Abstract
IL-7 signaling is required for thymocyte development and its loss has a severe deleterious effect on thymus function. Thymocyte–stromal cell interactions and other mechanisms tightly regulate IL-7 expression. We show that disruption of that regulation by over-expression of IL-7 inhibits T-cell development and promotes extensive B-cell lymphopoiesis in the thymus. Our data reveal that high levels of IL-7 negate Notch-1 function in thymocytes found in IL-7 transgenic mice and in co-culture with OP9-DL1 cells. While high levels of IL-7R are present on thymocytes, increased suppressor of cytokine signaling-1 expression blunts IL-7 downstream signaling, resulting in hypo-phosphorylation of proteins in the PI3K-Akt pathway. Consequently, GSK3β remains active and inhibits Notch-1 signaling as observed by decreased Hes-1 and Deltex expression in thymic progenitors. This is the first demonstration that high levels of IL-7 antagonize Notch-1 signaling and suggest that IL-7 may affect T- versus B-lineage choice in the thymus.
Keywords: CD127, IL-7, IL-7R, Notch-1, thymus
Introduction
A regular influx of bone marrow (BM)–derived precursor cells into the thymus is required for sustained T-cell development. While in transit and upon arrival at the thymus, accumulating effects of signals delivered in discrete environmental niches shape the developmental potential and direct the lineage choice of precursor cells. IL-7 receptor (IL-7R) signaling plays a critical role in T-cell development (1). Although intra-thymic early thymocyte precursor (ETP) cells do not express IL-7R, recent data based on a IL-7R reporter system indicate that about 85% of ETPs have expressed IL-7R prior to seeding the thymus (2), providing an opportunity for prior IL-7 exposure to influence responses to downstream developmental signals. By the double negative 2 (DN2) stage, IL-7R is expressed and IL-7 plays a critical role by providing survival and proliferation signals to T-cell precursors. The profound defect in T-cell development observed in IL-7 null mice is mainly due to loss of precursor cells at this stage (3). The dynamic regulation of IL-7 expression within the thymus underscores its importance in T-cell development. Thymic IL-7 expression occurs at varying levels in localized regions (4, 5). In addition, recent data indicate that TCRβ-selected thymocytes provide a negative feedback control to modulate thymic epithelial cell production of IL-7 (6).
Notch-1 plays an important role in T cell–lineage commitment and thymocyte proliferation (1). In experimental models that abrogate Notch-1 signaling, thymocytes are diverted into abnormal B-cell development in the thymus (7, 8), while the reciprocal Notch gain-of-function studies result in abnormal T-cell development at the expense of B-cell development in the BM (9). Therefore, precise regulation of Notch activity is required for efficient B and T lymphopoiesis. Notch function is inhibited when phosphorylated by GSK3β kinase (10, 11). Similarly, GSK3β kinase function is inactivated when Akt phosphorylates GSK3β following activation by IL-7 or other signals (11). Therefore, inactivation of GSK3β with lithium chloride or by other means leads to a dramatic increase in Notch signaling (10–12). Another negative regulator of Notch signaling is the proto-oncogene leukemia/lymphoma related factor (LRF) encoded by the Zbtb7a gene (13). LRF directs early lymphoid progenitors in mice to the B-cell lineage, and in its absence, B-cell development is blocked at the pre pro-B stage and T cells develop in the BM (14).
Given the importance of IL-7 on thymopoeisis and the recent clinical use of super-physiological levels to enhance T-cell reconstitution following BM transplant (15), we wished to determine the effect of chronic high-level exposure on thymus function. Because we were concerned about uncontrollable variables inherent in studies done in vivo using exogenous administration of IL-7, we produced a transgenic (Tg) mouse line that expresses IL-7 driven by the proximal lck promoter resulting in a high level of IL-7 expression. In this study, we show that high IL-7 levels, such as those found in the IL-7 Tg mice, actively inhibit Notch-1 signaling and result in defective αβ T-cell development, while abnormally high numbers of developing B cells are found in the thymus. These findings are confirmed in vitro using precursor cells and OP9-DL1 co-cultures (16). Therefore, high IL-7 levels within a clinically relevant range dysregulate T- and B-cell development in the thymus by opposing Notch-1 signaling.
Methods
Animal procedures
The IL-7 Tg mice were bred and housed at National Cancer Institute (NCI) facilities. All animal procedures were done according to National Institutes of Health and Animal Care and Use Committee guidelines.
Antibodies and analytical flow cytometry
Anti-IC STAT5 and biotinylated Lin Abs (Becton Dickinson, San Jose, CA). The Lin cocktail included anti-CD3 (2C11), anti-TCRβ (H57), anti-CD8α, anti-B220 (CD45R), anti CD19, anti-Mac-1 (2CD11b), anti-NK1.1 (PK136), anti-GR-1, anti-TCRγ (GL3), anti-TER 119 and anti-CD11c. All other antibodies were supplied by eBiosciences, San Diego, CA. For thymic common lymphoid precursor-2 (CLP-2) analysis, B220 and CD19 Abs were absent from the lineage cocktail and anti-CD25 (clone 7D4) antibody was added. For co-culture experiments, the stromal cells were excluded from the analysis by gating out GFP+ (FITC) and CD45− cells. Fluorescence data were displayed using Flowjo software (Tree Star, Ashland, OR).
Mixed BM chimeras
Three-month-old C57BL/6 mice were lethally irradiated (10 Gy) prior to intravenous injection of a 50/50 mixture of 10×106 T cell–depleted BM cells from IL-7 Tg (CD45.2)/or wild type (WT) (CD45.2) mixed with congenic Ly5.2 (CD45.1) mice. Thymocytes from individual lobe recipients were analyzed as indicated 7–12 weeks after the procedure.
IP injection of anti-IL-7 or isotype control Abs
As previously described (17), 1mg of anti-IL-7 mAb) (M25) or its isotype control (CC57) was injected three times weekly up to a total of 10mg into IL-7 Tg mice. The mice were sacrificed 2–3 days after the last injection.
Cell enrichment and cell sorting
All FACS gates were set using control staining where cells were stained with a complete antibody cocktail in which an isotype antibody was substituted for one antibody. For DN sort and culture, thymocytes were CD8 depleted (Miltenyi Biotec, Auburn, CA). For ETP sorting, the cells were Lin−CD44hiCD25−c-Kithi and IL-7R−/lo. For DN thymocyte analysis by western blot, ETPs or DN1 c-Kit+, DN2-c-Kit+ and DN3-c-Kit− cells were sorted from 10 freshly pooled IL-7 Tg mice and 10 normal littermates, or after culture on OP9-DL1, mixed with the same ratio from both groups. For LSK-Flt3+ isolation, the samples were enriched for Lin− cells using the Miltenyi lineage depletion cocktail Kit (Miltenyi Biotec, Auburn, CA) then sorted as Sca-1+, Flt3+, c-Kit+ and Lin− 7AAD−.
OP9 stromal cell co-culture
Sorted progenitors were seeded at 103 cells/well into 24-well tissue culture plates containing either OP9 control or OP9-DL1 cells to which 1ng ml−1 of IL-7 and 5ng ml−1 of Flt3 ligand were added (16, 18). As described, 1ng ml−1 of IL-6 and 25ng ml−1 of IL-15 were added to DN2 culture (18). For total lineage marker negative (Lin-), Sca+, c-Kit+ (LSK) cell culture, the plates were seeded at 100 cells/well, and 50ng ml−1 of c-kit-ligand were added for the first 10 days. Doses of IL-7 were applied at 1, 5, 10 or 40ngml−1 to WT progenitors. Stromal cells and cytokines were changed on a weekly basis.
Real time PCR
RNA was isolated from sorted cells (Pico Pure, Applied Biosystems, Foster City, CA) and amplified if needed (Arcturis RiboAmp HS plus kit, Applied Biosystems, Foster City, CA). Quantitation was performed using a LightCycler assay (Roche Diagnostics, Indianapolis, IN) with standard curves generated from serial dilutions of cDNA from C57BL/6 thymi. Hes-1 and Deltex data were normalized to RNA input. All other data were normalized to GAPDH.
Protein preparation and immunoblot analysis
For western blot analysis, the cells were lysed in 1× radioimmunoprecipitation assay buffer plus the 10 µg ml−1 complete protease inhibitor and 1mM One-Stop phosphatase inhibitors (Roche, Indianapolis, IN). All antibodies used were from Cell Signaling, Danvers, MA, except anti-DLL1 (Abcam, Cambridge, MA), anti-DLL4 (Rockland, Gilbertsville, PA) and anti-tubulin (Santa Cruz, Santa Cruz, CA).
Statistical analysis
Statistical analysis was performed using Statview 5.0.1 software. All studies were analyzed using Student’s t-test with a value of 0.05.
Results
IL-7 over-expression decreases αβ T-cell development and increases numbers of thymic B-lineage cells
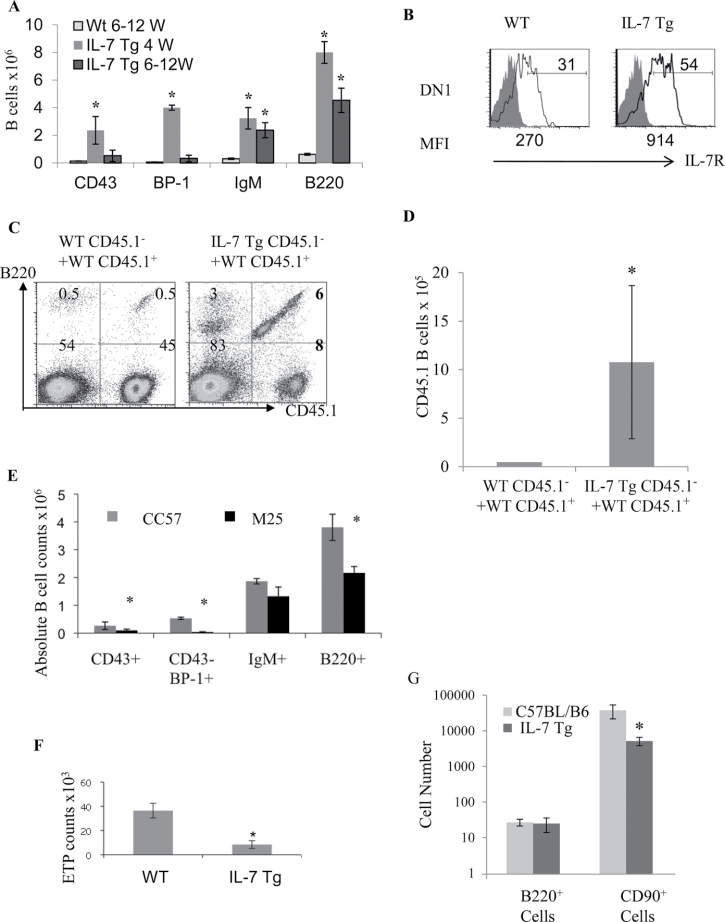
The IL-7 Tg mice used in this study (previously designated TgB) express 39-fold more IL-7 mRNA that blocks αβ T-cell lineage development (17). In parallel, abnormally robust B-cell development was taking place in the thymus of these animals (Fig. 1A). The peak of thymic B-cell production in IL-7 Tg mice was seen at 4 weeks, when significant increases in the numbers of cells in all B-cell developmental stages were observed (Fig. 1A). The percentages of thymic CD43+ (pro-B), BP1+ (late pro-B and pre-B) and IgM+ (immature/mature) B cells were 18-fold, 74-fold and 11-fold higher, respectively, in 4-week-old IL-7 Tg mice (Fig. 1A). At 6–12 weeks of age, while the size of the thymus in IL-7 Tg mice was dramatically decreased (5×106 to 10×106 cells/thymus) (17), the numbers of thymic mature B cells and B220+ cells were significantly increased, whereas numbers of immature CD43+ and BP1+ B cells were not increased (Fig. 1A). We did not detect circulating pre-B or pro-B cells in spleen, lymph nodes or peripheral blood in 4- to 12-week-old IL-7 Tg (unpublished results). There were no differences noted in BM B-cell development in the IL-7 Tg compared with control WT mice (Supplemental Figure 1A is available at International Immunology Online). Therefore, it seemed likely that elevated IL-7 levels promoted the development of B cells at the expense of thymocyte development in the IL-7 Tg thymus.
Fig. 1.
Analysis of αβ thymocytes and B cells in the thymi of IL-7 Tg mice. (A) Absolute counts of pro-B (CD43+), pre-B and late pro-B (BP-1+), immature/mature B cells (IgM+) and total B220+ in the thymus of 4-week- (black bars) or 6- to 12-week- (grey bars) old IL-7 Tg mice and 6- to 12-week- (outlined bar) old WT mice are shown as mean ± standard error (10 mice in each group, *P < 0.0001). (B) IL-7R expression on thymocytes comprising the DN1 population (Lin−CD44+CD25−c-Kit+) in IL-7 Tg and WT mice (open histograms) compared with the isotype control (filled histograms), the mean fluorescence intensity (MFI) is shown below the histograms. The full gating for the FACS is shown in Supplemental Figure 1B, available at International Immunology Online. Data are representative of five experiments with three mice for each group. (C) Analysis of thymic B cells in mixed BM chimeras. The plots show thymic B220+ cells derived from CD45.1+ and CD45.1− BM cells in both recipients. (D) Average numbers of thymic CD45.1+ B cells obtained when CD45.1+ WT BM were co-injected with IL-7 Tg or WT CD45.1− cells. n = 7 mice in each group (*P value of 0.022). (E) Absolute number of thymic B cells in IL-7 Tg mice that received neutralizing anti-IL-7 mAb M25 (black bars) versus control CC57 (grey bars) are shown. (*P values of 0.03, 0.005 and 0.008 for total B220+, pro-B (CD43+) and pre-B (CD43−BP-1+) in the M25-treated IL-7 Tg mice, respectively). Each group included four mice. (F) ETPs in the thymus of 4-week-old IL-7 Tg and WT mice (*P = 0.0019; n = 9–13 mice). (G) Average numbers of B220+ or CD90+ cells obtained from co-cultures of ETPs sorted from either WT or IL-7 Tg mice and OP9-DL1 cells are shown from quadruplicate wells (P < 0.001).
We next sought to demonstrate that IL-7 protein was responsible for the dysregulation of thymus function observed in IL-7 Tg thymus. We first examined the expression of IL-7R on thymocytes from the IL-7 Tg mice because it has been shown that exposure to IL-7 reduces IL-7R expression on WT thymocytes (19). However, IL-7R levels were increased in Lin− DN precursor populations DN1 (CD44+, c-Kit+), DN2 (CD44+, CD25+, c-Kit+) and DN3 (CD25+, c-Kit−) in IL-7 Tg mice relative to WT controls (Fig. 1B and Supplemental Figure 1C, available at International Immunology Online). Next, we used mixed BM chimeras in which WT mice were reconstituted with a one-to-one mixture of T cell–depleted BM from IL-7 Tg mice or WT littermates (expressing CD45.2) and CD45.1+ WT congenic mice (17). In recipients of IL-7 Tg BM cells, there were higher percentages and significantly higher numbers of WT CD45.1+ B220+ cells than in the recipients reconstituted only with WT BM cells (Fig. 1C and 1D). Development of CD45.1+ αβ T cells was reduced in recipients of IL-7 Tg BM cells compared with controls as described in our previous paper(17). Therefore, excess IL-7 expressed by IL-7 Tg cells altered the development of co-injected WT T-cell depleted BM cells. We also injected 4-week-old IL-7 Tg mice with a neutralizing anti-IL-7 mAb (M25) or with an isotype control mAb (CC57) (17). A significant decrease of pro-B and pre-B cell numbers was observed in the thymus of the group injected with anti-IL-7 mAb (Fig. 1E), in parallel with restored αβ T cell development (17). Thymic B220+IgM+ numbers were still significantly higher in the IL-7 Tg mice injected with an anti-IL-7 mAb, which likely correspond to mature B cells produced before the mAb injection. In summary, high IL-7 levels induced an increase in thymic B-cell production along with a specific disruption of αβ T-cell development in the thymus. Both abnormalities were observed when an excess of IL-7 protein was present and were reversed when IL-7 levels were reduced.
High IL-7 doses blunt the developmental potential of ETPs
We next investigated the potential precursor cell populations that could give rise to the B cells developing in IL-7 Tg thymi. We have shown that disruption of αβ T-cell development in IL-7 Tg mice occurs after the DN1 stage (17). The cells within the DN1 population comprise a heterogeneous collection of subpopulations based on their developmental potential and c-kit expression (20). ETPs are a discrete subset DN1 cells defined as Lin−CD44hiCD25−c-KithiIL-7R−/lo (21). ETPs are thought to be the intrathymic precursors responsible for canonical T-cell development and have been reported to lack robust B-cell potential (21). Therefore, we evaluated the ETP numbers of 2- to 3-week-old IL-7 Tg mice and WT littermates. We observed a dramatic decrease in the percentage and numbers (4.3-fold) of ETP in the IL-7 Tg thymus (Fig. 1F and Supplemental Figure 1D, available at International Immunology Online). To explore the T- and B-cell potential of sorted IL-7 Tg ETPs, we incorporated and cultured them in congenic re-aggregated fetal thymi. After 14 days, the WT thymocytes expanded by approximately 20-fold into CD4 and CD8 double positive (DP) and CD4 or CD8 single positive (SP) cells, while the IL-7 Tg ETPs barely increased 2-fold, without any distinct T- or B-cell development (Supplemental Fig. 2A and B, available at International Immunology Online). In addition, ETPs sorted from either WT or IL-7 Tg mice were equally inefficient in generating B220+ cells when they were co-cultured with OP9-DL1 cells (Fig. 1G). ETPs from WT mice did produce B cells at low efficiency when cultured on OP9 cells, while ETPs from IL-7 Tg mice did not (data not shown). In addition, we observed a 7-fold decrease in the number of CD90+ cells generated in OP9-DL1 co-cultures when ETPs from IL-7 Tg mice were compared with those from WT mice (Fig. 1G). Therefore, ETPs from the IL7 Tg mice are inefficient at producing normal numbers of thymocytes and do not appear to be the source of the robust B-cell development observed in the Tg mice.
We extended these results by analyzing cells obtained from OP9-DL1 co-cultures seeded with sorted DN1 c-Kit+ and DN2 c-Kit+ cells from WT and IL-7 Tg mice. No DP or B cells developed from the atypical IL-7 Tg DN1 cells (Supplemental Figure 2C, available at International Immunology Online, and unpublished data). In addition, DN2 progenitors from IL-7 Tg mice showed less DP than WT cells (Supplemental Figure 2C). Thus, progenitor cells (ETPs, DN1 c-Kit+, DN2 and DN3) from IL7 Tg mice are less efficient at producing thymocytes and do not appear to be the source of the B-cell development observed in the Tg thymus.
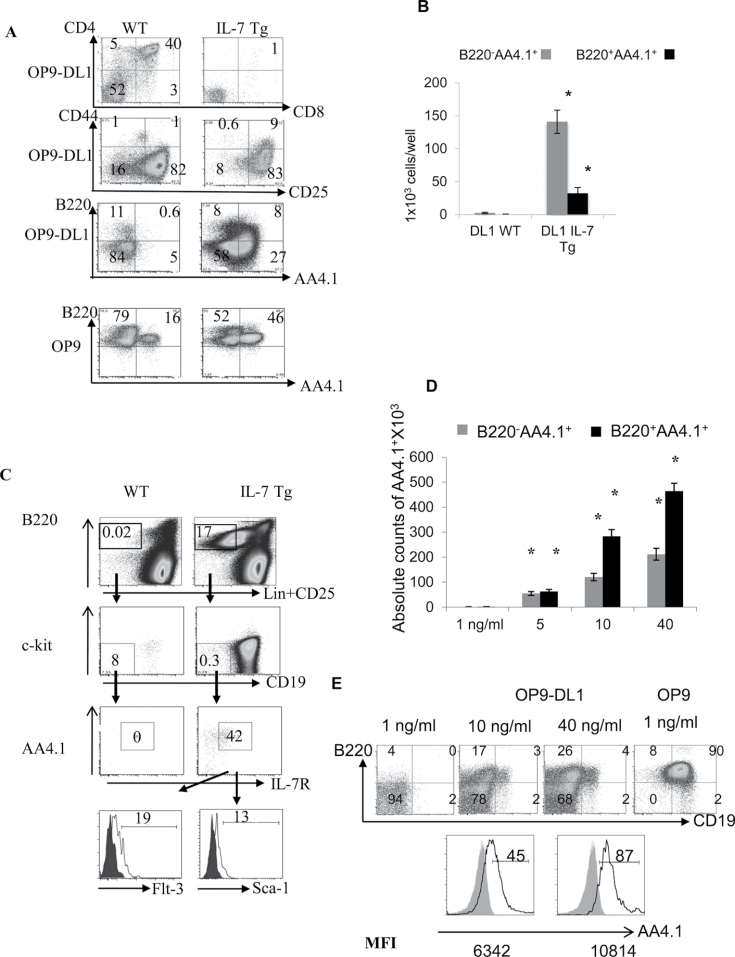
High IL-7 increases numbers of differentiated cells expressing B-cell markers in the thymus and in OP9-DL1 co-cultures
Because the B cells developing in the IL-7 Tg thymus did not appear to arise from ETPs, we evaluated other possible progenitor cell populations. We sorted BM-LSKs (Lin−, c-Kit+Sca-1+) from IL-7 Tg and WT littermate mice and co-cultured them (100 cells/well) on OP9 or OP9-DL1 stromal cells. After 3 weeks of co-culture, few T cells developed beyond the DN3 stage in the IL-7 Tg co-cultures (Fig. 2A and data not shown). However, we did observe an increase of both B220−AA4.1+ and B220+AA4.1+ cells from IL-7 Tg LSK cells compared with WT in the presence of the Notch-1 ligand DLL1 (Fig. 2A and B). Both of these populations expressed IL-7R (Supplemental Figure 3, available at International Immunology Online). The B220−AA4.1+CD19− IL-7R+ phenotype can define common lymphoid progenitors or CLP (22) and the B220+AA4.1+CD19−IL-7R+ phenotype corresponds to the CLP-2 cells (23) in an early stage of B-cell development. CLPs and CLPs-2 are lymphoid-restricted BM-progenitors with lower T- and higher B-cell potential than ETPs (21, 23, 24). Although CLP-2 progenitors have not been shown to be present in normal thymus, we observed a population of cells that were Lin−B220+CD19−c-Kit−AA4.1+IL-7R+ScaloFlt3lo compatible with the CLP-2 phenotype in the thymi of 2-week-old IL-7 Tg mice (Fig. 2C). The absolute numbers of thymic-CLP-2 cells ranged from 2700 to 6700 in the IL-7 Tg mice (n = 8), while they were absent from the thymus of WT mice. They were also found in the BM of both IL-7 Tg mice and their WT littermates (data not shown). When sorted from normal BM, CLP-2 progenitors developed into B and T cells on OP9 and OP9-DL1, respectively (data not shown). We also sorted BM-LSK-Flt3+ from WT mice and cultured them with increasingly high doses of IL-7 (Fig. 2D and E). We observed an increase in the number of B220+AA4.1+ and B220−AA4.1+ cells in cultures treated with high IL-7. At 10 or 40ng ml−1 IL-7, a small percentage of these cells became CD19+ (Fig. 2E, top panel) and AA4.1hi (Fig. 2E, bottom panel). Although the proportion of these cells was below 5%, the phenotype of these cells suggests that further B-cell maturation occurred even in the presence of the strong Notch-1 stimulation found in OP9-DL1 co-cultures. Therefore, high IL-7 levels inhibit thymocyte development of BM-progenitor and OP9-DL1 co-cultures while permitting a significant increase in the number of cells expressing the B-cell markers AA4.1, B220 and CD19.
Fig. 2.
Increase of cells with B-cell markers instead of DP cells on OP9-DL1 stromal cells in presence of high IL-7. (A) T- and B-cell development from WT or IL-7 Tg-LSKs co-cultured with OP9-DL1 cells. The percentages of DP, DN1-4 and B220/AA4.1 cells are shown in the corresponding quadrants. Representative of experiments repeated three times. (B) Numbers of B220−AA4.1+ (grey bars) and B220+AA4.1+ (black bars) obtained from IL-7 Tg-LSKs cultured on OP9-DL1 cells compared with WT-LSKs. (*P values for B220−AA4.1+ and B220+AA4.1+ are 0.0014 and 0.02 on OP9-DL1, respectively). The experiment was repeated three times. (C) Thymic-CLP-2 (T-CLP-2) in 2-week-old IL-7 Tg mice and WT littermates. The percentage of T-CLP-2 are shown, as well as the profiles of Flt-3 and Sca-1 markers on CLP-2 from IL-7 Tg thymus (open histograms) compared with isotype controls (shaded histograms). The results were confirmed in eight mice in each group included in three experiments. (D) Numbers of B220−AA4.1+ (grey bars) and B220+AA4.1+ (black bars) obtained from OP9-DL1 and LSK-Flt3+ WT progenitors co-cultures in proportion to the increasing doses of IL-7 (*P < 0.0001 in all groups). The experiment was repeated three times. (E) FACS analysis of B cells obtained from WT LSK-Flt3+ cells co-cultured with OP9 or OP9-DL1 cells and treated with different levels of IL-7. The open histograms show the profile of AA4.1+ cells on B220−CD19+ (left histogram) and B220+CD19− cells (right histogram) compared with the isotype control (shaded histograms) from the sample treated with 40ng ml−1 IL-7. The numbers below the histograms represent the MFI. The experiment was repeated twice.
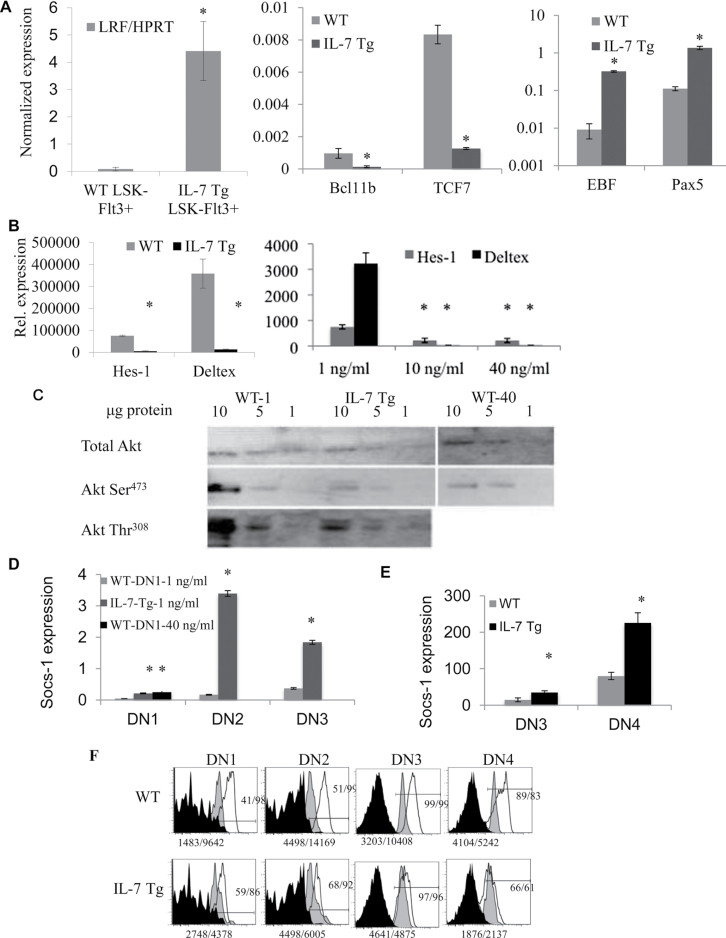
Thymocytes exposed to high levels of IL-7 adopt a B-lineage specific gene expression profile
A microarray comparison of gene expression profiles of DN1 c-kit+ and DN2 c-kit+ progenitors revealed a widespread change in gene expression patterns reflecting down-regulation of PI3K, JAK-STAT and Notch-1 signaling pathways in IL-7 Tg mice compared with WT controls (data not shown). To extend these findings, we performed q-RT-PCR assays for several genes that have been identified as critical mediators of T- and B-cell development. LRF has been described recently as having an important role in T- versus B-lineage decision through regulation of Notch signaling (14). Bcl11b and TCF7 are both functionally important markers for T-cell lineage choice and development (25, 26). On the other hand, early B cell factor (EBF) and Pax5 are critical mediators of B-cell development and they repress thymopoeisis (27). LRF expression was examined in sorted LSK-Flt3+ from WT mice and IL-7 Tg mice. Relative LRF mRNA levels normalized to hypoxanthine phosphoribosyltransferase 1 (HPRT) expression were increased 56-fold in LSK-Flt3+ progenitors from IL-7 Tg mice compared with WT mice (Fig. 3A). The analysis of the other genes was done using sorted c-Kit+ DN1 cells from 1- to 2-month-old IL-7 Tg and WT mice and normalized using GAPDH expression. We observed that the thymocyte markers Bcl11b and TCF-7 were dramatically decreased in IL-7 Tg DN1 cells in addition to a substantial increase in the expression of the B-cell restricted genes EBF and Pax5 in these cells (Fig. 3A).
Fig. 3.
Gene expression analysis of cells exposed to high IL-7 doses. (A) Panels compare the relative gene expression of IL-7 Tg and WT. Left panel, LRF expression in LSK-Flt3+ cells; middle panel, Bcl11b and TCF-7 expression in DN1 c-Kit+ cells; right panel, EBF and Pax5 expression in DN1 c-Kit+ cells (*P < 0.001). (B) Left panel, relative expression levels of Hes-1 and Deltex expression in DN1 c-Kit+ cells from WT and IL-7 Tg mice (*P < 0.0001 for Hes-1 and 0.0008 for Deltex). Right panel, relative expression levels of Hes-1 and Deltex expression in DN1 c-Kit+ cells sorted from WT LSK-Flt3+ and cultured with OP9-DL1 cells at 1, 10 or 40ng ml−1 of IL-7 (Hes-1 *P < 0.05, Deltex *P < 0.0001). Data shown in (A) n = 8–9 mice each group done in triplicate and (B) are from two experiments done in triplicate. (C) LSK-Flt-3+ progenitors were sorted from IL-7 Tg and WT mice. WT cells were cultured on OP9-DL1 stromal cells with 1ng ml−1 (WT-1) or 40ng ml−1 of IL-7 (WT-40). IL-7 Tg cells were cultured on OP9-DL1 stromal cells with 1ng ml−1 IL-7 (IL-7 Tg). After culture, DN1 c-Kit+, DN2 c-Kit+ and DN3 c-Kit− progenitors were separately sorted then mixed in the same proportions for each group. Total and phospho-Akt Ser473 are shown for all groups. Phospho-Akt-Thr308 is shown in IL-7 Tg and WT-derived cells. (D) Normalized SOCS-1 expression in sorted DN1-c-Kit+, DN2-ckit+ and DN3-c-Kit− from WT LSK-Flt-3+ and OP9-DL1 cell co-cultures treated with1ng ml−1 or 40ng ml−1 of IL-7. IL-7 Tg LSK-Flt3+ were cultured with 1ng ml−1 of IL-7 (*P < 0.0001). (E) Normalized SOCS-1 expression in sorted DN3-c-Kit− and DN4-c-Kit− thymocytes from IL-7 Tg and WT mice (*P = 0.0004; **P = 0.0001). (F) STAT5 intracellular staining in DN thymocytes from IL-7 Tg and WT mice. The black filled histograms show the isotype control for STAT5 staining; the grey histograms show STAT5 staining for unstimulated cells; the open histograms correspond to STAT5 staining in progenitors stimulated with IL-7. Gates were DN1-c-Kit+, DN2-ckit+, DN3-c-Kit− and DN4-c-Kit− within the Lin− population. The experiment was repeated three times.
Decreased Notch-1 signaling and attenuation of the PI3 kinase pathway in the IL-7 Tg thymus
We documented apparent thymic B-cell differentiation coupled with inhibition of T-cell development in the thymus of IL-7 Tg mice and in OP9-DL1 co-cultures treated with high levels of IL-7. Because both of these models contain ample Notch-1 ligand and because the response to high IL-7 is similar to that observed in Notch-1-deficient mice, we hypothesized that a down-regulation of Notch signaling in thymocytes might occur in high IL-7 conditions. First we confirmed that the expression of the Notch-1 and DLL1 and DLL4 were normal in IL-7 Tg thymi using qRT-PCR (Supplemental Figure 4A is available at International Immunology Online). Western blots confirmed that DLL1 and DLL4 protein levels in the IL-7 Tg thymus were similar to the WT control (Supplemental Figure 4B, available at International Immunology Online). In addition, Notch-1 mRNA levels in sorted DN1 and DN2 thymocytes from Tg or WT mice were not significantly different (unpublished data). We analyzed mRNA expression of the Notch-regulated genes Hes-1 and Deltex in c-Kit+ DN1 cells sorted from IL-7 Tg mice and normal littermates (Fig. 3B). In IL-7 Tg c-Kit+ DN1 cells, Hes-1 expression was decreased on average 9-fold (range, 7.1–14.5) and Deltex 28-fold (range, 11.4–35) (Fig. 3B). We also noted a significant decrease in the expression of both Hes-1 and Deltex in IL-7 Tg-DN2 progenitors compared with WT (P < 0.0001 for both genes, data not shown), as well as a decrease in Hes1 expression in IL-7 Tg-DN3 progenitors compared with WT (data not shown). The same results were obtained in c-Kit+ DN1 generated in vitro using OP9-DL stromal cells with WT LSK-Flt-3+ cells cultured with high doses of IL-7. Hes-1 expression was decreased 12-fold and Deltex was undetectable in cells exposed to 10 or 40ng ml−1 IL-7 compared with 1ng ml−1 IL-7–treated samples (Fig. 3B). Again, a similar decrease in Hes1 expression was observed in DN3 cells sorted from OP9-DL1 co-cultures treated with 10 or 40ng ml−1 IL-7 compared with controls treated with 1ng ml−1 IL-7 (data not shown). Therefore, we documented an overall reduction of Notch target genes in precursor cells exposed to high levels of IL-7 in culture or in our IL-7 Tg mice.
Total thymocytes from IL-7 Tg mice and WT matched controls were used to study PI3K signaling by western blot. Phosphorylated regulatory (p85α) PI3K subunit, phosphorylated PDK-1 and phosphorylated Akt at Ser473 and Thr308 were decreased in IL-7 Tg mice, while total Akt protein and tubulin content were similar (Supplemental Figure 4C is available at International Immunology Online). In order to confirm these results in vitro in the presence of high IL-7 levels, LSK-Flt3+ BM cells were isolated from WT and IL-7 Tg mice and cultured for 3–4 weeks on OP9-DL1 cells. WT cells were cultured with 1ng ml−1 or 40ng ml−1 of IL-7. After the culture period, DN1 c-Kit+, DN2 c-Kit+ and DN3 c-Kit− progenitor cells that developed in vitro were separately sorted and then pooled together in the same proportions for WT and IL-7 Tg to produce similar populations of cells that were analyzed by western blot. Again, phosphorylation of Akt at Ser473 and Thr308 was decreased in IL-7 Tg progenitors compared with WT (Fig. 3C, middle lanes). WT progenitors cultured with high dose of IL-7 also showed lower phospho-Akt at ser473 (Fig. 3C, right lanes). These in vitro data confirm our in vivo results that show disrupted PI3K-Akt signaling in cells exposed to high levels of IL-7.
These findings suggested that, in spite of availability of IL-7 and its receptor, the IL-7 signaling pathway was blunted. Therefore, we examined expression of the suppressor of cytokine signaling-1 (SOCS-1) protein that directly binds to JAKs and inhibits downstream IL-7-induced events (28). We analyzed the expression of SOCS-1 by RT-PCR in thymocyte progenitors treated with high IL-7 doses. In cells cultured on OP9-DL1 stromal cells, SOCS-1 expression was increased in DN1 c-Kit+ from IL-7 Tg (5-fold) and WT cells cultured with 40ng ml−1 of IL-7 (6-fold) compared with cells cultured with 1ng ml−1 of IL-7 (Fig. 3D). SOCS-1 expression was also increased in cultured DN2 c-Kit+ (20-fold) and DN3 c-Kit− cells (5-fold) (Fig. 3D), and in freshly sorted DN3 (2.5-fold) and DN4 (2.8-fold) cells from IL-7 Tg mice compared with WT mice (Fig. 3E). In accord with increased SOCS-1 expression, we observed decreased levels of STAT5 phosphorylation in thymocytes from IL-7 Tg compared with normal WT mice (Fig. 3F). Therefore, the increased levels of SOCS-1 expression observed in IL-7 Tg thymocytes likely explains why IL-7 downstream signaling is down-regulated in IL-7 Tg thymocytes. In addition, it might also explain why IL-7R is not down-regulated in the IL-7 Tg in spite of high levels of IL-7.
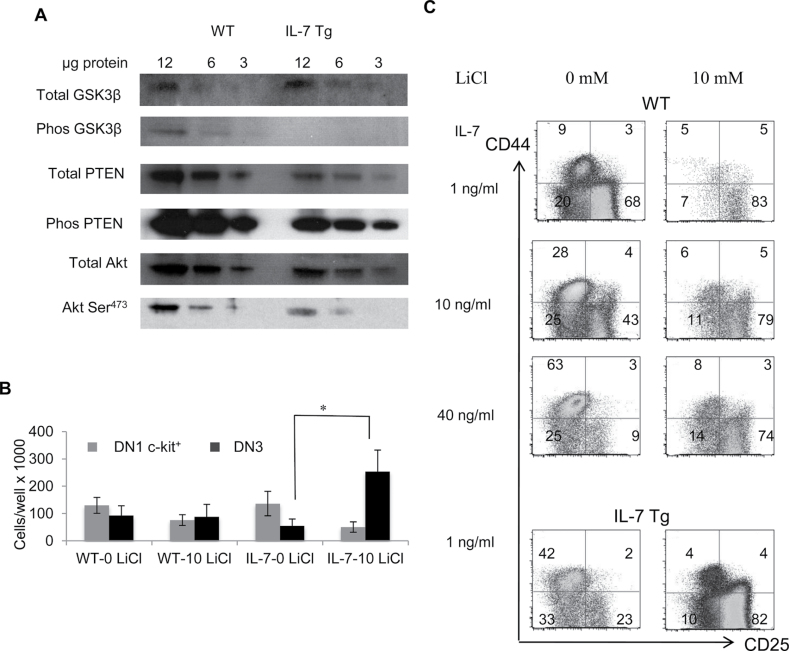
GSK3β mediates inhibition of Notch function in response to high IL-7 levels
The attenuation of IL-7 signaling measured by a reduced level of STAT5 phosphorylation was likely due to elevated SOCS-1 expression in IL-7 Tg thymocytes. Therefore, we examined another arm of IL-7R signaling. IL-7R also activates the PI3K pathway leading to AKT (also known as Protein Kinase B) phosphorylation and inactivation of GSK3β a negative regulator of Notch-1 (10, 11). Therefore, reduced GSK3β phosphorylation should inhibit Notch-1 signaling. We used western blots to examine PI3K-Akt-PTEN phosphorylation in sorted DN1-3 thymocytes from IL-7 Tg and control mice. Phospho-GSK3β was decreased in IL-7 Tg DN1-3 thymocytes, while phospho-PTEN was unchanged compared with WT cell thymocytes (Fig. 4A). We also confirmed that phospho-Akt at Ser473 was decreased in these samples (Fig. 4A). Therefore, these data suggest that the inhibition of the PI3K pathway we observed is associated with a reduction of GSK3β phosphorylation.
Fig. 4.
PI3K-Akt pathway controls Notch signaling through GSK3β. (A) Western blots for total and phospho-GSK3β, PTEN and Akt were performed on normalized populations of DN1 c-Kit+, DN2 c-Kit+, and DN3 c-Kit− progenitors freshly sorted from WT and IL-7 Tg mice. (B) Total counts of DN1 c-Kit+ (grey bars) and DN3 c-Kit− (black bars) cells after culture of LSKs in presence of normal or high IL-7 doses with or without lithium chloride. LSKs from WT mice were treated with 1, 10 or 40ng ml−1 while LSK from IL-7 Tg mice were cultured with 1ng ml−1 IL-7. Cell numbers from all cultures are compared with WT cultures with 1ng ml−1 of IL-7. The results represent three combined experiments (*P = 0.02). (C) DN1-4 profiles after culture of IL-7 Tg or WT progenitors with or without LiCl and different IL-7 doses. The percentages of all DN stages are shown from one out of three representative experiments.
Based on these results, we hypothesized that reduced Akt-dependent inactivation of GSK3β was responsible for Notch down-regulation. Therefore, we explored T- and B development in the presence of IL-7 and lithium chloride (LiCl) a GSK3β inhibitor, on OP9-DL stromal cells (11). We cultured LSK cells from IL7-Tg mice at 1ng ml−1 of IL-7 and LSK cells from WT mice at 1, 10 and 40ng ml−1 of IL-7 in the presence or absence of 10mM of LiCl added during the first week of culture. We analyzed T- and B-cell development after 2–4 weeks. The total cell counts were similar in cultures treated with or without LiCl. In the groups exposed to high IL-7 doses (IL-7 Tg or WT progenitors cultured with 10 and 40ng ml−1 of IL-7), a significant increase of the number of DN3 c-Kit− cells (4.6-fold) was observed in LiCl-treated cultures compared with non-treated cultures (Fig. 4B and C) (P = 0.02). The DN1 c-Kit+ cells also decreased in the same group by 2.7-fold, but the difference was not statistically significant (P = 0.09). In addition, a significant decrease in the percentage of B220+ cells was observed in IL-7 Tg or WT cells treated with high IL-7 when LiCl was present (P = 0.0002) (data not shown). These data suggest that LiCl inactivation of GSK3β restored Notch-dependent thymocyte development as evidenced by cells transiting beyond the DN1 block observed under high IL-7 conditions.
Discussion
Within the thymic microenvironment, integration of Notch-1 and the IL-7 signaling pathways are crucial to support T-cell development. IL-7 expression in thymic epithelial cells is regulated by location, ageing and thymocyte interactions (4–6,29,30). Given the importance of IL-7 and Notch signaling on thymus function, it seemed likely that efficient thymocyte development might depend on cross-talk between these pathways. Therefore, alterations in bioavailability of IL-7, e.g. during high dose clinical treatments, may affect T-cell development. Support for this concept was provided by a study showing that IL-7 enhances proliferation of early thymocytes while opposing Notch-1-dependent progression beyond the DN3 stage in OP9-DL1 co-cultures (31). Another example was the finding that Notch-1 signaling potentiates IL-7 signaling in human thymocytes grown on OP9-DL1 (32). Together, data from these in vitro studies suggest that regulatory circuits do exist between the IL-7 and Notch pathways.
To investigate the effect of clinically therapeutic levels of IL-7 in vivo, we created a Tg mouse that expresses about 40-fold higher than normal levels of IL-7 in the thymus (17). Unexpectedly, high IL-7 levels reduced αβ T-cell development (17), while promoting thymic B-cell development. Our findings differ from data using a different IL-7 Tg where B-lineage cells entered the thymus from the circulation due to very high levels of B lymphopoiesis in spleen, lymph nodes, and blood (33). In contrast, we propose that high levels of IL-7 inhibited Notch based on our observation of numerous phenotypic abnormalities observed in the IL-7 Tg thymus and known to be associated with decreased Notch-1 signaling: fewer T cells and more B cells in the thymus; (7, 8) reduced numbers of ETPs, DN2 and pre T cells and increases in γδ T cells and NK cells (18). This idea was strengthened when we observed decreased steady state levels of the Notch-1 target genes Hes-1 and Deltex in ETP cells. Together, our data strongly suggested that a major effect of high IL-7 on thymocytes is an active inhibition of Notch-1 signaling in spite of abundant DLL1 or DLL4 in the thymus and in our OP9-DL1 co-cultures.
We next examined signal transduction pathways to determine how Notch signaling was reduced and determined that PI3K, Akt and GSK3β are hypo-phosphorylated in IL-7 Tg thymocytes compared with controls. These data provided insight toward a potential mechanism because it is known that GSK3β phosphorylates and inhibits Notch (10, 11). Based on this idea, we showed that LiCl inactivation of GSK3β (10, 11, 34) in cells exposed to high IL-7 levels resulted in restoration of normal thymocyte development in OP9-DL co-cultures. Together, our data strongly suggested that a major effect of high IL-7 on thymocytes is an active inhibition of Notch-1 signaling.
Our results indicate that high levels of IL-7 also skew signaling networks and alter the developmental potential of thymocyte precursor cells. For example, we observed an increase in SOCS-1 expression in IL-7 Tg c-Kit+ DN1 cells. In accord with SOCS-1 ability to blunt cytokine responses (35), we also found that phospho-STAT5 levels were reduced in IL-7 Tg DN1-4 cells compared with WT controls. This finding distinguishes our work from others that showed that expression of constitutively active STAT5 drives thymic B-cell development (36).
In addition, we documented an increase in EBF and Pax5, master regulators of B-cell development in DN1 c-Kit+ cells sorted from IL-7 Tg mice. Conversely, we showed a dramatic decrease in Bcl11b and TCF-7, two genes critical for T-cell lineage commitment and development in DN1 c-Kit+ cells sorted from IL-7 Tg mice. Of particular interest is the recent finding that Bcl11b expression is repressed in culture conditions with high levels of IL-7 and Notch ligand (25). In these conditions, DN2 cells that retain myeloid and T-cell (but not B-cell) potential accumulate but are unable to undergo T-cell development until IL-7 levels are reduced (25). Although our data suggest that cells from an earlier developmental stage are diverted to thymic B-cell development in the IL-7 Tg, it seems likely that repression of Bcl11b by high IL-7 contributes to the inefficiency of T-cell production observed in vivo and in vitro. Together, these data are consistent with a diversion from T- to B-lineage development of IL-7 Tg thymic precursors.
One intriguing question is the nature of the cells that give rise to the abnormal thymic B-cell development in our Tg model. The majority of the earliest thymic precursor cells has a history of IL-7R expression (2) that may affect lineage choice in the thymus. Using the OP9-DL1 co-culture system, we tested different progenitors in the IL-7 Tg for B-cell potential. ETPs did not appear to be the source of the thymic B cells observed in the IL-7 Tg mice. However, we identified CLP-2 cells in the thymus of IL-7 Tg mice that was absent in WT mice, possibly representing a rare population of independent progenitor cells seeding the thymus (20). We also observed a distinctive population of B220+AA4.1+ CD19+cells that developed from LSK progenitors in the presence of high IL-7 and Notch-1 ligand (37). These cells are similar to the B220+CD19lo cells described in vivo as pro-T cells that convert to B cells upon Notch-1 deletion (38). CLP-2 cells normally die in the thymus in response to Notch signaling (39). Together, these findings suggest that the abnormal B-cell development in the IL-7 Tg thymus results from inhibition of Notch-1 signaling in lymphoid progenitors. This idea was supported by the reduced number and T-cell potential of IL-7 Tg ETP cells in spite of the presence of Notch ligand. In addition, TCRαβ differentiation in IL-7 Tg mice is blocked at the DN3 stage, a point where it has been reported that high Notch levels are required for survival and expansion (40). In addition, a down-regulation of Notch-1 signal may lead to accumulation of CLP-2 progenitors in the IL-7 Tg thymus and may constitute an alternative progenitor population giving rise to the B cells and thymocytes found in the IL-7 Tg thymus. The increase of LRF expression in LSK Flt3+ BM cells in IL-7 Tg mice could also explain the variability of thymic seeding progenitors in their T versus B potential leading to less T and more B cells in the thymus.
These findings demonstrate for the first time that high levels of IL-7 antagonize Notch signaling and dysregulate thymus function. It is likely that this information will lead to new understandings regarding lineage choice and expansion of precursor cells. For example, precursor cells are exposed to differing levels of IL-7 due to gradients formed by clusters of cells expressing higher levels of IL-7 in the thymus and other anatomical sites (5). Because our work suggests that IL-7 mediates an environmental regulation of Notch signaling, differences in localized IL-7 availability in niches such as BM and thymus may be an important component of B–T lineage development. It may also explain the functional importance of thymocyte down-regulation of IL-7 expression in thymic stromal cells (6). Our findings also present new considerations for patient treatment since IL-7 has been introduced in clinical settings (15, 41). Optimal clinical effects on peripheral expansion occurred at levels 1000-fold over baseline (15), which corresponds to the high levels of IL-7 where we observed negative effects on thymic function in our experimental models. Based on our data, we hypothesize that high IL-7 levels would diminish thymus function and may preclude simultaneous augmentation of T-cell populations by thymic dependent and peripheral expansion pathways by high dose IL-7 treatment. This may be an especially important consideration for pediatric patients because they often exhibit robust thymic function following hematopoietic stem cell transplantation.
Supplementary data
Supplementary data are available at International Immunology Online.
Funding
The Intramural Research Program of the National Cancer Institute at the National Institutes of Health (Bethesda, MD USA) provided funding for these studies that were performed solely at the NCI.
Acknowledgements
The authors thank Avinash Bandhoola, Remy Bosselut, and Alfred Singer for expert review of the manuscript. We acknowledge the expertise of Lionel Feigenbaum, William Teldford, Genevieve Sanchez and Amy James.
References
- 1. Rothenberg E. V. 2007. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity 26:690–702 [DOI] [PubMed] [Google Scholar]
- 2. Schlenner S. M.,, Madan V.,, Busch K., et al. 2010. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32:426–436 [DOI] [PubMed] [Google Scholar]
- 3. von Freeden-Jeffry U.,, Solvason N.,, Howard M., Murray R. 1997. The earliest T lineage-committed cells depend on IL-7 for Bcl-2 expression and normal cell cycle progression. Immunity 7:147–154 [DOI] [PubMed] [Google Scholar]
- 4. Mazzucchelli R. I.,, Warming S.,, Lawrence S. M., et al. 2009. Visualization and identification of IL-7 producing cells in reporter mice. PLoS ONE 4:e7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alves N. L.,, Richard-Le Goff O.,, Huntington N. D., et al. 2009. Characterization of the thymic IL-7 niche in vivo. Proc. Natl. Acad. Sci. U.S.A. 106:1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alves N. L.,, Huntington N. D.,, Mention J. J.,, Richard-Le Goff O., Di Santo J. P. 2010. Cutting Edge: a thymocyte-thymic epithelial cell cross-talk dynamically regulates intrathymic IL-7 expression in vivo. J. Immunol. 184:5949–5953 [DOI] [PubMed] [Google Scholar]
- 7. Koch U.,, Lacombe T. A.,, Holland D., et al. 2001. Subversion of the T/B lineage decision in the thymus by lunatic fringe-mediated inhibition of Notch-1. Immunity 15:225–236 [DOI] [PubMed] [Google Scholar]
- 8. Wilson A.,, MacDonald H. R., Radtke F. 2001. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194:1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pui J. C.,, Allman D.,, Xu L., et al. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11:299–308 [DOI] [PubMed] [Google Scholar]
- 10. Espinosa L.,, Inglés-Esteve J.,, Aguilera C., Bigas A. 2003. Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 278:32227–32235 [DOI] [PubMed] [Google Scholar]
- 11. McKenzie G.,, Ward G.,, Stallwood Y., et al. 2006. Cellular -otch responsiveness is defined by phosphoinositide 3-kinase-dependent signals. BMC Cell Biol. 7:10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O’Brien W. T., Klein P. S. 2009. Validating GSK3 as an in vivo target of lithium action. Biochem. Soc. Trans. 37(Pt 5):1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davies J. M.,, Hawe N.,, Kabarowski J., et al. 1999. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene 18:365–375 [DOI] [PubMed] [Google Scholar]
- 14. Maeda T.,, Merghoub T.,, Hobbs R. M., et al. 2007. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science 316:860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sportès C.,, Hakim F. T.,, Memon S. A., et al. 2008. Administration of rhIL-7 in humans increases in vivo TCR repertoire diversity by preferential expansion of naive T cell subsets. J. Exp. Med. 205:1701–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmitt T. M., Zúñiga-Pflücker J. C. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 17:749–756 [DOI] [PubMed] [Google Scholar]
- 17. El Kassar N.,, Lucas P. J.,, Klug D. B., et al. 2004. A dose effect of IL-7 on thymocyte development. Blood 104:1419–1427 [DOI] [PubMed] [Google Scholar]
- 18. Schmitt T. M.,, Ciofani M.,, Petrie H. T., Zúñiga-Pflücker J. C. 2004. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J. Exp. Med. 200:469–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park J. H.,, Yu Q.,, Erman B., et al. 2004. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity 21:289–302 [DOI] [PubMed] [Google Scholar]
- 20. Porritt H. E.,, Rumfelt L. L.,, Tabrizifard S.,, Schmitt T. M.,, Zúñiga- Pflücker J. C., Petrie H. T. 2004. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20:735–745 [DOI] [PubMed] [Google Scholar]
- 21. Allman D.,, Sambandam A.,, Kim S., et al. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4:168–174 [DOI] [PubMed] [Google Scholar]
- 22. Kondo M.,, Weissman I. L., Akashi K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91:661–672 [DOI] [PubMed] [Google Scholar]
- 23. Martin C. H.,, Aifantis I.,, Scimone M. L., et al. 2003. Efficient thymic immigration of B220+ lymphoid-restricted bone marrow cells with T precursor potential. Nat. Immunol. 4:866–873 [DOI] [PubMed] [Google Scholar]
- 24. Krueger A.,, Garbe A. I., von Boehmer H. 2006. Phenotypic plasticity of T cell progenitors upon exposure to Notch ligands. J. Exp. Med. 203:1977–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li L.,, Leid M., Rothenberg E. V. 2010. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weber B. N.,, Chi A. W.,, Chavez A., et al. 2011. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476:63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Northrup D. L., Allman D. 2008. Transcriptional regulation of early B cell development. Immunol. Res. 42:106–117 [DOI] [PubMed] [Google Scholar]
- 28. Yu Q.,, Park J. H.,, Doan L. L.,, Erman B.,, Feigenbaum L., Singer A. 2006. Cytokine signal transduction is suppressed in preselection double-positive thymocytes and restored by positive selection. J. Exp. Med. 203:165–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zamisch M.,, Moore-Scott B.,, Su D. M.,, Lucas P. J.,, Manley N., Richie E. R. 2005. Ontogeny and regulation of IL-7-expressing thymic epithelial cells. J. Immunol. 174:60–67 [DOI] [PubMed] [Google Scholar]
- 30. Park J. H.,, Adoro S.,, Lucas P. J., et al. 2007. ‘Coreceptor tuning’: cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat. Immunol. 8:1049–1059 [DOI] [PubMed] [Google Scholar]
- 31. Balciunaite G.,, Ceredig R.,, Fehling H. J.,, Zúñiga-Pflücker J. C., Rolink A. G. 2005. The role of Notch and IL-7 signaling in early thymocyte proliferation and differentiation. Eur. J. Immunol. 35:1292–1300 [DOI] [PubMed] [Google Scholar]
- 32. Magri M.,, Yatim A.,, Benne C., et al. 2009. Notch ligands potentiate IL-7-driven proliferation and survival of human thymocyte precursors. Eur. J. Immunol. 39:1231–1240 [DOI] [PubMed] [Google Scholar]
- 33. Ceredig R. 2002. The ontogeny of B cells in the thymus of normal, CD3 epsilon knockout (KO), RAG-2 KO and IL-7 transgenic mice. Int. Immunol. 14:87–99 [DOI] [PubMed] [Google Scholar]
- 34. O’Brien W. T.,, Huang J.,, Buccafusca R., et al. 2011. Glycogen synthase kinase-3 is essential for ß-arrestin-2 complex formation and lithium-sensitive behaviors in mice. J. Clin. Invest. 121:3756–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chong M. M.,, Cornish A. L.,, Darwiche R., et al. 2003. Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation. Immunity 18:475–487 [DOI] [PubMed] [Google Scholar]
- 36. Goetz C. A.,, Harmon I. R.,, O’Neil J. J.,, Burchill M. A.,, Johanns T. M., Farrar M. A. 2005. Restricted STAT5 activation dictates appropriate thymic B versus T cell lineage commitment. J. Immunol. 174:7753–7763 [DOI] [PubMed] [Google Scholar]
- 37. Li Y. S.,, Wasserman R.,, Hayakawa K., Hardy R. R. 1996. Identification of the earliest B lineage stage in mouse bone marrow. Immunity 5:527–535 [DOI] [PubMed] [Google Scholar]
- 38. Feyerabend T. B.,, Terszowski G.,, Tietz A., et al. 2009. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity 30:67–79 [DOI] [PubMed] [Google Scholar]
- 39. Nie L.,, Perry S. S.,, Zhao Y., et al. 2008. Regulation of lymphocyte development by cell-type-specific interpretation of Notch signals. Mol. Cell. Biol. 28:2078–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vicente R.,, Swainson L.,, Marty-Grès S., et al. 2010. Molecular and cellular basis of T cell lineage commitment. Semin. Immunol. 22:270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sportès C., Gress R. E. 2007. Interleukin-7 immunotherapy. Adv. Exp. Med. Biol. 601:321–333 [DOI] [PubMed] [Google Scholar]