Abstract
The current study comprehensively examined the association between common genetic variants of the kallikrein-kinin system (KKS) and blood pressure salt sensitivity. A 7-day low-sodium followed by a 7-day high-sodium dietary intervention was conducted among 1,906 Han Chinese participants recruited from 2003 to 2005. Blood pressure was measured by using a random-zero sphygmomanometer through the study. A total of 205 single nucleotide polymorphisms (SNPs) covering 11 genes of the KKS were selected for the analyses. Genetic variants of the bradykinin receptor B2 gene (BDKRB2) and the endothelin converting enzyme 1 gene (ECE1) showed significant associations with the salt-sensitivity phenotypes even after adjustment for multiple testing. Compared with the major G allele, the BDKRB2 rs11847625 minor C allele was significantly associated with increased systolic blood pressure responses to low-sodium intervention (P = 0.0001). Furthermore, a haplotype containing allele C was associated with an increased systolic blood pressure response to high-sodium intervention (P = 0.0009). Seven highly correlated ECE1 SNPs were shown to increase the diastolic blood pressure response to low-sodium intervention (P values ranged from 0.0003 to 0.002), with 2 haplotypes containing these 7 SNPs also associated with this same phenotype (P values ranged from 0.0004 to 0.002). In summary, genetic variants of the genes involved in the regulation of KKS may contribute to the salt sensitivity of blood pressure.
Keywords: blood pressure; genetics; kallikreins; kinins; polymorphism; sodium, dietary
Salt sensitivity of blood pressure (BP) is a complex trait documented to increase the risk of hypertension, cardiovascular disease, and premature death (1, 2). Although the heritability of this phenotype has been well established (3, 4), the genetic mechanisms underlying BP response to sodium intake remain relatively unknown. The kallikrein-kinin system (KKS) plays a key role in the regulation of BP and sodium homeostasis (5–7) and has been implicated in the pathogenesis of salt sensitivity. For example, animal models of kininogen-deficient Brown Norway Katholiek rats and kinin receptor knockout mice demonstrated greater BP responses to sodium loading and chronic high sodium intake, respectively, compared with controls (8, 9). Furthermore, observational epidemiologic data demonstrated an inverse association between urinary kallikrein excretion and BP responses to sodium loading (10). Despite strong evidence supporting a role for the KKS in salt sensitivity, the genetic mechanisms underlying this relation are not well understood.
In the KKS, kinins, which are derived from the enzymatic action of kallikrein on kininogen, cause vasodilation, diuresis, and natriuresis (11). Plasma and tissue kallikreins convert kininogens to vasoactive bradykinin and kallidin, the 2 major types of kinin peptides (12). The kinins act through G-protein-coupled B1 and B2 receptors (13). The B2 receptor is constitutively expressed and responsible for most physiologic actions of the KKS, whereas the B1 receptor is rarely expressed and induced by tissue injury (14). Carboxypeptidase N, also known as kininase I, and carboxypeptidase M remove arginine from the carboxyl terminus of the kinins and generate their des-Arg derivatives, which are agonists mainly of the B1 receptor. The kinins can also be inactivated by the action of kininase II (angiotensin-converting enzyme, ACE), neutral endopeptidase, and endothelin-converting enzyme which remove 2 amino acids from the carboxyl terminus (13).
The current study aimed to comprehensively examine common genetic variants from 11 candidate genes for their association with BP responses to dietary sodium interventions among a large and homogeneous sample of Han Chinese families. These candidate genes encode components of the KKS and include the bradykinin receptor B1 gene (BDKRB1), the bradykinin receptor B2 gene (BDKRB2), the carboxypeptidase M gene (CPM), the carboxypeptidase N, polypeptide 1 gene (CPN1), the carboxypeptidase N, polypeptide 2 gene (CPN2), the endothelin converting enzyme 1 gene (ECE1), the kallikrein 1 gene (KLK1); the kallikrein B, plasma (Fletcher factor) 1 gene (KLKB1), the kininogen 1 gene (KNG1), the membrane metalloendopeptidase gene (MME), and the serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 gene (SERPINA4).
MATERIALS AND METHODS
Study population
All study subjects were participants of the Genetic Epidemiology Network of Salt Sensitivity (GenSalt), a family-based dietary feeding study conducted in rural areas of northern China from 2003 to 2005 (15). A community-based BP screening was conducted among persons aged 18–60 years in the study villages to identify potential probands and their families. The proband was defined as an individual with a mean systolic blood pressure (SBP) between 130 and 160 mm Hg and/or a diastolic blood pressure (DBP) between 85 and 100 mm Hg and no use of antihypertensive medications. The probands, their spouses, siblings, and offspring were recruited for the dietary feeding study. All participants were of Han Chinese ethnicity. Individuals who had stage 2 hypertension, secondary hypertension, a history of clinical cardiovascular disease or diabetes, used antihypertensive medications, or were pregnant, heavy alcohol drinkers, or currently on a low-sodium diet were excluded from the study. Among 1,906 eligible participants for the dietary intervention, 1,871 (98.2%) and 1,860 (97.6%) completed the low-sodium and high-sodium interventions, respectively, and were included in the current analysis. Institutional review boards or ethics committees at all participating institutes approved the study protocol. Written, informed consents for the baseline observation and for the interventions were obtained from each participant prior to data collection or intervention, respectively.
Dietary intervention and BP measurements
After a 3-day baseline observation, the study participants received a 7-day low-sodium diet (3 g of sodium chloride or 51.3 mmol of sodium per day) followed by a 7-day high-sodium diet (18 g of sodium chloride or 307.8 mmol of sodium per day). All foods were cooked without salt, and prepackaged salt was added to the individual study participant's meal when it was served by the study staff. To ensure study participants' compliance to the intervention program, they were required to avoid consuming any foods and beverages that were not provided by the study. Dietary compliance by the participants was confirmed by measurements of 24-hour urinary excretion of sodium and potassium. The means of 24-hour urinary excretions of sodium and potassium were 242.4 (standard deviation (SD), 66.7) mmol and 36.9 (SD, 9.6) mmol at baseline, 47.5 (SD, 16.0) mmol and 31.4 (SD, 7.7) mmol during the low-sodium intervention, and 244.3 (SD, 37.7) mmol and 35.7 (SD, 7.5) mmol during the high-sodium intervention, respectively. Three sitting BP measurements were obtained each morning of the 3-day baseline observation and on days 5, 6, and 7 of each intervention period by the trained and certified observers using a random-zero sphygmomanometer according to a standard protocol (16).
Candidate gene and genetic variant selection and genotyping
To conduct a systematic analysis of the KKS, we selected 11 candidate genes involved in the formation, action, and degradation of kinins. They include KNG1, KLK1, KLKB1, BDKRB1, BDKRB2, SERPINA4, CPN1, CPN2, CPM, ECE1, and MME. We didn't include the angiotensin-converting enzyme gene (ACE) for one of the major inactivating enzymes for kinins, because we have previously reported the association between genetic variants of the renin-angiotensin-aldosterone system and salt sensitivity (17). We didn't observe significant association between ACE and salt sensitivity in the previous analysis. Table1 shows the physical location, number of genotyped single nucleotide polymorphisms (SNPs), and role for each gene in the KKS. Two sources of genotype information were available for the current study. Based on the International HapMap Project (referred to as “HapMap”; http://hapmap.ncbi.nlm.nih.gov/) linkage disequilibrium (LD) structure of the Chinese Han of Beijing, tag SNPs were selected from KLK1, BDKRB1, and BDKRB2 by using pairwise r2 ≥ 0.8. Genotyping was conducted by using SNPlex assays (Applied Biosystems, Foster City, California) based on an oligonucleotide ligation assay for capillary electrophoresis on ABI 3700 DNA analyzers (Applied Biosystems). For all 11 candidate genes as well as their 5,000 base pair (bp) flanking regions, we also included SNPs genotyped on the Affymetrix 6.0 platform (Affymetrix, Santa Clara, California). A total of 205 SNPs with minor allele frequencies ≥5% and an average call rate > 99% from both genotype sources were included in the analysis. Detailed information for these SNPs, including their physical positions, alleles, minor allele frequencies, and P values for the Hardy-Weinberg equilibrium test, is presented in Web Table 1, available at http://aje.oxfordjournals.org/.
Table 1.
Information on the Genes From the Kallikrein-Kinin System
| Official Symbol | Gene Name | Gene Location | No. of Genotyped SNPs | Role in the Kallikrein-Kinin System |
|---|---|---|---|---|
| KNG1 | Kininogen 1 | 3q27 | 18 | The precursor of vasoactive kinin peptides |
| KLK1 | Kallikrein 1 | 19q13.3 | 8 | Tissue kallikrein, a serine protease that generates kallidin by specific proteolysis of kininogen 1 |
| KLKB1 | Kallikrein B, plasma (Fletcher factor) 1 | 4q35 | 13 | Plasma kallikrein, cleaves high-molecular-weight kininogen to release bradykinin |
| BDKRB1 | Bradykinin receptor B1 | 14q32.1-q32.2 | 11 | B1 receptor of kinins, inducible |
| BDKRB2 | Bradykinin receptor B2 | 14q32.1-q32.2 | 26 | B2 receptor of kinins, constitutive |
| SERPINA4 | Serpin peptidase inhibitor, clade A, member 4 | 14q31-q32.1 | 12 | Also known as kallistatin, a specific inhibitor of tissue kallikreins |
| CPN1 | Carboxypeptidase N, polypeptide 1 | 10q24.2 | 11 | The small subunit of kininase-1, a plasma metalloprotease that removes arginine from the carboxyl terminus of the kinins and generates their des-Arg derivatives mainly activating B1 receptor |
| CPN2 | Carboxypeptidase N, polypeptide 2 | 3q29 | 12 | The large subunit of kininase-1 |
| CPM | Carboxypeptidase M | 12q14.3 | 36 | Removes arginine from the carboxyl terminus of the kinins |
| ECE1 | Endothelin converting enzyme 1 | 1p36.1 | 28 | Removes 2 amino acids from the carboxyl terminus of the kinins and inactivates them |
| MME | Membrane metalloendopeptidase | 3q25.1-q25.2 | 30 | A neutral endopeptidase that removes 2 amino acids from the carboxyl terminus of the kinins and inactivates them |
Abbreviation: SNP, single nucleotide polymorphism.
Statistical analysis
BP levels at baseline and during intervention were calculated as the mean of 9 measurements from each period. Mean arterial pressure was calculated as [(SBP − DBP)/3] + DBP. Salt sensitivity was defined continuously as the percent changes in mean SBP, DBP, and mean arterial pressure from baseline to low-sodium intervention and from low-sodium to high-sodium intervention. Salt-sensitivity phenotypes were adjusted for the effects of age and examination room temperature separately within sex–field center groups. The adjustment procedure included regressing each phenotype on the covariates in a stepwise manner, retaining only significant terms (P < 0.05). The residual variance was also examined by regressing the squared residual from the first regression on the same covariates (stepwise) and retaining significant terms. The final adjusted phenotype was computed as the residual from the first regression, divided by the square root of the predicted score from the second regression. A final standardization step was taken to ensure a mean of 0 and a standard deviation of 1.
Mendelian consistency and Hardy-Weinberg equilibrium for the SNP genotype data were assessed by PLINK, version 1.05, software (http://pngu.mgh.harvard.edu/~purcell/plink/) (18, 19). We used Haploview, version 4.2, software (http://www.broadinstitute.org/haploview) to estimate the extent of pairwise LD between SNPs and to define LD blocks.
The Family Based Association Test program, version 2.0.2 (http://www.biostat.harvard.edu/~fbat/default.html), was used to examine the association between each SNP and the adjusted BP responses. This method takes advantage of data from nuclear families, sibships, pedigrees, and any combination of familial data to provide unbiased tests of association. We conducted haplotype analyses to follow up on genes with significant results in single marker analyses. Additive genetic models were used for both single marker and haplotype analyses. The Family Based Association Test provides a z statistic with its corresponding P value for a tested allele or haplotype. In our study, a positive z statistic for a variant indicates a decreased response to low-sodium intervention and an increased response to high-sodium intervention. The false discovery rate method was used to adjust for multiple testing (20). The false discovery rate Q value represents the proportion of rejected null hypotheses that are erroneously rejected. We used the Proc Multtest procedure, along with the false discovery rate option, in SAS, version 9.2, software (SAS Institute, Inc., Cary, North Carolina) to calculate the Q value for all tested SNPs and haplotypes. A Q value less than 0.05 was considered statistically significant in this study.
RESULTS
The characteristics of 1,906 GenSalt participants included in the analysis are shown in Table 2. Overall, the participants' BP decreased from baseline to low-sodium intervention and increased from low-sodium to high-sodium intervention. BP levels were similar between baseline and high-sodium intervention. All of the BP responses to dietary sodium interventions were significantly different from zero.
Table 2.
Characteristics of 1,906 Han Chinese Participants, GenSalt Study, China, 2003–2005
| Variable | % | Mean (SD) |
|---|---|---|
| Age, years | 38.7 (9.6) | |
| Male | 53.0 | |
| Body mass indexa | 23.3 (3.2) | |
| Baseline blood pressure, mm Hg | ||
| Systolic blood pressure | 116.9 (14.2) | |
| Diastolic blood pressure | 73.7 (10.3) | |
| Mean arterial pressure | 88.1 (10.9) | |
| Blood pressure during low-sodium intervention, mm Hg | ||
| Systolic blood pressure | 111.4 (12.2) | |
| Diastolic blood pressure | 71.0 (9.7) | |
| Mean arterial pressure | 84.5 (9.7) | |
| Percentage blood pressure response to low sodium | ||
| Systolic blood pressure | −4.4 (5.5)* | |
| Diastolic blood pressure | −3.4 (7.5)* | |
| Mean arterial pressure | −3.9 (5.8)* | |
| Blood pressure during high-sodium intervention, mm Hg | ||
| Systolic blood pressure | 116.3 (13.6) | |
| Diastolic blood pressure | 72.9 (10.3) | |
| Mean arterial pressure | 87.4 (10.6) | |
| Percentage blood pressure response to high sodium | ||
| Systolic blood pressure | 4.4 (5.4)* | |
| Diastolic blood pressure | 2.9 (7.9)* | |
| Mean arterial pressure | 3.5 (6.0)* |
Abbreviations: GenSalt, Genetic Epidemiology Network of Salt Sensitivity; SD, standard deviation.
* P < 0.0001 (compared with no blood pressure change during sodium interventions).
a Body mass index: weight (kg)/height (m)2.
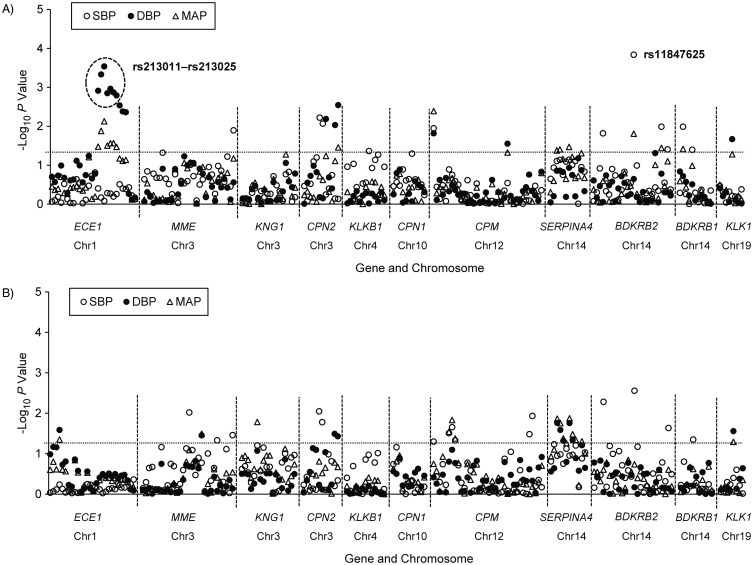
As shown in Figure 1, multiple SNPs from the 11 KKS genes were associated with BP responses to low-sodium and high-sodium interventions at an alpha threshold of 0.05 (−log10 P > 1.3). After adjustment for multiple testing, SNP rs11847625 of BDKRB2 remained significantly associated with the SBP response to low-sodium intervention (P = 1.4 × 10−4 and Q = 0.03). Similarly, 7 SNPs of ECE1, which were all in high LD (all pairwise r2 > 0.8), remained significantly associated with the DBP response to low-sodium intervention after accounting for multiple tests (P ranged from 2.9 × 10−4 to 1.6 × 10−3 and all Q = 0.047). Compared with its major G allele, the minor C allele of BDKRB2 rs11847625 was associated with an increased SBP response to low-sodium intervention (z = −3.798). A similar trend was observed for its association with the SBP response to high-sodium intervention (z = 2.992, P = 2.8 × 10−3) (Table 3). In addition, the minor alleles of the 7 ECE1 SNPs were associated with an increased DBP response to low-sodium intervention (all z < 0).
Figure 1.
−Log10 P values for the association between 205 SNPs in the kallikrein-kinin system genes and systolic blood pressure (SBP), diastolic blood pressure (DBP), and mean arterial pressure (MAP) responses to low (top) and high (bottom) sodium interventions, GenSalt Study, China, 2003–2005. Labeled SNPs are significant after adjustment for multiple testing, with a false discovery rate Q value less than 0.05. The dashed lines correspond to unadjusted P = 0.05. Chr, chromosome designation; GenSalt, Genetic Epidemiology Network of Salt Sensitivity; SNP, single nucleotide polymorphism. Genes symbols and their expanded names: bradykinin receptor B1 gene (BDKRB1); bradykinin receptor B2 gene (BDKRB2); carboxypeptidase M gene (CPM); carboxypeptidase N, polypeptide 1 gene (CPN1); carboxypeptidase N, polypeptide 2 gene (CPN2); endothelin converting enzyme 1 gene (ECE1); kallikrein 1 gene (KLK1); kallikrein B, plasma (Fletcher factor) 1 gene (KLKB1); kininogen 1 gene (KNG1); membrane metalloendopeptidase gene (MME); and serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 gene (SERPINA4).
Table 3.
Summary of Individual SNPs Significantly Associated With Blood Pressure Response to Sodium Interventions, GenSalt Study, China, 2003–2005
| SNP | Minor Allele | Minor Allele Frequency | Response to Low Sodium |
Response to High Sodium |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Systolic Blood Pressure |
Diastolic Blood Pressure |
Mean Arterial Pressure |
Systolic Blood Pressure |
Diastolic Blood Pressure |
Mean Arterial Pressure |
|||||||||||
| z Statistic | P Value | Q Valuea | z Statistic | P Value | Q Valuea | z Statistic | P Value | z Statistic | P Value | z Statistic | P Value | z Statistic | P Value | |||
| BDKRB2b | ||||||||||||||||
| rs11847625 | C | 0.20 | −3.798 | 1.4 × 10−4 | 0.03 | −1.210 | 0.23 | −2.420 | 0.02 | 2.992 | 2.8 × 10−3 | 0.415 | 0.68 | 1.332 | 0.18 | |
| ECE1b | ||||||||||||||||
| rs213011 | A | 0.37 | 0.846 | 0.40 | −3.232 | 1.2 × 10−3 | 0.047 | −2.127 | 0.03 | −0.607 | 0.54 | 0.613 | 0.54 | 0.454 | 0.65 | |
| rs169884 | A | 0.39 | 0.409 | 0.68 | −3.499 | 4.7 × 10−4 | 0.047 | −2.476 | 0.01 | −0.290 | 0.77 | 0.855 | 0.39 | 0.756 | 0.45 | |
| rs84853 | T | 0.39 | 0.218 | 0.83 | −3.623 | 2.9 × 10−4 | 0.047 | −2.675 | 7 × 10−3 | 0.069 | 0.95 | 0.992 | 0.32 | 0.980 | 0.33 | |
| rs213012 | T | 0.38 | 0.638 | 0.52 | −3.191 | 1.4 × 10−3 | 0.047 | −2.151 | 0.03 | −0.161 | 0.87 | 0.911 | 0.36 | 0.848 | 0.40 | |
| rs213014 | C | 0.40 | 0.615 | 0.54 | −3.265 | 1.1 × 10−3 | 0.047 | −2.205 | 0.03 | −0.315 | 0.75 | 1.012 | 0.31 | 0.853 | 0.39 | |
| rs213018 | A | 0.40 | 0.465 | 0.64 | −3.204 | 1.4 × 10−3 | 0.047 | −2.208 | 0.03 | −0.356 | 0.72 | 0.887 | 0.37 | 0.721 | 0.47 | |
| rs213025 | A | 0.38 | 0.594 | 0.55 | −3.154 | 1.6 × 10−3 | 0.047 | −2.120 | 0.03 | −0.318 | 0.75 | 0.993 | 0.32 | 0.841 | 0.40 | |
Abbreviations: GenSalt, Genetic Epidemiology Network of Salt Sensitivity; SNP, single nucleotide polymorphism.
a False discovery rate Q values that are less than 0.05 are provided.
b Gene symbols and their expanded names: BDKRB2, bradykinin receptor B2 gene; ECE1, endothelin converting enzyme 1 gene.
Seven and 4 LD blocks were identified in BDKRB2 and ECE1, respectively (Web Figures 1 and 2). A total of 25 common haplotypes with frequencies ≥ 5% were inferred within the BDKRB2 blocks. LD block 4, containing rs11847625, had 4 common haplotypes including T-T-T-A-G (26.2%), T-C-G-G-C (20.5%), C-C-G-G-G (35.8%), and T-T-G-G-G (15.1%). Similar to single-marker analysis, haplotype T-C-G-G-C that contained the minor C allele of rs11847625 was associated with a larger SBP response to low-sodium intervention (z = −3.089, P = 2.0 × 10−3, Q > 0.05) and high-sodium intervention (z = 3.314, P = 9.2 × 10−4, Q = 0.034) (Table 4). The 7 significant ECE1 SNPs were in the same LD block with 3 other SNPs. Because of the high LD between the SNPs, only 2 common haplotypes, A-A-T-T-C-A-A-A-T-T (35.8%) and G-G-C-C-T-C-C-G-C-C (58.7%), were observed within this 10-SNP block. As expected, the former haplotype, containing the minor alleles of the 7 significant SNPs, was associated with an increased DBP response to low-sodium intervention (z = −3.175, P = 1.5 × 10−3, Q = 0.028), and the latter one, containing the major alleles, was associated with a decreased DBP response (z = 3.575, P = 3.5 × 10−4, Q = 0.013) (Table 4). However, neither haplotype was more significant than the single marker finding of lead ECE1 SNP rs84853.
Table 4.
Summary of Haplotypes Significantly Associated With Blood Pressure Response to Sodium Interventions, GenSalt Study, China, 2003–2005
| Haplotype | Frequency | Response to Low Sodium |
Response to High Sodium |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Systolic Blood Pressure |
Diastolic Blood Pressure |
Mean Arterial Pressure |
Systolic Blood Pressure |
Diastolic Blood Pressure |
Mean Arterial Pressure |
||||||||||
| z Statistic | P Value | z Statistic | P Value | Q Valuea | z Statistic | P Value | z Statistic | P Value | Q Valuea | z Statistic | P Value | z Statistic | P Value | ||
| BDKRB2b | |||||||||||||||
| Block of rs8005195–rs11847625 | |||||||||||||||
| T-C-G-G-C | 0.21 | −3.089 | 2.0 × 10−3 | −0.55 | 0.58 | −1.759 | 0.08 | 3.314 | 9.2 × 10−4 | 0.034 | −0.262 | 0.79 | 0.946 | 0.34 | |
| ECE1b | |||||||||||||||
| Block of rs213011–rs213037 | |||||||||||||||
| G-G-C-C-T-C-C-G-C-C | 0.59 | −0.261 | 0.79 | 3.575 | 3.5 × 10−4 | 0.013 | 2.577 | 0.01 | 0.517 | 0.60 | −0.718 | 0.47 | −0.581 | 0.56 | |
| A-A-T-T-C-A-A-A-T-T | 0.36 | 0.394 | 0.69 | −3.175 | 1.5 × 10−3 | 0.02 | −2.273 | 0.02 | −0.409 | 0.68 | 0.707 | 0.48 | 0.645 | 0.52 | |
Abbreviations: GenSalt, Genetic Epidemiology Network of Salt Sensitivity.
a False discovery rate Q values that are less than 0.05 are provided.
b Genes symbols and their expanded names: BDKRB2, bradykinin receptor B2 gene; ECE1, endothelin converting enzyme 1 gene.
DISCUSSION
Using data from the largest dietary sodium feeding study to date, we identified multiple genetic variants of the KKS that contributed significantly to the salt sensitivity of BP. Specifically, several SNPs and haplotypes of the kinin receptor gene, BDKRB2, and the kinin degrading gene, ECE1, were significantly associated with BP responses to dietary sodium interventions. To our knowledge, this is one of only 2 reports examining genetic variants of KKS genes and salt sensitivity in humans (21). The novel findings reported here may help to enhance our knowledge of the role of the KKS in the development of hypertension.
Previous studies have indicated that salt sensitivity of BP could mediate the relation between KKS dysfunction and hypertension. Reports have documented that kinins are potent vasodilators and also promote diuresis and natriuresis (22). However, neither kininogen-deficient rats nor bradykinin receptor gene knockout mice showed elevated BP compared with their controls under normal sodium intake. On the contrary, when fed a high-sodium diet, both animal models exhibited higher BP levels than their corresponding controls (8, 9). High BP was accompanied by increased arteriolar sensitivity to vasopressor substances, such as angiotensin II, and sodium accumulation due to aldosterone release after salt loading (23). Despite the strong evidence of a mechanistic link between the KKS and salt sensitivity, only one variant of the KKS had been examined previously for association with BP response to sodium in humans, with Svetkey et al. (21) reporting a significant association between the nonsynonymous KLK1 Q121E (rs5516) variant and salt sensitivity. Although we didn't genotype rs5516 in the current study, genotype data were available for rs2659058, a variant highly correlated with rs5516 (r2 = 0.88 based on the HapMap data of the Chinese Han of Beijing). We did not observe a significant association between rs2659058 and salt sensitivity. Because of the inconsistent findings, future studies are warranted to clarify the association between KLK1 and salt sensitivity.
Although there is a paucity of data linking KKS genes to salt sensitivity, several genes of the KKS have been investigated for a potential relation with BP and hypertension. Genetic variants in KNG1 (24), KLK1 (25–27), KLKB1 (28), ECE1 (3, 29), BDKRB1 (30), and BDKRB2 (30–34) have been associated with BP levels or hypertension in numerous studies. Among them, 2 variants of BDKRB2, −58C/T and +9/−9 bp, have been studied extensively. However, their associations with hypertension are still unclear. A recent meta-analysis of the promoter polymorphism, −58C/T, concluded that the −58C allele showed a protective effect in African Americans and Asians but a risk effect in Caucasians (34). The BDKRB2 +9 polymorphism was linked to increased SBP in European Americans but not in African Americans in a paper by Pretorius et al. (32), whereas Freitas et al. (33) related the −9 bp allele with increased DBP in a Brazilian population. The +9/−9 bp variant was monomorphic in a Japanese population (35). These controversial findings may indicate that neither of the 2 polymorphisms is causally related to hypertension. On the other hand, it is also possible that failure to control environmental factors that interact with these variants leads to the inconsistency among studies. Although we did not include these 2 variants in our study, we did find that genetic variants in BDKRB2 were associated with salt sensitivity, interacting with sodium intake to influence BP.
We identified an intronic SNP of BDKRB2, rs11847625, and a haplotype including this SNP associated with SBP response to sodium interventions. Because this variant had not been reported previously, we used the National Institute of Environmental Health Sciences SNP Function Prediction Tool (http://snpinfo.niehs.nih.gov/snpfunc.htm) to explore its potential function. SNP rs11847625 is located at a transcription factor binding site of BDKRB2, suggesting that this variant could affect the level, location, or timing of gene expression. Specifically, this bioinformatics tool suggests that allelic variation at rs11847625 could result in differential activity at up to the 25-transcription factor binding site (36). However, experimental studies will be needed to confirm this inferred functional mechanism. Furthermore, the significant haplotype block appears to be in a regulatory region of the gene, containing at least one other variant (rs10132462) predicted to influence BDKRB2 transcription. Although we cannot directly infer that the variants reported here are causally related to salt sensitivity, our results highlight a promising region for future resequencing and functional studies.
The endothelin-converting enzyme is a multifunctional protease. It is not only the key enzyme in the production of the potent vasoconstrictor endothelin but also one of the main inactivating enzymes of the vasodilator bradykinin (13, 37). In previous studies, a genetic polymorphism located in the 5′-regulatory region of ECE1, C-338A (rs213045), was demonstrated to create a binding site for the E2F2 transcription factor and subsequently shown to influence promoter activity and BP levels (29, 38). Although we did not genotype this polymorphism, one of its proxies, rs212544 (r2 = 0.81), was included in our analyses. However, we did not observe any association between rs212544 and BP response to sodium interventions. In addition, an intronic SNP of ECE1, rs212528, was reported to associate with hypertension and BP levels exclusively in Japanese women (3). Again, we did not observe an association with BP response in our study. However, we identified 7 highly correlated intronic SNPs in an LD block of ECE1 associated with the DBP response to low-sodium intervention. Although none of these SNPs or their proxies (r2 > 0.80) had any predicted function, the strong association reported by us warrants further investigation to help clarify this novel relation.
The large and homogenous sample, excellent adherence of the GenSalt participants to the controlled feedings, and comprehensive analyses of common variants in the KKS highlight important strengths of the current study. Furthermore, we used rigorous quality control procedures to ensure high quality genotype and phenotype data, as well as stringent analytical methods that were immune to population structure and corrected for multiple testing. Despite these strengths, the contribution of low-frequency and rare variants in the KKS genes to salt sensitivity could have been missed by this study. Still, we provide 2 promising regions for future sequencing studies that will likely help to further elucidate the genomic architecture of salt sensitivity.
In summary, our study identified genetic variants in BDKRB2 and ECE1 strongly associated with BP response to dietary sodium intervention. These data provide some of the first empirical evidence linking genes encoding components of the KKS to salt sensitivity in human populations. Replication of these associations in independent samples, as well as sequencing and functional studies, will be necessary to follow up our research findings and to determine the true causal variants underlying the novel associations reported here.
ACKNOWLEDGMENTS
Author affiliations: Department of Evidence Based Medicine, Fuwai Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, and National Center for Cardiovascular Diseases, Beijing, China (Dongfeng Gu, Jie Cao, Jianxin Li, Jichun Chen); Department of Epidemiology, Tulane University School of Public Health and Tropical Medicine, New Orleans, Louisiana (Qi Zhao, Tanika N. Kelly, Jiang He); Human Genetics Center, University of Texas School of Public Health, Houston, Texas (James E. Hixson); Division of Biostatistics, Washington University School of Medicine, St Louis, Missouri (Dabeeru C. Rao); Department of Medicine, Tulane University School of Medicine, New Orleans, Louisiana (Jing Chen); Xinle Traditional Chinese Medicine Hospital, Hebei, China (Xu Ji); Division of Preventive Medicine, Shenzhen University School of Medicine, Guangdong, China (Dongsheng Hu); Ganyu Center for Disease Control and Prevention, Jiangsu, China (Xushan Wang); and Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China (De-Pei Liu).
The Genetic Epidemiology Network of Salt Sensitivity (GenSalt) is supported by research grants (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, Maryland.
GenSalt Study Steering Committee: Dongfeng Gu, Jiang He (Chair), James E. Hixson, Cashell E. Jaquish, Depei Liu, Dabeeru C. Rao, Paul K. Whelton, and Zhijian Yao. GenSalt Collaborative Research Group: Tulane University Health Sciences Center, New Orleans, Louisiana: Jiang He (Principal Investigator), Lydia A. Bazzano, Chung-Shiuan Chen, Jing Chen, Mei Hao, Lee Hamm, Tanika Kelly, Paul Muntner, Kristi Reynolds, Paul K. Whelton, Wenjie Yang, and Qi Zhao. Washington University School of Medicine, St Louis, Missouri: Dabeeru C. Rao (Principal Investigator), Matthew Brown, Charles Gu, Hongyan Huang, Treva Rice, Karen Schwander, and Shiping Wang. University of Texas Health Sciences Center at Houston, Houston, TX: James E. Hixson (Principal Investigator) and Lawrence C. Shimmin. National Heart, Lung, and Blood Institute, Bethesda, MD: Cashell E. Jaquish. Chinese Academy of Medical Sciences, Beijing, China: Dongfeng Gu (Principal Investigator), Jie Cao, Jichun Chen, Jingping Chen, Zhenhan Du, Jianfeng Huang, Hongwen Jiang, Jianxin Li, Xiaohua Liang, Depei Liu, Xiangfeng Lu, Donghua Liu, Qunxia Mao, Dongling Sun, Hongwei Wang, Qianqian Wang, Xigui Wu, Ying Yang, and Dahai Yu. Shandong Academy of Medical Sciences, Shandong, China: Fanghong Lu (Principal Investigator), Zhendong Liu, Shikuan Jin, Yingxin Zhao, Shangwen Sun, Shujian Wang, Qengjie Meng, Baojin Liu, Zhaodong Yang, and Chuanrui Wei. Shandong Center for Disease Control and Prevention, Shandong, China: Jixiang Ma (Principal Investigator), Jiyu Zhang, and Junli Tang. Zhengzhou University, Zhengzhou, Henan, China: Dongsheng Hu, Weidong Zhang, Hongwei Wen, Chongjian Wang, Minghui Shen, Jingjing Pan, and Liming Yang. Xinle Traditional Chinese Medicine Hospital, Hebei, China: Xu Ji (Principal Investigator), Rongyan Li, Haijun Zu, and Junwei Song. Ganyu Center for Disease Control and Prevention, Ganyu, Jiangsu, China: Delin Wu (Principal Investigator), Xushan Wang, and Xiaofeng Zhang. Xi'an Jiaotong University, Shanxi, China: Jianjun Mu (Principal Investigator), Enrang Chen, Fuqiang Liu, and Guanji Wu. Chinese National Human Genome Center, Beijing, China: Zhi-Jian Yao (Principal Investigator), Shufeng Chen, Dongfeng Gu, Hongfan Li, Laiyuan Wang, and Penghua Zhang.
Conflict of interest: none declared.
REFERENCES
- 1.Morimoto A, Uzu T, Fujii T, et al. Sodium sensitivity and cardiovascular events in patients with essential hypertension. Lancet. 1997;350(9093):1734–1737. doi: 10.1016/S0140-6736(97)05189-1. [DOI] [PubMed] [Google Scholar]
- 2.Weinberger MH, Fineberg NS, Fineberg SE, et al. Salt sensitivity, pulse pressure, and death in normal and hypertensive humans. Hypertension. 2001;37(2 part 2):429–432. doi: 10.1161/01.hyp.37.2.429. [DOI] [PubMed] [Google Scholar]
- 3.Banno M, Hanada H, Kamide K, et al. Association of genetic polymorphisms of endothelin-converting enzyme-1 gene with hypertension in a Japanese population and rare missense mutation in preproendothelin-1 in Japanese hypertensives. Hypertens Res. 2007;30(6):513–520. doi: 10.1291/hypres.30.513. [DOI] [PubMed] [Google Scholar]
- 4.Svetkey LP, McKeown SP, Wilson AF. Heritability of salt sensitivity in black Americans. Hypertension. 1996;28(5):854–858. doi: 10.1161/01.hyp.28.5.854. [DOI] [PubMed] [Google Scholar]
- 5.Sharma JN. Hypertension and the bradykinin system. Curr Hypertens Rep. 2009;11(3):178–181. doi: 10.1007/s11906-009-0032-7. [DOI] [PubMed] [Google Scholar]
- 6.Sharma JN. The kallikrein-kinin system: from mediator of inflammation to modulator of cardioprotection. Inflammopharmacology. 2005;12(5-6):591–596. doi: 10.1163/156856005774382760. [DOI] [PubMed] [Google Scholar]
- 7.Marcondes S, Antunes E. The plasma and tissue kininogen-kallikrein-kinin system: role in the cardiovascular system. Curr Med Chem Cardiovasc Hematol Agents. 2005;3(1):33–44. doi: 10.2174/1568016052773351. [DOI] [PubMed] [Google Scholar]
- 8.Majima M, Yoshida O, Mihara H, et al. High sensitivity to salt in kininogen-deficient brown Norway Katholiek rats. Hypertension. 1993;22(5):705–714. doi: 10.1161/01.hyp.22.5.705. [DOI] [PubMed] [Google Scholar]
- 9.Alfie ME, Yang XP, Hess F, et al. Salt-sensitive hypertension in bradykinin B2 receptor knockout mice. Biochem Biophys Res Commun. 1996;224(3):625–630. doi: 10.1006/bbrc.1996.1076. [DOI] [PubMed] [Google Scholar]
- 10.Bonner G, Thieven B, Rutten H, et al. Renal kallikrein is a determinant of salt sensitivity. J Hypertens Suppl. 1993;11(5):S210–S211. [PubMed] [Google Scholar]
- 11.Madeddu P, Varoni MV, Demontis MP, et al. Kallikrein-kinin system and blood pressure sensitivity to salt. Hypertension. 1997;29(1 pt 2):471–477. doi: 10.1161/01.hyp.29.1.471. [DOI] [PubMed] [Google Scholar]
- 12.Campbell DJ. The kallikrein-kinin system in humans. Clin Exp Pharmacol Physiol. 2001;28(12):1060–1065. doi: 10.1046/j.1440-1681.2001.03564.x. [DOI] [PubMed] [Google Scholar]
- 13.Kakoki M, Smithies O. The kallikrein-kinin system in health and in diseases of the kidney. Kidney Int. 2009;75(10):1019–1030. doi: 10.1038/ki.2008.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marceau F, Hess JF, Bachvarov DR. The B1 receptors for kinins. Pharmacol Rev. 1998;50(3):357–386. [PubMed] [Google Scholar]
- 15.GenSalt: rationale, design, methods, baseline characteristics of study participants. J Hum Hypertens. 2007;21(8):639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perloff D, Grim C, Flack J, et al. Human blood pressure determination by sphygmomanometry. Circulation. 1993;88(5):2460–2470. doi: 10.1161/01.cir.88.5.2460. [DOI] [PubMed] [Google Scholar]
- 17.Gu D, Kelly TN, Hixson JE, et al. Genetic variants in the renin-angiotensin-aldosterone system and salt sensitivity of blood pressure. J Hypertens. 2010;28(6):1210–1220. [PMC free article] [PubMed] [Google Scholar]
- 18.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 21.Svetkey LP, Harris EL, Martin E, et al. Modulation of the BP response to diet by genes in the renin-angiotensin system and the adrenergic nervous system. Am J Hypertens. 2011;24(2):209–217. doi: 10.1038/ajh.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma JN, Uma K, Noor AR, et al. Blood pressure regulation by the kallikrein-kinin system. Gen Pharmacol. 1996;27(1):55–63. doi: 10.1016/0306-3623(95)00028-3. [DOI] [PubMed] [Google Scholar]
- 23.Katori M, Majima M. A missing link between a high salt intake and blood pressure increase. J Pharmacol Sci. 2006;100(5):370–390. doi: 10.1254/jphs.crj06003x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao W, Wang Y, Wang L, et al. Gender-specific association between the kininogen 1 gene variants and essential hypertension in Chinese Han population. J Hypertens. 2009;27(3):484–490. doi: 10.1097/hjh.0b013e32831e19f9. [DOI] [PubMed] [Google Scholar]
- 25.Zhao W, Wang L, Lu X, et al. A coding polymorphism of the kallikrein 1 gene is associated with essential hypertension: a tagging SNP-based association study in a Chinese Han population. J Hypertens. 2007;25(9):1821–1827. doi: 10.1097/HJH.0b013e328244e119. [DOI] [PubMed] [Google Scholar]
- 26.Hua H, Zhou S, Liu Y, et al. Relationship between the regulatory region polymorphism of human tissue kallikrein gene and essential hypertension. J Hum Hypertens. 2005;19(9):715–721. doi: 10.1038/sj.jhh.1001875. [DOI] [PubMed] [Google Scholar]
- 27.Jiang S, Hsu YH, Venners SA, et al. Effects of protein coding polymorphisms in the kallikrein 1 gene on baseline blood pressure and antihypertensive response to irbesartan in Chinese hypertensive patients. J Hum Hypertens. 2011;25(5):327–333. doi: 10.1038/jhh.2010.70. [DOI] [PubMed] [Google Scholar]
- 28.Lu X, Zhao W, Huang J, et al. Common variation in KLKB1 and essential hypertension risk: tagging-SNP haplotype analysis in a case-control study. Hum Genet. 2007;121(3-4):327–335. doi: 10.1007/s00439-007-0340-4. [DOI] [PubMed] [Google Scholar]
- 29.Funalot B, Courbon D, Brousseau T, et al. Genes encoding endothelin-converting enzyme-1 and endothelin-1 interact to influence blood pressure in women: the EVA Study. J Hypertens. 2004;22(4):739–743. doi: 10.1097/00004872-200404000-00016. [DOI] [PubMed] [Google Scholar]
- 30.Cui J, Melista E, Chazaro I, et al. Sequence variation of bradykinin receptors B1 and B2 and association with hypertension. J Hypertens. 2005;23(1):55–62. doi: 10.1097/00004872-200501000-00013. [DOI] [PubMed] [Google Scholar]
- 31.Gainer JV, Brown NJ, Bachvarova M, et al. Altered frequency of a promoter polymorphism of the kinin B2 receptor gene in hypertensive African-Americans. Am J Hypertens. 2000;13(12):1268–1273. doi: 10.1016/s0895-7061(00)01215-2. [DOI] [PubMed] [Google Scholar]
- 32.Pretorius MM, Gainer JV, Van Guilder GP, et al. The bradykinin type 2 receptor BE1 polymorphism and ethnicity influence systolic blood pressure and vascular resistance. Clin Pharmacol Ther. 2008;83(1):122–129. doi: 10.1038/sj.clpt.6100250. [DOI] [PubMed] [Google Scholar]
- 33.Freitas SR, Pereira AC, Floriano MS, et al. Insertion/deletion polymorphism of the bradykinin type 2 receptor gene influence diastolic blood pressure. J Hum Hypertens. 2009;23(8):553–555. doi: 10.1038/jhh.2009.23. [DOI] [PubMed] [Google Scholar]
- 34.Niu W, Qi Y, Gao P, et al. A meta-analysis of the bradykinin B2 receptor gene −58C/T polymorphism with hypertension. Clin Chim Acta. 2010;411(5-6):324–328. doi: 10.1016/j.cca.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Fu Y, Katsuya T, Matsuo A, et al. Relationship of bradykinin B2 receptor gene polymorphism with essential hypertension and left ventricular hypertrophy. Hypertens Res. 2004;27(12):933–938. doi: 10.1291/hypres.27.933. [DOI] [PubMed] [Google Scholar]
- 36.Xu Z, Taylor JA. SNPinfo: integrating GWAS candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. (Web server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoang MV, Turner AJ. Novel activity of endothelin-converting enzyme: hydrolysis of bradykinin. Biochem J. 1997;327:23–26. doi: 10.1042/bj3270023. (pt 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Funke-Kaiser H, Reichenberger F, Kopke K, et al. Differential binding of transcription factor E2F-2 to the endothelin-converting enzyme-1b promoter affects blood pressure regulation. Hum Mol Genet. 2003;12(4):423–433. doi: 10.1093/hmg/ddg040. [DOI] [PubMed] [Google Scholar]