Abstract
Tetrahydrofolate cofactors are required for one carbon transfer reactions involved in the synthesis of purines, amino acids, and thymidine. Inhibition of tetrahydrofolate biosynthesis is a powerful therapeutic strategy in the treatment of several diseases, and the possibility of using antifolates to inhibit enzymes from Mycobacterium tuberculosis has been explored. This work focuses on the study of the first enzyme in tetrahydrofolate biosynthesis that is unique to bacteria, dihydroneopterin aldolase (MtDHNA). This enzyme requires no metals or cofactors, and does not form a protein mediated Schiff base with the substrate, unlike most aldolases. Here, we were able to demonstrate that the reaction catalyzed by MtDHNA generates three different pterin products, one of which is not produced by other wild type DHNAs. The enzyme-substrate complex partitions 51% in the first turnover to form the aldolase product, 24% to the epimerase product, and 25% to the oxygenase product. The aldolase reaction is strongly pH-dependent, and apparent pKa values were obtained for the first time for this class of enzyme. Furthermore, chemistry is rate-limiting for the aldolase reaction, and the analysis of solvent kinetic isotope effects in steady-state and pre-steady-state conditions, combined with proton inventory studies revealed that two protons and a likely solvent contribution are involved in formation and breakage of a common intermediate. This study provides information about the plasticity required from a catalyst that possesses high substrate specificity while being capable of utilizing two distinct epimers with the same efficiency to generate five distinct products.
Keywords: Folate biosynthesis; Aldehyde-Lyases; 7,8-dihydroneopterin; Tuberculosis; Kinetics; Solvent Kinetic Isotope Effects; Nuclear Magnetic Resonance; Oxygen chemistry; Oxygenases
INTRODUCTION
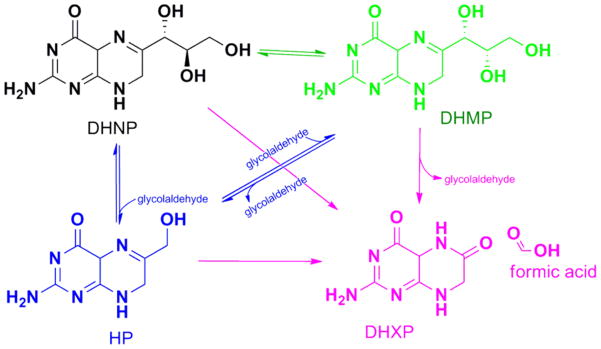
Dihydroneopterin aldolase (DHNA) catalyzes the conversion of 7,8-dihydroneopterin (DHNP), to 6-hydroxymethyl-7,8-dihydropterin (HP) in the biosynthesis of folate, a pathway currently targeted for the treatment of several distinct types of cancers and infections.1 It has been proposed that DHNA represents a promising new target for the development of antibacterials, underlying the importance of studying the catalytic and chemical mechanism of this enzyme in pathogenic bacteria such as Mycobacterium tuberculosis, the causative agent of tuberculosis.2,3 DHNAs are interesting enzymes since they catalyze the cleavage of a carbon-carbon bond without the requirement of metals or cofactors, being considered a unique type of aldolase in which a substrate imbedded imine assists in catalysis. In addition to their function as an aldolase, DHNAs catalyze an epimerization reaction in which the configuration of the C2′ carbon-oxygen bond is inverted, forming 7,8-dihydromonapterin (DHMP, Scheme 1).
Scheme 1.
Reactions catalyzed by MtDHNA
DHNAs from Escherichia coli and Staphylococcus aureus (EcDHNA and SaDHNA, respectively) have been characterized, and the reactions catalyzed by these enzymes were shown to possess distinct rate limiting steps and kinetic parameters. Additionally, mutational studies revealed that acid-base catalysis is probably crucial for the reaction, but pKa values for group(s) possibly involved in chemistry are lacking.
Here we report the kinetic characterization of the reactions catalyzed by the DHNA from M. tuberculosis (MtDHNA) using pH studies, solvent kinetic isotope effects, NMR, steady-state and pre-steady-state methods, and based on our findings conclude that MtDHNA catalyzes three distinct reactions with comparable proficiency. This is the first report of a wild type DHNA capable of catalyzing a cofactor-independent oxygenase reaction with significant product formation.
MATERIALS AND METHODS
Materials
All chemicals were of analytical or reagent grade and were used without further purification. All pterins were purchased from Schirck laboratories (Jona, Switzerland). Deuterium oxide (99.9 at. % D) was from Cambridge Isotope Laboratories.
Cloning of M. tuberculosis DHNA into plasmid pET28a(+)
The folB gene (Rv3607c) encoding DHNA from M. tuberculosis was amplified by PCR to generate a PCR product with NdeI and HindIII restriction sites at the ends. The oligonucleotide primers were 5′-GGAATTCCATATGGCTGACCGAATCGAAC-3′ and 5′-CCCAAGCTTTCATACCGCGCCGCCCGCCG-3′. The amplified DNA product was digested with NdeI and HindIII and the purified insert was ligated into pET28a(+) previously linearized with the same restriction enzymes. This recombinant plasmid was transformed into DH5α cells following the instructions supplied by the manufacturer (Invitrogen). Cloning into these restriction sites adds a thrombin-cleavable His6-tag at the N-terminus of the recombinant protein.
Purification of MtDHNA
Competent E. coli BL21 (DE3) cells (Novagen) were transformed with the recombinant plasmid pET28a(+):folB, and MtDHNA was expressed and purified as previously described.4 Protein concentration was determined by using the theoretical extinction coefficient per monomer, accounting for the addition of the sequence MGSSHHHHHHSSGLVPRGSH at the N-terminus of the protein (ε280 = 15,470 M−1cm−1). The Ni-NTA affinity purified enzyme was dialyzed against 100 mM HEPES containing 50 mM KCl, pH 7.5, and frozen at -80° C. Protein to be used for sedimentation equilibrium, equilibrium binding, single turnover and burst experiments was further purified using a gel filtration column (Superdex 200, GE) to make sure that no DHXP was bound to the protein after the purification. The apo-MtDHNA generated by this method was less stable and more prone to precipitation at μM concentrations than the DHXP-bound enzyme.
Mass-Spectrometry
Purified proteins had their molecular mass determined by LC-ESI-MS. Detailed methods can be found in the supporting information.
Ultracentrifugation
Sedimentation equilibrium experiments were conducted at 8,000 and 12,000 rpm using six channel centerpieces using an Optima XL-I analytical centrifuge (Beckman). Apo-MtDHNA (25 μM) alone or with equimolar concentrations of neopterin or DHXP were analyzed. The identity of scans taken at 22 and 24 h confirmed that the samples had reached equilibrium. The data was globally analyzed using HeteroAnalysis v1.1.33 (J. L. Cole and J. W. Lary, Analytical Ultracentrifugation Facility, Biotechnology Services Center, University of Connecticut, Storrs, CT 06269). The best-fit values and joint confidence intervals are reported.
Equilibrium dissociation constant
DHXP binding to MtDHNA was measured by fluorescence titration at 25°C using a Horiba Fluoromax 3 spectrofluorometer (Horiba Jobin Yvon). Excitation and emission wavelengths were 295 and 345 nm, respectively. Titrations were conducted by serial additions of 1 μL aliquots of ligand to a 1 cm2 quartz cuvette that contained 2 mL of 2 μM of MtDHNA in 100 mM HEPES with 50 mM NaCl pH 7.0, 1 mM EDTA, 5 mM DTT, and 1% BSA. Fluorescence readings were recorded 2 min after the addition of ligand to allow temperature equilibration, and the dissociation constants were estimated by nonlinear regression using eq. 1.5 A complete description of the equations fitted is available in the supporting information. The concentration of DHXP did not exceed 5 μM to avoid inner filter effects.
| (1) |
Where
a = Kd1 + Kd2 + S1t + S2t − Lt
b = Kd2*S1t + Kd1* S2t + Kd1*Kd2
c = − Kd1*Kd2*Lt
For eq.1, Kd1 and Kd2 are the equilibrium binding constants for sites 1 and 2, respectively, S1t and S2t are the total concentrations of sites 1 and 2, Lt is the total ligand (DHXP) concentration, Lb is the concentration of the MtDHNA-DHXP complex, and y0 is the initial y value.
Fluorescence steady-state assays
The aldolase reaction was measured as previously described,6 Briefly, HP formation was monitored with an excitation wavelength of 430 nm, followed by an increase in fluorescence emission at 535 nm due to HP formation. The slits were 2 nm and 10 nm for excitation and emission, respectively. The reaction mixture was kept at 25° for 2 minutes before adding enzyme to assure that the temperature was equilibrated. After the enzyme was added, there was a lag period of approximately 30 seconds when DHNP was used as substrate, which was excluded from the fitting to obtain initial rates of reaction. Standard curves for HP fluorescence were measured at all pH values and buffers utilized, and a strong pH-dependence of HP fluorescence was observed. Fluorescence emission at 535 nm was plotted as a function of HP concentration, and the slope of this plot was used as a scaling factor to obtain kcat values for the aldolase reaction. In an attempt to stabilize the enzyme for longer periods of time, 1% BSA was present in all assays and distinct monovalent and divalent metals were tested, demonstrating no influence on enzyme activity, and thus were excluded from all subsequent assays. Due to the tight binding to DHNP and DHMP and low turnover number, the concentration of MtDHNA was usually between 100 – 500 nM, comparable to substrate concentrations. Therefore, eq. 2 was used to fit initial velocity data, where Vmax is the maximum velocity, E0 is the initial total enzyme concentration, ST is the initial total substrate concentration, KM is the Michaelis constants for the substrate used.
| (2) |
NMR-based assays
Formation of products in the MtDHNA-catalyzed reaction was followed by 1H NMR. Reactions were performed in 100 mM phosphate, 50 mM NaCl, 1 mM EDTA, 5 mM DTT, 1% BSA, pH 7.0, made containing 90% H2O and 10% D2O with 8 mM 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) as internal standard. Spectra were collected before enzyme addition, and a control reaction with no MtDHNA was kept at room temperature for 1 hour, after which NMR spectra were collected to account for DHNP degradation. NMR spectra were analyzed using MestReNova, using changes in integration values relative to DSS over 1 hour. Reactions performed to obtain rates of substrate consumption/product formation had 1 mM DHNP and were initiated with the addition of 3 μM MtDHNA. Reactions to verify complete conversion of DHNP into products had 200 μM DHNP and 10 μM MtDHNA. All spectra were collected in a 14.1 Tesla magnet Bruker DRX600, using a sequence with water suppression. Standard spectra for DHNP, DHMP, DHXP, HP, and 6-formyl-7,8-dihydropterin (F-DHP) were obtained individually.
HPLC detection of products
Formation of products in the reverse direction was verified by mixing 200 μM HP and 200 mM glycolaldehyde with 5, 10, 15, or 20 μM MtDHNA. The reaction was stopped after 10 min or 1 hour by adding 1 N HCl, followed by neutralization with 1 N NaOH, and HPLC analysis. For the formation of products in the forward direction, 220 μM of DHNP or DHMP were mixed with 5, 10, 15, or 20 μM MtDHNA, and reaction was stopped after 10 min or 1 hour by adding 1 N HCl, followed by neutralization with 1 N NaOH, and HPLC analysis. For all HPLC separations, samples were loaded onto a C18 Gemini column (Phenomenex) coupled to a 1100 series HPLC (Agilent), where the mobile phase was 1% Methanol and 0.1% Acetic acid pH 3.3 (adjusted with NaOH), the flow was 1 mL min−1, and detection was at 330 nm.
Stopped-flow fluorescence assays
Pre-steady-state experiments were conducted on an Applied Photophysics model SX20 stopped-flow spectrofluorometer equipped with a Xenon lamp, which has a dead-time of 3 ms, a 1 cm path length for absorbance measurements and a 2 mm path length for fluorescence measurements. All experiments were performed at 25 °C in 100 mM HEPES, 50 mM NaCl, 1 mM EDTA, 5 mM DTT, and 1% BSA pH 7.0 in water or 90% deuterated water. For the 90% deuterated water experiments, buffers and substrates were prepared in D2O. For single turnovers in the forward direction using DHNP, 5 and 10 μM MtDHNA were mixed with 1 μM DHNP. For the measurement of solvent kinetic isotope effects under single-turnover conditions, 2, 3, 4, and 5 μM MtDHNA were mixed with 0.5 μM of DHMP in H2O or 90% D2O.
For the reverse reaction, 2 μM HP and 5, 11, 23, 34, or 69 μM MtDHNA were mixed. Typically, each observed rate constant was obtained after averaging 3–5 shots, and fitting the average to the appropriate exponential equation. The reported concentrations are final, after volumes of 60 μL were mixed from each syringe. Excitation was at 430 nm, and fluorescence emission above 495 nm was detected. Slits were 1 and 2 mm for excitation and emission, respectively.
Stopped-flow traces were fitted analytically using Microsoft Origin version 7, and the first 3 ms of data were discarded when fitting the data. Inspection of residuals provided an assessment of the quality of the fits. The fitting functions had the general form of eq. 3, where y(t) is the observed signal at time t, i is the number of transients, Ai is the amplitude of the ith transient, ki is the observed rate constant for the ith transient, and C is the offset.
| (3) |
Quench-flow assays
Burst experiments were conducted at 25 °C in a KinTek rapid quench apparatus (model RQF-3) equipped with a constant temperature circulating water bath. For this assay, 33 μM MtDHNA in 100 mM HEPES, 50 mM NaCl, 1 mM EDTA, 5 mM DTT, and 1% BSA, pH 7.0 were mixed with 1 mM DHNP in the same buffer. The reaction was quenched with 1 N HCl, rapidly neutralized with 1 N NaOH, flash frozen to avoid product degradation, and kept at -80 °C until HPLC analysis. A negative control reaction without MtDHNA addition was also collected. Samples were analyzed by HPLC using a C18 Gemini column (Phenomenex) and the protocol described above. Standard curves were collected at various concentrations of DHNP, DHMP, DHXP, and HP to obtain integration calibration values. Quenched reactions were thawed immediately before HPLC injection. The curves obtained were fitted to eq. 4, which is identical to eq. 3 but includes a linear phase, whose rate is represented by v.
| (4) |
Oxygen Consumption Assay
Oxygen consumption was measured using a Eutech ECDO6HANDY containing a galvanic dissolved oxygen probe, calibrated with 2 M sodium sulfite. Samples were 5mL containing 1mM DHNP, in 100 mM HEPES, pH 7.0 with 50 mM NaCl, 1mM EDTA, 5mM DTT, 1% BSA at 25 °C. A control for DHNP degradation was collected before adding enzyme, and the slope of this line was subtracted from the slope obtained in the enzyme-catalyzed reaction to calculate the turnover number. Reactions were started by adding 3 μM MtDHNA.
Coupled assay with formate dehydrogenase
To corroborate that formic acid was formed as one of the products of the oxygenase reaction, 100 μM HP was used as substrate for MtDHNA, and 10 μM formate dehydrogenase from Candida boidinii with 1 mM NAD+ were added to the reaction mixture. The total amount of NADH formed was calculated based on the total change in absorbance at 340 nm using ε340 = 6,220 M−1 cm−1. HPLC assays using methodology described above were used to demonstrate that in the presence of MtDHNA and HP, DHXP was the only product formed.
pH-Rate Profiles
To investigate the role of acid-base catalysis in the MtDHNA-catalyzed aldolase reaction, the pH dependence of the kinetic parameters was determined by measuring initial rates at varying concentrations of DHNP or DHMP. The experiments were conducted throughout the pH range of 4.5–9.5, using the following buffers in a final concentration of 100 mM with 50 mM NaCl, 1mM EDTA, 5 mM DTT, and 1% BSA: citrate (4.5–5.5), MES (5.5–6.5), HEPES (6.5–8.5), and TAPS (8.5–9.5). The pH stability of the enzyme was tested by incubating the enzyme in the desired buffer for 5 minutes and conducting the standard assay using HEPES (pH 7.0). pH-rate profiles with two ionizable groups in the acidic limb and one in the basic limb were fitted to eq. 5, where y is the kinetic parameter, C is the pH-independent value of y, H is the proton concentration, Ka is the apparent acid dissociation constant, and Kb is the apparent basic dissociation constant for ionizing groups.
| (5) |
Steady-state solvent kinetic isotope effects for the aldolase reaction
Solvent kinetic isotope effects (SKIEs) were measured with the fluorescence assay described above in either H2O or 93 % D2O, and initial velocities with DHNP or DHMP as substrates were obtained. The reaction was carried out in 100 mM HEPES, 50 mM NaCl, 1 mM EDTA, 5 mM DTT, and 1% BSA, pH 7.0, a plateau region in the pH-rate profiles to assure that no pH effects were influencing the SKIEs. Proton inventory studies using DHMP as substrate were conducted at 10 % increments of D2O. Data for the proton inventory on kcat/KDHMP were fitted to eq. 6, while data for the kcat proton inventories were fitted to eq. 7. For eq. 6 and 7, kn is the kcat or kcat/KM at each atom % D2O, k0 is the kcat or kcat/KM with H2O, n is the atom % D2O, φ1 and φ2 represent transition state fractionation factors, and φsolvent represents a fractionation factor arising from solvent contributions.
| (6) |
| (7) |
Global data fitting
Fluorescence stopped-flow data with more than one transient phase, and experiments with concentration series, were fit globally by regression analysis based upon numerical integration of the rate-equations using KinTek Global Kinetic Explorer version 2.2 (KinTek Corp. Austin, TX). For different ligand concentrations, scaling factors were included in the data fitting process, and estimates for errors on parameters were derived by nonlinear regression and by FitSpace confidence contour analysis.7,8
RESULTS AND DISCUSSION
General properties of the MtDHNA
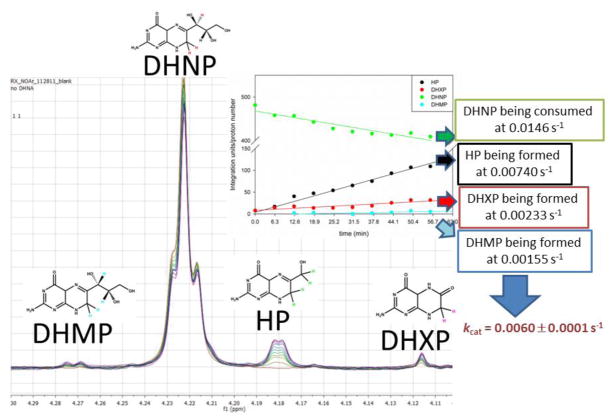
Purified MtDHNA in the DHXP-bound form and apo-MtDHNA were analyzed by mass spectrometry and sedimentation equilibrium experiments. A total monomeric mass of 16,585 Da was observed by ESI-MS, consistent with the loss of the N-terminal methionine (−131 Da from the predicted molecular weight of 16,716 per monomer Figure S1). The oligomeric state of purified proteins was determined by sedimentation equilibrium experiments, showing that apo-MtDHNA has a mass consistent with a tetramer (62,299 ± 25,715 Da), while the MtDHNA-NP and MtDHNA-DHXP forms possess the expected mass for octamers (117,853 ± 9,774 and 127,525 ± 4,105 respectively). Figure S2 shows the data and fits of sedimentation equilibrium experiments. Being both an aldolase and epimerase, DHNA can utilize both DHNP and DHMP as substrates to generate HP. This was confirmed by initial velocity fluorescence assays monitoring HP formation. Similar KM values were obtained with both epimers (KDHMP = 150 ± 30 nM, KDHNP = 170 ± 30 nM, Table 1, Figure S3), and the same turnover number was observed (kcat = 0.0060 ± 0.0001 s−1). This low kcat value could not be augmented by the addition of monovalent or divalent metals. Despite its low kcat value, the tight binding interaction with dihydropterin substrates gives MtDHNA a higher kcat/KM value than other DHNAs.9 Importantly, the kcat/KM value obtained for MtDHNA falls within the average of catalytic efficiencies obtained for intermediate metabolism enzymes.10 It is also important to note that there is a lag of HP formation when DHNP is the substrate, but the lag is absent when DHMP is the substrate. Further information on measured rates can be found in Table S1.
Table 1.
Comparison between kinetic parameters between different reactions and DHNAs from M. tuberculosis, S. aureus, and E. coli.
| substrate | kcat (s−1) | KM (μM) | kcat/KM (M−1s−1) | |
|---|---|---|---|---|
| MtDHNA-aldolase | DHNP | 0.0054 ± 0.0002 | 0.165 ± 0.026 | 3.6 × 104 ± 5.5 × 103 |
| DHMP | 0.0060 ± 0.0002 | 0.154 ± 0.027 | 3.8 × 104 ± 6.9 × 103 | |
| MtDHNA-epimerase | DHNP | 0.0015 ± 0.0003a | 0.154 ± 0.027 | 9.8 × 103 ± 2.5 × 103 |
| MtDHNA-oxygenase | DHNP | 0.0011± 0.0002a | 0.154 ± 0.027 | 7.3 × 103 ± 1.8 × 103 |
| EcDHNAb | DHNP | 0.045 | 4.6 | 9.8 × 103 |
| DHMP | 0.01 | 5.5 | 1.8 × 103 | |
| SaDHNAb | DHNP | 0.082 | 7.4 | 1.1 × 104 |
| DHMP | 0.089 | 8.0 | 1.1 × 104 |
calculated from linear phase of burst experiments
from 9.
Identification of Products by HPLC
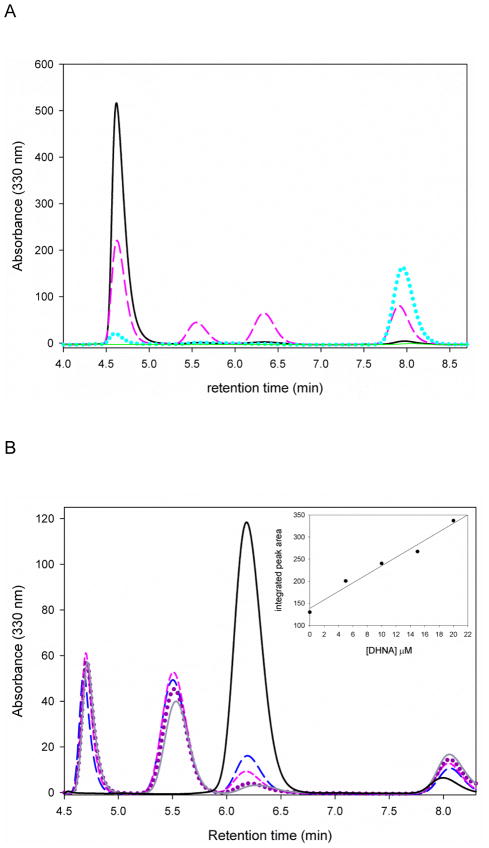
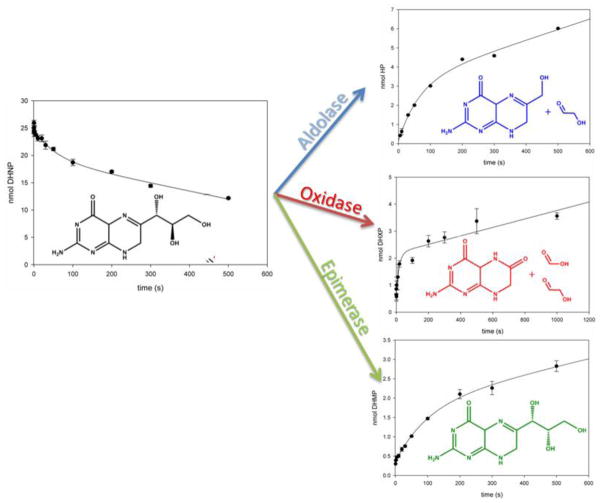
In order to test whether MtDHNA was capable of catalyzing both the epimerase and the aldolase reactions in a reversible manner, reactions were started by adding DHNP, DHMP, or glycolaldehyde and HP. As expected, MtDHNA catalyzes both reactions reversibly, and the reverse reaction has both DHNP and DHMP as products. Surprisingly, a peak with retention time at 8 minutes, not corresponding to DHNP, DHMP, or HP was observed in significant amounts, and its formation was directly proportional to the concentration of MtDHNA in the reverse reaction. When the forward reaction is started with either DHNP or DHMP, after one hour, only this unknown compound was observed as reaction product (Figure 1). This was not caused by degradation of any of the reactants or by contaminations in the enzyme preparation. In a recent work conducted with a tyrosine to phenylalanine mutant of Staphylococcus aureus and Escherichia coli DHNAs (SaDHNA and EcDHNA, respectively), the wild type aldolase/epimerase enzyme was converted to an oxygenase whose main product was 7,8-dihydroxanthopterin (DHXP), and smaller amounts of 6- formyl-7,8-dihydropterin (F-DHP).11 Both DHXP and F-DHP were used as HPLC standards, and DHXP had a retention time of 8 min under the same conditions. Additionally, DHXP production was confirmed by LC-MS/MS (Data not shown).
Figure 1.
HPLC-based assays showing the forward reaction with DHNP (A), and the reverse aldolase reaction with HP and GA. (B). The absorbance of all dihydropterins was observed at 330 nm. (A) Reaction with 10 μM MtDHNA, 220 μM DHNP in 100 mM HEPES pH 7.0 with 50 mM NaCl, 1mM EDTA, 5 mM DTT. (—, black) curve before adding MtDHNA, (---, pink) after 10 min of reaction, (…, blue) after 1 hour of reaction. (B) Reaction with 10 μM MtDHNA, 220 μM HP, 50 mM GA in 100 mM HEPES pH 7.0 with 50 mM NaCl, 1mM EDTA, 5 mM DTT. (—, black) curve before adding MtDHNA, (– –, blue) after 10 min of reaction with 5 μM MtDHNA, (---, pink) after 10 minutes of reaction with 10 μM MtDHNA, (…, purple) after 10 min of reaction with 15 μM MtDHNA, (—, gray) after 10 min of reaction with 20 μM MtDHNA. The inset represents the replot of the area under the peak with retention time equal to 8.1 min, increasing linearly with the increase in protfein concentration. Retention times, in minutes, are 4.7 (DHNP), 5.6 (DHMP), 6.4 (HP), and 8.1 (DHXP).
Identification of Products by NMR
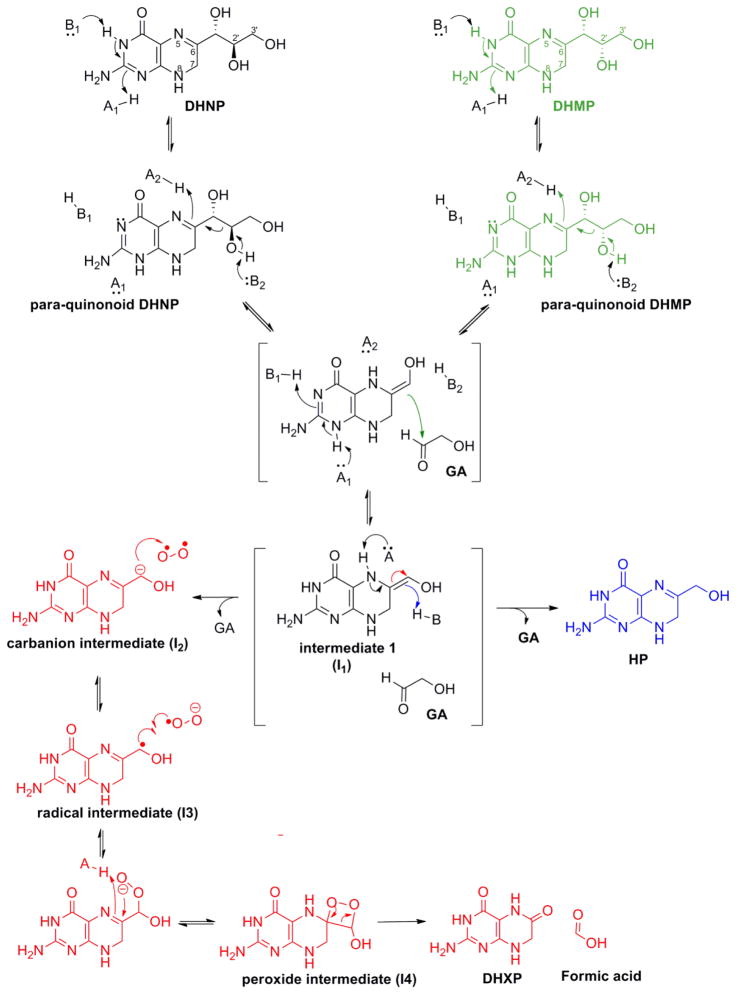
To further confirm that MtDHNA can produce DHXP as a reaction product, the reaction was monitored by NMR-based assays. Commercial standards of DHNP, DHMP, HP, DHXP, and F-DHP were used for comparison. Conveniently, in the region between 3.9–4.3 ppm unique peaks corresponding to each pterin can be observed, allowing the direct observation of each product formed/substrate consumed. Figure 2 shows the time dependence of DHNP consumption, resulting in HP, DHMP, and DHXP formation, and that no detectable FDHP was observed. Conversely, the reverse reaction with glycolaldehyde and HP formed DHNP, DHMP, and DHXP as products (Figure S4). Steady-state rates were obtained for DHNP consumption = 0.015 s−1, HP formation = 0.007 s−1, DHXP formation = 0.002 s−1, and DHMP formation = 0.001 s−1. Importantly, the sum of the steady-state rates of formation of DHMP, HP, and DHXP is comparable to the rate of DHNP depletion. For the reverse reaction, DHMP formation = 0.003 s−1, DHNP formation = 0.013 s−1, and HP consumption= 0.025 s−1, and the rate of DHXP formation could not be determined possibly due to the high concentrations of glycolaldehyde required. Despite faster rates obtained for the reverse reaction as compared to forward reaction, it is important to note that the Kd value for HP is higher than 100 μM, and could not be experimentally determined due to inner filter effects, and that the KM value for glycolaldehyde was shown to be in the mM range in DHNAs from other organisms.12 Finally, NMR-based assays confirmed that when the forward reaction is driven to completion the only product present was DHXP. The current chemical mechanism proposed for mutant DHNAs that form DHXP demonstrates that in order to form DHXP, glycolaldehyde and formic acid will be the other products (Scheme 2). Similar reactions catalyzed by other cofactor and metal independent oxygenases have been characterized, and are believed to be irreversible.13 Thus, despite the fact that HP formation over DHXP is kinetically favored, irreversible formation of DHXP would slowly result in its accumulation so that DHXP would build up until it is the only product present in the reaction mixture.
Figure 2.
NMR-based assays showing the forward reaction with 1 mM DHNP, and 3 μM MtDHNA in 100 mM sodium phosphate pH 7.0 with 5 mM DTT and 1% BSA. The area under the standard DSS was used as reference for the integration of peaks using MestReNova (MestreLab Research S.L.). Values for the integration of each peak corrected for the number of protons of each peak (shown in distinct colors for each molecule) as a function of time are shown in the inset. A negative control without enzyme was kept at 25°C for 1 hour and this spectrum was subtracted from the ones shown above.
Scheme 2.
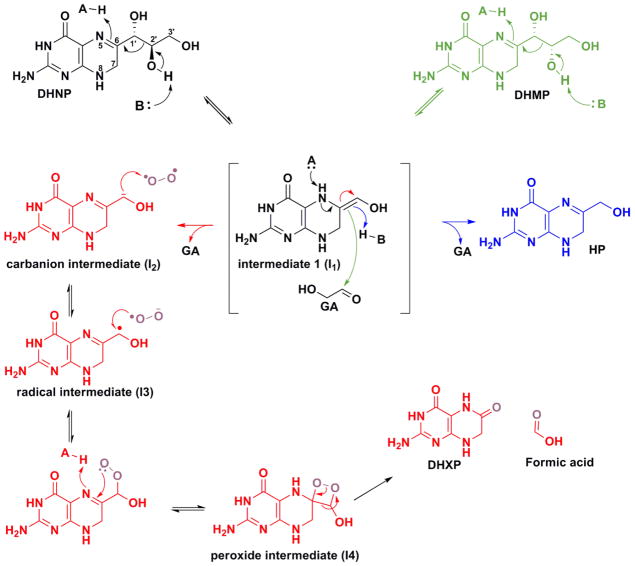
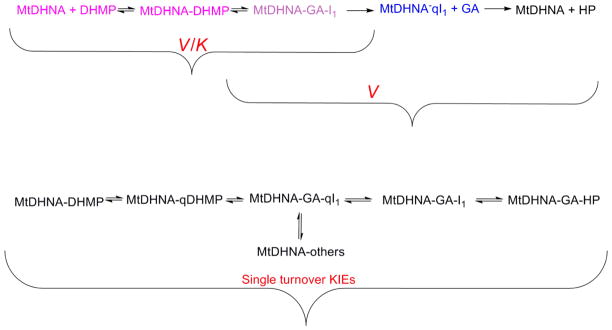
Proposed chemical mechanism for aldolase, epimerase and oxygenase. Intermediate 1 (I1) has three possible fates, represented by green, blue and red arrow. I1 partitions to form the epimerase (green arrow, attack on GA’s carbonyl forms DHMP), aldolase (blue arrow, abstraction of a proton from conjugated acid forms HP) or oxygenase (red arrow, formation of a carbanion intermediate which reacts with oxygen generates DHXP) reaction products11.
DHXP binding to MtDHNA
To further investigate the interaction between MtDHNA and DHXP, equilibrium binding experiments were performed, demonstrating that DHXP binds tightly to MtDHNA. Purified MtDHNA has an absorption maxima at 310 nm, indicating that the enzyme co-purified with bound DHXP. DHXP could be removed by gel filtration, but the apo-protein was unstable and prone to precipitation. The apo-protein was utilized for equilibrium binding experiments. Attempts to fit the data to a one site binding quadratic equation yielded a fitted curve that diverges considerably from experimental data, and better fits were obtained when using a hyperbolic equation for a two site binding interaction. Knowing that an equation accounting for ligand depletion was necessary since the concentrations of free DHXP was not equal to the total concentration of DHXP in the assay, an equation proposed by Wang et. al., which considers two binding sites and ligand depletion was used (eq. 1).5,14 Approximate equilibrium dissociation constants were obtained for the two sites, being Kd1 = 0.07 ± 0.07 μM and Kd2 = 6 ± 4 μM. Despite the high standard errors, this is the most realistic model to be fitted. It is possible that, because protein fluorescence was monitored, the contribution for fluorescence emission for the initial binding site is not the same as the one for the subsequent binding site, a situation not anticipated using this equation. Nevertheless, this fitting allowed an estimate of the affinity of DHXP binding, and the values of Kd1 and Kd2 indicate negative cooperativity (Figure S5). The advantages of negative cooperativity have been extensively discussed for several enzymatic reactions, and calculations suggest that enzymes that exhibit such behavior respond less sensitively to broader ranges of metabolite concentrations.15 Also, it was previously shown that DHNP was an activator of DHNA, inducing octamer formation.16 It might be advantageous for this particular enzyme to be regulated by distinct mechanisms depending on which metabolite is present, since DHNP is a potent antioxidant present at μM concentrations in sites of inflammation and free-radical damage.17–19
Pre-steady-state burst
To verify whether the MtDHNA-DHNP complex is capable of partitioning to generate the aldolase, epimerase and oxygenase products in the first turnover, burst experiments were performed using a quench-flow apparatus. Figure 4 shows that in the first turnover, DHNP is consumed at 0.018 s−1, to form HP (0.037 s−1), DHMP (0.009 s−1), and DHXP (0.065 s−1). Analysis of the burst amplitude reveals that 51% of the MtDHNA-DHNP complex is turned over to generate HP, 24% generates DHMP, and the other 25% produces DHXP. The rates for the linear phases of each of the reactions correlates well with the steady-state rates measured by NMR-based assays, demonstrating that in spite of the small kcat values for all three reactions, each can be independently and reproducibly determined. The oxygenase reaction has the fastest burst rate, but production of DHXP generates a stable octameric MtDHNA-DHXP complex (discussed above) that represents an inactive complex. In order to proceed to the next productive turnover, DHNP or DHMP need to compete for the pterin binding site, displacing DHXP. It is important to point out that in other DHNAs previously characterized, the aldolase reaction is by far the most significant to be catalyzed, and even though EcDHNA shows significant epimerase activity while SaDHNA shows almost none, neither of these wild type enzymes produced DHXP in a significant manner (less than 2% DHXP produced in comparison to the epimerase and aldolase products).12
Figure 4.
Multiple turnover showing bursts of substrate consumption and in the formation of the three dihydropterin products. 33 μM MtDHNA was mixed with 1 mM DHNP in 100 mM HEPES, pH 7.0, with 50 mM NaCl, 1mM EDTA, 5 mM DTT, 1% BSA, and the reaction was quenched at different time points with 1N HCl, neutralized with 1N NaOH and flash-frozen for posterior HPLC analysis. Standard curves with DHNP, DHMP, HP, and DHXP were used as standards for concentration. The values obtained were: DHNA consumption kburst-DHNP = 0.018 ± 0.006 s−1 and kcat-DHNP = 0.011 ± 0.008 s− 1, HP formation kburst-HP = 0.037 ± 0.031 s−1 and kcat-HP = 0.009 ± 0.001 s−1, DHMP formation kburst-DHMP = 0.010 ± 0.003 s−1, kcat-DHMP = 0.0015 ± 0.0006 s−1, and DHXP formation at kburst-DHXP = 0.07 ± 0.03 s−1, kcat-DHXP = 0.0011 ± 0.0002 s−1. Based on the burst amplitudes (total product formation = 6.0 nmol), the partition to form each product can be calculated, so that 51% of the MtDHNA-DHNP partitions to form HP, 24% to form DHMP, and 25% to form DHXP.
Oxygen Consumption Assay
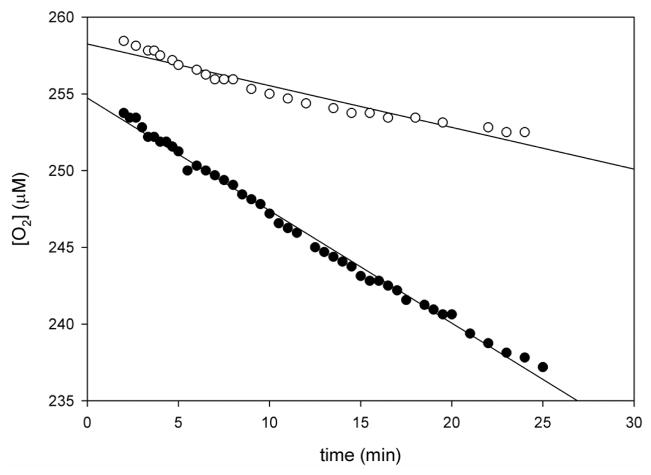
To test whether the oxygen source for the oxygenase reaction generating DHXP was dissolved oxygen, an oxygen consumption assay was conducted. Figure 5 compares the decrease in dissolved oxygen concentration before and after adding MtDHNA. Stability studies with reduced pterins demonstrated that dihydro and tetrahydro pterins slowly react with dissolved oxygen to generate oxidized pterins, and that DHXP is the major oxidation product of DHNP, 7,8- dihydrobiopterin, and 7,8-dihydrofolic acid.20 This spontaneous reaction is very slow (t1/2 = 60 hours), and significantly slower than the MtDHNA-catalyzed oxidation. Importantly, the corrected rate of oxygen consumption (0.0025 s−1) is equal, within experimental error, to the rate of DHXP formation determined by NMR-based assays (0.0023 s−1), further indicating that in order to produce DHXP dissolved oxygen is consumed. Both mutant SaDHNA and EcDHNA that were converted in oxygenases upon a tyrosine to phenylalanine substitution (Y54F and Y53F, respectively) consumed dissolved oxygen to convert DHNP into DHXP.11
Figure 5.
Oxygen consumption assay. The reaction was conducted with 1 mM DHNP in 100 mM HEPES, pH 7.0, with 50 mM NaCl, 1mM EDTA, 5 mM DTT, and 1% BSA. (○) Reaction mixture without enzyme, (●) reaction mixture after the addition of 3 μM MtDHNA. The slope of the curve without enzyme was subtracted from the one were MtDHNA was added, yielding a kcat of 0.0025 s−1.
Formation of formic acid
Formation of formic acid as one of the reaction products was probed using a coupled assay with formate dehydrogenase. When 100 μM HP was used as substrate for the oxygenase reaction, the formation of 80 μM NADH occurred. Knowing that every molecule of formic acid consumed is associated with the formation of one molecule of NADH, under the experimental conditions tested the majority of HP was converted in DHXP (Figure S6). It is possible that the combined Δε340 is smaller than the ε340 for NADH alone, since HP and DHXP have some residual absorbance at 340. The conversion of HP to DHXP in the presence of MtDHNA was confirmed by HPLC (Figure S7).
pH-rate profiles
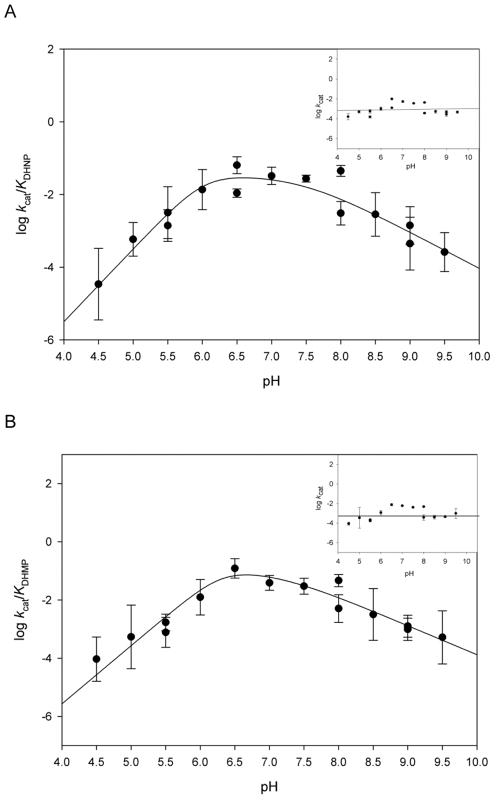
DHNAs are cofactor and metal independent, relying solely on acid-base catalysis to catalyze the cleavage of a carbon-carbon bond.21 Despite the availability of structural and kinetic information, pH studies are still lacking for all DHNAs characterized to date. We investigated the influence of pH on binding and catalysis of the aldolase reaction, using DHNP or DHMP as substrates. In these studies, two kinetic parameters were evaluated: kcat/KM, which reports on steps from free enzyme and free substrates up to and including the first irreversible step, in this case glycolaldehyde release, and kcat, which includes all steps from the formation of a productive enzyme-substrate complex until the release of the last product. Identical patterns were obtained for both epimers: for the kcat/KM profile, an apparent pKa of approximately 6.0 was assigned to two groups required to be unprotonated, and an apparent pKa of 7.4 was assigned to one group required to be protonated. No pH dependence was observed for either kcat profile, indicating that a pH-independent process is limiting enzyme turnover (Figure 6). The pH independence of kcat, while there is a strong pH dependence for kcat/KM, could also indicate that substrate binding can only occur to the correctly protonated form of the enzyme. Due to the fact that solvent KIEs under steady-state conditions (see below) presented considerably higher values in D2Okcat/KM than in D2Okcat, we believe the latter to be less likely than the presence of a pH independent process limiting turnover, which would increase reverse commitments and decrease the magnitude of the D2Okcat.
Figure 6.
pH dependence of (A) log kcat/KDHNP and log kcat-DHNP (inset) and (B) log kcat/KDHMP and log kcat-DHMP (inset). The lines represent fits to eq 7. The values obtained were 6.0 ± 0.5 with slope of two in the acidic limb, and 7.4 ± 0.3 with a slope of one in the basic region (A), and 6.1 ± 0.3 with slope of two in the acidic limb, 7.1 ± 0.3 with a slope of one in the basic region (B). The following buffers were utilized with 50 mM NaCl, 1 mM EDTA and 5 mM DTT: 100 mM citrate (pH 4.5–5.5), 100 mM MES (5.5–6.5), 100 mM HEPES (6.5–8.0), 100 mM TAPS (8.0–9.0), 100 mM CHES (9.0–9.5). Calibration curves for HP fluorescence emission at 535 nm were obtained for all buffers utilized. Lines are fits to eq. 5.
Mutational and structural studies conducted with the SaDHNA and EcDHNA showed that five residues in the active site could be playing catalytic roles. All these residues are conserved in DHNAs, including MtDHNA. For the SaDHNA and EcDHNA, mutations of E22, E74, and K99 (MtDHNA numbering) decreased kcat/KM for both pterin epimers by 3, 2 and 4 orders of magnitude, respectively.9 Interestingly, mutation of Y54 to a phenylalanine converted SaDHNA and EcDHNA into oxygenases whose main reaction product was DHXP.11. This residue is also conserved throughout species, being at the monomer-monomer interface such that Y54 from subunit 1 interacts with the active site of subunit 2, whose Y54 interacts with the active site of subunit 3, and so on. The role of Y54, although important, remains a subject of debate. It could be acting as a catalytic acid to protonate intermediate 1 (I1), or it could be modulating water accessibility to the active site, required to protonate N5. Based on the pK values reported here, another possibility is that proximity of Y54 to K99 lowers the pKa of the general base, which facilitates deprotonation and neutralization, avoiding charge repulsion.22 pKa values significantly lower than expected for primary amines were observed for a lysine acting as a general acid in the reaction catalyzed by acetoacetate decarboxylase, in which proximity to another lysine residue was shown to decrease the pKa values of both groups.23
Furthermore, the chemical mechanism proposed for the Y54F mutants indicates the formation of a carbanion intermediate (I2), which reacts with dioxygen to form DHXP as reaction product (Scheme 2). This type of mechanism seems to be common for cofactor independent dioxygenases; an initial proton abstraction that generates a carbanion intermediate, which then reacts with dioxygen. Additionally, several examples of enzymes that produce carbanionic intermediates and possess oxygenase side reactions are known, and it has been hypothesized that cofactor-independent oxygenases could be utilizing the reactivity of carbanions with electrophiles as a general catalytic strategy.13,24 As illustrated in Scheme 2, I1 has three possible fates: π electrons from the C6-C1′ double bond can abstract a proton from the conjugate acid of the general base (K99 in the Figure S8) to generate HP (blue arrow) or attack the carbonyl group of glycolaldehyde to form DHMP. Finally, deprotonation of N5 can generate the C1’ carbanion (I2) and after the formation of the three intermediates shown, generate DHXP and formic acid.11 The rate-limiting nature of the proton abstraction from the conjugate acid could be influencing the formation and stabilization of the carbanion intermediate and its reaction with dioxygen.
Single turnover studies on the reverse aldolase reaction
To evaluate the internal reversibility of the aldolase reaction, single turnover experiments were performed using 2 μM HP, 50 mM glycolaldehyde, and increasing concentrations of MtDHNA monitoring decrease in fluorescence emission above 495 nm due to HP disappearance. The observed rate constant for HP consumption showed a linear dependence with increasing enzyme concentration, further demonstrating the low affinity of HP for MtDHNA. Because binding was considerably rate limiting, thus contributing to the observed rate constant obtained under single turnover conditions, pseudo-first-order conditions were not satisfied, complicating the analysis by analytical fitting (Figure S9). In order to obtain estimates for the rates of HP binding and the rate of the reverse chemical step, data were analyzed by numerical integration using KinTek Global Explorer. A simple two step model was utilized (Scheme 4). Based on the boundaries obtained for k1 and k−1 (k1 between 0.07–0.17 μM−1s−1, k−1 between 5.2–19.3 s−1), a Kd between 30 and 250 μM can be estimated. Based on the fact that no saturation was observed in equilibrium binding experiments with up to 100 μM HP (data not shown), the true Kd value is more likely to approximate the upper boundary. It is important to point out that MtDHNA binds DHNP, DHMP, and DHXP very tightly, but possesses considerably lower affinity for HP, the pterin product of the aldolase reaction, a discrimination absent in previously characterized DHNAs in which binding of HP, DHNP and DHMP occurs with comparable affinity. The best fitted apparent value for the reverse chemical step was 0.52 s−1 (k2 boundaries: 0.31–0.81). The value for k−2 was kept fixed at 0.037 s−1, based on the value obtained in quench flow burst experiments for the conversion of MtDHNA-DHNP to MtDHNA-HP- glycolaldehyde. Interestingly, the on-enzyme reverse reaction is considerably faster than the forward reaction, but it probably rarely occurs since HP and glycolaldehyde bind extremely weakly. In the SaDHNA-catalyzed reaction, the rate of conversion of SaDHNA-HP-glycolaldehyde into I1 is very fast (100 s−1) if compared to the rates of conversion of both SaDHNA-DHNP and SaDHNA-DHMP into I1 (0.33 s−1 and 0.076 s−1, respectively).
Scheme 4.
Steady-state solvent kinetic isotope effects for the aldolase reaction
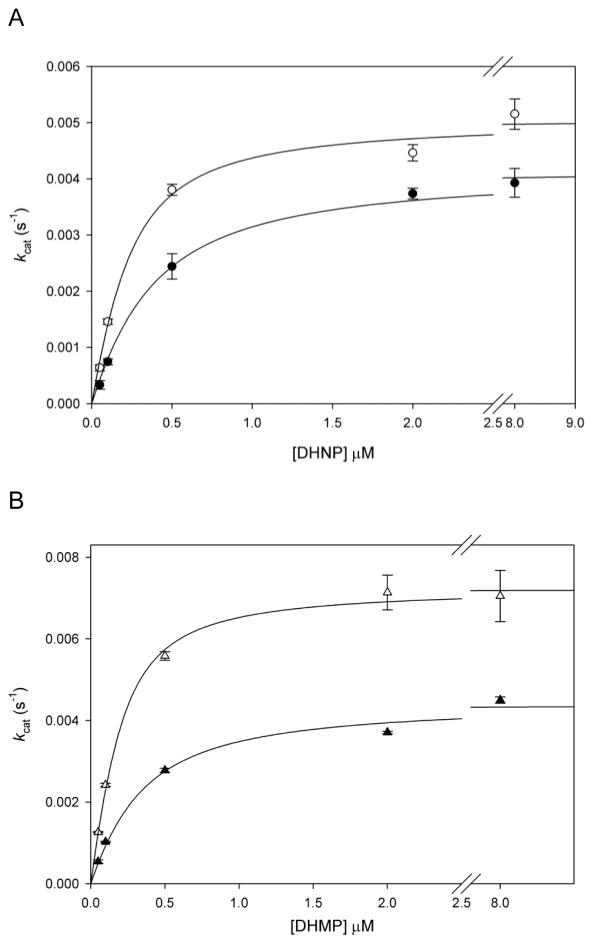
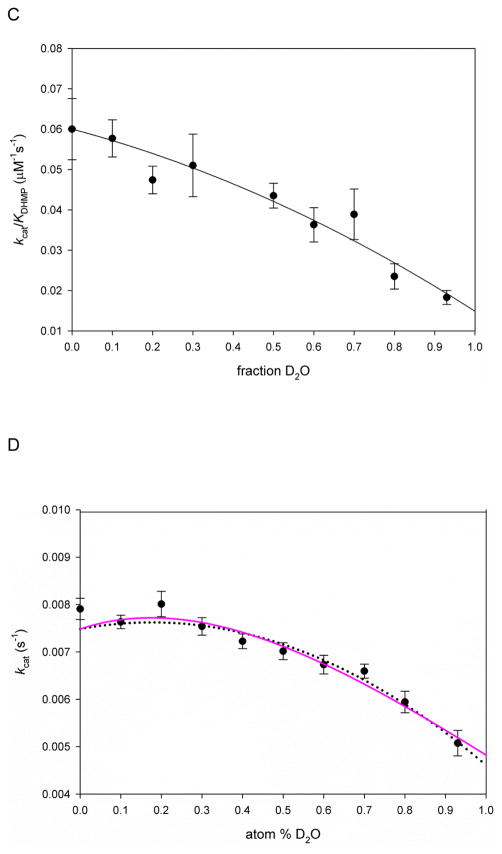
In order to investigate the rate-limiting nature of protonation steps, SKIEs and proton inventory studies were conducted for the aldolase reaction using DHNP or DHMP as substrates. When DHNP was the substrate, D2Okcat-DHNP = 1.2 ± 0.1, and D2Okcat/KDHNP = 2.6 ± 0.6 were obtained, while when DHMP was used as substrate, values were D2Okcat-DHMP = 1.6 ± 0.1 and D2Okcat/KDHMP = 4.1 ± 0.9 (Figures 7A and 7B, respectively). The difference in SKIE values when using distinct epimers can be explained based on the different orientation of the C2′-hydroxyl group, from which proton abstraction occurs. Based on the crystal structures of SaDHNA with bound substrate analogs monapterin (MP) and neopterin (NP), (Figure S8), it has been suggested that the complex SaDHNA-MP adopts an unfavorable orientation, and proton abstraction from the C2′-bound hydroxyl group requires the breakage of two hydrogen bonds and a 180° rotation to align the C2′-OH from MP with the catalytic lysine and for catalysis to occur (K99 in MtDHNA).25 The SKIEs determined here corroborate this hypothesis, since the D2Okcat/KDHMP is two times larger than D2Okcat//KDHMP, and D2Okcat-DHMP, despite being small, is significant and larger than D2Okcat-DHNP.
Figure 7.
Solvent kinetic isotope effects with (A) DHNP or (B) DHMP. The lines represent fits to eq. 2. The values obtained for kcat or kcat/KM in H2O were divided by the ones obtained at 93 % fraction of D2O. The reaction was carried out in 100 mM HEPES, 50 mM NaCl, 1 mM EDTA, 5 mM DTT, and 1% BSA, pH 7.0, a plateau region in the pH-rate profiles to assure that no pH effects were influencing the SKIEs, and the pH value was not altered by increasing fractions of D2O. Panels C and D show the proton inventory using DHMP as substrate. The fitted line in panel C is a fit to eq 8, indicating the contribution of two transition state protons, one with a normal fractionation factor (φ) and one with an inverse fractionation factors. The calculated D2Okcat/KDHMP using the relationship D2Okcat/KDHMP = (1/φTS1)(1/φTS2) was equal to 4.0 (where φTS1 and φTS2 are the fractionation factors for transition state protons 1 and 2). The continuous line in panel D is a fit to eq. 6, while the dotted line represents a fit to eq. 7, indicating the contribution of two transition state protons with an inverse and a normal fractionation factor each, yielding a normal isotope effect, and various solvent protons contributing with inverse isotope effects. For all data points, the values obtained for kcat or kcat/KM in H2O were divided by the ones obtained at different fractions of D2O, and the errors were propagated as described elsewhere32.
Because higher observed SKIE values were obtained when DHMP was the substrate, proton inventory studies using DHMP were carried out to obtain more information about the number of protons originating the observed SKIE. In the proton inventory on V/KDHMP, a better fit of the data was obtained with a model predicting contributions from two transition state protons than to a linear dependence (one transition state proton) model (further discussion about proton inventory data fitting is available in Table S2 in the supporting information). With this model, fractionation factors of 1.38 ± 0.23, and 0.18 ± 0.07 were obtained, yielding a calculated SKIE of 4.01, consistent with the experimental value (Figure 7C). It is possible that the protons generating this D2Okcat/KDHMP are involved in the deprotonation of C2′-OH group and protonation of N5. Considering the pKa of this group to be between 14–18 as expected for a secondary alcohol, the difference between the pKa values of proton donor and proton acceptor would be significant, rendering this proton transfer considerably rate limiting.26
The proton inventory on D2Okcat revealed a much more pronounced dome-shaped profile, indicating that both normal and inverse solvent contributions were giving rise to the observed SKIE (Figure 7D). To properly interpret the results, nine different equations for distinct models describing proton inventories were compared (Table S2). Three of these models fit the data and are in agreement with the proposed chemical mechanism for this reaction (Scheme 2). The first is equivalent to the case discussed above for the D2Okcat/KDHMP proton inventory, in which two transition state protons with distinct normal and inverse fractionation factors are causing the SKIE. The second possible scenario predicts contributions from two transition state protons with distinct fractionation factors accompanied by multiple inverse solvent contributions. Last, it is possible that two distinct transition states, contributing with two protons each, are responsible for the dome-shaped proton inventory. According to Scheme 2, base abstraction of the C2′-OH proton and protonation of N5 occur through transition state 1 (TS1), to form I1. Next, pi electrons from the C6-C1′ double bond abstract a proton from the conjugated acid of the base, and N5 is deprotonated, passing through transition state 2 (TS2) and forming HP. This “two distinct transition state” model requires the weighting factor to estimate the relative contributions of TS1 and TS2 to the observed KIE. The weighting factor revealed that one of the transition states is contributing for the majority of the effect observed (approximately 95% of the SKIE), and this illustrates why these three models fit equally well to the data. Despite the fact that the occurrence of two distinct transition states is likely, better fits and smaller errors were obtained when fitting data to a single transition state model, which possesses fewer unknowns. The fractionation factors obtained for kcat are φ1 = 1.88 ± 0.09, and φ2 = 0.33 ± 0.03 (two distinct transition state protons), φ1 = 0.49 ± 0.27, φ2 = 2.5 ± 0.7, and φSolv = 0.52 ± 0.40 (two distinct transition state protons, and a solvent contribution), both contributing to a calculated SKIE equal within experimental error to the experimentally determined SKIE value of 1.6. Previous work conducted on the dihydrofolate reductase from M. tuberculosis suggested that the N5 of 7,8-dihydrofolic acid has an inverse fractionation factor, and because both dihydrofolate and DHMP have identical pterin moieties, an analogous situation might be observed in the reaction studied here.27
It is important to point out that the first irreversible step in the reaction under steady-state conditions is glycolaldehyde release, so that the magnitude of the observed D2Okcat is diminished by subsequent events including HP release, and other steps that increase the reverse commitment. Thus, because D2Okcat/KDHMP includes steps up to and including glycolaldehyde release, reverse commitment factors are not decreasing its magnitude. This is corroborated by the presence of a burst of product formation (Figure 4), indicating that steps after the chemical step are limiting enzyme turnover (See upper portion of Scheme 6 for a comparison of steps that contribute to kcat and kcat/KM).
Scheme 6.
It is difficult, if not impossible, to rule out media contributions to the SKIEs and proton inventory experiments conducted here. Similar proton inventories were observed in the reactions catalyzed by the aryl acylamidase from Pseudomonas fluorescens and tryptophan indole-lyase from E. coli in which media contributions arising from conformational changes and high commitment factors were associated with dome-shaped proton inventories.28,29 Therefore, it is possible that the more pronounced deviation from linearity of the Dkcat proton inventory is due to increased commitment factors that are absent from Dkcat/KM. To summarize, the SKIEs discussed here clearly indicate the presence of offsetting normal and inverse contributions to the SKIE, and data are consistent with at least two transition state protons with opposite contributions, and possibly a solvent contribution to kcat.
Single turnover experiments for the aldolase reaction
To obtain more information about the rate of chemistry in the aldolase reaction, the increase in fluorescence due to HP formation was monitored under single turnover conditions using limiting concentrations of DHNP or DHMP, and increasing concentrations of MtDHNA. Three exponential phases with opposite amplitudes showing no dependence on MtDHNA concentration were observed when either epimer was the substrate (Figure 8). The first transient was characterized by a fast increase in fluorescence (kobs1-DHNP = 30 s−1, kobs1- DHMP = 6 s−1); the second transient showed decrease in fluorescence emission (kobs2-DHNP = 0.5 s−1, kobs2-DHMP = 0.4 s−1); the third exponential phase was characterized by a pronounced slow increase in fluorescence (kobs3-DHNP = 0.019 s−1, kobs3-DHMP = 0.025 s−1). The third transient most likely represents HP formation, since it has rates comparable to the ones obtained in burst experiments (kburst-DHNP = 0.037 s−1), demonstrating consistency as the burst rate must be larger than rates obtained under single turnover conditions since it can be extrapolated to the sum of the chemical steps in forward and reverse reactions and subsequent steps such as product release.
Figure 8.
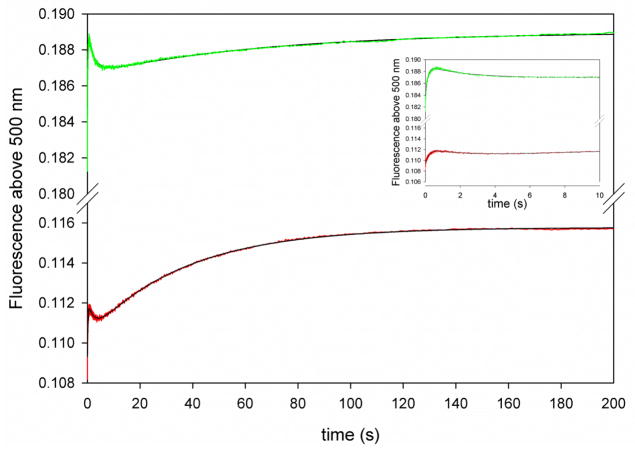
Solvent kinetic isotope effects under single turnover conditions. Data collected with 0.5 μM DHMP and 5 μM MtDHNA. The lines represent fits to eq. 3. The values obtained for k2 in H2O were divided by the ones obtained at 90 % fraction of D2O. The inset shows the first 10 s of the reaction, showing a clear increase in fluorescence followed by a slower decrease in fluorescence.
To investigate what could give rise to the first two transients observed, simulations were conducted using Kintek Global explorer testing distinct mechanistic models that could generate the observed signals. Because the three phases observed showed no dependence on enzyme concentration, bimolecular steps were fixed at diffusion limit. A model testing whether formation of I1 could be giving rise to kobs1 was incapable of describing the experimental data, and very fast rates for formation and consumption of I1 were obtained. This is inconsistent with the SKIEs observed because it would suggest that chemistry was several orders of magnitude higher than kcat, and if that was the case SKIEs close to unity would be observed. The best fitted model (bottom portion of Scheme 6) consisted of fast binding followed by an isomerization step causing an increase in fluorescence, after which this fluorescent species partitioned to form I1, and part of I1 was rapidly converted in DHXP. Both events (decay of the fluorescent species to form I1 and DHXP formation) caused decreases in fluorescence, after which protonation of I1 occurred causing an increase in fluorescence due to HP formation.
Dihydrobiopterin and other pterins have been shown to form oxidized quinonoid tautomers, which could possess a red-shifted maximum absorbance, if compared to non quinonoid pterins, whose maxima is around 330 nm 30,31. Therefore, it is possible that the first fast transient characterized by an increase in fluorescence represents a tautomerization step to form a para quinonoid form of DHNP. Scheme 5 shows an expansion of the chemical mechanism for this reaction, including the formation of the quinonoid species.
Scheme 5.
Aldolase reaction is shown in blue, epimerase reaction in green, and oxygenase reaction in red, including the formation of the proposed para-quinonoid species.
Pre-steady-state solvent kinetic isotope effects for the aldolase reaction
To better understand the role of rate limiting proton transfers in the aldolase reaction, solvent KIEs were measured under single turnover conditions. DHMP was used as substrate due to the higher expression of kinetic isotope effects as compared to DHNP. A limiting concentration of DHMP and increasing concentrations of MtDHNA where utilized to assure that experiments were conducted under pseudo-first order conditions (0.5 μM and 2, 3, 4, and 5 μM, respectively). Three concentration independent transients with identical appearance as observed for single-turnover experiments conducted with DHNP as substrate were observed both in H2O and D2O, indicating that only on-enzyme unimolecular steps were causing the changes in fluorescence. The first and fastest transient was characterized by an increase in fluorescence and an inverse solvent isotope effect of 0.6. As discussed above, the first transient is thought to be a tautomerization step, so that the observed effect is an equilibrium isotope effect instead of a kinetic effect.
The second transient was characterized by a decrease in fluorescence as a function of time, and no isotope effect was observed (KIE = 1.1). In contrast, the third slower transient corresponding to HP formation had a kinetic isotope effect of 1.7. Interestingly, the amplitude of this transient in D2O was two fold smaller than the amplitude of the same transient in the reaction measured in water. Because MtDHNA catalyzes three distinct reactions, we hypothesized that the partition between each one of the reactions was no longer the same when the reaction occurred in D2O (MtDHNA-DHNP or MtDHNA-DHMP partitions in three distinct reactions in the first turnover, and 51% becomes the aldolase product, as discussed above). To test this hypothesis, MtDHNA and DHMP were mixed at 10 and 3 μM, respectively, using standard reaction conditions in H2O or 90% D2O, the reaction was quenched after 200 s, and products were analyzed by HPLC. After separating DHMP and products, it was observed that the reaction in D2O partitioned differently between the aldolase/epimerase/oxygenase reactions than the reaction occurring in water. In the reaction carried out in H2O, the majority of product formed corresponds to HP, while in the D2O reaction, less DHMP is consumed, less HP is formed, and more DHNP and DHXP are generated (Figure S10). We propose that the magnitude of the oxygenase reaction is related to the capability of the enzyme to protonate I1 to generate HP, so that any reduction in I1 protonation would allow this intermediate to react with molecular oxygen and form DHXP. It is likely that I1 protonation is slowed in D2O, causing the aldolase reaction to occur less frequently.
Often, kinetic isotope effects are measured under single-turnover conditions to decrease kinetic complexity since steps such as product release which usually decrease the magnitude of observed KIEs by increasing reverse commitments are not observed under these conditions. Nevertheless, the assumption that single turnover studies decrease kinetic complexity must be considered on a case-by-case basis. In the kinetic mechanism analyzed here, under steady-state conditions, release of glycolaldehyde is fast, causing the reverse chemical step to occur very rarely. Under single-turnover conditions, however, since only on-enzyme events contribute to the rates observed, the internal reversibility of the chemical reaction has a more significant contribution, therefore decreasing the magnitude of the KIEs observed under single-turnover conditions. Scheme 6 compares steps that contribute for KIEs under steady-state or single-turnover conditions.
Relevance of the oxygenase reaction
MtDHNA is the first wild type form of this class of enzymes to concurrently catalyze the epimerase and oxygenase reactions to a significant extent. Because the enzyme co-purifies with bound DHXP, we conclude that the oxygenase reaction occurs in vivo, despite the fact that studies identifying and quantifying pterin molecules in M. tuberculosis are lacking. The fact that DHXP induces octamerization of the protein could have regulatory implications. It has been proposed that the substrate of the reaction, DHNP, is an antioxidant that could exert a protective effect on M. tuberculosis against host defenses, so that it would be advantageous for the bacteria to maintain the enzyme in the inactive MtDHNA-DHXP form until enough DHNP was present to compete with DHXP and generate a productive MtDHNA-DHNP complex.
Supplementary Material
Figure 3.
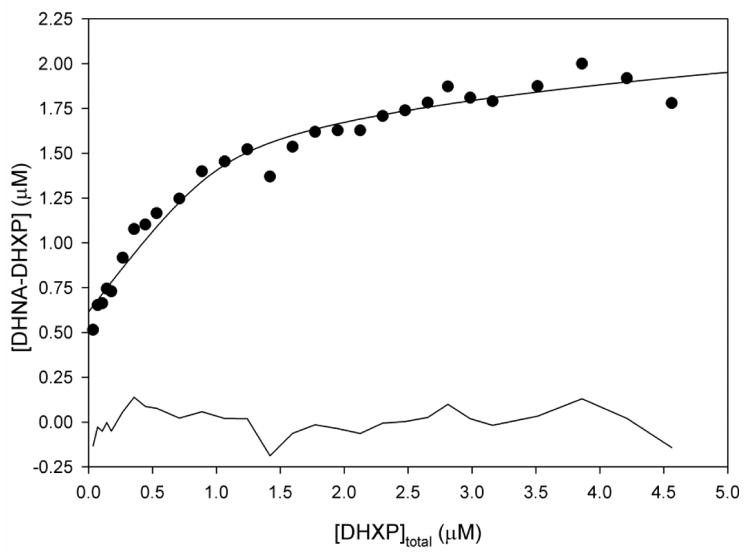
DHXP binding to MtDHNA measured by fluorescence titration. Excitation and emission wavelengths were 295 and 345 nm, respectively. Serial additions of 1 μL aliquots of ligand were made to a 1 cm2 quartz cuvette that contained 2 mL of 2 μM of MtDHNA in 100 mM HEPES with 50 mM NaCl pH 7.0, 1 mM EDTA, 5 mM DTT, and 1% BSA. Dissociation constants were estimated by nonlinear regression using eq. 1, giving Kd1= 0.07 ± 0.07 μM and Kd2 = 6 ± 4 μM. More details about other equations fitted are available in the supporting information. The line in the bottom of the graph represents the residuals of the fit.
Scheme 3.
Acknowledgments
This work was supported by the NIH (AI33696), the Einstein-Montefiore CFAR (NIH AI-515109).
We thank Dr. Bruce Palfey (University of Michigan) for help discussing tSKIEs, and Dr. Richard Schowen (University of Kansas) for discussions about proton inventory data fitting. We also thank Dr. Luiz Pedro S. de Carvalho for critical reading of the manuscript, and Dr. Rafael G. da Silva for insightful discussions.
Abbreviations
- TB
tuberculosis
- MDR-TB
multi-drug resistant tuberculosis
- XDR-TB
extensively drug resistant tuberculosis
- DHNA
dihydroneopterin aldolase
- MtDHNA
dihydroneopterin aldolase from Mycobacterium tuberculosis H37Rv
- EcDHNA
DHNA from Escherichia coli
- SaDHNA
DHNA from Staphylococcus aureus
- DHNP
7,8- dihydroneopterin
- DHMP
7,8-dihydromonapterin
- HP
7,8-dihydro-6-methylpterin
- DHXP
7,8-dihydroxanthopterin
- kobs
observed rate constant
- kcat
steady state turnover number
- kcat-rev
steady state turnover number for the reverse reaction
- D2Okcat-DHNP or D2Okcat-DHMP
solvent kinetic isotope effect on kcat using DHNP or DHMP, respectively, as substrate
- D2Okcat/KDHNP or D2Okcat/KDHMP
solvent kinetic isotope effect on kcat/KM using DHNP or DHMP, respectively, as substrate
- kburst
rate of the burst phase
- kchem-rev
rate constant of chemistry for the reverse reaction
- KIE
kinetic isotope effect
- SKIE
solvent kinetic isotope effect
Footnotes
An extended description of proton inventory data fitting, ESI-MS spectra, and additional figures, are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wright DL, Anderson AC. Expert Opin Ther Pat. 2011;21:1293. doi: 10.1517/13543776.2011.587804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermingham A, Derrick JP. Bioessays. 2002;24:637. doi: 10.1002/bies.10114. [DOI] [PubMed] [Google Scholar]
- 3.Walsh C. Nat Rev Microbiol. 2003;1:65. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 4.Goulding CW, Parseghian A, Sawaya MR, Cascio D, Apostol MI, Gennaro ML, Eisenberg D. Protein Sci. 2002;11:2887. doi: 10.1110/ps.0219002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang ZX, Jiang RF. FEBS Lett. 1996;392:245. doi: 10.1016/0014-5793(96)00818-6. [DOI] [PubMed] [Google Scholar]
- 6.Sanders WJ, Nienaber VL, Lerner CG, McCall JO, Merrick SM, Swanson SJ, Harlan JE, Stoll VS, Stamper GF, Betz SF, Condroski KR, Meadows RP, Severin JM, Walter KA, Magdalinos P, Jakob CG, Wagner R, Beutel BA. J Med Chem. 2004;47:1709. doi: 10.1021/jm030497y. [DOI] [PubMed] [Google Scholar]
- 7.Johnson KA, Simpson ZB, Blom T. Anal Biochem. 2009;387:20. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 8.Johnson KA, Simpson ZB, Blom T. Anal Biochem. 2009;387:30. doi: 10.1016/j.ab.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Li Y, Yan H. Biochemistry. 2006;45:15232. doi: 10.1021/bi060949j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bar-Even A, Noor E, Savir Y, Liebermeister W, Davidi D, Tawfik DS, Milo R. Biochemistry. 2011;50:4402. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Scherperel G, Roberts KD, Jones AD, Reid GE, Yan H. J Am Chem Soc. 2006;128:13216. doi: 10.1021/ja063455i. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Li Y, Wu Y, Yan H. FEBS J. 2007;274:2240. doi: 10.1111/j.1742-4658.2007.05761.x. [DOI] [PubMed] [Google Scholar]
- 13.Fetzner S, Steiner RA. Appl Microbiol Biotechnol. 2010;86:791. doi: 10.1007/s00253-010-2455-0. [DOI] [PubMed] [Google Scholar]
- 14.Stein RA, Wilkinson JC, Guyer CA, Staros JV. Biochemistry. 2001;40:6142. doi: 10.1021/bi002817a. [DOI] [PubMed] [Google Scholar]
- 15.Koshland DE., Jr Curr Opin Struct Biol. 1996;6:757. doi: 10.1016/s0959-440x(96)80004-2. [DOI] [PubMed] [Google Scholar]
- 16.Goulding CW, Apostol MI, Sawaya MR, Phillips M, Parseghian A, Eisenberg D. J Mol Biol. 2005;349:61. doi: 10.1016/j.jmb.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 17.Gieseg SP, Amit Z, Yang YT, Shchepetkina A, Katouah H. Antioxid Redox Signal. 2010;13:1525. doi: 10.1089/ars.2009.3065. [DOI] [PubMed] [Google Scholar]
- 18.Firth CA, Crone EM, Flavall EA, Roake JA, Gieseg SP. Biochim Biophys Acta. 2008;1783:1095. doi: 10.1016/j.bbamcr.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Duggan S, Rait C, Gebicki JM, Gieseg SP. Redox Rep. 2001;6:188. doi: 10.1179/135100001101536175. [DOI] [PubMed] [Google Scholar]
- 20.Dantola ML, Vignoni M, Capparelli AL, Lorente C, Thomas AH. Helv Chim Acta. 2008;91:411. [Google Scholar]
- 21.Mathis JB, Brown GM. J Biol Chem. 1970;245:3015. [PubMed] [Google Scholar]
- 22.Harris TK, Turner GJ. IUBMB Life. 2002;53:85. doi: 10.1080/15216540211468. [DOI] [PubMed] [Google Scholar]
- 23.Highbarger LA, Gerlt JA, Kenyon GL. Biochemistry. 1996;35:41. doi: 10.1021/bi9518306. [DOI] [PubMed] [Google Scholar]
- 24.Abell LM, Schloss JV. Biochemistry. 1991;30:7883. doi: 10.1021/bi00246a002. [DOI] [PubMed] [Google Scholar]
- 25.Blaszczyk J, Li Y, Gan J, Yan H, Ji X. J Mol Biol. 2007;368:161. doi: 10.1016/j.jmb.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyte J. Mechanism in Protein Chemistry. Garland Publishing, Inc; 1995. [Google Scholar]
- 27.Czekster CM, Vandemeulebroucke A, Blanchard JS. Biochemistry. 2010;50:367. doi: 10.1021/bi1016843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiick DM. J Am Chem Soc. 1991;113:8499. [Google Scholar]
- 29.Stein RL. Biochemistry. 2002;41:991. doi: 10.1021/bi0118198. [DOI] [PubMed] [Google Scholar]
- 30.Lyudnikova TA, Dashina OA, Telegina TA, Kritsky MS. Appl Biochem and Microbiol. 2009;45:104. [Google Scholar]
- 31.Eberlein G, Bruice TC, Lazarus RA, Henrie R, Benkovic SJ. J Am Chem Soc. 1984;106:7916. [Google Scholar]
- 32.Skoog D, Holler J, Crouch S. Principles of Instrumental Analysis. 6. Thomson Brooks/Cole; Belmont: 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.