Abstract
Rationale: Extracellular matrix (ECM) is a dynamic tissue that contributes to organ integrity and function, and its regulation of cell phenotype is a major aspect of cell biology. However, standard in vitro culture approaches are of unclear physiologic relevance because they do not mimic the compositional, architectural, or distensible nature of a living organ. In the lung, fibroblasts exist in ECM-rich interstitial spaces and are key effectors of lung fibrogenesis.
Objectives: To better address how ECM influences fibroblast phenotype in a disease-specific manner, we developed a culture system using acellular human normal and fibrotic lungs.
Methods: Decellularization was achieved using treatment with detergents, salts, and DNase. The resultant matrices can be sectioned as uniform slices within which cells were cultured.
Measurements and Main Results: We report that the decellularization process effectively removes cellular and nuclear material while retaining native dimensionality and stiffness of lung tissue. We demonstrate that lung fibroblasts reseeded into acellular lung matrices can be subsequently assayed using conventional protocols; in this manner we show that fibrotic matrices clearly promote transforming growth factor-β–independent myofibroblast differentiation compared with normal matrices. Furthermore, comprehensive analysis of acellular matrix ECM details significant compositional differences between normal and fibrotic lungs, paving the way for further study of novel hypotheses.
Conclusions: This methodology is expected to allow investigation of important ECM-based hypotheses in human tissues and permits future scientific exploration in an organ- and disease-specific manner.
Keywords: extracellular matrix, lung fibrosis, fibroblast
At a Glance Commentary
Scientific Knowledge on the Subject
Extracellular matrix is important in driving cellular phenotypic responses in fibrotic and other lung disorders. Typical investigation using planar, rigid dishes coated with isolated matrix proteins may not accurately reflect the impact of matrix on cells in vivo.
What This Study Adds to the Field
We define a new method to prepare acellular lung matrix that can be used to study the impact of extracellular matrix on lung cell phenotype. This process results in a more physiologic substrate that retains normal stiffness, architecture, and matrix composition compared with native lungs. By using human lungs and cells, this new system provides a novel way to culture cells in vitro to investigate matrix contributions to lung pathology in a disease-specific manner.
The extracellular matrix (ECM), long believed to simply serve as a structural support for cells, is now recognized as an important mediator in health and disease; ECM is a highly dynamic complex that varies in composition according to its tissue localization and physiological circumstances. In addition to providing structural integrity, ECM delivers important spatial and contextual cues to drive varied behaviors such as proliferation, migration, epithelial-to-mesenchymal transition, apoptosis, and production of various secreted mediators (as reviewed in References 1–3, among others). ECM also acts as a reservoir for growth factors and cytokines that, during tissue injury and repair, help to localize and direct the wound healing response. The ECM is critical during embryogenesis, dictating key developmental events such as branching morphogenesis in the lung (4, 5) as well as cardiovascular and neural development (6, 7). Thus, ECM is an integral contributor to tissue homeostasis and development.
In the human lung, ECM consists primarily of collagen I (which provides tensile strength), elastins (which provide elasticity), and laminins and collagen IV (which constitutes the bulk of the alveolar and airway basement membranes). Other ECM proteins (e.g., fibronectin and glycosaminoglycans) serve to maintain lung cell polarity and survival. Although the groundwork for our understanding of individual ECM component functions has been established using standard culture techniques, mechanistic studies of human lung ECM have been hampered by lack of a biologically relevant model. Although reductionist studies using cells grown on planar ECM-coated plastic dishes have informed our understanding of cell signaling and behavior, this approach does not account for the complexity of a three-dimensional (3D), deformable microenvironment in the lung where cells encounter a variety of different proteins simultaneously in a unique spatial geometry. To address these issues, some investigators have used human fibroblast-derived 3D matrices to determine the effects of ECM on cell phenotype, often with strikingly disparate results compared with planar matrices (8). This approach, although more appropriate than culturing cells on planar ECM-coated dishes, does not recapitulate an organ-specific ECM structure and composition. More recently, newer technologies involving artificial polymeric or hydrogel scaffolds (9) or bioengineered cantilever posts that deform under stress (10) have been used to evaluate cell–ECM interactions, but these studies are often limited to single ECM proteins rather than the complex mixture found in situ and lack the topographic variability of the organs they are designed to mimic. Notably, none of these models allows for ECM analysis in an organ- or disease-specific manner.
Traditional tissue culture approaches are inadequate to study disease states that are themselves defined by disordered ECM. Idiopathic pulmonary fibrosis (IPF) is a progressive disorder of the lungs characterized by deposition and remodeling of the lung ECM, which ultimately leads to chronic respiratory failure and death. At present, no effective therapeutic options exist to slow progression of IPF (11). This has primarily been attributed to the fact that no animal model faithfully recapitulates all features of IPF (12), and in vitro investigation typically relies on standard culture techniques.
Because ECM is a critical regulator of health and disease, some investigators have relied on acellular matrices for organ regeneration. In experimental rodent models, we and others have demonstrated an ability to regenerate functional lungs (13, 14) and heart (15) by repopulating acellular matrices with various cell types. Perhaps more importantly, this approach has been successfully used in humans to correct large airway defects (16, 17).
To better study the influences of human lung ECM on cell behavior in vitro, we developed a biologically relevant in vitro model using decellularized human lung matrices. Our method results in a lung matrix devoid of cellular and nuclear material. Structural and ultrastructural analysis confirms the normal architecture of the matrix, with normally aligned matrix fibers and intact, homogeneous basement membranes. In contrast, acellular IPF lung matrix reveals marked ECM deposition in the lung interstitium with disorganized alignment and disrupted basement membranes. Proteomic analysis of acellular lung matrices identifies significant differences in ECM composition between normal and IPF lung. Atomic force microscopy confirms that acellular normal lung matrices exhibit an elastic modulus similar to that expected in vivo, whereas IPF acellular matrices demonstrated increased but heterogeneous stiffness. Finally, when cultured in normal or in IPF acellular lung matrices, normal human fibroblasts adopted starkly different phenotypes, confirming the active role of the ECM in shaping cell behavior. Taken together, these data highlight the feasibility of using an entirely human in vitro system to study lung ECM biology in a manner that takes into account tissue stiffness, architecture, and matrix composition. We believe this new approach will enable biologically relevant, mechanistic study of ECM in a fully humanized system that can be adapted to any disease state. Moreover, our data provide novel compositional information regarding the human lung ECM, opening new potential avenues of research.
Some of these results have been previously presented in abstract form (18).
Methods
Detailed methods can be found in the online supplement.
Cells and Reagents
Primary human lung fibroblasts were isolated as we have described previously (19). Written informed consent was obtained from all subjects in accordance with the University of Michigan Institutional Review Board, and cell lines were derived from patients in a blinded fashion, without regard to clinical data except diagnosis. Cells were maintained in Dulbecco’s modified Eagle medium with 10% fetal calf serum, antibiotics, glutamine, and N-2-hydroxyethylpiperazine-N′-ethane sulfonic acid. The ALK5 inhibitor A83-01 (20) was from Tocris Bioscience (Minneapolis, MN) and was used at a concentration of 20 nM in dimethyl sulfoxide.
Human Lung Procurement
Human control lungs deemed to be unsuitable for lung transplantation were obtained from beating-heart (or warm autopsy) donors through Gift of Life Michigan. Human deidentified IPF lung samples were obtained from explants of patients with IPF undergoing lung transplantation at the University of Michigan. The University of Michigan Institutional Review Board has deemed these approaches exempt from oversight as all subjects were considered deceased.
Decellularization of Lung Matrices
Decellularization was achieved based on modifications of a previously published protocol (21). A detailed protocol used for these studies is included in the online supplement.
Preparation of Acellular Matrix Slices for Tissue Culture
Cylinders of acellular lung matrices were obtained with a sterile 12-mm biopsy punch (Acuderm, Inc., Fort Lauderdale, FL), embedded in 2% warm ultra-low melt agarose (SeaPrep; Lonza, Rockland, ME), and cooled at 4°C. Sections 1,000 μm thick were cut on a standard Vibratome (Leica Biosystems, Richmond, IL). Sections were gently warmed to remove the agarose surrounding and within tissue sections. Before culture, slices were sterilized in 0.18% peracetic acid and 4.8% ethanol solution and rinsed extensively in phosphate-buffered saline (PBS). Slices were stored at 4°C in PBS containing antibiotics until use in culture. Using this technique with human lungs, a minimum of 60 cylinders can be obtained per lung lobe (depending on lobe size), and each cylinder can be sliced into 6 to 10 uniform slices.
Reseeding Acellular Matrices
Acellular matrices were placed in individual wells of a 24-well dish without media. Fibroblasts were harvested from culture, resuspended in media at 1 × 107 cells/ml, and 10 μl of cells were seeded dropwise into slices. After 30 minutes, tissue slices were transferred to individual wells of a Teflon-coated 24-well plate and media slowly added to the side of the well. Slices were cultured for indicated time points, changing media every other day.
Immunohistochemistry
Immunohistochemistry on paraffin-embedded sections was performed as previously described (22), with standard hematoxylin and eosin, Masson trichrome, Alcian blue, or Verhoeff elastic stains. Anti–α-smooth muscle actin (SMA) (clone 1A4; Sigma, St. Louis, MO) was used at 1:200. A Nikon Eclipse 50i microscope fitted with an Evolution MP Color CCD camera (Media Cybernetics, Bethesda, MD) was used to visualize and photograph sections.
Electron Microscopy
Scanning and transmission electron microscopy were performed by the University of Michigan Microscopy and Image Analysis Core.
Atomic Force Microscopy
A detailed protocol describing atomic force microscopy is available in the online supplement.
Sample Preparation and Mass Spectrometry
A detailed protocol describing the method used for mass spectrometry is available in the online supplement.
Western Blot
Western blot analysis of cellular lysates was performed as previously described (22). Antibodies against the EDA domain of fibronectin (clone 3E2) and α-SMA (clone 1A4) was from Sigma. Antibodies to β-tubulin, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), PTEN (phosphatase and tensin homolog deleted on chromosome 10), and E cadherin were from Cell Signaling Technologies (Danvers, MA). Antibodies to vimentin were from Santa Cruz Biotechnology (Santa Cruz, CA); antibodies to cytokeratin-19 were from Novus Biologicals (Littleton, CO).
RNA Extraction and Quantitative Reverse Transcriptase–Polymerase Chain Reaction
RNA extraction from cells cultured in acellular matrices was performed using a previously published method (23). Quantitative reverse transcriptase–polymerase chain reaction was performed on an Applied Biosystems (Foster City, CA) 7300 thermocycler as previously described (22), using SYBR Green for detection. All housekeeping genes were assayed using a gene array (catalog # PAHS-000; SA Biosciences/Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Results
Decellularization Effectively Removes Cellular and Nuclear Material
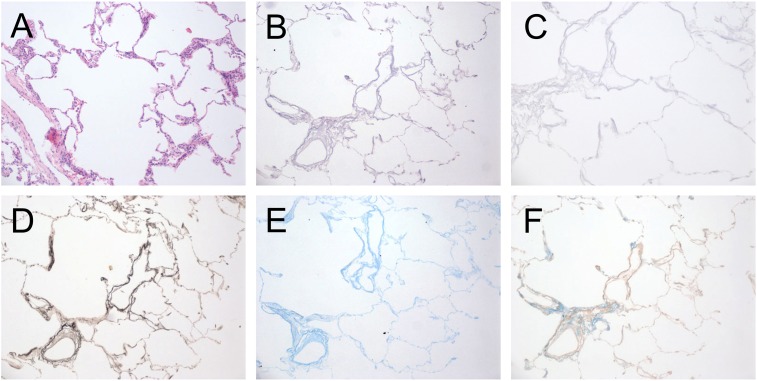
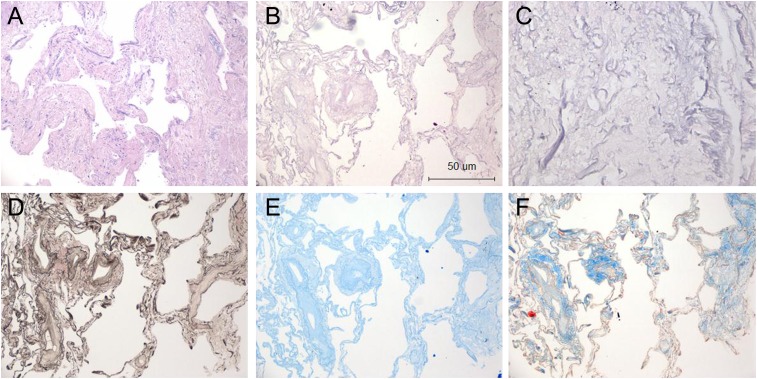
Culturing cells in acellular human lungs to study effects of ECM on target cell behavior requires they first be devoid of endogenous cellular material that might influence experimental results. Thus, we first confirmed that the decellularization process resulted in extraction of all cellular and nuclear material. Native or decellularized normal human donor lung samples (n = 3) were fixed in 10% buffered formalin and paraffin embedded. Sections were prepared and stained with hematoxylin and eosin (H&E), Verhoeff elastic (to identify elastic fibers), Alcian blue (to identify glycosaminoglycans), and Masson trichrome stains (to identify collagens). Figure 1A shows the typical appearance of normal human lung before decellularization, with cells lining alveolar, airway, and vascular structures (H&E stain). Acellular normal human lung maintained a similar architecture, albeit without evidence of cells (Figure 1B, H&E stain). Additional stains indicated the presence of elastin fibers, glycosaminoglycans, and collagens (Figures 1C–1E). Given the dense nature of fibrotic tissue, we also wished to confirm that the decellularization procedure was capable of removing cellular material from IPF lungs as well. Figure 2 reveals the absence of cellular material in densely fibrotic regions, suggesting that the decellularization protocol is capable of removing cells from denser tissues and is consistent with the literature (16).
Figure 1.
Histologic evaluation of native and acellular human normal lungs. (A) Native human normal lung, hematoxylin and eosin (H&E) stain. (B and C) Decellularized human lung stained with H&E at 10× (B) and 40× (C) magnification. (D) Verhoeff elastin stain, (E) Alcian blue, and (F) Masson trichrome stain (all 10× magnification). The decellularization process leaves the lung architecture intact.
Figure 2.
Histologic evaluation of native and acellular human idiopathic pulmonary fibrosis (IPF) lungs. (A) Native human IPF lung, hematoxylin and eosin (H&E) stain. (B and C) Decellularized human IPF lung stained with H&E at 10× (B) and 40× (C) magnification. (D) Verhoeff elastin stain, (E) Alcian blue, and (F) Masson trichrome stain (all 10× magnification). The decellularization process leaves the lung architecture intact.
To confirm that cellular products were absent in the matrices, we next performed Western blot on radioimmunoprecipitation assay (RIPA) extracts from native or acellular human lungs (n = 2 each) for cellular proteins; as shown in Figure 3A, native lungs expressed all cellular proteins investigated to various degrees. In contrast, none were detected in extracts of acellular lungs, with the exception of a minimal amount of collagen I(αI) chain, which was likely leached from the matrix. Finally, to ensure RNA was effectively removed from the acellular lung, we also extracted RNA from native and acellular lung matrices. Quantitative real-time polymerase chain reaction for a panel of human housekeeping genes on extracted samples showed that mRNA expression in acellular matrices was reduced greater than 99% compared with native matrices (Figure 3B). These data suggest effective clearance of cells and nuclear material using this method and are consistent with prior approaches (16, 21).
Figure 3.
Confirmation of cellular protein and DNA removal. (A) Western blot analysis for cellular proteins indicated in right margin before (lanes 1 and 3) and after (lanes 2 and 4) decellularization; n = 2 separate lung samples. Results are representative of three Western blots. (B) Real-time semiquantitative polymerase chain reaction of various housekeeping genes in non-decellularized lung (closed bars) and decellularized lung matrix (open bars). Relative expression of each gene was less than 99.95% in decellularized matrix compared with native lung. Results are pooled from two separate experiments. α-SMA = α-smooth muscle actin; β-2-MG = β-2-microglobulin; β-glc = β-glucuronidase; CD71 = transferrin receptor; CycA = cyclophilin A; G3PDH = glyceraldehyde-3-phosphate dehydrogenase; HPRT1 = hypoxanthine phosphoribosyltransferase 1; HSP90 = heat shock protein 90 kD α (cytosolic); L13a = ribosomal protein L13a; P0 = ribosomal protein, large; PTEN = phosphatase and tensin homolog deleted on chromosome 10. *P < 0.005, **P < 0.01, ***P < 0.001.
Dimensionality and Stiffness of Normal and Fibrotic Decellularized Lung
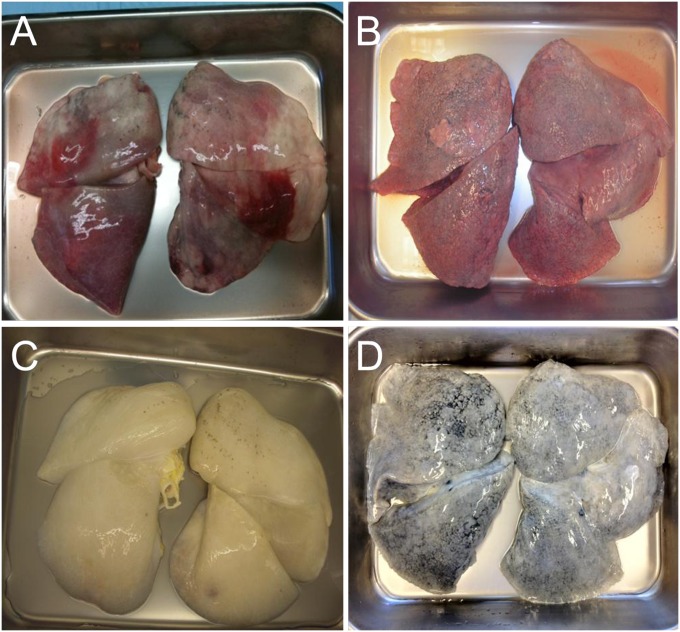
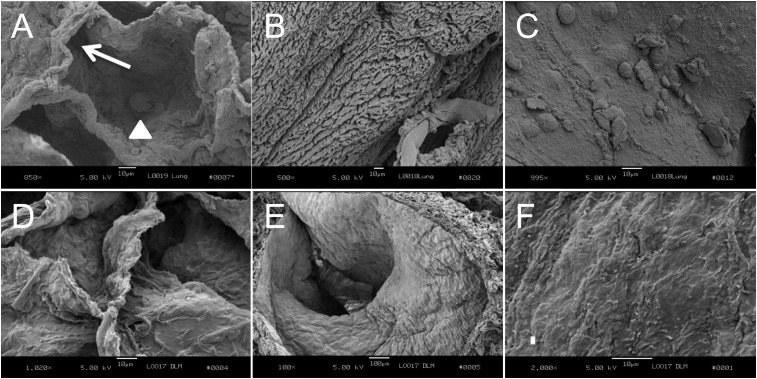
Grossly, acellular lung maintains a 3D morphology similar in appearance to normal lungs (Figure 4), comparable to results achieved in the rodent (13). However, it was critical to ensure that the normal microscopic architecture was preserved after decellularization. Thus, sections of lung were subjected to scanning electron microscopy. Figure 5 shows alveoli (Figures 5A and 5D), airway (Figures 5B and 5E), and vessel (Figures 5C and 5F) before and after decellularization, respectively. Notably, the remaining ECM basement membranes appeared intact, with maintenance of the normal 3D morphology of lung.
Figure 4.
Gross appearance of human lungs before and after decellularization. Normal lungs deemed unsuitable for transplantation and idiopathic pulmonary fibrosis lungs before (A and B, respectively) and after (C and D, respectively) decellularization. Lungs retain their normal gross structure and anatomic relationships.
Figure 5.
Scanning electron microscopy of normal lung matrices before (A–C) and after (D–F) decellularization. (A) Normal alveolus. White arrow shows normal-appearing alveolar wall. Arrowhead shows an alveolar macrophage; 850× magnification. (B) Ciliated bronchus; 500× magnification. (C) Pulmonary blood vessel. Blood cells are adherent to the wall; 995× magnification. (D) Acellular alveoli; 1,020× magnification. (E) Acellular bronchus without ciliated lining cells; 100× magnification. (F) Acellular vessel without lining cells; 2,000× magnification. Scale bar = 10 μm.
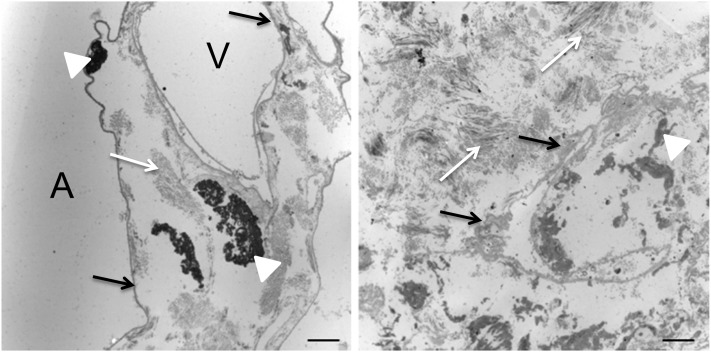
Ultrastructurally, transmission electron microscopy highlights the abnormal appearance of IPF lung compared with normal lung (Figure 6). Normal lung revealed collagen fibers and elastin arranged in discrete organized bundles traversing interstitial spaces and what appear to be intact, homogeneous alveolar epithelial and endothelial basement membranes lining alveolar spaces and vessels, respectively, whereas IPF lungs showed a markedly expanded interstitial space with heterogeneous, partially disrupted basement membrane and disorganized ECM organization. Importantly, these findings (coupled with those in Figures 1 and 2) support our conclusion that cells were completely removed from the tissues.
Figure 6.
Transmission electron microscopy of normal and idiopathic pulmonary fibrosis (IPF) acellular lung matrices. In normal matrix (left panel), alveolar epithelial and vascular endothelial basement membranes (black arrows) are of uniform thickness, and the interstitial space has organized fibers of collagen (white arrows) and discrete bundles of elastin (white arrowheads). In contrast, IPF matrix (right panel) shows a markedly thickened, heterogeneous basement membrane (black arrows) and haphazardly located, disorganized collagen fibers (white arrows) with disorganized elastin fragments (white arrowhead). No cellular material is observed. A = alveolar space. V = vascular space. Both panels, 8,520× magnification. Scale bar = 2 μm.
To ensure that decellularization did not result in significant ECM protein loss, we performed proteomic analysis on the effluent of the various wash steps during decellularization. More than 500 protein peptides were identified in the wash effluent; manual review of these revealed that the large majority of expressed peptides were non–matrix-associated (see Table E1 in the online supplement). Moreover, paired proteomic analyses of lungs before and after decellularization (n = 3 each) showed very few proteins that were significantly reduced after decellularization (Table E2). The observations suggest that the decellularization procedure results in the loss of few structural matrix proteins.
Tissue stiffness (measured as Young’s elastic modulus) varies depending on the organ (24), ranging from distensible (as in lung, around 1 kPa [25]) to very rigid (as in bone, 6.9–31.6 GPa; reviewed in [26]). However, standard cell culture techniques often use plastic dishes, which do not account for this variability in stiffness to which cells are exposed in vivo. Indeed, tissue culture plastic possesses a Young’s modulus ranging from 2 to 4 GPa (24), suggesting that lung cells will experience a much stiffer surface, and bone cells a much more deformable surface, when cultured on plastic dishes in vitro.
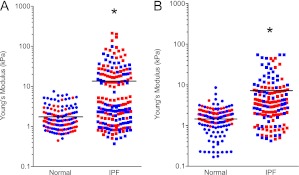
To ensure the acellular lung matrix retains its physiologic stiffness, we subjected native and acellular normal (n = 2 separate donors) and IPF (n = 2 separate donors) lung matrices to atomic force microscopy as outlined in Methods. Consistent with prior estimates, native normal human lung possessed a mean (± SEM) Young’s modulus of 1.96 ± 0.13 kPa (24) arranged in a relatively homogeneous pattern. In contrast, native IPF lung displayed a bimodal distribution of stiffness, with each sample exhibiting stiffness comparable to normal tissue as well as much higher stiffness representing fibrotic tissue (Figure 7A). Combined, mean (± SEM) stiffness of IPF tissue was 16.52 ± 2.25 kPa, significantly higher than normal tissue (P < 0.0001 by Mann-Whitney test) but still markedly less stiff than standard tissue culture plastic.
Figure 7.
Stiffness of native and acellular normal and idiopathic pulmonary fibrosis (IPF) lung matrices as determined by atomic force microscopy (n = 2 samples per condition; each color represents a different individual). (A) Native normal human lung possessed a mean (± SEM) Young’s modulus of 1.96 ± 0.13 kPa, whereas native IPF lung possessed a mean (± SEM) stiffness of 16.52 ± 2.25 kPa, significantly higher than normal tissue (P < 0.0001 by Mann-Whitney test). (B) The mean (± SEM) elastic (Young’s) modulus of decellularized normal lung is approximately 1.6 (± 0.08) kPa. By comparison, the mean (± SEM) Young’s modulus of decellularized IPF lung was 7.34 ± 0.6 kPa. Each point represents a single measurement of a 30 × 30 μm2 area. *P < 0.0001.
Interestingly, we found that after decellularization, normal lung matrices exhibited a mean (± SEM) Young’s modulus of 1.606 ± 0.08 kPa, which is no different than native lung (Figure 7B). In contrast, the mean (± SEM) Young’s modulus of acellular IPF lung matrices was 7.34 ± 0.6 kPa (P < 0.0001 compared with normal acellular lung by Mann-Whitney test) and somewhat more homogeneous in distribution (Figure 7B). We conclude that decellularization of IPF lungs may result in a relaxation of matrix proteins resulting in tissue that is slightly less stiff but still significantly stiffer than normal lung.
Proteomic Analysis of Acellular Lung Matrix
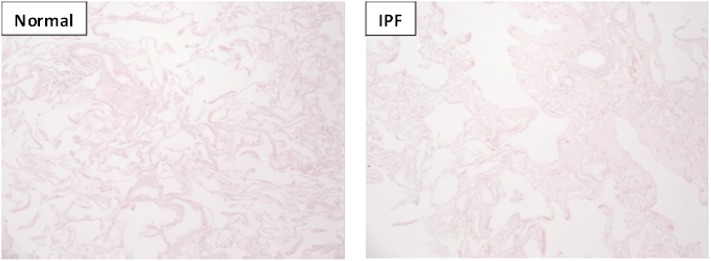
To better understand the composition of lung ECM, samples of acellular lung matrix were next evaluated using liquid chromatography/tandem mass spectrometry (LC-MS/MS). To ensure appropriate sampling, matrix for mass spectrometry was obtained from regions located adjacent to histologically confirmed representative samples (Figure 8). We identified 94 proteins in the normal lung “matrisome” (as defined in [27]), comprising 61 core ECM proteins (glycoproteins, collagens, and proteoglycans) and 33 ECM-related proteins (Table 1). As expected, we identified collagens (VI, XIV, XII, and IV), laminins, fibronectin, and various proteoglycans and their associated core proteins in normal lung. Notably, elastin and its peptides (desmosine, isodesmosine) were not identified, likely due to the inability of trypsin (used in the preparation for LC-MS/MS) to cleave this hydrophobic protein (28). As evidence of the presence of elastin, however, we did identify EMILIN-1 (elastin microfibril interfacer-1) and fibrillin-1 (both of which are necessary for formation of elastin fibers) in our samples. Moreover, our immunohistochemistry (Figure 1) and electron microscopy (Figure 6) studies clearly demonstrate the presence of elastin. In contrast, IPF matrices expressed 85 matrisome proteins (64 core ECM proteins and 21 ECM-related proteins) with a different protein signature than normal lungs (for example, collagens I, V, and XV were only expressed in IPF lungs) (Table 1). Despite the relatively equal numbers of various ECM proteins between normal and IPF lungs, the data also showed marked differences in the relative quantities of important proteins (Table 2). For example, IPF lungs were enriched for matrisome-associated proteins that might be predicted, such as glycosaminoglycans and latent transforming growth factor (TGF)-β binding protein 1, as well as relatively deficient in others, such as laminin α-3, β-3, and γ-2 chains, all components of the alveolar basement membrane. Moreover, we identified other proteins enriched in IPF lung, such as matrix Gla protein and nephronectin, that have not previously been associated with human IPF, although evidence suggests that they may have a role in experimental fibrosis models (29, 30). Similarly, collagens III and VI and periostin (a matricellular protein downstream of IL-13 signaling and implicated in lung fibrosis [31]) were enriched in IPF matrices.
Figure 8.
Hematoxylin and eosin stain (10× magnification) of normal and idiopathic pulmonary fibrosis lung tissue immediately adjacent to tissue assessed by liquid chromatography–tandem mass spectrometry reported in Table 1.
TABLE 1.
HUMAN NORMAL AND IPF LUNG MATRISOME AS DETERMINED BY MASS SPECTROMETRY OF ACELLULAR MATRICES
| Spectral Counts |
Spectral Counts |
||||
| Identified Proteins | Normal | IPF | Identified Proteins | Normal | IPF |
| Collagen α-3(VI) chain | 3,936 | 6140 | Dermatopontin | 32 | 84 |
| Basement membrane-specific heparan sulfate proteoglycan core protein | 2,707 | 1898 | Annexin A4 | 111 | 8 |
| Collagen α-1(VI) chain | 2,042 | 2,241 | Nidogen-2 | 67 | 37 |
| Collagen α-2(VI) chain | 1,471 | 1,891 | Tubulointerstitial nephritis antigen-like | 46 | 30 |
| Fibrillin-1 | 1,399 | 1,837 | Matrix metalloproteinase-9 | 70 | 0 |
| Fibronectin | 1,294 | 1,072 | Microfibrillar-associated protein 2 | 31 | 35 |
| Laminin subunit α-5 | 1,793 | 416 | Collagen α-1(XVIII) chain | 43 | 37 |
| Laminin subunit β-2 | 1,527 | 456 | von Willebrand factor | 49 | 20 |
| Fibrinogen γ chain | 1,136 | 450 | Fibulin-1 | 39 | 0 |
| Collagen α-1(XIV) chain | 145 | 1,268 | Galectin-3 | 32 | 16 |
| Fibrinogen β chain | 949 | 313 | Fibulin-2 | 21 | 26 |
| Laminin subunit γ-1 | 995 | 256 | Inter-α-trypsin inhibitor heavy chain H4 | 28 | 24 |
| Laminin subunit α-3 | 933 | 70 | Collagen α-1(VIII) chain | 38 | 14 |
| Periostin | 145 | 777 | Coagulation factor XIII A chain | 23 | 25 |
| Protein-glutamine γ-glutamyltransferase 2 | 401 | 444 | Metalloproteinase inhibitor 3 | 43 | 0 |
| Fibrinogen α chain | 565 | 254 | Plasma protease C1 inhibitor | 23 | 22 |
| Annexin A2 | 459 | 295 | Annexin A3 | 33 | 0 |
| Transforming growth factor-β–induced protein ig-h3 | 118 | 552 | Serpin B3 | 7 | 23 |
| Tenascin-X | 342 | 328 | Pulmonary surfactant-associated protein B | 38 | 0 |
| Decorin | 265 | 340 | Microfibrillar-associated protein 5 | 21 | 8 |
| Biglycan | 427 | 135 | Annexin A1 | 25 | 6 |
| Collagen α-1(XII) chain | 408 | 159 | Nephronectin | 32 | 4 |
| Collagen α-6(VI) chain | 332 | 163 | Collagen α-2(I) chain | 0 | 35 |
| Nidogen-1 | 285 | 211 | Neutrophil elastase | 31 | 0 |
| Laminin subunit α-4 | 297 | 57 | Antithrombin-III | 19 | 5 |
| EMILIN-1 | 235 | 168 | Laminin subunit α-2 | 18 | 5 |
| Mimecan | 145 | 178 | Collagen α-3(IV) chain | 16 | 0 |
| α-1-antichymotrypsin | 279 | 30 | Deleted in malignant brain tumors 1 protein | 16 | 9 |
| Laminin subunit β-1 | 266 | 67 | Inter-α-trypsin inhibitor heavy chain H1 | 12 | 6 |
| Collagen α-2(IV) chain | 274 | 108 | Hyaluronan and proteoglycan link protein 1 | 6 | 11 |
| Collagen α-1(IV) chain | 257 | 92 | Collagen α-1(I) chain | 0 | 13 |
| Laminin subunit γ-2 | 250 | 30 | Target of Nesh-SH3 | 10 | 5 |
| Laminin subunit β-3 | 250 | 65 | Serpin B12 | 8 | 12 |
| α-1-antitrypsin | 188 | 92 | Galectin-1 | 13 | 0 |
| Asporin | 36 | 219 | Plasminogen | 10 | 0 |
| Vitronectin | 117 | 90 | Latent-transforming growth factor β-binding protein 4 | 0 | 8 |
| Collagen α-5(VI) chain | 84 | 153 | Fibrillin-2 | 3 | 2 |
| Versican core protein | 97 | 156 | Serpin B6 | 12 | 3 |
| Agrin | 207 | 13 | Annexin A7 | 11 | 0 |
| Microfibril-associated glycoprotein 4 | 121 | 59 | Mucin-1 | 14 | 0 |
| Lumican | 124 | 70 | Protein-glutamine γ-glutamyltransferase E | 5 | 5 |
| Tenascin | 126 | 88 | Collagen α-1(XV) chain | 0 | 11 |
| Pulmonary surfactant-associated protein A1 | 216 | 10 | Multimerin-2 | 8 | 0 |
| Fibulin-5 | 54 | 129 | Latent-transforming growth factor β-binding protein 2 | 4 | 4 |
| EGF-containing fibulin-like extracellular matrix protein 1 | 103 | 85 | Galectin-7 | 0 | 5 |
| Aggrecan core protein | 6 | 139 | Annexin A11 | 5 | 0 |
| Protein AMBP | 124 | 45 | von Willebrand factor A domain-containing protein 5A | 2 | 0 |
| Annexin A5 | 99 | 20 | Pulmonary surfactant-associated protein D | 5 | 0 |
| Prolargin | 78 | 34 | Protein-glutamine γ-glutamyltransferase K | 0 | 3 |
| Annexin A6 | 57 | 30 | Collagen α-1(V) chain | 0 | 4 |
| Disintegrin and metalloproteinase domain-containing protein 10 | 3 | 0 | |||
Definition of abbreviation: IPF = idiopathic pulmonary fibrosis.
TABLE 2.
RELATIVE EXPRESSION OF EXTRACELLULAR MATRIX PROTEINS IN ACELLULAR IPF LUNG COMPARED TO ACELLULAR NORMAL LUNG
| Protein | Fold Change (IPF:Normal) |
| Hyaluronan and proteoglycan link protein 1 | 21.20 |
| Matrix Gla protein | 19.33 |
| Latent-transforming growth factor β-binding protein 1 | 15.83 |
| Microfibrillar-associated protein 2 | 8.08 |
| Nephronectin | 5.67 |
| Fibrillin-2 | 4.73 |
| Periostin | 3.49 |
| EGF-containing fibulin-like extracellular matrix protein 1 | 3.25 |
| Fibrillin-1 | 3.18 |
| Fibulin-5 | 3.11 |
| Collagen α-1(III) chain | 2.72 |
| Fibulin-2 | 2.60 |
| Deleted in malignant brain tumors 1 protein | 2.56 |
| Dermatopontin | 2.53 |
| Versican core protein | 2.47 |
| Metalloproteinase inhibitor 3 | 2.37 |
| Collagen α-1(VIII) chain | 2.29 |
| Tubulointerstitial nephritis antigen-like | 1.72 |
| Collagen α-2(VI) chain | 1.72 |
| Asporin | 1.71 |
| Transforming growth factor-β–induced protein ig-h3 | 1.62 |
| Microfibrillar-associated protein 5 | 1.57 |
| Collagen α-1(VI) chain | 1.54 |
| Collagen α-1(XIV) chain | 1.54 |
| Annexin A1 | 1.51 |
| Microfibril-associated glycoprotein 4 | 1.46 |
| Aggrecan core protein | 1.39 |
| Collagen α-3(VI) chain | 1.31 |
| Collagen α-5(VI) chain | 1.30 |
| Cartilage acidic protein 1 | 1.28 |
| Inter-α-trypsin inhibitor heavy chain H4 | 1.26 |
| Collagen α-2(IV) chain | 1.20 |
| Vitronectin | 1.18 |
| α1-antitrypsin | 1.10 |
| Collagen α-1(I) chain | 1.02 |
| Mimecan | 1.01 |
| Collagen α-1(IV) chain | 0.99 |
| Basement membrane-specific heparan sulfate proteoglycan core protein | 0.92 |
| Nidogen-1 | 0.89 |
| Protein AMBP | 0.86 |
| Collagen α-6(VI) chain | 0.86 |
| Annexin A2 | 0.81 |
| Tenascin | 0.79 |
| Fibrinogen γ chain | 0.76 |
| Elastin | 0.73 |
| Fibronectin | 0.72 |
| Annexin A7 | 0.71 |
| Annexin A5 | 0.70 |
| EMILIN-1 | 0.67 |
| Fibrinogen β chain | 0.67 |
| Fibrinogen α chain | 0.67 |
| Collagen α-2(I) chain | 0.66 |
| Decorin | 0.62 |
| Neutrophil elastase | 0.61 |
| Laminin subunit γ-1 | 0.61 |
| Fibulin-1 | 0.60 |
| Collagen α-1(XII) chain | 0.58 |
| Laminin subunit α-5 | 0.57 |
| Laminin subunit β-2 | 0.46 |
| Lumican | 0.44 |
| Nidogen-2 | 0.44 |
| Tenascin-X | 0.43 |
| Laminin subunit β-1 | 0.43 |
| Inter-α-trypsin inhibitor heavy chain H1 | 0.43 |
| Biglycan | 0.37 |
| Leukocyte elastase inhibitor | 0.34 |
| Collagen α-1(XV) chain | 0.33 |
| Laminin subunit γ-2 | 0.32 |
| Laminin subunit β-3 | 0.31 |
| Collagen α-1(XVIII) chain | 0.29 |
| Laminin subunit α-4 | 0.28 |
| Prolargin | 0.25 |
| α1-antichymotrypsin | 0.25 |
| Latent-transforming growth factor β-binding protein 2 | 0.24 |
| Annexin A4 | 0.21 |
| Laminin subunit α-2 | 0.17 |
| Laminin subunit α-3 | 0.14 |
| EGF-containing fibulin-like extracellular matrix protein 2 | 0.13 |
| Agrin | 0.06 |
| Annexin A6 | 0.58 |
| Fibromodulin | 0.05 |
Definition of abbreviation: IPF = idiopathic pulmonary fibrosis.
ECM Dictates Cellular Phenotype
To determine whether acellular lung matrices would influence the phenotype of exogenously added cells, we next cultured primary human lung fibroblasts (1 × 105) in acellular normal and IPF ECM slices (prepared as in Methods). Cells were allowed to adhere for 30 minutes before adding media to the wells and were then cultured for 48 hours before harvesting cell lysates for Western analysis.
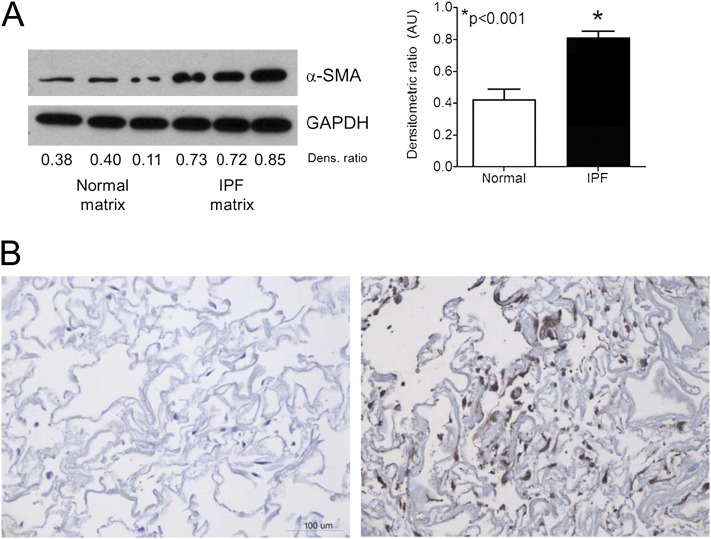
Fibroblasts cultured in such a manner migrate into the 3D matrix (Figure E1). Importantly, we observed a significant phenotypic difference in cells plated on the two different matrices. Whereas fibroblasts in normal lung ECM retained a phenotype characterized by low α-SMA production, cells in the IPF lung ECM demonstrated a significant increase in the production of α-SMA, consistent with myofibroblast differentiation (22) (Figure 9A). Immunohistochemistry corroborated this finding (Figure 9B).
Figure 9.
Idiopathic pulmonary fibrosis (IPF) matrices induce α-smooth muscle actin (α-SMA) expression in normal human lung fibroblasts. (A) Western blot analysis of normal human lung fibroblasts cultured within human acellular normal or IPF lung matrix. The same cells at the same passage were cultured in matrices for 48 hours before harvesting. IPF lung matrix significantly induced α-SMA. Bar graphs depict the mean (± SEM) densitometric ratio of α-SMA normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The blot is representative of triplicate experiments using three different primary cell lines and matrices derived from three separate individuals. (B) α-SMA immunohistochemistry of normal fibroblasts seeded within normal (left panel) or IPF (right panel) matrices for 48 hours; 20× magnification.
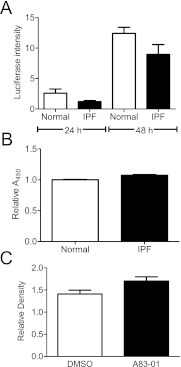
To determine whether the profibrotic mediator TGF-β could account for these findings, we next cultured primary normal human lung fibroblast-seeded acellular matrices in transwells opposite mink lung epithelial cells stably expressing the firefly luciferase gene under the control of the PAI-1 promoter (a reporter system to identify TGF-β activity [32]). Surprisingly, we observed no difference in TGF-β activity between normal and IPF tissues (Figure 10A), a finding that was corroborated by both enzyme immunoassay (EIA) measurement of TGF-β in acid-activated ECM slices showing no difference in TGF-β levels (Figure 10B) and the lack of effect on fibroblast α-SMA expression in fibroblasts cultured in IPF ECM in the presence of A83-01, an ALK-5 inhibitor (Figure 10C). Together, these data indicate that IPF lung matrix is capable of inducing myofibroblast differentiation in part through TGF-β–independent mechanisms and is consistent with recent literature (33).
Figure 10.
Enhanced α-smooth muscle actin expression in fibroblasts in idiopathic pulmonary fibrosis (IPF) matrices is transforming growth factor (TGF)-β–independent. (A) Normal human lung fibroblasts were cultured in normal or IPF matrices for 24 hours in the upper well of a transwell system, and 1 × 105 mink lung epithelial cells (MLECs) were cultured in the lower chamber. Lysates of MLECs were assayed by a luminometer and relative intensity was plotted above. There was no difference in TGF-β activity released from normal or IPF matrices. Results are pooled from three separate experiments each performed in triplicate tissue slices. (B) Intact acellular matrices were acid activated and the resultant media was assayed for TGF-β levels by enzyme immunoassay. No difference in total TGF-β was observed between normal and IPF slices. Data are the pooled mean (± SD) of samples from three separate normal and five separate IPF tissues. (C) Normal lung fibroblasts (n = 2) were seeded within triplicate IPF matrices from two separate donors for 48 hours in the presence of the ALK5 inhibitor A83-01 or dimethyl sulfoxide (vehicle control) for 48 hours. Lysates were assessed for α-SMA induction by Western blot. Pooled densitometric evaluation revealed no differences between groups, suggesting α-SMA induction was TGF-β–independent.
Discussion
The ECM is dynamic tissue providing structural support and spatial and temporal contextual cues to cells during many processes, including embryonic development, wound repair, and homeostasis. A great deal has been learned about the role of ECM in health and disease using standard cell culture techniques, although it is clear that these approaches mimic neither the compositional heterogeneity of ECM in vivo nor the dimensionality of tissues. As a result, data gleaned using traditional approaches may not accurately reflect cell behavior that occurs in vivo. Along the same lines, experimental animal models using transgenic approaches have helped to elucidate the role of many ECM proteins in various disease states (34–36). Unfortunately, animal models do not always recapitulate human disease, raising questions about the ability to translate results from animal to human. For example, anatomic differences in the lung between humans and rodents, such as lobar distribution, branching patterns, presence of bronchial submucosal glands, and goblet cells (37), likely substantially influence results. Indeed, these factors may account for the lack of effective therapies for patients with idiopathic pulmonary fibrosis; numerous compounds that are efficacious in rodent models of lung fibrosis are ineffective in the human disease (11).
To overcome some of these limitations, we have developed a fully humanized novel model for culturing cells in vitro in an organ- and disease-specific manner. Our approach retains the 3D spatial orientation, relative stiffness, complex ECM composition, and architecture found in the human lung; it is likely that these features will be maintained regardless of the organ tested. In fact, prior studies using decellularized heart (15), bone (38), and murine lung (39) as matrices to support regeneration imply these features remain true to the native organ, although this has not been directly shown previously. Our model also provides physiologically relevant tissue stiffness and ECM composition, as compared with the standard planar rigidity of tissue culture plastic. Moreover, it allows investigators to study the roles of ECM and cellular processes in a disease-specific manner, which is a major advance over currently used techniques.
In addition to defining a new model culture system, our data also show marked differences between the normal and (IPF) disease state, which may provide insight into disease pathogenesis. For example, we observed that IPF lung ECM markedly expresses glycosaminoglycans, matrix Gla protein, and microfibrillar-associated proteins. These observations are notable in that, although heparan sulfate– and chondroitin sulfate–containing glycosaminoglycans are known to be produced by various lung cell types (40), their presence in IPF has only been indirectly shown. Similarly, matrix Gla proteins, although identified in TGF-α–induced rodent lung fibrosis (29), have not previously been implicated in human lung fibrosis.
Studies of tissue fibrosis in general, and IPF specifically, clearly implicate the profibrotic effects of TGF-β (41–44). Thus, there has been an intense effort to delineate the role of TGF-β in IPF and other fibrosing disorders. These efforts suggest that TGF-β promotes myofibroblast differentiation (45), the fibroblast antiapoptotic phenotype (46), epithelial–mesenchymal transition (47), and ECM production (48). However, our current work suggests that myofibroblast differentiation (as measured by expression of α-SMA and cellular fibronectin) occurs at least partially independent of TGF-β in our system. Whether this finding applies to the true in vivo condition is not known, because our data show (Table E2) that latent TGF-β–binding proteins-2 and -4 are stripped away during the decellularization process. Regardless, evidence supports the notion that myofibroblast differentiation in the absence of TGF-β signaling can be induced in fibroblasts merely by altering the stiffness of the underlying substrate (49). Our current work is highly concordant with a seminal finding by Liu and colleagues (49), namely that fibroblasts seeded on polyacrylamide-crosslinked hydrogels maintained a quiescent phenotype or underwent apoptosis at low stiffness levels (0.1 to ∼ 3 kPa), whereas higher stiffness levels (20–50 kPa) induced an appearance of activation characterized by accumulation of cells aligned in parallel clusters (49). This is very similar to our own findings (Figure 9), in which fibroblasts cultured in stiffer IPF matrices clearly adopted an activated myofibroblast phenotype.
The pathogenesis of IPF remains incompletely understood. In part, this is due to the lack of an animal model that faithfully recapitulates the human disease (12). However, the bleomycin model (in which a dose of intratracheal bleomycin is instilled in rodents and fibrosis is measured 21–35 d later) remains one of the most widely used to address mechanistic questions about IPF. Although this model shares some features with human fibrosis, its complete reliance on the preceding inflammatory injury makes it a less-than-ideal model to study IPF. As proof of this shortcoming, many therapeutic agents that are shown to be successful in bleomycin-induced lung fibrosis are often ineffective in patients with IPF (50, 51). Thus, new models that more closely resemble the human disease are necessary to further our understanding. The in vitro model described herein provides such a tool to begin to dissect, mechanistically, the contribution of the ECM to IPF pathogenesis. Moreover, we believe this model can be adapted to other organs and diseases as a means to enhance the translatability of biomedical research.
Supplementary Material
Acknowledgments
The authors thank the patients and families who donated lung tissue for this study.
Footnotes
Supported by National Institutes of Health grants R01 HL085083 (E.S.W.) and U01 HL111016 (E.S.W. and L.E.N.), the Drews Sarcoidosis Research Fund (E.S.W.), the Martin Edward Galvin Fund for Idiopathic Pulmonary Fibrosis Research (E.S.W.), and the Health Effects Institute at Georgia Institute of Technology (T.H.B.). A.J.B. and S.A.M. are supported by National Institutes of Health training grant T32 HL07749.
Author Contributions: A.J.B. performed experiments and analyzed data. R.H., A.M.C., A.A.D., S.A.M., J.L.T., and K.W. performed experiments and provided intellectual input. V.F.F. and T.H.B. performed atomic force microscopy experiments and analyzed data. B.B.M., F.J.M., J.C.H., and L.E.N. provided critical intellectual guidance of experiments and assisted with data analysis. E.S.W. conceptualized the project, performed experiments, analyzed data, and wrote the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201204-0754OC on August 30, 2012
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Hardie WD, Glasser SW, Hagood JS. Emerging concepts in the pathogenesis of lung fibrosis. Am J Pathol 2009;175:3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mutsaers SE, Bishop JE, McGrouther G, Laurent GJ. Mechanisms of tissue repair: from wound healing to fibrosis. Int J Biochem Cell Biol 1997;29:5–17 [DOI] [PubMed] [Google Scholar]
- 3.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol 2008;214:199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature 2003;423:876–881 [DOI] [PubMed] [Google Scholar]
- 5.Schuger L, Skubitz AP, de las Morenas A, Gilbride K. Two separate domains of laminin promote lung organogenesis by different mechanisms of action. Dev Biol 1995;169:520–532 [DOI] [PubMed] [Google Scholar]
- 6.George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development 1993;119:1079–1091 [DOI] [PubMed] [Google Scholar]
- 7.Li DY, Brooke B, Davis EC, Mecham RP, Sorensen LK, Boak BB, Eichwald E, Keating MT. Elastin is an essential determinant of arterial morphogenesis. Nature 1998;393:276–280 [DOI] [PubMed] [Google Scholar]
- 8.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science 2001;294:1708–1712 [DOI] [PubMed] [Google Scholar]
- 9.Matsusaki M, Yoshida H, Akashi M. The construction of 3D-engineered tissues composed of cells and extracellular matrices by hydrogel template approach. Biomaterials 2007;28:2729–2737 [DOI] [PubMed] [Google Scholar]
- 10.Lemmon CA, Chen CS, Romer LH. Cell traction forces direct fibronectin matrix assembly. Biophys J 2009;96:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, Colby TV, Cordier JF, Flaherty KR, Lasky JA, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 2011;183:788–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore BB, Hogaboam CM. Murine models of pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2008;294:L152–L160 [DOI] [PubMed] [Google Scholar]
- 13.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, Gavrilov K, Yi T, Zhuang ZW, Breuer C, et al. Tissue-engineered lungs for in vivo implantation. Science 2010;329:538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, Kotton D, Vacanti JP. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med 2010;16:927–933 [DOI] [PubMed] [Google Scholar]
- 15.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med 2008;14:213–221 [DOI] [PubMed] [Google Scholar]
- 16.Macchiarini P, Jungebluth P, Go T, Asnaghi MA, Rees LE, Cogan TA, Dodson A, Martorell J, Bellini S, Parnigotto PP, et al. Clinical transplantation of a tissue-engineered airway. Lancet 2008;372:2023–2030 [DOI] [PubMed] [Google Scholar]
- 17.Delaere P, Vranckx J, Verleden G, De Leyn P, Van Raemdonck D. Tracheal allotransplantation after withdrawal of immunosuppressive therapy. N Engl J Med 2010;362:138–145 [DOI] [PubMed] [Google Scholar]
- 18.White ES, Booth AJ, Coley CJ. Decellularized human lung slices implicate the extracellular matrix in directing fibroblast phenotypes. Am J Respir Crit Care Med 2011;183:A2699 [Google Scholar]
- 19.White ES, Thannickal VJ, Carskadon SL, Dickie EG, Livant DL, Markwart S, Toews GB, Arenberg DA. Integrin alpha4beta1 regulates migration across basement membranes by lung fibroblasts: a role for phosphatase and tensin homologue deleted on chromosome 10. Am J Respir Crit Care Med 2003;168:436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tojo M, Hamashima Y, Hanyu A, Kajimoto T, Saitoh M, Miyazono K, Node M, Imamura T. The ALK-5 inhibitor A-83–01 inhibits Smad signaling and epithelial-to-mesenchymal transition by transforming growth factor-beta. Cancer Sci 2005;96:791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AP, England KA, Matson AM, Blazar BR, Panoskaltsis-Mortari A. Development of a decellularized lung bioreactor system for bioengineering the lung: the matrix reloaded. Tissue Eng Part A 2010;16:2581–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White ES, Atrasz RG, Hu B, Phan SH, Stambolic V, Mak TW, Hogaboam CM, Flaherty KR, Martinez FJ, Kontos CD, et al. Negative regulation of myofibroblast differentiation by PTEN (Phosphatase and Tensin Homolog Deleted on chromosome 10). Am J Respir Crit Care Med 2006;173:112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bracete AM, Fox DK, Simms D. Isolation and long term storage of RNA from ribonuclease-rich pancreas tissue. Focus 1998;20:82 [Google Scholar]
- 24.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer 2009;9:108–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goss BC, McGee KP, Ehman EC, Manduca A, Ehman RL. Magnetic resonance elastography of the lung: technical feasibility. Magn Reson Med 2006;56:1060–1066 [DOI] [PubMed] [Google Scholar]
- 26.Thurner PJ. Atomic force microscopy and indentation force measurement of bone. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2009;1:624–649 [DOI] [PubMed] [Google Scholar]
- 27.Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics 2012;11:M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Getie M, Schmelzer CE, Neubert RH. Characterization of peptides resulting from digestion of human skin elastin with elastase. Proteins 2005;61:649–657 [DOI] [PubMed] [Google Scholar]
- 29.Hardie WD, Korfhagen TR, Sartor MA, Prestridge A, Medvedovic M, Le Cras TD, Ikegami M, Wesselkamper SC, Davidson C, Dietsch M, et al. Genomic profile of matrix and vasculature remodeling in TGF-alpha induced pulmonary fibrosis. Am J Respir Cell Mol Biol 2007;37:309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu M, Munger JS, Steadele M, Busald C, Tellier M, Schnapp LM. Integrin alpha8beta1 mediates adhesion to LAP-TGFbeta1. J Cell Sci 2002;115:4641–4648 [DOI] [PubMed] [Google Scholar]
- 31.Okamoto M, Hoshino T, Kitasato Y, Sakazaki Y, Kawayama T, Fujimoto K, Ohshima K, Shiraishi H, Uchida M, Ono J, et al. Periostin, a matrix protein, is a novel biomarker for idiopathic interstitial pneumonias. Eur Respir J 2011;37:1119–1127 [DOI] [PubMed] [Google Scholar]
- 32.Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 1999;96:319–328 [DOI] [PubMed] [Google Scholar]
- 33.Huang X, Yang N, Fiore VF, Barker TH, Sun Y, Morris SW, Ding Q, Thannickal VJ, Zhou Y. Matrix stiffness-induced myofibroblast differentiation is mediated by intrinsic mechanotransduction. Am J Respir Cell Mol Biol 2012;47:340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Astrof S, Crowley D, George EL, Fukuda T, Sekiguchi K, Hanahan D, Hynes RO. Direct test of potential roles of EIIIA and EIIIB alternatively spliced segments of fibronectin in physiological and tumor angiogenesis. Mol Cell Biol 2004;24:8662–8670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dietz HC, Mecham RP. Mouse models of genetic diseases resulting from mutations in elastic fiber proteins. Matrix Biol 2000;19:481–488 [DOI] [PubMed] [Google Scholar]
- 36.Cosgrove D, Meehan DT, Grunkemeyer JA, Kornak JM, Sayers R, Hunter WJ, Samuelson GC. Collagen COL4A3 knockout: a mouse model for autosomal Alport syndrome. Genes Dev 1996;10:2981–2992 [DOI] [PubMed] [Google Scholar]
- 37.Dixon D, Herbert RA, Sills RC, Boorman GA. Lungs, pleura, and mediastinum. In: Maronpot R, Boorman G, Gaul B, editors. Pathology of the mouse. Vienna: Cache River Press; 1999. p. 293–332.
- 38.Yates P, Thomson J, Galea G. Processing of whole femoral head allografts: validation methodology for the reliable removal of nucleated cells, lipid and soluble proteins using a multi-step washing procedure. Cell Tissue Bank 2005;6:277–285 [DOI] [PubMed] [Google Scholar]
- 39.Cortiella J, Niles J, Cantu A, Brettler A, Pham A, Vargas G, Winston S, Wang J, Walls S, Nichols JE. Influence of acellular natural lung matrix on murine embryonic stem cell differentiation and tissue formation. Tissue Eng Part A 2010;16:2565–2580 [DOI] [PubMed] [Google Scholar]
- 40.David G, Lories V, Heremans A, Van der Schueren B, Cassiman JJ, Van den Berghe H. Membrane-associated chondroitin sulfate proteoglycans of human lung fibroblasts. J Cell Biol 1989;108:1165–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson MS, Wynn TA. Pulmonary fibrosis: pathogenesis, etiology and regulation. Mucosal Immunol 2009;2:103–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Booth AJ, Csencsits-Smith K, Wood SC, Lu G, Lipson KE, Bishop DK. Connective tissue growth factor promotes fibrosis downstream of TGFbeta and IL-6 in chronic cardiac allograft rejection. Am J Transplant 2010;10:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bissell DM. Chronic liver injury, TGF-beta, and cancer. Exp Mol Med 2001;33:179–190 [DOI] [PubMed] [Google Scholar]
- 44.Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J. Adenovector-mediated gene transfer of active transforming growth factor-beta1 induces prolonged severe fibrosis in rat lung. J Clin Invest 1997;100:768–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serini G, Bochaton-Piallat ML, Ropraz P, Geinoz A, Borsi L, Zardi L, Gabbiani G. The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. J Cell Biol 1998;142:873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horowitz JC, Rogers DS, Sharma V, Vittal R, White ES, Cui Z, Thannickal VJ. Combinatorial activation of FAK and AKT by transforming growth factor-beta1 confers an anoikis-resistant phenotype to myofibroblasts. Cell Signal 2007;19:761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willis BC, Liebler JM, Luby-Phelps K, Nicholson AG, Crandall ED, du Bois RM, Borok Z. Induction of epithelial-mesenchymal transition in alveolar epithelial cells by transforming growth factor-beta1: potential role in idiopathic pulmonary fibrosis. Am J Pathol 2005;166:1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuhn C, McDonald JA. The roles of the myofibroblast in idiopathic pulmonary fibrosis. Ultrastructural and immunohistochemical features of sites of active extracellular matrix synthesis. Am J Pathol 1991;138:1257–1265 [PMC free article] [PubMed] [Google Scholar]
- 49.Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, Tschumperlin DJ. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol 2010;190:693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daniels CE, Lasky JA, Limper AH, Mieras K, Gabor E, Schroeder DR. Imatinib treatment for idiopathic pulmonary fibrosis: randomized placebo-controlled trial results. Am J Respir Crit Care Med 2010;181:604–610 [DOI] [PubMed] [Google Scholar]
- 51.Daniels CE, Wilkes MC, Edens M, Kottom TJ, Murphy SJ, Limper AH, Leof EB. Imatinib mesylate inhibits the profibrogenic activity of TGF-beta and prevents bleomycin-mediated lung fibrosis. J Clin Invest 2004;114:1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.