Abstract
Clinicians generally record medical narrative data, such as current complaints, physical examination, and progress notes, as free text in paper-based medical records. The medical narrative involves heterogeneous and detailed data that include the description of (multiple) occurrences of medical findings or symptoms that may progress over time. Structured, electronic recording of narrative data would facilitate the use of these data for research. The authors' OpenSDE application supports clinicians with the structured recording of narrative data in both research and care settings. Data entry is enabled using forms that are generated using domain-specific trees of medical concepts. For data storage, the authors have expanded the traditional row modeling methodology with additional columns that allow structured representation of medical narratives including descriptions of findings, multiple occurrences of findings, and the progression of findings over time.
The medical narrative section of the patient record comprises the medical history, physical examination, progress notes, and reports on additional tests and interventions. Medical narrative data vary per discipline, per patient, and over time. Besides the heterogeneity of the data, the level of detail in recording varies greatly among clinicians. The unruliness and large variation in the collected data have made it difficult to support structured recording of the medical narrative.1 Clinicians convinced of the potential benefit of electronically available data (e.g., greater availability, data sharing, data analysis, or use of decision support) have launched efforts to develop dedicated systems to accommodate their data needs. Such attempts are far from ideal2; over time, adaptation and expansion of databases result in haphazard collections of tables and data. New tables will make older tables (partially) obsolete, and data redundancy is frequent. Performing research on one or more such databases is on the verge of being unmanageable especially for clinicians or researchers who are relatively unfamiliar with database management.2
Our objective is to support structured recording of narrative data in the form of an application that allows tailoring to specific medical domains and individual preferences without the need for technical adaptation.3 Furthermore, we want to support structured recording of data with a high degree of expressiveness. We developed an application called OpenSDE4 (SDE: Structured Data Entry) that supports structured data entry in a variety of settings, thus facilitating the use of data for both care and research. OpenSDE supports data entry using customizable entry forms based on domain-specific trees. In this report we describe how we implemented row modeling to enable structured recording of medical narrative data.
Row Modeling
Row modeling is a methodology that is suitable for storing heterogeneous and evolving data sets.5 In essence, row modeling involves a column-to-row transformation; the attributes (or column headings) of the conventional column-modeled table are stored as data in the row-modeled table. A column-modeled table contains a column for every attribute. A row-modeled table contains one column that holds all attributes and one column that holds the values of the attributes. In a column-modeled table, one record holds a set of facts about a patient, whereas in a row-modeled table, every record holds one particular fact about a patient.6 A row-modeled table only holds those attributes for which a value actually has been recorded.
In row modeling, the data definition is not defined in the data tables themselves. The data definitions are stored separately and often are referred to as “metadata.” The advantage of separating the metadata from the physical data schema is that one eliminates the need to change the physical data structure when the data set changes: only the metadata content needs change. In a conventional column-modeled approach, metadata are held in table definitions and relations between tables. Changes to a column-modeled table would involve adding or removing columns from tables, i.e., changing the database structure.
Row modeling can be used as a generic structuring technique for diverse and changing data sets. Metadata hold the information necessary for the correct semantic interpretation of the data held in the row-modeled table. Metadata, therefore, need to be edited and adapted for different disciplines and constitute an important area of research.7
Method
In OpenSDE, metadata are represented as discipline-specific domain models. The domain model defines the content of the medical narrative in a specific discipline. Domain models vary in content but not in structure. The content consists of concepts and constraints organized in a rooted tree structure. The nodes of the tree structure represent the concepts and are connected to each other via one-directional arcs; a node at the end of an arc represents a descriptor of the node at the beginning of an arc. For every node, one path extends from the root to the particular node.
We developed a toolset that uses a graphic interface to define domain models; using this toolset, clinicians can define their own domain models.8
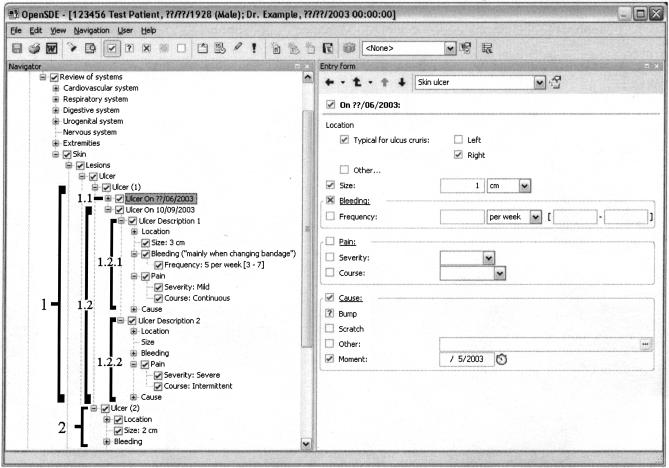
OpenSDE uses the domain models to generate an interface for data entry. ▶is a screen capture of OpenSDE. The domain model tree (metadata) is presented on the left of the figure, while the right shows the dynamically generated entry form with all nodes detailing the node selected in the domain model. The forms can be customized by clinicians themselves.
Figure 1.
Screen capture of the OpenSDE data entry application. The left-hand side shows the domain model tree, which contains medical concepts. On the right is the form on which data are entered. The form is associated with the selected node, in this case, “skin ulcer.” The brackets on the left (included in this figure as examples) indicate that two different ulcers are described: ulcer 1 on the right shin (see entry form) and ulcer 2 on the left shin (location is hidden in this view). Ulcer 1 consists of two descriptions over time (progress descriptions); the first description (1.1; shown on entry form) is of June 2003, describing the probable cause of the ulcer in May 2003; progress description 2 (bracket 1.2) shows the progression of the ulcer on September 10, 2003. Progress description 2 contains two descriptions of pain to indicate that pain is continuously mild (Ulcer Description 1/bracket 1.2.1) and intermittently severe (Ulcer Description 2/bracket 1.2.2).
To accommodate expressiveness for the recording of medical narratives, OpenSDE supports a number of general items that can be recorded for each concept in the domain model. Every instance of a concept has a “presence state,” which states whether a concept is present, absent, or unknown. Numeric values can be a single value (with a deviation), a range, or a date/time value; each value may have a unit. Domain models, however, have their boundaries; clinicians may encounter narrative that cannot be expressed using the domain model. To deal with this limitation of the domain model, clinicians may add free text to any node in the tree, i.e., each recorded finding may be supplemented by free text.
OpenSDE uses an extended row-modeled table to support the complexity of the medical narrative. The example shown in ▶ illustrates that complexity; the patient reports that he has several skin ulcers; one of the ulcers is located on the right shin and the other on the left shin. The ulcer on the right shin was possibly caused by bumping into a table several months earlier; in the past few weeks, this skin ulcer has grown, started to bleed, and is increasingly painful. In OpenSDE, the row-modeled table has been extended with columns for multiple instances, progress descriptions, and multiple descriptions. Multiple instances represent findings that can occur more than once (in ▶, the patient describes two skin ulcers: one on the left shin and one on the right shin). Progress descriptions represent findings that evolve over time (in ▶, the patient describes that as of September 10, 2003, the skin ulcer on the right shin has started bleeding, mainly when the bandage is changed). Multiple descriptions represent findings that present themselves differently under different circumstances (in ▶ the patient complains that the ulcer is always a little painful, but that the pain is sometimes severe).
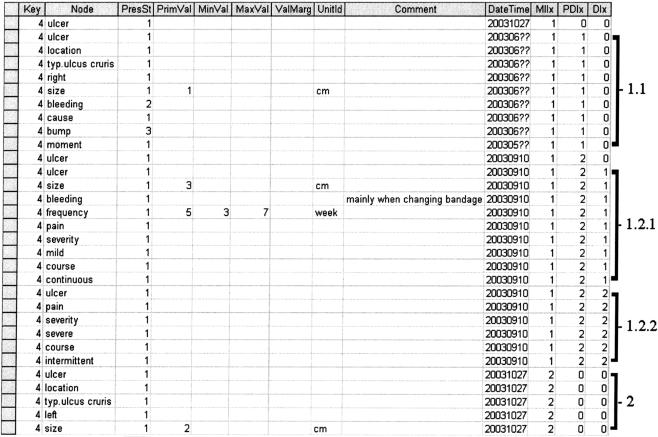
The data presented in ▶ are represented in ▶ . Every concept for which data have been entered (both in the tree and on the form in ▶) corresponds to one record in ▶.
Figure 2.
An excerpt from the row-modeled table that we use to store data collected using OpenSDE. The first row contains the column headings. The following 31 rows contain patient data. The first “key” column is shortened for this example; it normally consists of a reference to the patient, the event, and the domain model version and discipline. The column “Node” is actually a code, but for this example we have used the associated text. “Node” refers to the node in the domain model associated with the recorded data. The following 11 columns are the data items. PresSt, Presence state (1, present, 2, absent, 3, unknown). The columns that include “Val” are used to represent the values (primary value, min, max, and margin) and unitId refers to the unit of the value. The “Comment” column holds free-text values, and “DateTime” refers to date applicable, i.e., data entry date unless otherwise specified by clinician. The last three columns are index columns: MIIx for multiple instances, PDIx for progress descriptions, and DIx for description index. The brackets at the right side of Figure 2 correspond to the brackets in ▶.
Discussion
Row modeling is a technique frequently used for representing heterogeneous data sets. In a row-modeled table, every record ideally holds one particular fact about a patient.6 Although applying the same underlying principle, different researchers have developed alternative approaches. Salgado and Gouveia-Oliveira9 use a combination of conventional and row-modeled tables for their clinical trials information system, COATI. Their approach was to create a row-modeled table per separate entity for those entities that are either trial specific or have attributes that vary between trials. Nadkarni et al.10 use an entity-attribute-value model with classes and relationships (EAV/CR) for the Human Brain Project and clinical trials data. In addition, many researchers6,11 have separate tables for each data type; a change, for example, in data type from free text to a numeric value implies that from then on the attribute will be stored in a different table. This relocation of attributes is not necessary when hybrid data types are allowed in one column. In general, the use of multiple tables requires a decision about where to store which data, which implies the possible need for changes to the data structure when the data set changes. In OpenSDE, all items are stored in a single table. That is, in OpenSDE we use an extended row-modeled table to hold extra data items in preassigned columns rather than introducing new tables. A row in our row-modeled table, therefore, corresponds to one fact about a patient but allows more detail about this fact to be described in one row.
A difference between the extended tables in the model by Friedman et al.12 and OpenSDE is that Friedman represents context of data using nested rows, i.e., internal row reference. OpenSDE represents the context of each row with a reference to a unique node in the domain model.
The extensions we made to the row model fall in two categories. The first category deals with data types. Other researchers introduce different tables to deal with different data types, OpenSDE extends the row model with additional columns to reflect the data type. The second category deals with the complexity of the medical narrative (e.g., repeated descriptions over time of multiple lesions). OpenSDE extends the row model to represent descriptions of (multiple) occurrences of findings or symptoms that may progress over time.
OpenSDE does not model an ontology. At first sight, modeling an ontology in, for example, Protégé may seem similar to domain modeling in OpenSDE. Protégé, however, supports modeling for various purposes, such as decision support and data entry.13,14 OpenSDE domain models currently are used only to support structured data entry; to use the domain models for inference would require adding more knowledge to our domain models. Investigating whether the expressiveness of OpenSDE can be achieved using Protégé, would be an interesting study.
OpenSDE currently is being used in several pilot projects within the Erasmus MC University Medical Center and is used by several commercial vendors of hospital information systems. OpenSDE is used in different disciplines including neurology, radiology, immunology, and pediatrics. OpenSDE, written in Delphi, is available in open source.4
References
- 1.Tange HJ, Hasman A, de Vries Robbe PF, Schouten HC. Medical narratives in electronic medical records. Int J Med Inf. 1997;46(1):7–29. [DOI] [PubMed] [Google Scholar]
- 2.Pierik FH, van Ginneken AM, Timmers T, Stam H, Weber RF. Restructuring routinely collected patient data: ORCA applied to andrology. Methods Inf Med. 1997;36:184–90. [PubMed] [Google Scholar]
- 3.van Ginneken AM, de Wilde M. A New Approach to Structured Data Collection. In Waegemann CP (ed). Proc of TEPR 2000. San Francisco: Med Rec Inst, 2000, pp 627–35.
- 4.OpenSDE. Available at: <http://sourceforge.net/projects/opensde>. Accessed Nov 19, 2003.
- 5.Nadkarni PM. Available at: <http://ycmi.med.yale.edu/nadkarni/db_course/>. Accessed: Oct 20, 2003.
- 6.Nadkarni PM, Brandt C. Data extraction and ad hoc query of an entity-attribute-value database. J Am Med Inform Assoc. 1998;5:511–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Ginneken AM. Considerations for the representation of meta-data for the support of structured data entry. Methods Inf Med. 2003;42:226–35. [PubMed] [Google Scholar]
- 8.Doupi P, van Ginneken AM. Structured physical examination data: a modeling challenge. Medinfo. 2001;10(pt 1):614–8. [PubMed] [Google Scholar]
- 9.Salgado NC, Gouveia-Oliveira A. Towards a common framework for clinical trials information systems. Proc AMIA Symp. 2000:754–8. [PMC free article] [PubMed]
- 10.Miller PL, Nadkarni P, Singer M, Marenco L, Hines M, Shepherd G. Integration of multidisciplinary sensory data: a pilot model of the human brain project approach. J Am Med Inform Assoc. 2001;8:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganslandt T, Mueller M, Krieglstein CF, Senninger N, Prokosch HU. A flexible repository for clinical trial data based on an entity-attribute-value model. Proc AMIA Symp. 1999:1064–7.
- 12.Friedman C, Hripcsak G, Johnson SB, Cimino JJ, Clayton PD. A generalized relational schema for an integrated clinical patient database. Proc 14th Symp Comput Appl Med Care. 1990:335–9.
- 13.Musen MA. Modern architectures for intelligent systems: reusable ontologies and problem-solving methods. Proc AMIA Symp. 1998:46–52. [PMC free article] [PubMed]
- 14.The Protégé website. Available at: <http://protege.stanford.edu/>. Accessed Nov 19, 2003.