Abstract
To identify genes influencing blood pressure response to an angiotensin II receptor blocker, single nucleotide polymorphisms identified by genome-wide association analysis of the response to candesartan were validated by opposite direction associations with the response to a thiazide diuretic, hydrochlorothiazide. 198 White and 193 African Americans with primary hypertension were sampled from opposite tertiles of the race-sex-specific distributions of age-adjusted diastolic blood pressure response to candesartan. 285 polymorphisms associated with the response to candesartan at p<10−4 in Whites. 273 of the 285 polymorphisms, which were available for analysis in a separate sample of 196 Whites, validated for opposite direction associations with the response to hydrochlorothiazide (Fisher’s X2 1-sided p=0.02). Among the 273 polymorphisms, those in the chromosome 11q21 region were the most significantly associated with response to candesartan in Whites (e.g., rs11020821 near FUT4, p=8.98×10−7), had the strongest opposite direction associations with response to hydrochlorothiazide (e.g., rs3758785 in GPR83, p=7.10×10−3), and had same direction associations with response to candesartan in the 193 African Americans (e.g., rs16924603 near FUT4, p=1.52×10−2). Also notable among the 273 polymorphisms was rs11649420 on chromosome 16 in the amiloride-sensitive sodium channel subunit SCNN1G involved in mediating renal sodium reabsorption and maintaining blood pressure when the renin-angiotensin system is inhibited by candesartan. These results support the utility of genomewide association analyses to identify novel genes predictive of opposite direction associations with blood pressure responses to inhibitors of the renin-angiotensin and renal sodium transport systems.
Keywords: Hypertension, pharmacogenetics, diuretic, blood pressure, genome
INTRODUCTION
The purpose of this study was to scan the genome for common variants, i.e., single nucleotide polymorphisms (SNPs), that predict blood pressure (BP) response to an angiotensin II receptor blocker (ARB) in White and African Americans with primary hypertension. In previous studies, the only consistently identified predictors of BP responses to antihypertensive drug treatment have been pretreatment BP level, plasma renin activity, race, and age, which together explain less than 50% of inter-individual variation in BP responses.1, 2 While higher pretreatment BP level is nonspecifically associated with greater BP response to all antihypertensives regardless of pharmacological class, the other established predictors of BP response are drug-class specific.3 Notably, the associations of renin, race, and age with BP response to ARBs and other inhibitors of the renin-angiotensin system (RAS) are directionally opposite to their associations with BP response to thiazide and other diuretics.1, 2
Our specific objective was to identify single nucleotide polymorphisms (SNPs) that not only predict BP response to the ARB candesartan but also have directionally opposite associations with BP response to the thiazide diuretic, hydrochlorothiazide, the most commonly prescribed antihypertensive.4 We reasoned that such SNPs with directionally opposite effects on BP responses to drugs from different pharmacological classes would have greatest utility in individualizing antihypertensive drug therapy based on patients’ genotypes. Moreover, such SNPs could identify novel components of the vasoconstriction and volume-regulating systems that maintain BP and reciprocally oppose declines in BP when either system is inhibited by single-drug therapy.5
MATERIALS AND METHODS
Phenotypic data and DNA samples were collected in the Genetic Epidemiology of Responses to Antihypertensives (GERA) study between 1997 and 2007 (please see http://hyper.aha.journals.org, Supplementary Methods). Standardized drug-treatment protocols were conducted in the Centers for Transitional Science Activities (CTSA) at Emory University and the Mayo Clinic after approval by the respective institutional review committees and subjects gave informed consent.6, 7 A case-control sampling design was employed for genome-wide association (GWA) analyses,8 which contrasted good responders with poor responders sampled from opposite tertiles of the distributions of diastolic BP response to the ARB, candesartan and the thiazide diuretic, hydrochlorothiazide. For each protocol, we enrolled 300 Whites (in Rochester MN) and 300 African Americans (in Atlanta GA), who had uncomplicated primary hypertension, stage 1–2, and who were 30–59.9 years of age. Participants were instructed to discontinue previous antihypertensive medications for ≥4 weeks. Once stable elevation of the BP was achieved (diastolic BP ≥90 mmHg), the study drug was administered orally: hydrochlorothiazide 25 mg daily for four weeks or candesartan 16 mg daily for two weeks followed by 32 mg daily for four additional weeks. At the end of the drug-free and drug-treatment periods, three readings of BP were made by a trained assistant after the participant had been seated quietly for at least 5 minutes. The difference between averages of the second and third diastolic BP readings taken before and at the end of drug treatment was calculated as the primary measure of BP response.6, 7 A case-control sampling design was employed for genome-wide association (GWA) analyses, which contrasted good responders with poor responders sampled from opposite tertiles of the distributions of diastolic BP response to each drug.8 Genotypes from the Affymetrix GeneChip® Human Mapping 6.0 Array Sets were suitable for statistical analyses in the 198 White and 193 African Americans treated with candesartan. Genotypes from the Affymetrix GeneChip® Human Mapping 500K Array Sets were suitable for statistical analyses in 196 White and 194 African Americans treated with hydrochlorothiazide (please see http://hyper.aha.journals.org, Supplementary Methods). The GWA analyses conducted within each race-drug treatment group tested each SNP for association with the diastolic BP response categories by applying the Cochran-Armitage test for trend in the proportion of good or poor responders with increase in one of the two alleles at each locus.9, 10 For the SNPs most significantly associated with BP response to candesartan (defined by p-values <10−4 in the primary GWA analysis in each race), we tested for validation of the SNP associations with opposite direction BP response to hydrochlorothiazide using Fisher’s method of combining the separate 1-sided p-values (for hydrochlorothiazide) in the same race.11 Because of linkage disequilibrium among the SNPs (and non-independence of the p-values), we generated the probability distribution of the X2 test statistic empirically by repetitively permuting the BP response categories for the second drug, recalculating the X2 after each permutation, and thereby generating a distribution of p-values for the X2 statistic under the null hypothesis (i.e., no SNP associations with BP response). Departure from the null hypothesis was tested using a 1-sided X2 test statistic in which 1-sided p-values from the Cochran-Armitage test for an opposite trend in the proportion of good responders to hydrochlorothiazide with increases in the number of alleles associated with good response to candesartan. In the validation setting, 1-sided p-values were used because of the a priori direction for each SNP effect dictated by results of the primary GWA analysis.
RESULTS
Sample descriptions
By design, the 198 Whites and 193 African Americans treated with candesartan were nearly equally divided between good and poor responders and, within each response group, between men and women (Tables 1 and 2). Also consistent with the sampling design, the good and poor responders differed significantly in mean systolic and mean diastolic BP responses to candesartan. After race and sex-specific adjustments for age and baseline diastolic BP, the mean (±standard deviation) diastolic BP response in White good responders was −21.9±3.9 versus −4.7±5.6 mmHg in poor responders; and in African American good responders was −16.0±7.1 versus 0.3±4.4 mmHg in poor responders. Mean plasma renin activity prior to candesartan treatment was significantly greater in the good than poor responders (p<0.0001).7
Table 1.
Hypertensive White Americans treated with candesartan or hydrochlorothiazide
| Characteristic | Candesartan treatment (N=198) | Hydrochlorothiazide treatment (N=196) | ||||
|---|---|---|---|---|---|---|
| Good BP responders (N=99) | Poor BP responders (N=99) | Contrast P | Good BP responders (N=98) | Poor BP responders (N=98) | Contrast P | |
| Women/Men | 49/50 | 49/50 | 1.00 | 42/56 | 42/56 | 1.00 |
| Age, years | 49 ±7 | 49 ±7 | 0.84 | 48 ±8 | 49 ±7 | 0.84 |
| BMI, kg·m−2 | 29 ±4 | 30 ±3 | 0.07 | 31 ±5 | 32 ±6 | 0.35 |
| Pretreatment SBP, mmHg | 145 ±13 | 149 ±12 | 0.05 | 142 ±12 | 143 ±13 | 0.61 |
| Pretreatment DBP, mmHg | 95 ±5 | 95 ±5 | 1.00 | 96 ±5 | 96 ±6 | 1.00 |
| SBP response, mmHg | −26.8 ±12.9 | −9.9 ±11.2 | <0.0001 | −17.8 ±11.0 | −3.9 ±10.9 | <0.0001 |
| DBP response, mmHg | −22.0 ±5.2 | −4.7 ±5.5 | <0.0001 | −13.7 ±4.2 | 1.1 ± 5.3 | <0.0001 |
| Adjusted SBP response, mmHg | −27.7 ±9.3 | −9.3 ±10.3 | <0.0001 | −17.9 ±8.3 | −3.5 ±9.9 | <0.0001 |
| Adjusted DBP response, mmHg | −21.9 ±3.9 | −4.7 ±5.6 | <0.0001 | −13.6 ±3.1 | 1.20 ±4.4 | <0.0001 |
| Serum potassium, mmol·L−1 | 4.5 ±0.3 | 4.3 ±0.4 | 0.003 | 4.0 ±0.3 | 4.0 ±0.5 | 0.90 |
| Plasma renin activity, ng·ml−1·hr−1 | 1.4 ±1.0 | 0.7 ±0.9 | <0.0001 | 1.3 ±0.9 | 1.6 ±1.4 | 0.07 |
| Serum aldosterone, ng·dL−1 | 12.1 ±6.3 | 12.4 ±6.5 | 0.70 | 14.5 ±6.8 | 15.4 ±10.2 | 0.48 |
| Urine aldosterone, μg·(24hr)−1 | 10.4 ±6.3 | 11.9 ±6.7 | 0.11 | 8.7 ± 4.7 | 11.0 ±7.7 | 0.01 |
| Urine sodium, mmol·(24hr)−1 | 178 ±77 | 181 ±89 | 0.83 | 159 ±66 | 160 ±72 | 0.92 |
| Urine potassium, mmol·(24hr)−1 | 73 ±25 | 74 ±34 | 0.93 | 61 ±26 | 71 ±29 | 0.01 |
Table entries are means ± standard deviation. SBP, systolic blood pressure; DBP, diastolic blood pressure. Adjusted SBP and DBP responses after adjustment for pretreatment blood pressure levels. P is significance of contrast between good and poor responders within drug-treatment group.
Table 2.
Hypertensive African Americans treated with candesartan or hydrochlorothiazide
| Characteristic | Candesartan treatment (N=193) | Hydrochlorothiazide treatment (N=194) | ||||
|---|---|---|---|---|---|---|
| Good BP responders (N=99) | Poor BP responders (N=94) | Contrast P | Good BP responders (N=97) | Poor BP responders (N= 97) | Contrast P | |
| Women/Men | 49/50 | 48/46 | 0.83 | 50/47 | 50/47 | 1.00 |
| Age, years | 49 ±7 | 49 ±7 | 0.58 | 47 ±6 | 48 ±7 | 0.52 |
| BMI, kg·m−2 | 30 ±4 | 31 ±5 | 0.10 | 31 ±6 | 32 ±7 | 0.06 |
| Pretreatment systolic BP, mmHg | 147 ±13 | 147 ±12 | 0.94 | 148 ±14 | 154 ±15 | 0.004 |
| Pretreatment diastolic BP, mmHg | 96 ±5 | 96 ±5 | 0.86 | 97 ±5 | 97 ±6 | 0.76 |
| Systolic BP response, mmHg | −20.9 ±9.9 | 0.4 ±10.5 | <0.0001 | −26.0 ±12.1 | −9.5 ±10.2 | <0.0001 |
| Diastolic BP response, mmHg | −16.0 ±7.2 | 0.4 ±4.4 | <0.0001 | −18.3 ±4.9 | −0.3 ±4.4 | <0.0001 |
| SBP response, adjusted | −20.9 ±8.5 | 0.4 ±9.9 | <0.0001 | −26.6 ±9.6 | −8.3 ±9.6 | <0.0001 |
| DBP response, adjusted | −16.0 ±7.1 | 0.3 ±4.4 | <0.001 | −18.3 ±4.2 | −0.18 ±4.3 | <0.0001 |
| Serum potassium, mmol·L−1 | 4.1 ±0.4 | 4.0 ±0.4 | 0.03 | 3.8 ±0.3 | 3.7 ±0.3 | 0.03 |
| Plasma renin activity, ng·ml−1·hr−1 | 1.0 ±2.7 | 0.4 ±1.1 | <0.0001 | 0.70 ±0.71 | 1.12 ±1.29 | 0.007 |
| Serum aldosterone, ng·dL−1 | 7.8 ±4.5 | 7.6 ±4.5 | 0.80 | 15.8 ±8.6 | 19.1 ±10.7 | 0.02 |
| Urine aldosterone, μg·(24hr)−1 | 9.2 ±8.2 | 10.3 ±9.9 | 0.41 | 8.2 ±5.0 | 10.7 ±6.9 | 0.004 |
| Urine sodium, mmol·(24hr)−1 | 141 ±86 | 147 ±72 | 0.59 | 167 ±61 | 161 ±71 | 0.55 |
| Urine potassium, mmol·(24hr)−1 | 47 ±26 | 47 ±20 | 0.97 | 49 ±19 | 51 ±19 | 0.35 |
Table entries are means ± standard deviation. SBP, systolic blood pressure; DBP, diastolic blood pressure. Adjusted SBP and DBP responses after adjustment for pretreatment blood pressure levels. P is significance of contrast between good and poor responders within drug-treatment group.
The good and poor responders to hydrochlorothiazide were similar in number, gender distribution, mean age, body mass index, and pretreatment systolic and diastolic BPs to the candesartan-treated groups8 (shown for comparison in Tables 1 and 2). Mean plasma renin activity prior to hydrochlorothiazide treatment was lower in the good than poor responders (p≤0.07).6 In Whites, BP responses were greater to candesartan than to hydrochlorothiazide (Table 1); and in African Americans, BP responses were greater to hydrochlorothiazide than to candesartan (Table 2).6, 7
Genome-wide association analyses of BP response to candesartan
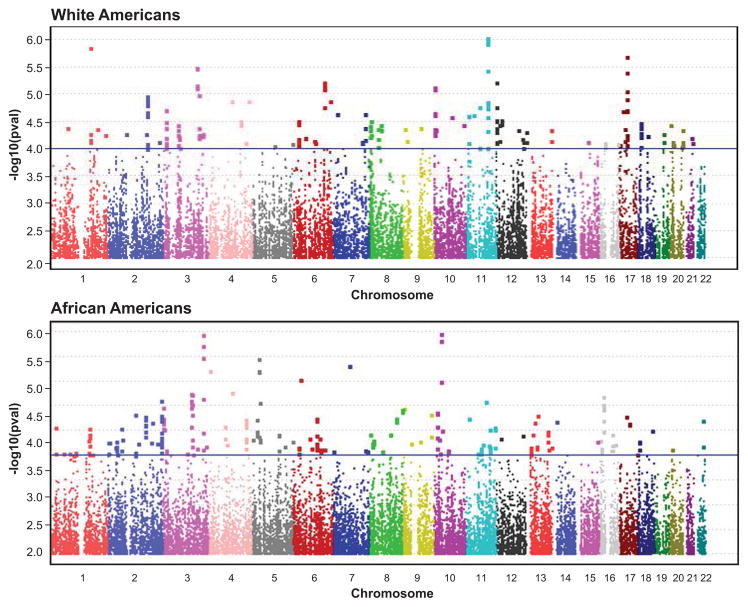
We used ≈2.3 million SNPs in Whites and ≈2.1 million SNPs in African Americans for the primary GWA analyses of categorically defined good versus poor BP response to candesartan in each race separately (Figure 1). While no SNP achieved the threshold of p<5×10−8 conventionally used for genome-wide significance,12 the Q-Q plots in both races indicated that the number of SNP p-values <10−4 deviated above the diagonal, i.e., the expectation under the null hypothesis (H0) of no relationship between SNPs and BP response (please see http://hyper.aha.journals.org, Figure S1). The observed number of SNP p-values <10−4 was 285 in Whites (versus 229 expected under H0) and 272 in African Americans (versus 214 expected under H0). The SNPs with p-values <10−4, their chromosomal locations, and other descriptive information are in Tables S1 and S2 (please see http://hyper.aha.journals.org).
Figure 1.
Race-specific Manhattan plots of single nucleotide polymorphism p-values from genome-wide association of diastolic blood pressure response to candesartan.
Two hundred and seventy-three of the 285 SNPs identified in Whites and 264 of the 272 SNPs identified in African Americans were available for validation of opposite-direction associations with BP response to hydrochlorothiazide. In Whites, the X2 test statistic for directionally opposite SNP associations with BP response to hydrochlorothiazide had a nominal 1-sided p-value of 3.36×10−10 and an empirically determined 1-sided p-value of 0.015 (upper 95% confidence limit of 0.038) based on 200 permutations of the hydrochlorothiazide BP response categories. The X2 test statistic in African Americans had a nominal 1-sided p-value of 0.81 and an empirically determined 1-sided p-value of 0.69. Therefore, only the 273 SNPs identified and validated in Whites are considered below.
Regional SNP associations with BP responses to candesartan and hydrochlorothiazide
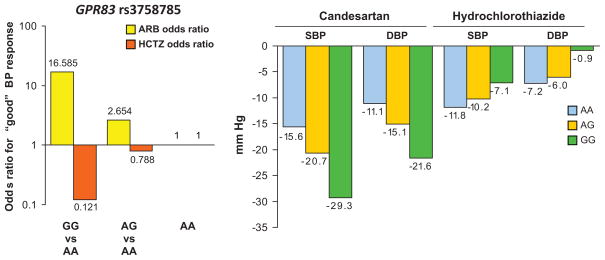
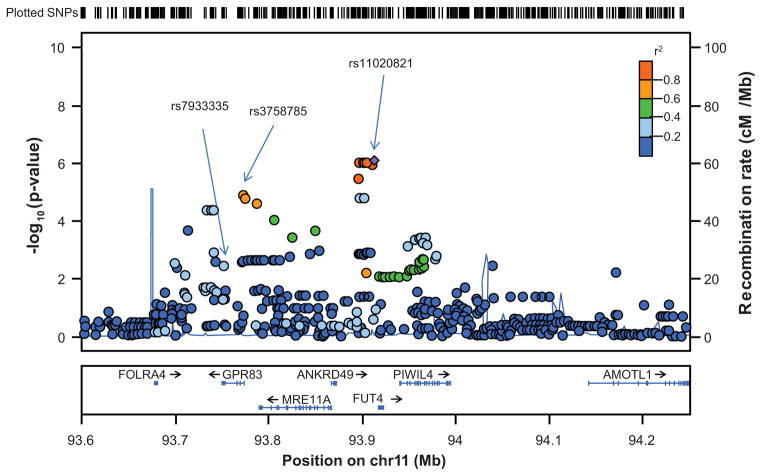
For the chromosomal regions identified in the primary GWA analyses of BP response to candesartan in Whites (by SNP p-values <10−4), lead SNPs were selected that had the most significant opposite direction associations with BP response to hydrochlorothiazide (i.e., 1-sided p<0.05) (Table 3). Adjustment for plasma renin activity slightly reduced statistical significance of the SNP associations with BP response to candesartan (Table 3). Two of the six lead SNPs were on chromosome 11, of which rs3758785 (A→G) in the G protein-coupled receptor 83 gene (GPR83) on 11q21 had the most significant opposite direction association with BP response to hydrochlorothiazide (1-sided p=7.10×10−3). For the rs3758785 GG genotype, the odds of good BP response to candesartan was more than 16-fold greater than for the AA genotype and the odds of good BP response to hydrochlorothiazide was almost 8-fold less than for the AA genotype (Figure 2). The mean adjusted systolic BP/diastolic BP responses to candesartan were 13.7/10.5 mmHg greater for the GG than for the AA genotype, and the mean systolic BP/diastolic BP responses to hydrochlorothiazide were 4.7/6.3 mmHg less for the GG than for the AA genotype (Figure 2). While statistical significance of the opposite direction association with BP response to hydrochlorothiazide was maximal at rs7933335 in GPR83 (1-sided p=3.33×10−3) (please see http://hyper.aha.journals.org, Figure S2, panel b), statistical significance of the association With BP response to candesartan was maximal at rs11020821 (p=8.98 ×10−7) near the fucosyltransferase 4 gene (FUT4) (Figure 3). Adjustment for rs11020821 near FUT4 markedly reduced statistical significance of the association of rs3758785 in GPR83 with BP response to candesartan (2-sided p=0.02) and its opposite direction association with BP response to hydrochlorothiazide (1-sided p= 0.05, please see http://hyper.aha.journals.org, Figure S3).
Table 3.
Lead single nucleotide polymorphisms associated with good versus poor blood pressure response to candesartan and opposite direction association with blood pressure response to hydrochlorothiazide in White Americans
| SNP | Nearest Gene | Candesartan treatment | Hydrochlorothiazide treatment | Alleles | Frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | β | ±SE | P* | Padj | β | ±SE | P† | 0 | 1 | C | H | ||
| rs13059339 | 3 | SLC9A9 | 0.99 | 0.23 | 3.30E-06 | 5.553E-05 | −0.37 | 0.21 | 3.88E-02 | G | T | 0.42 | 0.39 |
| rs3758785 | 11 | GPR83 | −1.11 | 0.27 | 1.39E-05 | 2.044E-04 | 0.58 | 0.24 | 7.10E-03 | A | G | 0.75 | 0.7 |
| rs7113572 | 11 | EHF | −1.45 | 0.38 | 2.41E-05 | 9.725E-05 | 0.7 | 0.37 | 2.65E-02 | C | T | 0.88 | 0.9 |
| rs2113653 | 2 | MYO3B | 1.00 | 0.25 | 2.57E-05 | 3.743E-05 | −0.36 | 0.20 | 3.49E-02 | A | C | 0.49 | 0.49 |
| rs12313639 | 12 | SLCO1B1 | −6.95 | 9.00 | 3.22E-05 | 2.225E-04 | 1.71 | 0.66 | 1.76E-03 | C | G | 0.03 | 0.04 |
| rs11649420 | 16 | SCNN1G | −1.09 | 0.30 | 9.41E-05 | 7.818E-04 | 0.57 | 0.28 | 2.00E-02 | A | G | 0.21 | 0.22 |
SNP, single nucleotide polymorphism; Chr, chromosome; β, logistic regression coefficient; SE, standard error of the logistic regression coefficient.
Alleles: 0, reference allele; 1, alternative allele. Frequency of reference allele: C, among candesartan treated group; H, among hydrochlorothiazide treated group.
, 2-sided p-value;
Padj, adjusted for plasma renin activity;
1-sided p-value.
The logistic regression coefficients (βs) represent log-of-the-odds of good blood pressure response associated with each copy of the alternative allele.
Figure 2.
GPR83 rs3758785 genotype dependence of the odds ratios for good BP response and the mean systolic and diastolic blood pressure declines in response to each drug.
Figure 3.
Chromosome 11q21 plot of p values for association of single nucleotide polymorphisms with blood pressure response to candesartan in White Americans.
Additional SNPs in the chromosome 11q21 region identified in Whites also validated for same direction associations with BP response to candesartan in the African American sample (1-sided p<0.05). Two of these SNPs were in GPR83 (rs3758786 and rs3758789); one was near MRE11A (rs12270338); and two near FUT4 (rs16924603 and rs16912567), of which rs16924603 was most significantly associated (1-sided p=1.52×10−2) (please see http://hyper.aha.journals.org, Figure 2, panel c). In the previous African American sample of good and poor BP responders to hydrochlorothiazide,8 the chromosome 11q21 region was independently identified by a lead SNP rs2851582 in the angiomotin like 1 gene (AMOTL1) that was primarily associated with BP response to hydrochlorothiazide (2-sided p=5.37×10−5) and also validated for directionally opposite association with BP response to candesartan in the present African American sample (1-sided p=3.61×10−3) (please see http://hyper.aha.journals.org, Figure S2, panel d). In both races, there was linkage disequilibrium among the SNPs in the GPR83-to-FUT4 region but not with SNPs in AMOTL1 (please see http://hyper.aha.journals.org, Figure S4).
Other lead SNPs discovered and validated in Whites included rs11649420 in the gene encoding the γ-subunit (SCNN1G) of the amiloride-sensitive sodium channel (Table 3). For the rs11649420 GG genotype, the odds of good BP response to candesartan was three-fold greater than for the combined AA+AG group and the odds of good BP response to hydrochlorothiazide was two-fold less than for the combined AA+AG group (please see http://hyper.aha.journals.org, Figure S5). The mean adjusted systolic BP/diastolic BP responses to candesartan were 7.0/5.5 mmHg greater for the GG than for the AA+AG group, and the mean systolic BP/diastolic BP responses to hydrochlorothiazide were 4.5/2.5 mmHg less for the GG than for the AA genotype.
DISCUSSION
We sought to identify SNPs associated with directionally opposite BP responses to candesartan and hydrochlorothiazide because of their greater potential utility in personalizing antihypertensive drug therapy than SNPs that only predict response to a single drug or the same response to drugs from multiple classes. Pursuit of this objective was also motivated by the knowledge that established predictors of antihypertensive drug responses have directionally opposite associations with BP responses to diuretics and inhibitors of the renin-angiotensin system.10, 11 These inverse relationships reflect the complementary effects of angiotensin-mediated arterial constriction and intra-arterial volume to maintain BP, which reciprocally oppose declines in BP when either system is inhibited.5 Accordingly, we hypothesized that alternative alleles of SNPs associated with BP response to candesartan may have directionally opposite associations with BP response to hydrochlorothiazide.
Our analytical approach was also guided by the opposite direction associations of known predictors with BP responses to candesartan and hydrochlorothiazide.6, 7 Since none of the large number of SNPs tested for associations with BP response to candesartan achieved the p-value threshold for genome-wide significance,12 we guarded against false positives by requiring that the SNPs discovered to be most strongly associated with BP response to candesartan also validated for opposite direction associations with BP response to hydrochlorothiazide in separate samples of each race. In lieu of replicate same-race, same-drug-treated samples, this validation approach was employed because of differences between races in frequencies of the marker SNPs,8 linkage disequilibrium of the markers with functional variants,13 and differences in variances of the BP responses between races.6, 7 Within each race, the number of SNPs strongly associated with BP response to candesartan exceeded what was expected by chance alone, but the identified SNPs differed between races. Only the SNPs discovered in Whites also validated for opposite direction associations with BP response to hydrochlorothiazide.
Obvious candidates to influence BP response to a drug are genes that encode components of the BP-regulating systems targeted by the drug.14 Additional logical candidates are genes encoding components of the counterregulatory systems opposing the decline in BP following drug administration. From this perspective, notable among the SNPs discovered and validated in Whites were SNPs in the gene encoding the the γ-subunit (SCNN1G) of the amiloride-sensitive sodium channel involved in distal renal tubular sodium reabsorption. Gain-of-function mutations in SCNN1G cause Liddle’s syndrome, a rare familial form of low-renin, sodium-volume dependent hypertension that responds to diuretic therapy with amiloride.15 In contrast, treatment of low-renin, sodium-volume dependent hypertension with inhibitors of the renin-angiotensin system has been associated with opposite-direction pressor responses.16 We have previously reported that SCNN1G SNPs (rs5723 and rs5729) in strong linkage disequilibrium with the lead SNP, rs11649420 (r2=0.87), were associated with BP response to hydrochlorothiazide.17 Our present findings that SCNN1G SNPs were among those most strongly associated with BP response to candesartan and had opposite direction associations with BP response to hydrochlorothiazide is consistent with the counterregulatory effect of renal sodium reabsorption to maintain intra-arterial volume and oppose the decline in BP following inhibition of the renin-angiotensin system with candesartan.18
Only three previous studies have reported polymorphisms in candidate genes to predict BP responses to ARBs.19–21 In the Swedish Irbesartan Left Ventricular Hypertrophy Investigation versus Atenolol (SILVHIA) trial, 49 adults with stage I–II hypertension were treated with the irbesartan for three months and 74 SNPs were genotyped in 25 genes encoding regulators of volume, vasoconstriction, and drug metabolism.22 The SNPs found to be associated with BP response were in the genes encoding aldosterone synthase20, 23 (CYP11B2), angiotensinogen,22 angiotensin converting enzyme,19, 22 endothelin receptor type B,22 endothelial nitric oxide synthase,22 apolipoprotein(a),22 hepatic lipase C,22 and a cytochrome P450 enzyme21, 24 (CYP2C9). In the other candidate gene studies of ARB responses, associations were also reported for the same CYP11B2 polymorphism (−344 C/T) with BP response to candesartan20 and the same CYP2C9 polymorphism (*1/*2/*3) with BP response to losartan.21 However, the alleles associated with greater BP lowering different from those found in the SILVHIA trial. Associations of polymorphisms in several of the candidate genes in the SILVHIA trial have also been reported with BP response to diuretic therapy.25–29 However, the alleles associated with greater BP response to diuretic therapy were not consistent across studies, nor were the alternative alleles consistently associated with greater BP response to inhibitors of the renin-angiotensin system.30, 31 Thus, none of the previously reported candidate gene associations with BP responses was replicated across independent samples. Of the previously investigated candidate genes, in the present study only SNPs in SCNN1G were strongly associated with BP response to candesartan and validated for opposite direction associations with BP response to hydrochlorothiazide.
While GWA analyses provide an unbiased opportunity to discover novel variants not previously implicated or suspected to influence BP responses, biologic mechanisms that may account for the other identified SNP associations with BP response to candesartan are not readily apparent. Variation in three of the five other identified genes has been associated with hypertension-related phenotypes: rs1371924 in SLC9A9, which encodes a sodium/hydrogen exchanger,32 was associated with arterial stiffness in the Framingham Heart Study 100K Project;33 rs4149014 in the promoter of SLCO1B1, which encodes an organic anion transporter implicated in statin-induced myopathy,34 was associated with increased risk for primary hypertension in Chinese Uyghurs;35 and MYO3B, which encodes a class III myosin, actin-dependent motor protein expressed in the kidney,36 has been implicated as a potential candidate for a rare syndrome including renal abnormalities and hypertension.37 The other two identified genes, EHF and GPR83, are 64Mbp apart on chromosome 11 and have no reported associations with hypertension-related phenotypes. EHF encodes a transcriptional activator implicated in epithelial cell differentiation and proliferation;38 and GPR83 encodes an orphan G protein-coupled receptor 83 that may be a neuropeptide Y receptor. 39 Variations in plasma renin activity did not account for the lead SNPs’ associations with BP response to candesartan. Considerable additional work will be necessary to replicate the identified novel associations of variation in these genes with opposite direction BP responses to candesartan and hydrochlorothiazide.
Our approach to validation of SNPs associated with BP response to candesartan by requiring opposite direction associations with BP response to hydrochlorothiazide has limitations. Although pretreatment plasma renin activity, race, and age conform to this pattern of inverse associations, other characteristics, e.g., pretreatment BP level, predict same direction BP response to all drugs40 or may only predict BP response to a single class of drugs.2 In Whites, our analyses may not have identified SNPs with same-direction associations with BP response to both candesartan and hydrochlorothiazide or SNPs only associated with BP response to one of the drugs. In African Americans, some of the SNPs that failed to validate for opposite direction associations with BP response to hydrochlorothiazide may still be true predictors of BP response to candesartan.
Perspective
Replication across multiple, well-powered, independent samples has become the gold standard for reliability of genetic associations.41 Because predictors of greater BP response to candesartan are expected also to be associated with lesser BP response to hydrochlorothiazide, we validated the SNPs most strongly associated with BP response to candesartan by testing for opposite direction associations with BP response to hydrochlorothiazide in another sample from the same race. This approach to validating reliability of antihypertensive pharmacogenetic associations is not only feasible in lieu of multiple same-race, same-drug-treated samples but also widely applicable since the majority of antihypertensive drugs target the renin-angiotensin or renal sodium transport systems. Moreover, genetic predictors associated with opposite direction BP responses to drugs that inhibit vasoconstriction and volume-regulating systems maintaining BP hold promise for individualizing antihypertensive drug therapy and identifying novel targets for antihypertensive drug therapy.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What is new?
Genome-wide association analysis of BP to an angiotensin II receptor blocker
SNPs associated with BP response to candesartan validate for opposite direction association with BP response to hydrochlorothiazide
What is relevant?
Such SNPs can identify components of the renin-angiotensin and renal sodium transport systems that reciprocally oppose BP declines when either system is inhibited.
Predictors of directionally opposite BP responses to drugs from different pharmacological classes have greatest utility in individualizing antihypertensive drug therapy.
Summary
SNPs in novel genes predictive of opposite direction associations with BP responses to the vasoconstriction and volume-retaining systems maintaining BP may provide new targets for antihypertensive drug therapy.
Acknowledgments
Technical assistance of Zhiying Wang, Meagan Grove, Jodie Van De Rostyne, Jeremy Palbicki.
SOURCES OF FUNDING
Supported by HL074735, HL053335, and Mayo Foundation.
Footnotes
DISCLOSURES
None.
References
- 1.Preston RA, Materson BJ, Reda DJ, Williams DW, Hamburger RJ, Cushman WC, Anderson RJ. Age-race subgroup compared with renin profile as predictors of blood pressure response to antihypertensive therapy. Department of veterans affairs cooperative study group on antihypertensive agents. JAMA. 1998;280:1168–1172. doi: 10.1001/jama.280.13.1168. [DOI] [PubMed] [Google Scholar]
- 2.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-DeHoff RM, Boerwinkle E, Johnson JA, Bailey KR. Plasma renin activity predicts blood pressure responses to beta-blocker and thiazide diuretic as monotherapy and add-on therapy for hypertension. Am J Hypertens. 2010;23:1014–1022. doi: 10.1038/ajh.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laragh JH, Lamport B, Sealey J, Alderman MH. Diagnosis ex juvantibus. Individual response patterns to drugs reveal hypertension mechanisms and simplify treatment. Hypertension. 1988;12:223–226. doi: 10.1161/01.hyp.12.3.223. [DOI] [PubMed] [Google Scholar]
- 4.Reilly RF, Peixoto AJ, Desir GV. The evidence-based use of thiazide diuretics in hypertension and nephrolithiasis. Clin J Am Soc Nephrol. 2010;5:1893–1903. doi: 10.2215/CJN.04670510. [DOI] [PubMed] [Google Scholar]
- 5.Laragh JH, Sealey JE. The plasma renin test reveals the contribution of body sodium-volume content (v) and renin-angiotensin (r) vasoconstriction to long-term blood pressure. Am J Hypertens. 2011;24:1164–1180. doi: 10.1038/ajh.2011.171. [DOI] [PubMed] [Google Scholar]
- 6.Chapman AB, Schwartz GL, Boerwinkle E, Turner ST. Predictors of antihypertensive response to a standard dose of hydrochlorothiazide for essential hypertension. Kidney Int. 2002;61:1047–1055. doi: 10.1046/j.1523-1755.2002.00200.x. [DOI] [PubMed] [Google Scholar]
- 7.Canzanello VJ, Baranco-Pryor E, Rahbari-Oskoui F, Schwartz GL, Boerwinkle E, Turner ST, Chapman AB. Predictors of blood pressure response to the angiotensin receptor blocker candesartan in essential hypertension. Am J Hypertens. 2008;21:61–66. doi: 10.1038/ajh.2007.24. [DOI] [PubMed] [Google Scholar]
- 8.Turner ST, Bailey KR, Fridley BL, Chapman AB, Schwartz GL, Chai HS, Sicotte H, Kocher JP, Rodin AS, Boerwinkle E. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension. 2008;52:359–365. doi: 10.1161/HYPERTENSIONAHA.107.104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochran WG. Some methods for strengthening the common chi-square tests. Biometrics. 1954;10:417–451. [Google Scholar]
- 10.Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- 11.Fisher RA. Statistical methods for research workers. Edinburgh, London: Oliver and Boyd; 1925. [Google Scholar]
- 12.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 13.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 14.Turner ST, Schwartz GL, Chapman AB, Hall WD, Boerwinkle E. Antihypertensive pharmacogenetics: Getting the right drug into the right patient. J Hypertens. 2001;19:1–11. doi: 10.1097/00004872-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 15.Hansson JH, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton RP. Hypertension caused by a truncated epithelial sodium channel gamma subunit: Genetic heterogeneity of liddle syndrome. Nat Genet. 1995;11:76–82. doi: 10.1038/ng0995-76. [DOI] [PubMed] [Google Scholar]
- 16.Alderman MH, Cohen HW, Sealey JE, Laragh JH. Pressor responses to antihypertensive drug types. Am J Hypertens. 2010;23:1031–1037. doi: 10.1038/ajh.2010.114. [DOI] [PubMed] [Google Scholar]
- 17.Turner ST, Schwartz GL, Chapman AB, Boerwinkle E. Wnk1 kinase polymorphism and blood pressure response to a thiazide diuretic. Hypertension. 2005;46:758–765. doi: 10.1161/01.HYP.0000186240.81996.57. [DOI] [PubMed] [Google Scholar]
- 18.Gavras H, Brunner HR, Laragh JH, Sealey JE, Gavras I, Vukovich RA. An angiotensin converting enzyme inhibitor to identify and treat vasoconstrictor and volume factors in hypertensive patients. N Engl J Med. 1974;291:817–821. doi: 10.1056/NEJM197410172911603. [DOI] [PubMed] [Google Scholar]
- 19.Kurland L, Melhus H, Karlsson J, Kahan T, Malmqvist K, Ohman KP, Nystrom F, Hagg A, Lind L. Angiotensin converting enzyme gene polymorphism predicts blood pressure response to angiotensin ii receptor type 1 antagonist treatment in hypertensive patients. J Hypertens. 2001;19:1783–1787. doi: 10.1097/00004872-200110000-00012. [DOI] [PubMed] [Google Scholar]
- 20.Ortlepp JR, Hanrath P, Mevissen V, Kiel G, Borggrefe M, Hoffmann R. Variants of the cyp11b2 gene predict response to therapy with candesartan. Eur J Pharmacol. 2002;445:151–152. doi: 10.1016/s0014-2999(02)01766-1. [DOI] [PubMed] [Google Scholar]
- 21.Sekino K, Kubota T, Okada Y, Yamada Y, Yamamoto K, Horiuchi R, Kimura K, Iga T. Effect of the single cyp2c9*3 allele on pharmacokinetics and pharmacodynamics of losartan in healthy japanese subjects. Eur J Clin Pharmacol. 2003;59:589–592. doi: 10.1007/s00228-003-0664-5. [DOI] [PubMed] [Google Scholar]
- 22.Liljedahl U, Karlsson J, Melhus H, Kurland L, Lindersson M, Kahan T, Nystrom F, Lind L, Syvanen AC. A microarray minisequencing system for pharmacogenetic profiling of antihypertensive drug response. Pharmacogenetics. 2003;13:7–17. doi: 10.1097/00008571-200301000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Kurland L, Melhus H, Karlsson J, Kahan T, Malmqvist K, Ohman P, Nystrom F, Hagg A, Lind L. Aldosterone synthase (cyp11b2) −344 c/t polymorphism is related to antihypertensive response: Result from the swedish irbesartan left ventricular hypertrophy investigation versus atenolol (silvhia) trial. Am J Hypertens. 2002;15:389–393. doi: 10.1016/s0895-7061(02)02256-2. [DOI] [PubMed] [Google Scholar]
- 24.Hallberg P, Karlsson J, Kurland L, Lind L, Kahan T, Malmqvist K, Ohman KP, Nystrom F, Melhus H. The cyp2c9 genotype predicts the blood pressure response to irbesartan: Results from the swedish irbesartan left ventricular hypertrophy investigation vs atenolol (silvhia) trial. J Hypertens. 2002;20:2089–2093. doi: 10.1097/00004872-200210000-00030. [DOI] [PubMed] [Google Scholar]
- 25.Jiang X, Sheng HH, Lin G, Li J, Lu XZ, Cheng YL, Huang J, Xiao HS, Zhan YY. Effect of renin-angiotensin-aldosterone system gene polymorphisms on blood pressure response to antihypertensive treatment. Chin Med J (Engl) 2007;120:782–786. [PubMed] [Google Scholar]
- 26.Li Y, Zhou Y, Yang P, Niu JQ, Wu Y, Zhao DD, Wu SL. Interaction of ace and cyp11b2 genes on blood pressure response to hydrochlorothiazide in han chinese hypertensive patients. Clin Exp Hypertens. 2011;33:141–146. doi: 10.3109/10641963.2010.531838. [DOI] [PubMed] [Google Scholar]
- 27.Wu SL, Li Y, Liu KJ, Hou GS, Wang JJ, Wu YT, Song SM. [association of polymorphisms in ace and cyp11b2 genes with antihypertensive effects of hydrochlorothiazide] Zhonghua Xin Xue Guan Bing Za Zhi. 2005;33:595–598. [PubMed] [Google Scholar]
- 28.Sciarrone MT, Stella P, Barlassina C, Manunta P, Lanzani C, Bianchi G, Cusi D. Ace and alpha-adducin polymorphism as markers of individual response to diuretic therapy. Hypertension. 2003;41:398–403. doi: 10.1161/01.HYP.0000057010.27011.2C. [DOI] [PubMed] [Google Scholar]
- 29.Turner ST, Chapman AB, Schwartz GL, Boerwinkle E. Effects of endothelial nitric oxide synthase, alpha-adducin, and other candidate gene polymorphisms on blood pressure response to hydrochlorothiazide. Am J Hypertens. 2003;16:834–839. doi: 10.1016/s0895-7061(03)01011-2. [DOI] [PubMed] [Google Scholar]
- 30.Turner ST, Schwartz GL. Gene markers and antihypertensive therapy. Curr Hypertens Rep. 2005;7:21–30. doi: 10.1007/s11906-005-0051-y. [DOI] [PubMed] [Google Scholar]
- 31.Mellen PB, Herrington DM. Pharmacogenomics of blood pressure response to antihypertensive treatment. J Hypertens. 2005;23:1311–1325. doi: 10.1097/01.hjh.0000173510.52987.68. [DOI] [PubMed] [Google Scholar]
- 32.Ohgaki R, van ISC, Matsushita M, Hoekstra D, Kanazawa H. Organellar na+/h+ exchangers: Novel players in organelle ph regulation and their emerging functions. Biochemistry (Mosc) 2011;50:443–450. doi: 10.1021/bi101082e. [DOI] [PubMed] [Google Scholar]
- 33.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, Vasan RS, Mitchell GF. Framingham heart study 100k project: Genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8 (Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. Slco1b1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 35.Lin R, Wang X, Zhou W, Fu W, Wang Y, Huang W, Jin L. Association of polymorphisms in the solute carrier organic anion transporter family member 1b1 gene with essential hypertension in the uyghur population. Ann Hum Genet. 2011;75:305–311. doi: 10.1111/j.1469-1809.2010.00622.x. [DOI] [PubMed] [Google Scholar]
- 36.Dose AC, Burnside B. A class iii myosin expressed in the retina is a potential candidate for bardet-biedl syndrome. Genomics. 2002;79:621–624. doi: 10.1006/geno.2002.6749. [DOI] [PubMed] [Google Scholar]
- 37.Waters AM, Beales PL. Bardet-biedl syndrome. 1993. [Google Scholar]
- 38.Kleinbaum LA, Duggan C, Ferreira E, Coffey GP, Buttice G, Burton FH. Human chromosomal localization, tissue/tumor expression, and regulatory function of the ets family gene ehf. Biochem Biophys Res Commun. 1999;264:119–126. doi: 10.1006/bbrc.1999.1493. [DOI] [PubMed] [Google Scholar]
- 39.De Moerlooze L, Williamson J, Liners F, Perret J, Parmentier M. Cloning and chromosomal mapping of the mouse and human genes encoding the orphan glucocorticoid-induced receptor (gpr83) Cytogenet Cell Genet. 2000;90:146–150. doi: 10.1159/000015650. [DOI] [PubMed] [Google Scholar]
- 40.Gill JS, Zezulka AV, Beevers DG, Davies P. Relation between initial blood pressure and its fall with treatment. Lancet. 1985;1:567–569. doi: 10.1016/s0140-6736(85)91219-x. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.