spines. We observed that Pcdh10 was present in dendritic spines where PSD-95 was also enriched (Figure 1H). To validate the specificity of the antibody used for immunohistochemistry, we performed staining on hippocampal neurons transfected with two independent shRNAs against Pcdh10 (Figure S1C). The results showed similar knock-down efficiency (>50%) by shRNAs as observed with Western blot (Figures 3F and S3B), validating the fluorescence signal of Pcdh10 seen in Fig. 1H. Therefore, Pcdh10 is present in spines at the site of excitatory synapses and may function in MEF2 regulation of excitatory synapse number.
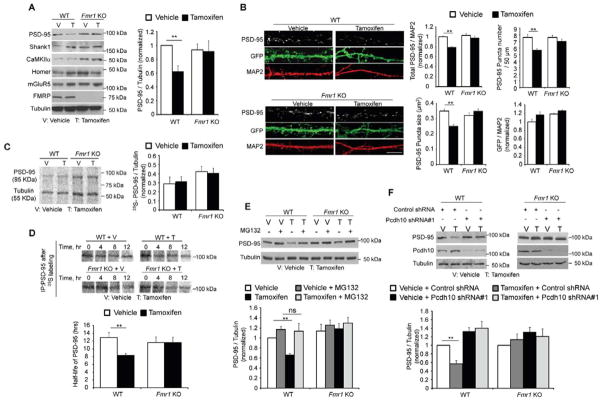
Figure 3. MEF2 induces Pcdh10-dependent degradation of PSD-95.
(A) Western blots of postsynaptic proteins from dissociated WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm and treated with either vehicle or Tamoxifen for 6 hours. Quantification of PSD-95 is shown on the right. (n = 3 cultures) (B) Immunohistochemistry of PSD-95 from dissociated WT or Fmr1 KO hippocampal neurons transfected with MEF2-VP16ERtm and treated with either vehicle or Tamoxifen for 6 hours. Bicistronic GFP from MEF2-VP16ERtm plasmid and endogenous MAP2 serve as transfection control and quantification control, respectively. Scale bars: 5 μm. Quantifications of total PSD-95 intensity, PSD-95 puncta number, PSD-95 puncta size and GFP intensity are shown on the right. (n = 21cells/condition). (C) 35S-Met/Cys metabolic labeling from vehicle- or Tamoxifen -treated WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm followed by immunoprecipitation of PSD-95 and Tubulin. Plotted is average of Pcdh10/Tubulin. (n = 3 cultures) (D) 35S-Met/Cys pulse-chase assay of Pcdh10 from vehicle- or Tamoxifen -treated WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm. Plotted is average half-life of Pcdh10. (n = 3 cultures) (E) Western blots of PSD-95 and Tubulin from dissociated WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm and treated with vehicle, Tamoxifen and/or proteasome inhibitor, MG132, as indicated for 6 hours. Quantification is shown below. (n = 4 cultures) (F) Western blots of PSD-95, Pcdh10 and Tubulin from dissociated cortical neurons of WT or Fmr1 KO mice transfected with MEF2-VP16ERtm and Pcdh10 shRNA. Quantification is shown below. (n = 3 cultures). In all figures, error bars represent SEM. (**p < 0.01) See also Figure S3.
Pcdh10 is required for MEF2-induced synapse elimination
Since the expression of Pcdh10 is regulated by both MEF2 and FMRP, we hypothesized that Pcdh10 is required for MEF2 and FMRP-dependent synapse elimination (Pfeiffer et al., 2010). We first determined if manipulation of Pcdh10 alone was sufficient to affect excitatory synaptic transmission. To do this, we overexpressed mouse Pcdh10, together with mCherry (as a transfection marker), in WT organotypic hippocampal slice cultures using biolistic transfection (Figure S2A and S2B) (Pfeiffer et al., 2010). 16–30 hours after transfection, we performed simultaneous whole cell patch clamp recordings from Pcdh10 transfected and neighboring untransfected CA1 pyramidal neurons. Pcdh10 overexpression had no effect on excitatory synaptic transmission as measured by the frequency or amplitude of miniature (m) EPSCs or evoked EPSC amplitude (Figure S2B and Table S1). Similarly, there was no effect of Pcdh10 on paired-pulse facilitation (PPF) of evoked EPSCs, an indicator of presynaptic release probability (Table S1). In the converse experiment, we knocked down endogenous Pcdh10 in WT hippocampal slice cultures using either one of two shRNAs targeting different regions of Pcdh10 mRNA. After 40–60 hours transfection, neither shRNA against Pcdh10, nor a control, nontarget shRNA, had any effect on the aforementioned measurements of excitatory synaptic function (Figures S2C, S2D, S2E and Table S1). Similarly, transfection of dissociated hippocampal cultures with either Flag-Pcdh10 or shRNA against Pcdh10 (Figure S2F and S2G) had no effect on synapse number as measured by co-clusters of the pre- and post-synaptic proteins, Synapsin-1 and PSD-95 or PSD-95 puncta number and size. These results indicate that acute changes in Pcdh10 levels are not sufficient to affect excitatory synaptic function.
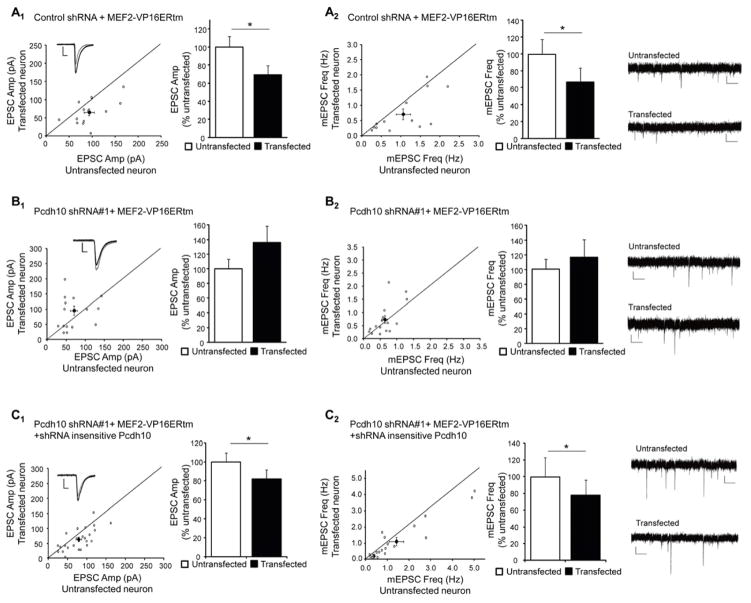
To test if Pcdh10 is required for MEF2-induced synapse elimination, we co-transfected plasmids generating either one of two shRNAs against Pcdh10 together with MEF2-VP16ERtm into WT hippocampal slice cultures. Sister cultures were co-transfected with a control shRNA together with MEF2-VP16ERtm. After 24 hours, Tamoxifen was applied to the slices to activate MEF2 for another 16–32 hours followed by simultaneous recordings of transfected and neighboring untransfected CA1 neurons. As shown, neurons transfected with MEF2-VP16ERtm and control shRNA had a decrease in mEPSC frequency and evoked EPSC amplitude, but no change in PPF (Figure 2A, and Table S1). These changes are consistent and correlate with structural synapse elimination that we previously demonstrated with MEF2-VP16ERtm in this preparation (Flavell et al., 2006; Pfeiffer et al., 2010). However, MEF2-induced functional synapse elimination was blocked in neurons transfected with shRNA against Pcdh10 (Figure 2B, Figure S2H and Table S1) and rescued by cotransfection of a shRNA-insensitive Pcdh10 (Figure 2C and Table S1) together with the Pcdh10 shRNA. shRNA-mediated knockdown of Pcdh10 did not affect MEF2 induced transcription of Nurr77 as measured with real-time RT-PCR (Figure S2I) suggesting that Pcdh10 functions in synapse elimination downstream of MEF2 activated transcription.
Figure 2. Pcdh10 is required for MEF2-induced functional synapse elimination in WT neurons.
(A) Average evoked EPSC amplitude (A1) and mEPSC frequency (A2) from untransfected and control shRNA with MEF2-VP16ERtm transfected CA1 neurons in slice culture (n = 13 for both E1 and E2). (B) Average evoked EPSC amplitude (B1) and mEPSC frequency (B2) from untransfected and Pcdh10 shRNA with MEF2-VP16ERtm transfected cells (n = 16 for both F1 and F2). (C) Average evoked EPSC amplitude (C1) and mEPSC frequency (C2) from untransfected and Pcdh10 shRNA with MEF2-VP16ERtm and shRNA-insensitive Pcdh10 construct transfected cells (n = 24 for both C1 and C2). In all figures, error bars represent SEM. *p < 0.05. Representative evoked EPSC and mEPSC traces are shown with the dot plots of individual cell pairs and on the right of each figure, respectively. See also Figure S2 and Table S1.
MEF2 induces Pcdh10 -dependent degradation of PSD-95
To determine the cellular mechanism by which Pcdh10 mediates MEF2-induced synapse elimination, we turned to dissociated cortical neuron cultures which are amenable to biochemical methods. We hypothesized that MEF2 activation causes rapid synapse elimination by the regulated degradation of synaptic scaffolding proteins. To investigate this possibility, we monitored the level of several postsynaptic scaffolding proteins in MEF2-VP16ERtm transfected cortical neuron cultures after 6 hours of Tamoxifen treatment. As shown (Figure 3A), PSD-95 levels were significantly decreased in WT neurons while other synaptic proteins (mGluR5, Homer and CaMKIIα) remained unchanged. Consistent with the deficit in MEF2-induced synapse elimination in Fmr1 KO neurons, PSD-95 levels were unchanged upon MEF2 activation in Fmr1 KO neurons. Similar results were observed with immunohistochemistry for PSD-95 in dissociated hippocampal neurons. PSD-95 puncta number, size and total PSD-95 intensity were decreased upon MEF2 activation in WT but not Fmr1 KO neurons (Figure 3B). Bicistronic GFP expression from the MEF2-VP16ERtm plasmid did not differ among all groups indicating similar transfection and expression efficiency of the construct across conditions. This result also suggests that the down-regulation of PSD-95 is a common consequence of MEF2 activation in both cortical and hippocampal neurons.
To determine whether the down-regulation of PSD-95 resulted from reduced synthesis or increased degradation, we performed metabolic labeling with 35S-Met/Cys in dissociated cortical neuron cultures. After 6-hours of vehicle or Tamoxifen treatment and 35S-Met/Cys label on MEF2-VP16ERtm transfected cells, there was no change in newly synthesized PSD-95 from either WT or Fmr1 KO neurons (Figure 3C). However, using the pulse-chase 35S-Met/Cys labeling assay the half-life of PSD-95 was reduced by ~35% after MEF2-activation in WT neurons (Figure 3D). Like PSD-95 protein levels (Figures 3A and 3B), PSD-95 half-life was unaffected by MEF2 activation in Fmr1 KO neurons. To investigate whether MEF2-induced degradation of PSD-95 in WT neurons is proteasome-dependent, we applied one of two proteasome inhibitors, MG132 (5 μM) or lactacystin (5 μM), together with Tamoxifen. Both MG132 and lactacystin blocked MEF2-induced down-regulation of PSD-95 in WT neurons and no changes in PSD-95 were observed under any condition in Fmr1 KO neurons (Figure 3E and S3A).
To determine whether MEF2-induced degradation of PSD-95 is mediated by Pcdh10, we used lentivirus to deliver MEF2-VP16ERtm along with one of two different shRNAs against Pcdh10 into dissociated cortical neuron cultures. Knocking down Pcdh10 with either shRNA inhibited MEF2-induced PSD-95 degradation (Figures 3F and S3B). PSD-95 degradation is rescued by lentiviral co-expression of shRNA-insensitive Pcdh10 together with the Pcdh10 shRNA (Figure S3C). These findings indicate that Pcdh10 is required for MEF2-induced degradation of PSD-95 and suggest a molecular mechanism for Pcdh10 in MEF2-induced synapse elimination.
Pcdh10 facilitates the proteasomal deposition of ubiquitinated PSD-95
To better determine the role of Pcdh10 in MEF2-induced regulation of PSD-95, we investigated whether Pcdh10 physically associated with PSD-95 and if this changed with MEF2 activation. Using co-immunoprecipitation of Pcdh10 with PSD-95 from cortical neuron cultures (in the presence of MG132), we found Pcdh10 could associate with PSD-95 (Figure 4A). Activation of MEF2 by treating MEF2-VP16ERtm transfected cultures with Tamoxifen for 6 hours, drastically increased the association between Pcdh10 and PSD-95 in WT neurons, but not in Fmr1 KO neurons (Figure 4A). Similar results were obtained in hippocampal neuron cultures (Figure S4A). Given that MEF2 increases the expression of Pcdh10 in WT cultures (Figure 1D), we considered whether the MEF2-induced association of Pcdh10 and PSD-95 is solely due to enhanced production of Pcdh10 or if MEF2 enhances their interaction independent of changes in Pcdh10 levels. To differentiate between these possibilities, we used lentivirus to transfect WT or Fmr1 KO cortical cultures with a Flag-tagged Pcdh10 driven by the cytomegalovirus (CMV) promoter whose transcription is not regulated by MEF2. Although total Flag-Pcdh10 levels were unchanged upon MEF2 activation, immunoprecipitation of PSD-95 revealed an increase in Flag-Pcdh10 association with PSD-95 (Figure 4B, in the presence of MG132). This suggests that MEF2 promotes the association of Pcdh10 with PSD-95 independent of regulating Pcdh10 levels. In Fmr1 KO cultures, MEF2 activation did not stimulate an interaction of PSD-95 with Flag-Pcdh10, suggesting that the deficit in PSD95 degradation in Fmr1 KO neurons is not due to the lack of MEF2-triggered Pcdh10 translation but instead on a distinct function of FMRP.
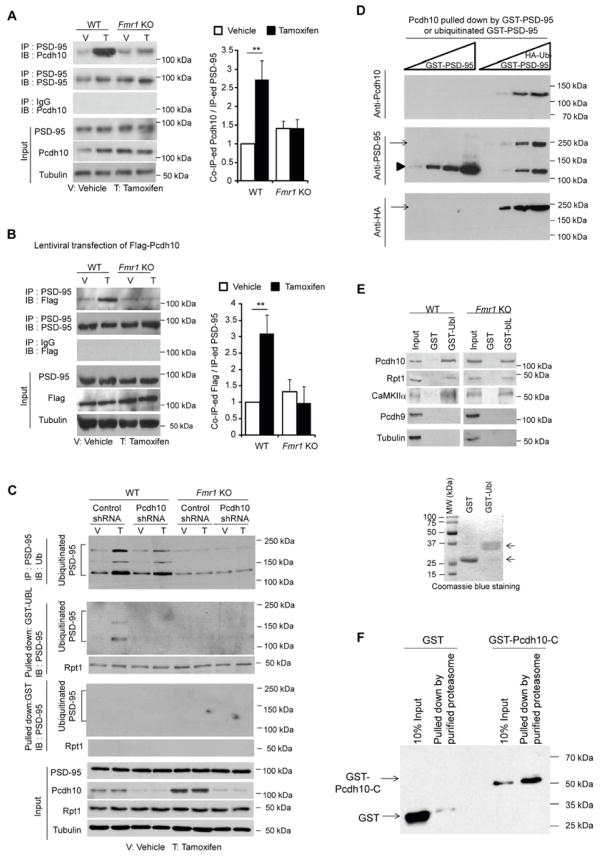
Figure 4. Pcdh10 facilitates the proteasomal degradation of ubiquitinated PSD-95.
(A and B) Western blots of (A) endogenous Pcdh10 or (B) Flag-Pcdh10 co-immunoprecipitated by anti-PSD-95 antibody. Protein samples were from dissociated WT or Fmr1 KO cortical neurons transfected with (A) MEF2-VP16ERtm or (B) MEF2-VP16ERtm plus Flag-Pcdh10, and treated with either vehicle or Tamoxifen for 6 hours. Inputs and quantification results are shown on the bottom and on the right. (n = 4 cultures) (C) Western blots of Ubiquitin after immunoprecipitation of PSD-95 (top); Western blots of PSD-95 and Rpt1 after pull down with GST-Ubl (second and third panels from the top); Western blots of PSD-95 and Rpt1 after the pull down with GST alone (fourth and fifth panels from the top). Protein samples were from dissociated WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm with either control shRNA or Pcdh10 shRNA. Treatment is as indicated and inputs are shown on the bottom. (D) Western blots of Pcdh10 from WT brain lysates pulled down by increasing amount of native GST-PSD-95 or in vitro ubiquitinated GST-PSD-95. GST-PSD-95 used for the assay was detected by anti-PSD-95 antibody shown in the middle panel while ubiquitinated GST-PSD-95 was detected by anti-HA antibody shown on the bottom. (E) Western blots of Pcdh10, Rpt1, CaMKIIα, Pcdh9 and Tubulin from WT or Fmr1 KO mice brains after pull down with either GST or GST-Ubl. GST proteins used are shown on the bottom after Coomassie blue staining on a SDS-PAGE gel. (F) Western blots of GST or GST-Pcdh10-C-terminus pulled down by purified proteasome from Flag-Rpt6 stably expressing HEK 293 cells. (Experiments in A, B and C were done in the presence of MG132). (**p < 0.01) See also Figure S4.
To determine the mechanism by which Pcdh10 regulates MEF2-dependent PSD-95 proteasomal degradation, we examined ubiquitination of PSD-95. MEF2 activation in WT cultures resulted in PSD-95 ubiquitination as observed by immunoprecipitation of PSD-95 followed by Western blotting with an anti-Ubiquitin (Ub) antibody (in the presence of MG132, Figure 4C, upper panel). Similar to previous reports (Colledge et al., 2003; Rezvani et al., 2007), polyubiquitinated PSD-95 was observed as multiple, distinct high molecular weight bands (115–200 kDa). Knockdown of Pcdh10 with a lentiviral expressed shRNA had a slight effect on MEF2-induced PSD-95 ubiquitination. However, using GST-Ubl (ubiquitin-like domain from Rad23) (Bingol et al., 2010) to pull down the proteasome and proteasome-interacting proteins (in the presence of MG132), we could only detect ubiquitinated PSD-95 in neurons transfected with MEF2 plus control shRNA, but not Pcdh10 shRNA (Figure 4C, second panel from the top, and S4B). These results suggest that the predominant role of Pcdh10 is to associate ubiquitinated PSD-95 with the proteasome.
Since Pcdh10 is important for association of ubiquitinated PSD-95 with the proteasome, we next asked whether PSD-95 ubiquitination or proteasome association is deficient in Fmr1 KO neurons. In contrast to WT cultures, MEF2 activation in Fmr1 KO cultures failed to increase PSD-95 ubiquitination (Figure 4C). Consistent with these results, basal ubiquitination of PSD-95 is reduced in vivo in Fmr1 KO hippocampi (Figure S4C). Taken together, these findings suggest that FMRP and Pcdh10 have distinct roles in MEF2-induced PSD-95 degradation: (1) FMRP is required for MEF2-induced ubiquitination of PSD-95, and (2) Pcdh10 is primarily required for linking ubiquitinated PSD-95 to the proteasomal machinery.
Based on results in WT cultures, we hypothesize that Pcdh10 associates ubiquitinated PSD-95 with the proteasome. To test this idea, we determined whether Pcdh10 interacts with ubiquitinated PSD-95 in vitro. We prepared native and ubiquitinated GST-PSD-95 by in vitro ubiquitination and observed that Pcdh10 from WT brain lysates preferentially interacts with ubiquitinated, but not native, GST-PSD-95 (Figure 4D). Ubiquitinated GST-PSD-95 failed to interact with in vitro translated Pcdh10 (Figure S4D) suggesting that posttranslational modifications of Pcdh10 or other interacting proteins are necessary for its interaction with ubiquitinated PSD-95. To test whether Pcdh10 interacts with the proteasome, we used GST-Ubl to pull down the proteasome, and find that Pcdh10, the proteasome component, Rpt1 (Demartino and Gillette, 2007), as well as the proteasome interacting protein, CaMKIIα (Bingol et al., 2010), were pulled down from both WT and Fmr1 KO mouse brain (Figure 4E). In contrast, another protocadherin-δ family member, Pcdh9 (Morishita and Yagi, 2007), was not pulled down by GST-Ubl. The interaction of Pcdh10 and the proteasome was also confirmed by the demonstration that the GST-tagged C-terminus of Pcdh10 interacts with a purified proteasome preparation from Flag-Rpt6 stably expressing HEK293 cells (Kim and DeMartino) (Figure 4F and S4E). Together, these results suggest that Pcdh10 interacts with ubiquitinated PSD-95 and the proteasome, and this interaction may not be conserved in other protocadherin family members.
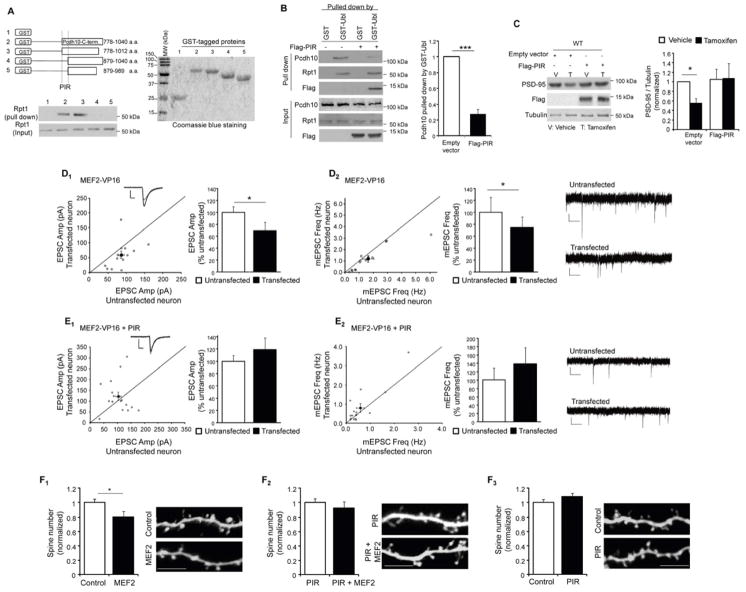
We next determined whether MEF2-induced PSD-95 degradation and synapse elimination required Pcdh10 association with the proteasome. Towards this goal, we first mapped the minimum region of Pcdh10 that is required for interaction with the proteasome. We focused on the C-terminus of Pcdh10 (266 amino acid) because it is predicted to be a cytoplasmic domain (Morishita and Yagi, 2007), is variable among protocadherin members. We made various deletion constructs of the C-terminus of Pcdh10 tagged with GST and tested their ability to pull down proteasomal complexes from brain lysates as determined by association with the essential proteasome protein component, Rpt1 (Bingol et al., 2010) (Figure 5A). Using this assay, we identified a 102 amino acid region (amino acids 778–879) of Pcdh10 required for the interaction with the proteasome. Further deletions of this region abolished any proteasome pull down (data not shown). Amino acids 778–879 are highly conserved in Pcdh10 across species (Figure S5A) and we termed this sequence the Proteasome Interacting Region, (PIR).
Figure 5. Blocking interaction of Pcdh10 and proteasome inhibits MEF2-induced PSD-95 degradation and synapse elimination.
(A) Western blots of Rpt1 from WT mouse brain after pull down by GST alone or different GST-Pcdh10 carboxyl-terminus proteins. Schematic diagram of Pcdh10-C-terminal deletions were shown on top. GST proteins used were shown on the right with Coomassie blue staining on a SDS-PAGE gel. (B) Western blots of Pcdh10, Rpt1 and Flag from dissociated WT cortical neurons transfected with Flag-PIR after pull down with either GST or GST-Ubl. Input of each protein and quantification are shown on the bottom and right respectively. (n = 3 cultures) (C) Western blots of PSD-95, Flag and Tubulin from dissociated WT cortical neurons transfected with MEF2-VP16ERtm and Flag-PIR with vehicle or Tamoxifen treatment as indicated. Quantification of PSD-95 level was shown on the right. (n = 4 cultures) (D and E) Average evoked EPSC amplitude (D1) and mEPSC frequency (D2) from untransfected and control vector plus MEF2-VP16 transfected cells (n = 14 for both D1 and D2). (E) Average evoked EPSC amplitude (E1) and mEPSC frequency (E2) from untransfected and PIR plus MEF2-VP16 transfected cells (n = 16 for both E1 and E2). Representative evoked EPSC and mEPSC traces are shown with the dot plots and on the right of each figure, respectively. (F) Quantification and representative images of apical, secondary, dendrites from WT CA1 pyramidal neurons transfected with GFP plus other plasmids as indicated. (n = 10 to 20 cells in each condition; Scale bar: 5 μm) In all figures, error bars represent SEM. (*p < 0.05, ***p < 0.001). See also Figure S5 and Table S1.
We aimed to determine if Pcdh10 association with the proteasome is necessary for MEF2-induced PSD-95 degradation and synapse elimination. In preparation for these experiments, we expressed a Flag-tagged PIR, in WT cultured neurons, and found that it distributes in dendrites and spines, but did not alter general proteasome activity (Figures S5B and S5C). Importantly, Flag-PIR expression greatly reduced the association of endogenous Pcdh10 with the proteasome in neurons as assessed by GST-Ubl pulldown (Figure 5B). This result suggests that exogenous expression of PIR can effectively compete with Pcdh10 for the proteasome. Coexpression of Flag-PIR and MEF2-VP16ERtm in WT cortical neuron cultures effectively blocked MEF2-induced degradation of PSD-95 (Figure 5C). Similarly, biolistic cotransfection of Flag-PIR and MEF2-VP16 into CA1 neurons of WT mouse hippocampal slice cultures blocked MEF2-induced functional synapse elimination as assessed by recordings of mEPSC frequency and evoked EPSC amplitude (Figure 5D, 5E and Table S1), and MEF2-induced reduction in dendritic spine density (Figure 5F). Expression of Flag-PIR alone did not alter synaptic function (Figure S5D), spine density (Figure 5F3) or MEF2-induced transcription (Figure S2I). These data demonstrate that Pcdh10 association with the proteasome is necessary for MEF2-induced PSD-95 degradation, as well as functional and structural synapse elimination. Together with our biochemical data (Figure 4) we propose that one role of Pcdh10 in synapse elimination is to deliver ubiquitinated PSD-95 to the proteasome for degradation.
The ubiquitin E3 ligase for PSD-95, Mdm2, is dysregulated in Fmr1 KO neurons
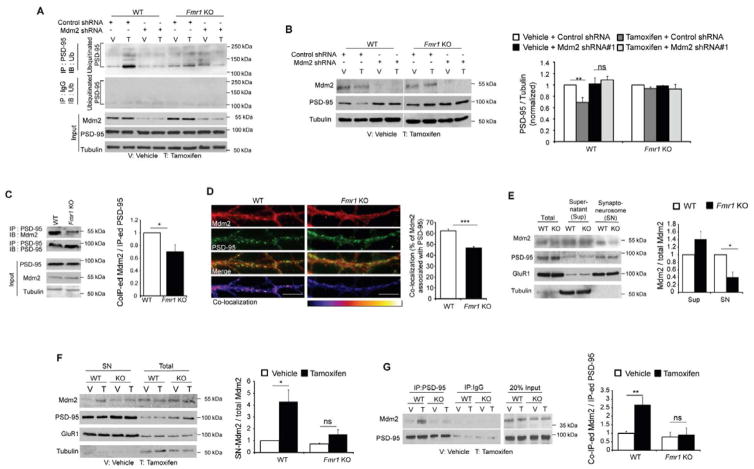
In Fmr1 KO neurons, MEF2-induced ubiquitination of PSD-95 is absent; a process that is independent of Pcdh10. To investigate the deficit in PSD-95 ubiquitination in Fmr1 KO neurons, we examined the substrate-specific step of the ubiquitination process which involves the ubiquitin E3 ligase for PSD-95, Mdm2 (Colledge et al., 2003). Using shRNAs against Mdm2, we observed that Mdm2 is required for MEF2-induced PSD-95 ubiquitination (Figure 6A) and degradation (Figure 6B, S6A, S6B and S6C) in WT cortical neurons, thus confirming that Mdm2 is the relevant E3 ligase for PSD-95. To investigate whether Mdm2 is dysregulated in Fmr1 KO neurons, we performed co-immunoprecipitation and observed reduced interaction of Mdm2 and PSD-95 in Fmr1 KO hippocampus in contrast to WT hippocampus (Figure 6C). Using immunohistochemistry for Mdm2 in dissociated hippocampal neurons, we also observed lower co-localization of Mdm2 with PSD-95 in dendrites of Fmr1 KO neurons compared to WT neurons (Figure 6D). Although total Mdm2 levels were normal in Fmr1 KO hippocampus (Figure 6C), which is consistent with the fact that Mdm2 has not been identified as an FMRP target mRNA (Darnell et al., 2011), Mdm2 levels in synaptoneurosome fractions of Fmr1 KO hippocampi were reduced to about 40% of WT levels (Figure 6E). We considered the possibility that MEF2 may enhance the synaptic localization of Mdm2 and this may be deficient in Fmr1 KO neurons. To address this possibility, we activated MEF2 in dissociated cortical neuron cultures using MEF2-VP16ERtm transfection and Tamoxifen and then examined Mdm2 levels in synaptoneurosome fractions and in co-immunoprecipitates with PSD-95. In WT neurons MEF2 activation increased Mdm2 levels in synaptoneurosomes (Figure 6F) as well as the interaction between PSD-95 and Mdm2 (Figure 6G). In contrast, in Fmr1 KO neurons, MEF2 failed to increase Mdm2 levels in synaptoneurosomes or the association of Mdm2 with PSD-95. MEF2 activation did not alter total Mdm2 levels in either WT or Fmr1 KO cultures (Figures 6B and 6F). Together, these results indicate that MEF2 stimulates the localization of Mdm2 to the synaptic compartment and to PSD-95 and this process is defective in Fmr1 KO neurons.
Figure 6. Mdm2 and Mdm2-mediated degradation of PSD-95 are dysregulated in Fmr1 KO neurons.
(A) Western blots of Ubiquitin after immunoprecipitation with PSD-95 (top) or IgG (second panel from the top). Protein samples were from dissociated WT or Fmr1 KO cortical neurons transfected with either control shRNA or Mdm2 shRNA together with MEF2-VP16ERtm. Treatment was as indicated and input was shown on the bottom. (B) Western blots of PSD-95, Mdm2 and Tubulin from dissociated WT or Fmr1 KO cortical neurons transfected with either control shRNA or Mdm2 shRNA together with MEF2-VP16ERtm. Quantification is shown on the right. (n = 3 cultures). (C) Western blots of Mdm2 immunoprecipitated with PSD-95 from WT or Fmr1 KO hippocampi. (n = 4 mice per genotype) (D) Immunohistochemistry of dendritic Mdm2 with PSD-95 from dissociated WT or Fmr1 KO hippocampal neurons. Co-localization images are shown on the bottom. Scale bars: 5 μm. Quantification of co-localization is shown on the right. (n = 21 cells) (E) Western blots of Mdm2, PSD-95, GluR1 and Tubulin in synaptoneurosomes prepared from WT or Fmr1 KO hippocampi. Quantification of Mdm2 in supernatant or synaptoneurosome is shown on the right. (n = 4 mice per genotype) (F) Western blots of Mdm2, PSD-95, GluR1 and Tubulin after synaptoneurosome preparation of dissociated WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERTm. Quantification of Mdm2 in synaptoneurosome is shown on the right. (n = 3 cultures) (G) Western blots of Mdm2 immunoprecipitated with PSD-95 from dissociated WT or Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm. Quantification is shown on the right. (n = 3 cultures). In all figures, error bars represent SEM. (*p < 0.05, **p < 0.01, ***p < 0.001) (Experiments in A, F and G were done in the presence of MG132) See also Figure S6.
Elevated EF1α inhibits Mdm2 from interacting with PSD-95 in Fmr1 KO neurons
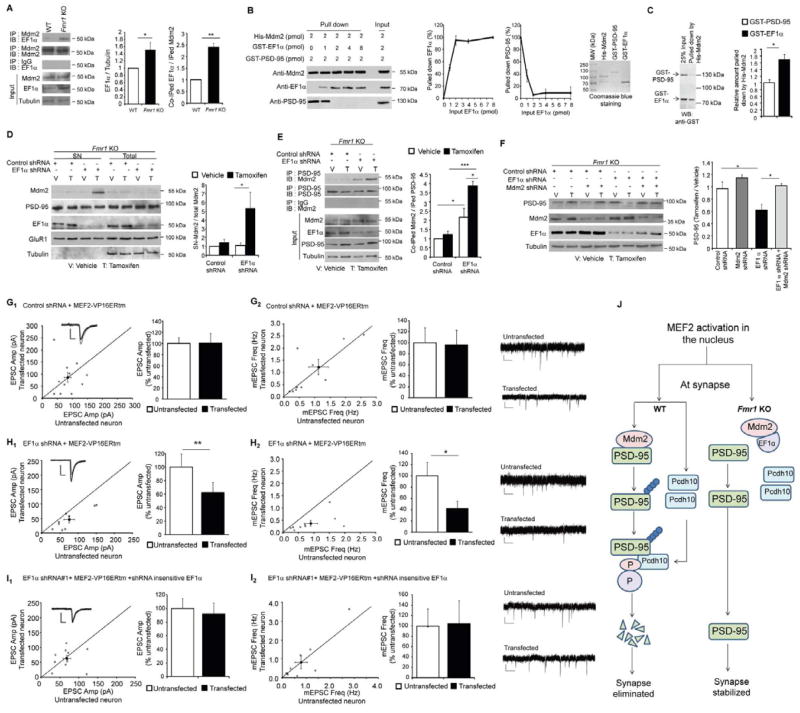
In Fmr1 KO neurons, Mdm2 localization at the synapse is reduced and MEF2-induced redistribution of Mdm2 to synapses is deficient (Figure 6). We hypothesized that Mdm2 may have an abnormal interactome in Fmr1 KO neurons. A previous proteomic study to identify human Mdm2 (Hdm2) -interacting proteins discovered a robust interaction of eukaryotic translation elongation factor 1-alpha (EF1α) and Hdm2 (Frum et al., 2007). EF1α is an FMRP-target mRNA and EF1α protein levels are up-regulated in Fmr1 KO brain (Darnell et al., 2011; Sung et al., 2003). We hypothesized that elevated EF1α in Fmr1 KO neurons sequesters Mdm2 and prevents it from ubiquitinating PSD-95. In support of this hypothesis immunoprecipitation of Mdm2 from hippocampal lysates revealed an increased interaction of EF1α and Mdm2 in vivo in Fmr1 KO mice (Figure 7A). This increased interaction may be due to the observed increase in total EF1α protein in Fmr1 KO (Figure 7A). To determine if EF1α and PSD-95 directly compete for interactions with Mdm2, we obtained recombinant His-Mdm2, GST-EF1α and GST-PSD-95. To Ni-beads with bound His-Mdm2, we first added increasing amounts of GST-EF1α. After washing off unbound GST-EF1α, we incubated the complex with GST-PSD-95. Because a possible degraded product of His-Mdm2 runs at the same position of GST- EF1α on a SDS-PAGE, we used Western blotting to detect the composition of the complex. As shown (Figure 7B), the binding of Mdm2 to EF1α occur with a stoichiometry of approximately 1:1 and EF1α-bound Mdm2 lost the ability to bind PSD-95. His-Mdm2 also runs at a position approximately doubled to its predicted molecular weight, suggesting formation of homodimer (Dolezelova et al., 2012). The GST tag does not interact with His-Mdm2 nor interfere with the binding of His-Mdm2 to GST-EF1α or GST-PSD-95 (Figure S7A). To test the affinity of Mdm2 toward binding EF1α or PSD-95, we pre-mixed GST- EF1α (1 pmol) and GST-PSD-95 (1 pmol) followed by adding Ni-beads with bound His-Mdm2 (0.5 pmol). As shown (Figure 7C), Mdm2 has higher affinity toward binding EF1α over PSD-95. Together, these results indicate that EF1α can effectively compete with PSD-95 for binding to Mdm2.
Figure 7. Elevated EF1α in Fmr1 KO neurons inhibits Mdm2 from interacting with PSD-95 and subsequent PSD-95 degradation.
(A) Western blots of EF1α from WT or Fmr1 KO hippocampus after immunoprecipitation by anti-Mdm2 antibody or IgG. Quantifications on total and immunoprecipitated EF1α with Mdm2 are shown on the right. (n = 3 cultures) (B) Western blots of Mdm2, EF1α and PSD-95 after in vitro, sequential, binding with recombinant proteins. Quantifications of binding and competition are shown in the middle (n = 3) while the purity of recombinant proteins is shown on the right with Coomassie blue stained SDS-PAGE gel. (C) Western blots of GST- EF1α and GST-PSD-95 after in vitro binding with His-Mdm2. Quantification of relative amount of GST proteins pulled down by His-Mdm2 is on the right. (n = 4) (D) Western blots of Mdm2, PSD-95, EF1α, GluR1 and Tubulin after synaptoneurosome preparation of dissociated Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm and either control shRNA or EF1α shRNA. Quantification of Mdm2 in synaptoneurosome is shown on the right. (n = 3 cultures) (E) Western blots of Mdm2 immunoprecipitated with PSD-95 from dissociated Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm and either control shRNA or EF1α shRNA. Quantification is shown on the right. (n = 3 cultures) Experiments in D and E were done in the presence of MG132. (F) Western blots of PSD-95, Mdm2, EF1α and Tubulin from dissociated Fmr1 KO cortical neurons transfected with MEF2-VP16ERtm and either control shRNA, EF1α shRNA and/or Mdm2 shRNA as indicated. Treatment with vehicle or Tamoxifen is also as indicated. Quantification is shown on the bottom. (n = 4 cultures). (G) Average evoked EPSC amplitude (G1) and mEPSC frequency (G2) from untransfected and control shRNA with MEF2-VP16ERtm transfected Fmr1 KO cells (n = 11 for both G1 and G2). (H) Average evoked EPSC amplitude (H1) and mEPSC frequency (H2) from untransfected and EF1α shRNA with MEF2-VP16ERtm transfected Fmr1 KO cells (n = 9 for both F1 and F2). (I) Average evoked EPSC amplitude (I1) and mEPSC frequency (I2) from untransfected and EF1α shRNA with MEF2-VP16ERtm and shRNA-insensitive EF1α transfected Fmr1 KO cells (n = 13 for both I1 and I2). In all figures, error bars represent SEM. Representative evoked EPSC and mEPSC traces are shown with the dot plots and on the right of each figure, respectively. (*p < 0.05, **p < 0.01). (J) Working model of MEF2-induced synapse elimination in WT neurons and the molecular basis of the deficit in synapse elimination in Fmr1 KO neurons. See also Figure S7 and Table S1.
EF1α competes with PSD-95 for interactions with Mdm2, and EF1α is elevated in Fmr1 KO brains. Therefore, we hypothesized that reducing EF1α levels in Fmr1 KO neurons may release Mdm2, allow interactions with PSD-95 and rescue MEF2-induced PSD-95 degradation. To test this possibility, we applied a lentivirus expressing a shRNA against EF1α that reduced EF1α in Fmr1 KO neurons to WT levels, but did not affect Pcdh10 protein levels (Figure S7B). Knocking down EF1α in Fmr1 KO neurons did not affect MEF2 activation (Figure S7C) but restored MEF2-induced synaptic localization of Mdm2 (Figure 7D) and the interaction between Mdm2 and PSD-95 (Figure 7E). Knocking down EF1α with shRNAs also restored MEF2-induced PSD-95 degradation in Fmr1 KO neurons which required Mdm2 (Figures 7F, and S7D). Coexpression of a shRNA-insensitive EF1α together with the EF1α shRNA inhibited MEF2-induced synaptic accumulation of Mdm2 (Figure S7E), the interaction between PSD-95 and Mdm2 (Figure S7F), and PSD-95 degradation (Figure S7G) in Fmr1 KO neurons demonstrating the specificity of the EF1α shRNA. These results demonstrate that elevated EF1α in Fmr1 KO neurons inhibits Mdm2 from interacting with PSD-95 and blocks the subsequent degradation of PSD-95.
To determine whether knocking down EF1α in Fmr1 KO neurons could also restore MEF2-induced synapse elimination, we biolistically transfected Fmr1 KO hippocampal slice cultures with a plasmid encoding a shRNA for EF1α or a control shRNA. Expression of either shRNA alone had no effect on synaptic function as measured by mEPSCs and evoked EPSCs (Figure S7H, S7I and Table S1). We then co-transfected Fmr1 KO slice cultures with either control shRNA or EF1α shRNA together with MEF2-VP16ERtm and subsequently treated the cultures with Tamoxifen. Fmr1 KO neurons transfected with control shRNA had a deficit in functional synapse elimination in response to MEF2 activation (Figure 7G) consistent with our previous report (Pfeiffer et al., 2010). As observed with PSD-95 degradation, EF1α shRNA transfection rescued robust functional synapse elimination in response to MEF2 activation (Figure 7H). Again, coexpression of a shRNA-insensitive EF1α together with the EF1α shRNA blocked MEF2-induced functional synapse elimination (Figure 7I). The shRNA-insensitive EF1α on its own did not affect excitatory synaptic function (Figure S7J). Taken together, our results indicate that elevated EF1α in Fmr1 KO neurons inhibits Mdm2 from triggering PSD-95 degradation and subsequent synapse elimination by MEF2 in the Fmr1 KO neurons. By knocking down EF1α, we were able to rescue the following in response to MEF2 activation: (1) synaptic re-distribution of Mdm2, (2) the interaction between Mdm2 and PSD-95, (3) PSD-95 degradation and (4) synapse elimination.
Discussion
MEF2 triggers synapse elimination by promoting Pcdh10-dependent degradation of a synaptic scaffolding protein
Here we identified a novel mechanism by which the activity-dependent transcription factor, MEF2, promotes synapse elimination as well as the molecular basis of the deficit in synapse elimination observed in Fragile X Syndrome, a prevalent cognitive and autistic disorder. In wild type neurons, MEF2 activation induces transcription of Pcdh10, which mediates degradation of the synaptic scaffold, PSD-95, and synapse elimination by associating ubiquitinated PSD-95 with the proteasome (Figure 7J). Although translation of Pcdh10, an FMRP target mRNA, is dysregulated in Fmr1 KO neurons, we find a deficit in MEF2-stimulated ubiquitination of PSD-95, a step that is upstream and independent of Pcdh10. The deficit in PSD-95 ubiquitination in Fmr1 KO neurons is caused by elevated levels of EF1α protein, an FMRP target mRNA, that sequesters Mdm2 from the synapse and PSD-95 (Figure 7J). In support of this conclusion, knockdown of EF1α levels rescues MEF2-induced PSD-95 degradation and synapse elimination in Fmr1 KO neurons.
Protocadherins and synapse elimination
The protocadherins are ancestors of the classical cadherins, the more well known members of the Ca2+ dependent cell adhesion molecule family (Giagtzoglou et al., 2009). Although the extracellular domains of protocadherins share homology with the traditional cadherins, they lack of a hydrophobic pocket within extracellular domain 1 that is important for strong homophilic adhesion (Morishita et al., 2006; Morishita and Yagi, 2007). Consistent with this prediction, Pcdh10, when expressed in cell lines, clusters at points of cell contact and stimulates cell aggregation, but less robustly than classical cadherins (Hirano et al., 1999). A recent study demonstrated that another δ2 non-clustered protocadherin family member, Pcdh8 or Arcadlin, is induced in neurons in response to seizures, and interacts in cis with N-cadherin (a classical cadherin) in postsynaptic membranes. Homophilic interactions of Pcdh8, in cis or trans, recruit p38 MAPK to its C-terminus which, in turn, stimulates the co-endocytosis of Pcdh8 and N-cadherin and decreases dendritic spine density (Yasuda et al., 2007). An interesting possibility is that the induction of the δ2 Pcdhs, such as Pcdh8 and Pcdh10, functions to increase the membrane concentration and probability of homophilic Pcdh interactions, which activates key signaling pathways and their endocytosis. Such endocytosis could facilitate interactions of Pcdh10 with ubiquitinated PSD-95 and the proteasome.
MEF2 promotes degradation of a postsynaptic scaffold to eliminate synapses
MEF2 activation is associated with a rapid reduction of synapse number suggesting that MEF2 eliminates pre-existing synapses (Flavell et al., 2006; Pfeiffer et al., 2010). However, MEF2 may also inhibit synapse formation rates. In support of an active elimination mechanism, MEF2 activation caused a rapid and selective degradation of PSD-95 and decrease in functional synapse number; effects that both required the MEF2 transcript, Pcdh10. Disrupting the association of Pcdh10 with the proteasome using the PIR peptide, blocked MEF2-induced PSD-95 degradation as well as functional and structural synapse elimination suggesting that ubiquitination and Pcdh10-dependent degradation of PSD-95 are key steps that couple MEF2-driven transcription to synapse elimination. PSD-95 stabilizes synaptic AMPARs, increases synaptic function, and results in larger and more stable spines (Keith and El-Husseini, 2008; Opazo et al., 2011). Conversely, reductions in PSD-95 can lead to diffusion and endocytosis of synaptic AMPARs, weaker synapses, smaller spines and spine elimination (Colledge et al., 2003; Haas et al., 2007; Keith and El-Husseini, 2008; Opazo et al., 2011; Woods et al., 2011). Specifically, in response to NMDAR activation, PSD-95 is ubiquitinated by Mdm2, degraded by the proteasome, removed from spines, and required for endocytosis of AMPARs (Colledge et al., 2003; Sturgill et al., 2009). Therefore, MEF2 triggered degradation PSD-95 may cause destabilization and removal of synaptic AMPARs that precedes elimination of the structural synapse. Of note, knockdown of Pcdh10 prevented MEF2-induced functional synapse elimination, but not ubiquitination of PSD-95. This suggests that ubiquitination of PSD-95 is not sufficient to mediate functional synapse elimination, but Pcdh10-dependent targeting of ubiquitinated PSD-95 to the proteasome is also required.
MEF2 triggered synaptic accumulation of Mdm2 is blocked by elevated EF1α in Fmr1 KO neurons
MEF2 activation triggered the synaptic accumulation of Mdm2, its association with PSD-95 and subsequent ubiquitination of PSD-95 by Mdm2, independently of Pcdh10. Therefore, another MEF2-activated transcript may stimulate Mdm2 movement to the synapse and association with PSD-95. In Fmr1 KO neurons, elevated levels of EF1α sequestered Mdm2 and prevented its synaptic accumulation and association with PSD-95 in response to MEF2. Whether MEF2 regulates the interaction of EF1α and Mdm2 to regulate PSD-95 degradation in WT neurons is unknown. Interestingly, local sequestration of an E3 ubiquitin ligase complex mediates elimination of specific synaptic connections during development in C. Elegans (Ding et al., 2007). Localized translational suppression of EF1α by FMRP may function to locally regulate Mdm2, PSD-95 and synapse number.
Recapitulating synapse elimination in Fragile X syndrome
Fragile X syndrome patients and Fmr1 KO mice display elevated dendritic spine number in mature cortical neurons which may contribute to the neuronal hyperexcitability, hypersensitivity to sensory stimuli and epileptic seizures observed in patients and the mouse model (Berry-Kravis, 2002; Dolen et al., 2010). A deficit in activity and MEF2-dependent, synapse elimination could lead to excess spines. In support of this idea, experience-dependent spine elimination in the somatosensory barrel cortex is deficient in Fmr1 KO mice (Pan et al., 2010). The molecular mechanisms that lead to elevated dendritic spine number or the deficits in synapse elimination associated with Fragile X are unknown. Current evidence suggests that FMRP functions as a translational switch of its target mRNAs. At steady state, FMRP suppresses translation of its mRNA targets in dendrites and upon synaptic stimulation dephosphorylation and/or degradation of FMRP derepresses or stimulates their translation (Bassell and Warren, 2008; Niere et al., 2012). Consistent with this model, we observed elevated Pcdh10 translation rates in Fmr1 KO neurons which were not stimulated by MEF2, despite induction of Pcdh10 mRNA. Alternatively, Pcdh10 translation rates in Fmr1 KO neurons may be saturated and cannot be further increased by elevated mRNA. We would expect that translational control of other FMRP target mRNAs that are induced in response to MEF2 would be similarly affected in Fmr1 KO neurons.
Although translational control of Pcdh10 is abnormal in Fmr1 KO neurons, our results suggest that this does not underlie the deficit in MEF2-induced synapse elimination. Knocking down EF1α restores MEF2-induced synapse elimination in Fmr1 KO mice without affecting Pcdh10 protein levels. Therefore, one requirement of FMRP in MEF2-induced synapse elimination is to suppress steady state translation of EF1α and allow appropriate MEF2-regulated synaptic targeting of Mdm2 and PSD-95 ubiquitination. This highlights the importance of translational suppression and regulation of protein concentration in dendrites for proper synaptic plasticity. Loss of function mutations in the MEF2 isoform, MEF2C, Fmr1 and Pcdh10 are all associated with autism, with or without intellectual disability (Abrahams and Geschwind, 2008; Morrow et al., 2008; Novara et al., 2010). Here we have described distinct roles for all three genes in regulated degradation of PSD-95 and synapse elimination. This work suggests a common deficit in activity-dependent synapse elimination among different genetic causes of autism.
Experimental Procedures
Organotypic slice culture and dissociated neuron culture
Organotypic hippocampal slice cultures were made from postnatal day (P) 6–7 WT or Fmr1 KO mice bred from the congenic C57BL/6 mouse strain and transfected as described (Pfeiffer et al., 2010) (Supplementary Experimental Procedures). Dissociated hippocampal or cortical cultures were prepared from P0-1 WT and Fmr1 KO mice as described (Niere et al., 2012).
Electrophysiology
Dual whole-cell patch recordings were obtained from CA1 pyramidal neurons in slice cultures using IR-DIC and GFP fluorescence to identify non-transfected and transfected neurons as described (Pfeiffer et al., 2010)(Supplemental Experimental Procedures).
Synaptoneurosome preparation
Hippocampal synaptoneurosomes were prepared as previously described (Waung et al., 2008). For synaptoneurosomes of dissociated cultures, cells were scraped off in buffer: 118 mM NaCl, 4.7 mM KCl, 1.2 mM MgSO4, 2.5 mM CaCl2, 1.53 mM KH2PO4, 212.7 mM glucose, 1 mM DTT (pH 7.4), and protease inhibitor cocktail (Calbiochem) followed by same procedures as described (Waung et al., 2008).
Statistical analysis
For multiple comparisons, a 2 way- ANOVA and Bonferroni post-hoc test were performed. Paired t-tests were used for electrophysiology assays. Independent t-test was used for Figures 1D, 5B, 5F, 6C and 7A. In all figures, error bars represent SEM and *p < 0.05, **p < 0.01, ***p < 0.001.
Supplementary Material
Highlights.
The transcription factor MEF2 induces Pcdh10 that is required for synapse elimination
MEF2 stimulates ubiquitination of PSD-95 by the ubiquitin E3 ligase, Mdm2
Pcdh10 chaperones ubiquitinated PSD-95 to proteasome
Elevated EF1α in Fmr1 KO neuron blocks Mdm2 and MEF2-mediated synapse elimination
Acknowledgments
This research was supported by the grants from the National Institutes of Health NS045711, HD052731 (K.M.H.), F32HD069111 (N.P.T.), F32HD062120 (J.R.W.) and the Simons Foundations (K.M.H. and C.W.C.). We would also like to thank L. Ormazabal, N. Cabalo, F. Niere, K. Loerwald, T. Zang, D. Thompson, X. Li and B. Chen for technical assistance, Dr. M. Rosen for the GST-Pcdh10 constructs and Dr. J. Gibson for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis E. Epilepsy in fragile X syndrome. Dev Med Child Neurol. 2002;44:724–728. doi: 10.1017/s0012162201002833. [DOI] [PubMed] [Google Scholar]
- Bianchetta MJ, Lam TT, Jones SN, Morabito MA. Cyclin-dependent kinase 5 regulates PSD-95 ubiquitination in neurons. J Neurosci. 2011;31:12029–12035. doi: 10.1523/JNEUROSCI.2388-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Wang CF, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demartino GN, Gillette TG. Proteasomes: machines for all reasons. Cell. 2007;129:659–662. doi: 10.1016/j.cell.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Ding M, Chao D, Wang G, Shen K. Spatial regulation of an E3 ubiquitin ligase directs selective synapse elimination. Science. 2007;317:947–951. doi: 10.1126/science.1145727. [DOI] [PubMed] [Google Scholar]
- Dolen G, Carpenter RL, Ocain TD, Bear MF. Mechanism-based approaches to treating fragile X. Pharmacol Ther. 2010;127:78–93. doi: 10.1016/j.pharmthera.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Dolezelova P, Cetkovska K, Vousden KH, Uldrijan S. Mutational analysis of Mdm2 C-terminal tail suggests an evolutionarily conserved role of its length in Mdm2 activity toward p53 and indicates structural differences between Mdm2 homodimers and Mdm2/MdmX heterodimers. Cell Cycle. 2012;11:953–962. doi: 10.4161/cc.11.5.19445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum R, Busby SA, Ramamoorthy M, Deb S, Shabanowitz J, Hunt DF, Deb SP. HDM2-binding partners: interaction with translation elongation factor EF1alpha. J Proteome Res. 2007;6:1410–1417. doi: 10.1021/pr060584p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giagtzoglou N, Ly CV, Bellen HJ. Cell adhesion, the backbone of the synapse: “vertebrate” and “invertebrate” perspectives. Cold Spring Harb Perspect Biol. 2009;1:a003079. doi: 10.1101/cshperspect.a003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas KF, Miller SL, Friedman DB, Broadie K. The ubiquitin-proteasome system postsynaptically regulates glutamatergic synaptic function. Mol Cell Neurosci. 2007;35:64–75. doi: 10.1016/j.mcn.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech Dev. 1998;76:79–90. doi: 10.1016/s0925-4773(98)00093-8. [DOI] [PubMed] [Google Scholar]
- Hanson JE, Madison DV. Presynaptic FMR1 genotype influences the degree of synaptic connectivity in a mosaic mouse model of fragile X syndrome. J Neurosci. 2007;27:4014–4018. doi: 10.1523/JNEUROSCI.4717-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S, Yan Q, Suzuki ST. Expression of a novel protocadherin, OL-protocadherin, in a subset of functional systems of the developing mouse brain. J Neurosci. 1999;19:995–1005. doi: 10.1523/JNEUROSCI.19-03-00995.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith D, El-Husseini A. Excitation Control: Balancing PSD-95 Function at the Synapse. Front Mol Neurosci. 2008;1:4. doi: 10.3389/neuro.02.004.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Kim SY, Yasuda S, Tanaka H, Yamagata K, Kim H. Non-clustered protocadherin. Cell Adh Migr. 2011;5:97–105. doi: 10.4161/cam.5.2.14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, DeMartino GN. C termini of proteasomal ATPases play nonequivalent roles in cellular assembly of mammalian 26 S proteasome. J Biol Chem. 2011;286:26652–26666. doi: 10.1074/jbc.M111.246793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Franke TF, Gan WB. Opposite effects of fear conditioning and extinction on dendritic spine remodelling. Nature. 2012 doi: 10.1038/nature10792. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- Morishita H, Umitsu M, Murata Y, Shibata N, Udaka K, Higuchi Y, Akutsu H, Yamaguchi T, Yagi T, Ikegami T. Structure of the cadherin-related neuronal receptor/protocadherin-alpha first extracellular cadherin domain reveals diversity across cadherin families. J Biol Chem. 2006;281:33650–33663. doi: 10.1074/jbc.M603298200. [DOI] [PubMed] [Google Scholar]
- Morishita H, Yagi T. Protocadherin family: diversity, structure, and function. Curr Opin Cell Biol. 2007;19:584–592. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, Mukaddes NM, Balkhy S, Gascon G, Hashmi A, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–223. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- Niere F, Wilkerson JR, Huber KM. Evidence for a fragile X mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered Arc translation and long-term depression. J Neurosci. 2012;32:5924–5936. doi: 10.1523/JNEUROSCI.4650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novara F, Beri S, Giorda R, Ortibus E, Nageshappa S, Darra F, Dalla Bernardina B, Zuffardi O, Van Esch H. Refining the phenotype associated with MEF2C haploinsufficiency. Clin Genet. 2010;78:471–477. doi: 10.1111/j.1399-0004.2010.01413.x. [DOI] [PubMed] [Google Scholar]
- Opazo P, Sainlos M, Choquet D. Regulation of AMPA receptor surface diffusion by PSD-95 slots. Curr Opin Neurobiol. 2011 doi: 10.1016/j.conb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- Pan F, Aldridge GM, Greenough WT, Gan WB. Dendritic spine instability and insensitivity to modulation by sensory experience in a mouse model of fragile X syndrome. Proc Natl Acad Sci U S A. 2010;107:17768–17773. doi: 10.1073/pnas.1012496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Zang T, Wilkerson JR, Taniguchi M, Maksimova MA, Smith LN, Cowan CW, Huber KM. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66:191–197. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, et al. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani K, Teng Y, Shim D, De Biasi M. Nicotine regulates multiple synaptic proteins by inhibiting proteasomal activity. J Neurosci. 2007;27:10508–10519. doi: 10.1523/JNEUROSCI.3353-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill JF, Steiner P, Czervionke BL, Sabatini BL. Distinct domains within PSD-95 mediate synaptic incorporation, stabilization, and activity-dependent trafficking. J Neurosci. 2009;29:12845–12854. doi: 10.1523/JNEUROSCI.1841-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YJ, Dolzhanskaya N, Nolin SL, Brown T, Currie JR, Denman RB. The fragile X mental retardation protein FMRP binds elongation factor 1A mRNA and negatively regulates its translation in vivo. J Biol Chem. 2003;278:15669–15678. doi: 10.1074/jbc.M211117200. [DOI] [PubMed] [Google Scholar]
- Tian X, Kai L, Hockberger PE, Wokosin DL, Surmeier DJ. MEF-2 regulates activity-dependent spine loss in striatopallidal medium spiny neurons. Mol Cell Neurosci. 2010;44:94–108. doi: 10.1016/j.mcn.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. OL-Protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat Neurosci. 2007;10:1151–1159. doi: 10.1038/nn1960. [DOI] [PubMed] [Google Scholar]
- Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods GF, Oh WC, Boudewyn LC, Mikula SK, Zito K. Loss of PSD-95 enrichment is not a prerequisite for spine retraction. J Neurosci. 2011;31:12129–12138. doi: 10.1523/JNEUROSCI.6662-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Yu X, Perlik AJ, Tobin WF, Zweig JA, Tennant K, Jones T, Zuo Y. Rapid formation and selective stabilization of synapses for enduring motor memories. Nature. 2009;462:915–919. doi: 10.1038/nature08389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Pan F, Gan WB. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462:920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, Takemiya T, Mizoguchi A, Yagita Y, Sakurai T, et al. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 2007;56:456–471. doi: 10.1016/j.neuron.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang Q, Huang Y. Fragile X mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize Nxf1 mRNA in neuronal cells. Proc Natl Acad Sci U S A. 2007;104:10057–10062. doi: 10.1073/pnas.0700169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.