Abstract
Numerous studies have shown that resistance to oxidative stress is crucial to stay healthy and to reduce the adverse effects of aging. Accordingly, nutritional interventions using antioxidant food-grade compounds or food products are currently an interesting option to help improve health and quality of life in the elderly. Live lactic acid bacteria (LAB) administered in food, such as probiotics, may be good antioxidant candidates. Nevertheless, information about LAB-induced oxidative stress protection is scarce. To identify and characterize new potential antioxidant probiotic strains, we have developed a new functional screening method using the nematode Caenorhabditis elegans as host. C. elegans were fed on different LAB strains (78 in total) and nematode viability was assessed after oxidative stress (3 mM and 5 mM H2O2). One strain, identified as Lactobacillus rhamnosus CNCM I-3690, protected worms by increasing their viability by 30% and, also, increased average worm lifespan by 20%. Moreover, transcriptomic analysis of C. elegans fed with this strain showed that increased lifespan is correlated with differential expression of the DAF-16/insulin-like pathway, which is highly conserved in humans. This strain also had a clear anti-inflammatory profile when co-cultured with HT-29 cells, stimulated by pro-inflammatory cytokines, and co-culture systems with HT-29 cells and DC in the presence of LPS. Finally, this Lactobacillus strain reduced inflammation in a murine model of colitis. This work suggests that C. elegans is a fast, predictive and convenient screening tool to identify new potential antioxidant probiotic strains for subsequent use in humans.
Introduction
Aerobic metabolism leads to the production of harmful byproducts. Organisms can only stay healthy by reducing natural by-products of oxygen metabolism, such as reactive oxygen species (ROS), which are mainly produced by mitochondria [1] and damage proteins, lipids and DNA on accumulating in cells [2], [3], [4], [5]. Oxidative stress plays an important role in the rate of aging processes, often referred to as the Mitochondrial Free Radical Theory of Aging [6], [7], [1]. This process is also a key factor in aging-associated degenerative diseases such as certain types of cancer, diabetes and heart failure, among others [4], [7]. Oxidative stress also plays an obvious role in the pathogenesis of a number of gastrointestinal diseases, including: gastric and duodenal ulcer disease, pancreatitis, IBD; gastric, esophageal, and colon cancers [8], [9]. Although the causal role of ROS in aging remains controversial, recent reports suggest that ROS mediate a stress response to age-related damage, rather than being the underlying cause of aging [10]. Moreover, ROS are shown to have protective effects in model organisms such as Saccharcomyces cerevisae, Caenorhabditis elegans and Drosophila melanogaster [11].
The search for new nutritional antioxidant compounds able to counterbalance the effects of oxidative stress poses an exciting research challenge. There is increasing pre-clinical and clinical evidence that nutritional interventions, using food products or food-grade compounds, can protect against oxidative stress [12], [13], [14], [15]. Probiotics, defined by FAO/WHO in 2001 as live micro-organisms which, when administered in adequate amounts, confer a health benefit to the host [16], can be good candidates for providing such antioxidant effects. While various methodologies have been developed to identify natural LAB strains with intrinsic antioxidant properties [17], [18], there are insufficient methods available for the fast and reliable massive screening of bacterial culture collections in vivo using complex multicellular organisms. Indeed, only a few natural antioxidant lactic acid bacteria (LAB) have been characterized in animals so far [19]. In systems mimicking colon fermentation, Lactobacillus paracasei Fn032, Lactobacillus rhamnosus GG and Lactobacillus spp Fn 001 have been shown to prevent hydroxyl radical production [20]. Moreover, it has been shown that orally-administered live recombinant LAB (Lactococcus lactis or Lactobacillus plantarum strains) producing bacterial SOD can improve TNBS-induced colitis in rats [21], [22] and mice (Foligné et al., unpublished data).
However, laboratory animals cannot be used in the first steps of screening large sets of strains and, thus, there is the need for more convenient and predictable screening tools for natural antioxidant LAB. In this respect, the use of less evolved animals may provide an attractive alternative [23], [24].
To identify new natural LAB strains with antioxidant effect, we have developed a completely new and highly predictive method that uses Caenorhabditis elegans as an in vivo screening model. Caenorhabditis elegans is an extremely powerful and well-studied biological system, which has been used as a model to study aging and related diseases [25], [10], [26]. This nematode is a good biological model for genetic studies [27] since many of its pathways are conserved in humans. Oxidative damage and its effects on aging have been studied in a C. elegans model system using a nematode mutant strain exhibiting hyper-resistance to oxidative stress, as compared to its parental strain [28]. Moreover, C. elegans proved an appropriate model to screen potential anti-Salmonella infection bacteria [29] and antibacterial compounds [30]. Furthermore, C. elegans longevity is known to be related to the insulin pathway and to orthologous genes in humans, involved in the insulin-like growth factor and diabetes [31], [32]. So far, little is known about the potential use of C. elegans as a screening tool for probiotic bacteria inducing resistance against oxidative stress or improving longevity.
Here, we have used the nematode Caenorhabditis elegans as a new preclinical model to carry out preliminary antioxidant screenings to identify potential probiotic strains and to provide insights into the mechanisms by which these strains lower oxidative stress. This animal model has enabled us to identify a new strain of Lactobacillus rhamnosus, designated CNCM I-3690, which exerted a strong antioxidant effect and extended nematode lifespan through the insulin-like pathway DAF-2/DAF-16. Furthermore, since inflammation can be associated with generation of ROS leading to oxidative stress [33], we observed that CNCM I-3690 strain might have the ability to protect against oxidative stress through an anti-inflammatory effect. These findings suggest that C. elegans can be a good predictive screening tool for new potential probiotic strains.
Materials and Methods
Cultures of Lactic Acid Bacteria (LAB) and Bifidobacteria
We included 78 bacterial strains from a Danone Research collection in the in vivo antioxidant screening methodology, using the model organism C. elegans, adapted from a previously reported protocol performed to measure the antioxidant activity of plant extracts [12]. This collection is composed by 62 Lactobacillus strains belonging to acidophilus, bulgaricus, casei, paracasei, plantarum and rhamnosus species, 9 Streptococcus thermophilus isolates and 6 Bifidobacterium strains belonging to the animalis, breve and longum species. See Table S1 for genera and species specifications of each strain used in this study. The strains belonging to Bifidobacterium, Lactobacillus and Streptococcus genera were grown in MRS with cysteine, MRS and Elliker media, respectively. As the bioassay of the in vivo antioxidant activity is to be carried out with samples of live cells of different LAB, cells must be recovered in the logarithmic phase growth. After growth curve analysis, the optimal time for cell recovery was established to be after 15 h of incubation at OD600 = 1, 1.5 and 1.7 for Streptococcus, Lactobacillus and Bifidobacterium, respectively. The different LAB cultures were added to the mixture at a final concentration of 4×106 cells/mL. See Supplementary Material S1 for growth curves of representative strains for each genus and detailed protocol.
In addition, we analyzed the sensitivity of bacterial strains to kanamycin, an antibiotic used to inhibit Escherichia coli growth. A concentration of 30 µg/mL was sufficient to inhibit E. coli OP50.
Cultures of Caenorhabditis Elegans
Experiments were carried out with a C. elegans mutant strain BA17 fem-1(hc17) (Caenorhabditis Genetics Center at the University of Minnesota, USA) which is infertile at 25°C. BA17 worms were synchronized by isolating eggs from gravid adults at 20°C, hatching the eggs overnight in M9 buffer and isolating L1-stage worms in the wells of a microtiter plate. The worms were grown without shaking for three days at 25°C and 80–85% relative humidity. After this incubation period, adult worms were subjected to oxidative stress with H2O2 (3 mM and 5 mM) or without H2O2 (no stress control). Two controls were used during this experiment: wells with Escherichia coli instead of LAB as the control of bacterial feeding and wells with E. coli and 3 mM or 5 mM H2O2 as the control for oxidative stress. Worms were incubated in these conditions for 5 h. In terms of scoring for antioxidant capacity, we considered paralyzed worms to be dead (stressed).
Oxidative Stress Assays on Agar Plates
In these experiments, we used the C. elegans wild type strain N2 (Caenorhabditis Genetics Center at the University of Minnesota, USA) to validate the potential antioxidant activity of a group of selected bacterial strains. For this purpose, worms were grown in NG medium (Nematode Growth medium: Agar 17.5 g/l, Sodium Chloride 3.0 g/l, Peptone 2.5 g/l, Cholesterol 0.005 g/l) on agar plates with a lawn of E.coli OP50 (control media) and NG with a lawn of each LAB. Worms were incubated at 20°C for 5 days, and then transferred to a medium with 3 mM of H2O2. After 5 h of incubation, worm viability was scored.
Longevity Assays in C. elegans
To measure the lifespan of C. elegans, synchronized worms of the wild-type strain (N2) and the mutant strains LG333 (skn-1), GR1307 (daf-16) and CB1370 (daf-2) were grown at 20°C until they reached the young adult stage. All mutant strains of C. elegans were obtained from the Caenorhabditis Genetics Center at the University of Minnesota (USA). Worms were then transferred to NGM agar plates covered with lawns of E. coli OP50 or the corresponding LAB (CNCM I-3690 or CNCM I-4317). The plates were incubated at 20°C and the numbers of live and dead worms were scored every two days. Parents were moved periodically to new plates to separate them from their progeny. A worm was considered as dead if it failed to respond to a platinum wire. Three independent assays were carried out with each strain.
Transcriptomic Analysis in C. elegans
Gene expression in C. elegans wild-type strain (N2) was analyzed in worm populations fed with E. coli OP50 (control condition) or the corresponding LAB (Lactobacillus rhamnosus CNCM I-3690 or Lactobacillus rhamnosus CNCM I-4317). Three days feeding period was analyzed (young-adult worms). Synchronized populations were obtained from embryos isolated from gravid adults in the different feeding conditions. After feeding period (3 days), samples of worms were collected with M9 buffer, washed three times and collected in eppendorf tubes for worm disruption by sonication (3 pulses at 10 W, 20 s/pulse). Total RNA isolation was performed with RNAasy Kit (Qiagen, Barcelona, Spain). RNA samples were processed for hybridization using the GeneChip® C. elegans Genome Array of Affymetrix (UCIM, University of Valencia). These chips contain oligonucleotide probesets designed to asses over 22500 transcripts from the C. elegans genome. Four biological replicates were examined per condition using Bioinformatics (CIPF, Valencia, Spain). Raw data obtained from Affymetrix arrays were background corrected using RMA methodology [34]. Signal intensity was standardized across arrays via quantile normalizaton algorithm. Differential gene expression assessment between control and treated conditions was carried out using limma moderated t-statistics. The p-values obtained for each gene were adjusted with multiple testing p-value correction procedures [35]. Finally, gene set analysis was carried out for each comparison using logistic regression models ner [36].
Microarray Data Analysis
Data analysis was accomplished in “R” (R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org) mainly through packages in the “Bioconductor” suite [37]. Normalization was performed using rma (robust multi-array average expression measure) in “affy” [38] and differential expression was assayed via “limma” [39]. Genes were considered differentially expressed when the multiple testing adjusted P-value <0,01. Ontology analysis was performed with “GeneAnswers” (R package version 1.8.0).
The microarray data produced and discussed in this work have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE42192 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE42192).
RT-qPCR Experiments and Analysis
Total RNA was extracted using RNeasy (Qiagen), and reverse transcribed using RevertAid H Minus Reverse Transcriptase (Fermentas). The resulting cDNA was subjected to RT-qPCR analysis using SYBR Green detection (LightCycler® FastStart DNA MasterPLUS SYBR Green, Roche) on LightCycler (Roche). Oligonucleotides for RT-qPCR were designed using Primer3Plus. All CT values were normalized against the control gene Y45F10D.4, which did not vary under the conditions being tested. All experiments were repeated at least three times (biological replicates) and were internally controlled (technical replicate). Expression changes were obtained by calculating the relative expression levels using the 2−ΔΔCT method. For expression ratio and comparison between microarray data and RT-qPCR data see Supplementary Material S8.
Nile Red Staining
The effect of the selected lactic bacteria strains on C. elegans body fat reduction was studied via fluorescence measurement in Red Nile (0.05 µg/mL, Sigma, St. Louis, USA) stained nematodes. Worms of the wild type strain N2 and the strain BX153, mutant in the 9 stearoyl-CoA fatty acid desaturase FAT-7, were cultured in NGM plates in the presence of E. coli OP50, Lactobacillus rhamnosus CNCM I-3690 or Lactobacillus rhamnosus CNCM I-4317. Fluorescence of age-synchronized young-adult worms (3-days-old) was quantified in a VersaFluorTM Fluorometer System (Bio-Rad, Hercules, USA). A total of 180 worms per condition were analyzed. Experiments were carried out in triplicate.
Epithelial Cell Co-cultures
HT-29 (HTB-38, ATCC) cells were cultured in DMEM supplemented with 10% fetal bovine serum. Cells were routinely propagated in 25 or 75 cm2 tissue culture flasks at 37°C, 5% CO2 in a humidified incubator until reaching 80–90% confluence. Subsequently, cells were trypsinized, adjusted the cellular concentration and used for different experiments. The initial passage number of the HT-29 wild-type cells was 5. Cells used for each assay were cultured for less than 15–20 passages.
HT-29 cells were seeded in a 12 well plate at a cell density of 2×105/well using a final volume of 2 mL and incubated at 37°C, 5% CO2 for 24 h. Cells were transfected with the pIgK-luciferase plasmid using a DNA: Lipofectamine 2000 ratio of 1∶2.5 (µg:µL). The NF-κB reporter system by using the pIgK-luciferase plasmid was kindly provided by Philippe Sansonetti’s group at Institut Pasteur (Paris, France), and is referred in [40].
Bacteria were grown overnight at 37°C in MRS broth (Oxoid, Cambridge, UK), then subcultured, and harvested by centrifugation (5 min at 3000 g). On co-culture day, bacteria were washed twice with PBS buffer and re-suspended in DMEM. A correlation curve between A570 nm and colony forming units was performed for each strain and A570 nm values were used for calculating the bacterial number used in each experiments.
Twenty four hours post-transfection, media was renewed and cells were pre-incubated for 2 hours with each bacterial strain using a MOI of 100 bacteria per cell. The MOI was determined taking into account the initial cell density of 2×105 cells/well. After 2 hours of pre-incubation with bacteria, an inflammatory stimulus was added to the cell without washing bacteria. The pro-inflammatory cytokine cocktail consisted of: TNF-α, IL-1β and IFN-γ; 50 ng, 2.5 ng and 7.5 ng/well, respectively and was applied for 6 hours. Viability of epithelial cells was controlled by trypan blue exclusion method. After this incubation time, supernatants were collected, cleared by centrifugation at 3000 g for 5 minutes, transferred to a new tube and finally frozen at −80°C for further cytokine quantification by Flow Cytometry and/or Luminex. Cells were rinsed with PBS and immediately lysated with Luciferase Cell Culture Lysis Reagent 1X (Promega E1500) according to the manufacturer instructions. Cell lysates were frozen at −80°C for further determination of I-κB.
Heterotypic Co-culture Systems
Non-polarized HT-29-NF-κB-luciferase cells (passage 25–35) were grown in the upper chamber of a transwell filter (3 µm diameter of pores; Costar, USA) and incubated for 2 days in RPMI supplemented with 10% heat-inactivated FCS. Monocyte-derived DCs were generated according to a modified protocol described by Dauer et al. [41]. Human PBMC were obtained after Ficoll-Hypaque density-gradient centrifugation of leukoreduction system chambers of plateletpheresis from healthy donors. Briefly, monocytes from PBMC were purified by plastic adherence. For DC differentiation, monocytes were incubated for 2 days in RPMI supplemented with 10% (v/v) heat-inactivated FBS, 800 U/mL GM-CSF (Clausen, Uruguay) and IL-4 (1% of conditioned supernatant from IL-4 transfected J588L cell line). Immature DC were harvested and cultured in twelve-well tissue culture plates (1.25×105 cells/well) for experiments.
Bacteria were grown overnight at 37°C in MRS broth (Oxoid, Cambridge, UK), then subcultured, and harvested by centrifugation (5 min at 3000×g). On co-culture day, bacteria were washed twice with PBS buffer and re-suspended in RPMI. A correlation curve between A570 nm and colony forming units was performed for each strain and A570 nm values were used for calculating the bacterial number used in each experiments. Bacteria were added apically in a 25∶1 relation (bacteria: DC). In addition to the correlation curve, the same day of each experiment, bacterial cells were inoculated in MRS plate, cultured ON and the colony number was determined. We considered valid only those experiments where a ratio of 25∶1 was achieved. Viability of epithelial cells and DC was controlled by trypan blue exclusion method.
LPS from E. coli O26:B6 (Sigma, Saint Louis, US) was added apically to a final concentration of 0.5 µg/mL per well. The co-culture was incubated for 18 h at 37°C in a 5% CO2 humidified atmosphere. HT-29-NF-κB-luciferase cells were harvested and lysed for protein and luminiscence determination and DCs surface markers were stained and analysed by flow cytometry. Co-culture supernatant was collected, cleared by centrifugation at 3000 g for 5 minutes, transferred to a new tube and finally frozen at −80°C for further cytokine quantification.
Protein and Luminescence Determination
Protein content of the cell lysate was determined by BCA method (SIGMA, Saint Louis, US) according to the manufacturer’s instructions. For the luminescence assay, luciferase kit was used according to the manufacturer’s instructions (Promega, Madison, USA). Luciferase was quantified in a luminometer with 0.5 second of measurement per well and 0.5 second delay, at maximum gain (LUMIstar Optima, BMG).
Cytokine Measurements
IL-8, IL-10, IL-6, TNF-α, and IL-12 concentrations were determined by FlowCytomix™ technology (Bender MedSystems, Austria) and analyzed by flow cytometry. For results calculation BMS FlowCytomix Software version 2.2.1 was used.
Flow Cytometry and Antibodies
The following antibodies (all purchased from BD PharMingen) were used for flow cytometry stainings: B-ly6 (anti-human CD11c, allophycocyanin-conjugated), 2331 (anti-human CD86, phycoerythrin (PE)-conjugated), TU36 (anti-human HLA-DR fluorescein isothiocyanate (FITC)-conjugated), HIB19 (anti-human CD19 PE-conjugated), HIT3a (anti-human CD3 FITC-conjugated), M5E2 (anti-human CD14 FITC-conjugated), all with matched isotype controls. The level of expression was analyzed as the median of the fluorescence intensity (MFI). DCs were stained and then analyzed using a CyAn™ ADP (DAKO) flow cytometer and Summit 4.3 software. For each analysis 10,000 counts were recorded, gated on FSC vs SSC dot plot. For results comparison the following equation was used:
In vivo Colitis Murine Mice Model
Ethics statement
Animal experiments were performed in an accredited establishment (number A59107; animal facility of the Institut Pasteur de Lille, France) and carried out in accordance with the guidelines of laboratory animal care published by the regional ethical committee (CREEA Nord-Pas de Calais) and subject to the rules of the European Union Normatives (number 86/609/EEC).
The name of the Institutional Animal Care and Use Committee (IACUC) is: “CEEA Nord Pas de Calais” as the regional ethical commitee for animal experiments, headed by Dr Dombrowicz (President), (mail: ceea.npdc@univ-lille2.fr). It has approved the study with the acceptance number CEEA/AF 019/2009.
All animal experiments were performed following the guidelines of the Institut Pasteur de Lille animal study board, which conforms to the Amsterdam Protocol on animal protection and welfare, and Directive 86/609/EEC on the Protection of Animals Used for Experimental and Other Scientific Purposes, updated in the Council of Europe's Appendix A complied with the French law (n° 87-848 dated 19-10-1987) and the European Communities: Amendment of Cruelty to Animals Act 1976. All manipulations involving animals were carried out by qualified personnel. The animal house was placed under the direct control of the Institut Pasteur de Lille director who is the “designated responsible person” under French law. The study has been approved by Ethical Committee for experiments on animals of the Région Nord-Pas-de-Calais (ceea.npdc@univ-lille2.fr), approval number AF 19/2009.
BALB/c mice (female, 8- week-old females) were obtained from Charles River (St Germain sur l’Arbresle, France). A standardized murine TNBS colitis model was used to induce acute levels of inflammation [42]. Briefly, anesthetized mice received an intra-rectal administration of 50 ml solution of 2,4,6-trinitrobenzene sulfonic acid (TNBS, Sigma-Aldrich Chemical, France) (100 mg/kg) dissolved in 0.9% NaCl/ethanol (50/50 v/v). Following the onset of colitis, animals were observed and weighted daily according to the animal welfare standards. No excessive inflammation was depicted during this study, as observed and measured by limit points, i.e. i) body weight loss over 20% of initial ii) prominent haunchbacked posture and iii) ruffled fur. Mice were euthanized by prompt cervical dislocation performed by trained individuals. Colons were removed at sacrifice, 72 h after administration, washed and opened. Inflammation grading was performed blindly using the Wallace scoring method [43], reflecting both the intensity of inflammation and the extent of the lesions. The protective effect of LAB was studied following 5-day s-intragastric administration of 108 CFU live bacteria in physiologic buffer or vehicle. A commercial preparation of prednisone (Cortancyl, Sanofi Aventis, France) was used as positive control of protection and was also orally administered for 5 subsequent days at 10 mg/kg. Histological analysis was performed on May-Grünwald-Giemsa stained 5 µm tissue sections from colon samples fixed in 10% formalin and embedded in paraffin and tissue lesions were scored according to the Ameho criteria [44].
Statistical Analysis
Data are expressed as the mean+/−standard deviation. Statistical analysis was determined using unpaired Student t test and One-Way ANOVA using GraphPad Prism Sofware version 5.00 Demo (San Diego, CA). Differences were considered statistically significant if p<0.05. Survival curves were compared using the log rank survival significance test, provided by GraphPad Prism 4 statistical software package.
Epithelial co-cultures were performed 3 times independently (n = 3). For each set of experiments, an internal control of HT-29 cells stimulated by pro-inflammatory cytokines without bacteria was used. Results were expressed in comparison to this internal control.
For heterotypic co-cultures data were expressed as the mean+/−standard deviation of triplicates. Experiments were performed with two different donors. Statistical analysis was determined using unpaired Student t test and One-Way ANOVA using GraphPad Prism Sofware version 5.00 Demo (San Diego, CA). Differences were considered statistically significant if p<0.05.
For in vivo colitis experiments, comparisons between the different animal groups were analyzed by the non-parametric one–way analysis of variance, Mann-Whitney U test. Differences were considered to be statistically significant when the p-value was <0.05.
Results
Antioxidant Screening and C. elegans Lifespan
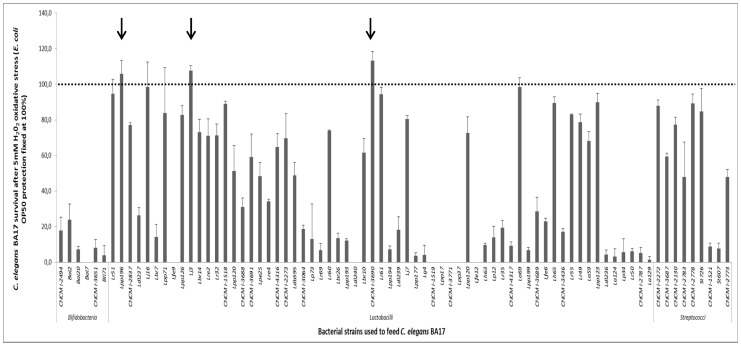
A pre-selection was made of 78 strains of lactic acid bacteria (LAB) and Bifidobacteria, taking into account their resistance to in vitro assays mimicking gastro-intestinal stresses, food safety and growth properties in acidic conditions. As a preliminary step, we verified that C. elegans exhibits similar growth on LAB strains as on E. coli OP50 strains, routinely used for feeding. Next, we fed worms with each of the 78 bacterial strains and we tested C. elegans survival rates upon H2O2-induced stress. Survival upon oxidative stress was determined in C. elegans BA17 by measuring worm viability after a 5-hour-long treatment at the previously determined optimal dose of 5 mM H2O2 [12]. To better determine LAB strain-induced protective effects, we compared them to the protective effect exerted by the control E. coli OP50, set at 100% (Figure 1). Under liquid experimental conditions and without any oxidative stress, LAB and Bifidobacteria didn’t affect worm viability (Figure S1). Upon oxidative stress, Bifidobacteria and Streptococci strains did not confer resistance to oxidative stress and only very few strains of Lactobacillus (strains Lpp196, Lj3 and CNCM I-3690, corresponding to L. casei, L. jonhsonii and L. rhamnosus, respectively) were able to confer a slight resistance to oxidative stress at a higher level than that conferred by the control strain (more than 100%). To validate the previous results, a second antioxidant screening was performed, in which the three potentially antioxidant strains identified in the first analysis were fed to C. elegans wild type N2 on agar plates. In this assay, the viability of worms was determined after being subjected to a treatment with 3 mM of H2O2 for 5 h. In these solid medium conditions, only the L. rhamnosus CNCM I-3690 proved significantly effective against oxidative stress with a level of protection of 171.3% (+/−8.1%) versus 67.6% (+/−2.1%) for Lpp196. The strain Lj3 didn’t show any protection in agar plates.
Figure 1. Screening for antioxidant bacteria in C. elegans.
Survival upon a 5 mM H2O2 treatment for 5 h of C. elegans BA 17 strain fed for 3 days with Bifidobacteria, Lactobacilli and Streptococci strains in liquid medium. Worms were fed with E. coli OP 50, for larvae 1st stage synchronization. E. coli OP 50 strain was inhibited with antibiotics prior of LAB and Bifidobacteria feeding. Protection by E. coli OP50 is fixed at 100%.
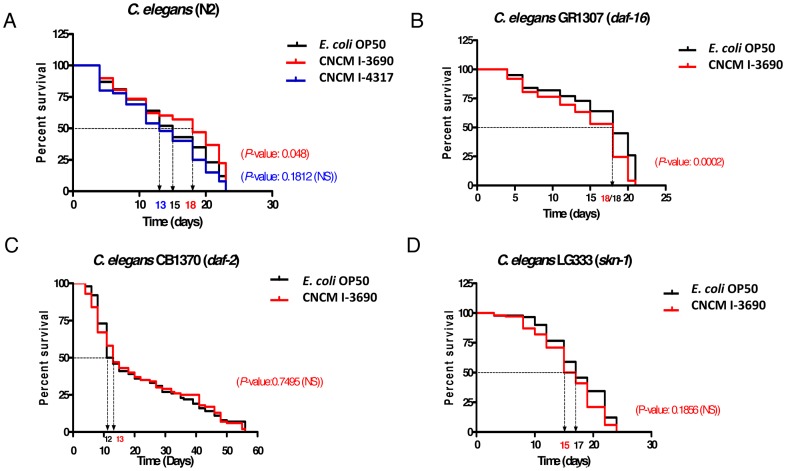
To further confirm the antioxidant properties of L. rhamnosus CNCM I-3690, and to analyze its potential anti-aging effect, we fed C. elegans wild type N2 with this strain and compared the lifespan of these worms with those fed on E. coli OP50. Figure 2A shows that feeding with strain CNCM I-3690 improved the average lifespan with 3 days compared to worms fed with the OP50 (lifespan of 18 versus 15 days). The protective effect of L. rhamnosus CNCM I-3690 became evident in worms after 7 days of feeding and a 20% increase in worm survival was recorded after 15 days.
Figure 2. Lactobacillus rhamnosus CNCM I-3690 increases lifespan of C. elegans wild-type N2 strain and this effect is at least partially dependent of the DAF2-DAF16 pathway.
A. C. elegans N2 wild-type strain was fed with L. rhamnosus CNCM I-3690, CNCM I-4317 and E. coli OP 50 and survival of worms was followed for 21 days. Mean lifespan, indicating the time in days where half of the worm population is still alive, is shown on the X-axis for the three situations. Curves comparisons are indicated (P-values) CNCM I-3690/CNCM I-4317 vs E. coli OP50 (red) and CNCM I-4317 vs E. coli OP50 (blue). NS: Non statistical difference. B, C, D. C. elegans daf-16 (GR1307), daf-2 (CB1370) and skn-1 (LG333) loss-of-function mutant strains were fed with L. rhamnosus CNCM I-3690 and E. coli OP 50. Survival of worms was followed up for 21 days. B. Curves comparisons vs E. coli OP50 are indicated in red (P-values).
Altogether, these results show that, among the 78 LAB strains analyzed; only Lactobacillus rhamnosus CNCM I-3690 strain induced a strong resistance to oxidative stress in C. elegans.
Comparative Transcriptomic Analysis in C. elegans Reveals a Specific Profile for CNCM I-3690 Strain
To better understand the mechanisms underlying the antioxidant effect and longevity in C. elegans induced by CNCM I-3690 consumption, we performed a comparative transcriptomic analysis. To do so, transcriptomes of CNCM I-3690 were compared to those of a control L. rhamnosus strain, CNCM I-4317, unable to reverse oxidative stress in the previous screening (9.3% of protection, see Figure 1), or to produce a statistically significant increase in C. elegans lifespan (Figure 2A and Table S2). Worms were fed on either L. rhamnosus strains (CNCM I-3690 or CNCM I-4317) or on E. coli (OP50) for 3 days and antioxidant effects were monitored. It confirmed that L. rhamnosus CNCM I-3690 was the only strain with an antioxidative phenotype in worms in agar plates (data not shown).
A comparative transcriptomic analysis of the three strains was carried out using the young developmental stage. In young C. elegans fed on CNCM I-3690, microarrays revealed 25 differentially expressed genes compared to CNCM I-4317-fed worms, and 1278 differentially expressed genes compared to OP50-fed worms (see Supplementary Material S2 for a detailed table of these genes). By comparing the two set of differentially expressed genes, we found 18 of the 25 genes differentially expressed in worms fed with CNCM I-3690 vs CNCM I-4317 also differentially expressed in worms fed with CNCM I-3690 vs E. coli OP50 (see Supplementary Material S3 for the details of these 18 genes). On the other hand, 7 genes were not found among the 1278 genes. (The details of these 7 genes are presented in Supplementary Material S4). Focusing our attention on the differences between the two L. rhamnosus strains, two genes were down-regulated and 23 genes were up-regulated (Table 1). Most of the up-regulated genes in young worms are involved in the insulin-like pathway, lipid metabolism and stress response. Conversely, in adult worms (fed with the strains for 10 days) fewer genes were differentially expressed for the three treatments, being 4, 113 and 196 for CNCM I-3690 versus CNCM I-4317, CNCM I-3690 versus OP50 and CNCM I-4317 versus OP50, respectively (see Supplemental Material S5, S6 and S7 for the details of these 4, 133 and 196 genes). Interestingly, transcriptional factor DAF-16 is involved in all three pathways modulated upon ingestion of L. rhamnosus CNCM I-3690. Moreover, downstream DAF-16 up-regulation could also explain the observed increase in lifespan after ingestion of strain CNCM I-3690 (see above). Finally, we confirmed the microarray data by RT-qPCR for the arf-1.1 and gst-22 genes (see Supplementary Material S8 for experimental details and results).
Table 1. Transcriptomic analysis of C. elegans N2 fed with CNCM I-3690 vs CNCM I-4317.
| Up regulated genes | |||||||
| Gene Symbol | Description | Fold change | p-Value | adjusted p-Value | |||
| lec-11 | gaLECtin | 2,7 | 1,27E–06 | 0,02866 | |||
| ZK593.3 | hypothetical protein | 2,4 | 4,57E–06 | 0,05164 | |||
| F23F12.12 | hypothetical protein | 7,7 | 1,66E–06 | 0,05462 | |||
| cpr-3 | hypothetical protein | 2,3 | 3,07E–05 | 0,05462 | |||
| Y71H2AM.16 | hypothetical protein | 2,3 | 2,61E–05 | 0,05462 | |||
| spp-16 | SaPosin-like Protein family | 1,8 | 3,13E–05 | 0,05462 | |||
| R08E5.3 | hypothetical protein | 2,7 | 2,39E–05 | 0,05462 | |||
| C06A12.3 | hypothetical protein | 1,6 | 1,94E–05 | 0,05462 | |||
| clc-1 | CLaudin-like protein | 2,4 | 2,59E–05 | 0,05462 | |||
| gst-22 | Glutathione S-Transferase | 3,4 | 3,14E–05 | 0,05462 | |||
| fat-7 | FATty acid desaturase | 4,4 | 1,66E–05 | 0,05462 | |||
| T04F3.1 | hypothetical protein | 2,2 | 2,62E–05 | 0,05462 | |||
| F44E7.5 | hypothetical protein | 1,8 | 4,48E–05 | 0,07109 | |||
| acl-12 | ACyLtransferase-like | 2,5 | 6,88E–05 | 0,07786 | |||
| Y34F4.2 | hypothetical protein | 2,1 | 6,20E–05 | 0,07786 | |||
| ric-3 | hypothetical protein | 1,4 | 6,71E–05 | 0,07786 | |||
| hsp-12.3 | Heat Shock Protein | 1,9 | 6,24E–05 | 0,07786 | |||
| Peroxidase | hypothetical protein | 1,7 | 7,33E–05 | 0,07897 | |||
| F49E2.5 | hypothetical protein | 2,3 | 7,78E–05 | 0,08005 | |||
| pqn-60 | Prion-like-(Q/N-rich)-domain-bearing protein | 2,1 | 9,14E–05 | 0,08833 | |||
| D2092.1 | hypothetical protein | 1,6 | 9,37E–05 | 0,08833 | |||
| W03D8.8 | hypothetical protein | 2,4 | 0,00011 | 0,09700 | |||
| cpr-3 | Cysteine Protease related | 2,5 | 0,00011 | 0,09816 | |||
| arf-1.1 | ADP-Ribosylation Factor related | 3,6 | 0,00012 | 0,09816 | |||
| Down regulated genes | |||||||
| Gene Symbol | Description | Fold change | p-Value | adjusted p-Value | |||
| T10B10.3 | hypothetical protein | −1,8 | 1,58E-05 | 0,05462 | |||
| F33A8.6 | hypothetical protein | −1,6 | 4,71E-05 | 0,07109 | |||
L. rhamnosus CNCM I-3690 Modulates DAF2/DAF-16 in C. elegans
To elucidate the role of DAF-16, we studied the antioxidant activity and effects on lifespan of L. rhamnosus CNCM I-3690 in C. elegans daf-2 (CB1370), daf-16 (GR1307) and skn-1 (LG333) mutant backgrounds. Unfortunately, both H2O2 doses of 5 mM and 3 mM proved toxic for these mutant strains and therefore the anti-oxidative phenotype could not be evaluated. Notwithstanding, effects on lifespan could be assessed. Data indicated that the increase in survival after 15 days observed in wild-type strain N2 was absent in GR1307, CB1370 and LG333 mutant worms; furthermore, there was no statistical difference in mean lifespan after consumption of CNCM I-3690 or OP50 bacterial strains (Figure 2B, C and D and Table S2). This would indicate that the increased lifespan observed after CNCM I-3690 consumption in wild-type N2 is dependent (at least partially) on the DAF-2/DAF-16 signaling pathway.
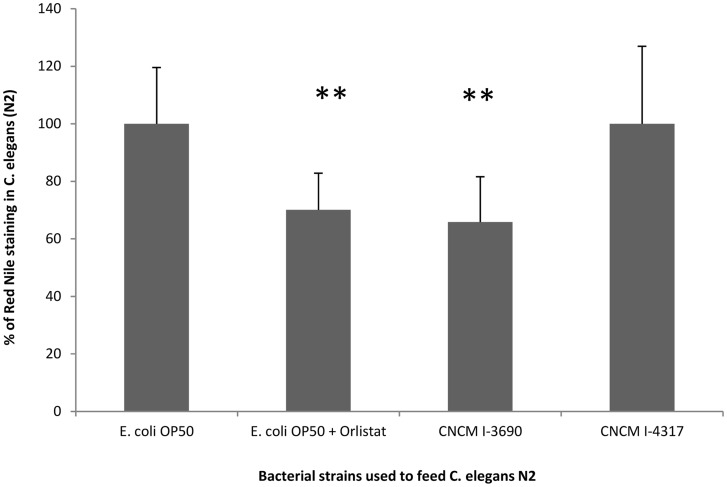
Long-lived worms are reported to display increased fat accumulation and altered metabolism [45]. Adult loss-of-function mutants in daf-2, corresponding to the C. elegans insulin-receptor, accumulate fat; indeed, DAF-16 transcriptional factor has been shown to control fat storage-related pathways [45]. For this reason, we studied lipid inclusion in worms fed on L. rhamnosus CNCM I-3690 and CNCM I-4317, and on E. coli OP50. We observed that worms fed with strain CNCM I-3690 showed a reduction of 34% in lipid inclusion when compared with worms fed on OP50 or CNCM I-4317 (Figure 3). Remarkably, this phenotype is partially dependent on fat-7 gene encoding delta-9 fatty acid desaturation enzyme, since in a fat-7 mutant (BX153) the decrease in lipid inclusion was reduced to 9% vs 34% in wild-type N2 (p-value<0.001). Interestingly, fat-7 was one of the lipid metabolism pathways up-regulated by L. rhamnosus CNCM I-3690 according to the transcriptomic analysis (Table 1).
Figure 3. Lactobacillus rhamnosus CNCM I-3690 inhibits total fat deposit in C. elegans wild-type N2.
Fluorescence of Red Nile stained C. elegans wild-type N2 strain fed with L. rhamnosus CNCM I-3690, CNCM I-4317, E. coli OP50 and E. coli OP50 with orlistat as a positive control. **p-value≤0,001. Fat deposit was measured by fluorescence counting.
In conclusion, transcriptome data indicate that the L. rhamnosus CNCM I-3690 strain diferentially modulates the DAF-16 dependent insulin-like pathway in C. elegans. The transcriptional activation of the DAF-16 pathway by the strain is in agreement with its phenotype of improved resistance to H2O2, its longevity and decreased lipid inclusion. Interestingly, these phenotypes are all associated with a reduction of inflammatory processes. In addition there is evidence in mammalian systems for a direct link between signaling via longevity factors, such as FoxOs or SIRT-1, and inhibition of NF-κB signaling [46], [47]. Thus, we hypothesized that a strain providing antioxidant protection in the C. elegans model through the DAF-16 transcriptional factor may also exhibit an anti-inflammatory profile in mammals. Therefore we decided to focus further experimentation on the analysis of anti-inflammatory properties of both strains CNCM I-3690 and the control CNCM I-4317 in in vitro and in vivo models.
In vitro Anti-inflammatory Profile of L. rhamnosus CNCM I-3690
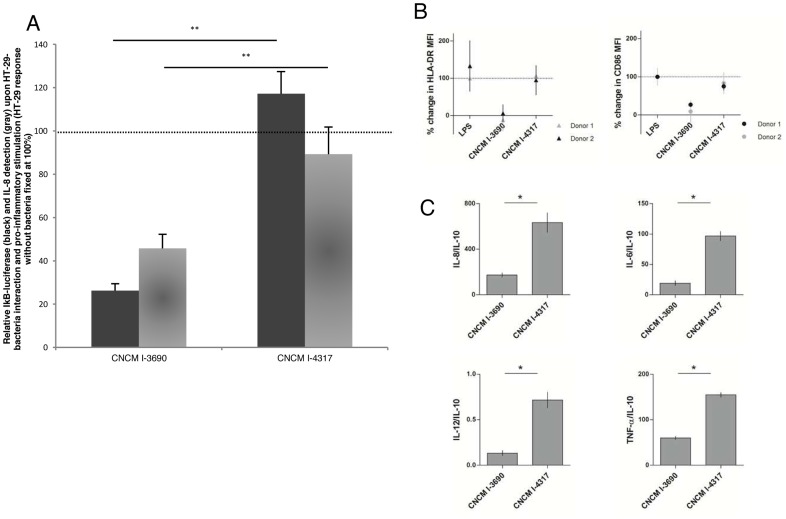
To validate the potential anti-inflammatory phenotype of the L. rhamnosus CNCM I-3690 strain in mammals, we studied the effect of a transient co-culture of CNCM I-3690 and CNCM I-4317 with HT-29 intestinal epithelial cell-line, after 6 h of pro-inflammatory stimulation with 50 ng TNF-α, 2.5 ng IL-1β and 7.5 ng IFN-γ. We observed that a two-hour pre-incubation of epithelial cells with L. rhamnosus CNCM I-3690 induced a significant reduction in NF-κB signaling observed by an I-κB reporter system and IL-8 after 6 h of pro-inflammatory stimulation when compared with CNCM I-4317 (Figure 4A). We also studied the effect of co-culturing L. rhamnosus CNCM I-3690 and CNCM I-4317 in a transwell-filter co-culture system with HT-29-NF-κB-luciferase epithelial cells in the apical side and human dendritic cells (DCs), obtained from human blood donors, in the basal side. A clear anti-inflammatory profile was induced by strain CNCM I-3690 in comparison with CNCM I-4317 (Figures 4B and 4C). In fact, phenotype of DCs was different when co-cultured with strains CNCM I-3690 or CNCM I-4317 in direct contact with epithelial cells. When the system was co-cultured with CNCM I-3690, we observed a 90% reduction in CD86 and HLA expression, which was similar to the LPS control for the co-culture with CNCM I-4317 (Figure 4B). We also observed that only CNCM I-3690 was able to reduce the pro-inflammatory cytokine ratios IL-12/IL-10, IL-6/IL-10, IL-8/IL-10 and TNFα/IL-10 measured in the co-culture system (Figure 4C). Moreover, we confirmed the reduction in NF-κB signaling in the context of the co-culture system with DCs, as previously shown with a single bacteria/epithelial cell interaction.
Figure 4. Lactobacillus rhamnosus CNCM I-3690 has an anti-inflammatory effect in vitro.
A. Relative IL-8, production and I-κB-luciferase detection from the HT-29-bacteria interaction assays. Each bar represent the mean value of three replicate samples and and error bars depict corresponding standard deviation. Black bars: I-kB-luciferase, grey bars: IL-8. **p-value ≤0.05. B. Phenotypical analysis of monocyte-derived DCs co-cultured with HT-29-NF-κB-luciferase cells. Cells were incubated with LPS or LPS+bacteria. CNCM I-3690 down-regulated the expression of HLA-DR and CD86 surface markers. Results were expressed according to the following equation [(bacteria+LPS)-LPS]/LPS-basal*100. Experiments were performed with two different donors. C. Cytokine ratios (IL-8/IL-10, IL-6/IL-10, IL-12/IL-10 and TNF-α/IL-10) of DCs from donor 2 in co-culture with HT-29-NF-κB-luciferase cells and bacteria w or w/o LPS. Experiments were performed with two different donors (data shown for donor 2). *p-value≤0.05.
L. rhamnosus CNCM I-3690 Alleviates Colitis Symptoms in a TNBS-induced Recto-colitis Murine Model
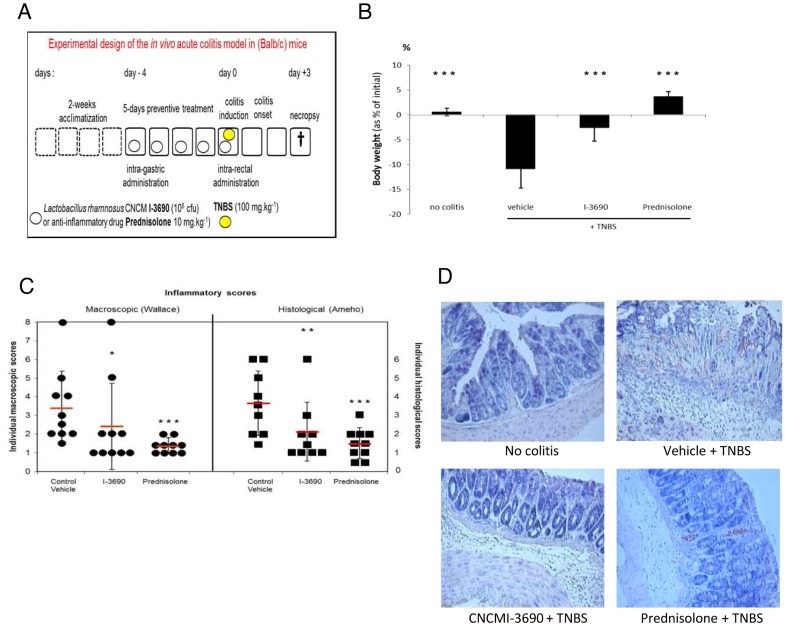
As the anti-oxidative and anti-inflammatory potential of L. rhamnosus CNCM I-3690 was confirmed by both the C. elegans model and the in vitro eukaryotic cell co-culture experiments, we went on to analyze the effects of this strain in a mouse model of intestinal inflammation. Accordingly, the effect of intra-gastric administration of 108 CFU/mL of strain CNCM I-3690 was evaluated over a 5-day period in an acute TNBS-induced recto-colitis gold-standard model, and compared with administration of an anti-inflammatory drug (Figure 5A). In comparison with the vehicle-TNBS-treated group, the 3-day post colitis body-weight losses were significantly lower when mice were fed strain CNCM I-3690 (respectively 11% +/−3.7 and 2.6% +/−3.7, p<0.001), Figure 5B. Similarly, there was nearly a 30% reduction in macroscopic lesions, as recorded by the mean Wallace scores, in the group that received strain CNCM I-3690, compared to the placebo (non-LAB) control group (from 3.4+/−0.8 to 2.4+/−0.77, p<0.01). Notably, the anti-inflammatory effect of strain CNCM I-3690 was similar to the prednisolone positive control, which showed a mean score of 1.5+/−0.13 and provided 58% of protection (p<0.001) (Figure 5C). Histological features confirmed L. rhamnosus CNCM I-3690 ability to prevent inflammation in this model (Figure 5C and D), the latter showing considerably fewer inflammatory cell infiltrates (neutrophils) and mucosal and submucosal lesions.
Figure 5. Protective effect of Lactobacillus rhamnosus CNCM I-3690 in a TNBS-induced murine model of colitis.
A. Design of the interventional animal study. B. 3-day post colitis body-weight losses (% of initial) for healthy control mice, vehicle-TNBS-treated animal and either L. rhamnosus CNCM I-3690-fed TNBS-treated mice or Prednisolone-TNBS-treated mice. Data represent the mean+/−SEM, (number of mice n = 10); ***: P<0.001. C: Individual inflammatory macroscopic (Wallace score, left panel), and histological damage scores (Ameho score, right panel) in Control, probiotic and drug treatment conditions respectively, means +/− SD are indicated, **: P<0.01, ***: P<0.001. D: Representative histological colon sections from mice treated in various conditions after induction of TNBS colitis; May-Grünwald and Giemsa-stainings of 5 µm paraffin sections, original magnification×40.
Taken together, these data confirm that, in the C. elegans model, the antioxidant effect exerted by this strain, selected from among 78 potential candidates, is associated with the anti-inflammatory profile, which was observed in host-bacteria interactions in vitro and in pre-clinical assays of TNBS-induced colitis in mice. These results clearly demonstrate that C. elegans is an efficient model for screening strains for potential probiotic properties and suggest that L. rhamnosus CNCM I-3690 is a novel anti-inflammatory potential probiotic strain.
Discussion
Caenorhabditis elegans was previously shown to be a useful tool for probiotic screening and identification. The potential antitumor activity and the growth inhibition of Lactobacillus salivarius, Lactobacillus reuteri and Pediococcus acidilactici were reported [48]. Some probiotic characteristics of a newly isolated Lactobacillus spp. were studied in terms of their impact on C. elegans longevity [49], while C. elegans was also used to preselect Lactobacillus isolates potentially able to control Salmonella typhimurium infection [50]. Our study describes an original approach, which uses the nematode C. elegans as a preclinical model not only to screen potential antioxidant probiotic strains, but also to identify the underlying molecular mechanisms of action triggered by the selected strains in the host. Interestingly, most of the pathways involved in protection against oxidative stress are highly conserved in higher organisms. Indeed, C. elegans and human genomes have ca 35% homology [51] and it is noteworthy that the number of human disease-related genes sharing at least modest homology with C. elegans can range from 40–75% [52]. Moreover, components of insulin signaling pathways have been conserved, such as kinases, nuclear hormone receptors, F-box proteins and transcription factors [53]. Here, we exploit the technical advantages of the nematode model to obtain new mechanistic insights on potential probiotic strains and to avoid the excessive use of mammals. Similar approaches have been used to identify conserved mechanisms in neuronal disorders [54] and to screen new antimicrobial or antifungal compounds in vivo upon infection of C. elegans and study its innate immune response [55], [56], [57].
Using the strategy described above, a Lactobacillus rhamnosus strain, CNCM I-3690, was identified among 78 bacterial strains as providing protection and increasing longevity in C. elegans. The transcriptome analysis showed that the consumption of this strain by worms activates genes downstream of the DAF-16/FOXO pathway, controlling the insulin-like pathway, lipid metabolism in C. elegans and increasing mean lifespan of worms. These results suggest that CNCM I-3690 strain has a health-aging effect in worms which needs to be confirmed in higher organisms. Transcriptomic analysis also showed that worms fed on CNCM I-3690 for 3 days, over-expressed the fatty acid Δ9 stearoyl-CoA desaturase FAT-7. In mammals, desaturases play a key role in maintaining appropriate fatty acid desaturation levels, which are important for membrane functioning and lipid metabolism control. In C. elegans, FAT-7, FAT-6 and FAT-2 desaturases also catalyze the first step in the synthesis of Poly-Unsaturated Fatty Acids (PUFAs) [45]. Several studies show that FAT-7 expression is controlled by “Nuclear Hormone Receptors” (NHRs), especially NHR-8, NHR-49 and NHR-80, which in turn controls lipid metabolism and nematode lifespan [58]–[59], [60]. Interestingly, it was shown that PUFAs can act as regulators of the insulin-like IGF-II pathway, and DAF-16, in particular, could be one of the transcriptional factors involved in fatty acid homeostasis in C. elegans by controlling the expression of FAT-2, FAT-6 and FAT-7 desaturases and elongases [61]. In this work, we observed a FAT-7 dependent lipid inclusion in worms fed with I-3690 alone, which supports the hypothesis that this strain directly mediates DAF-16-controlled desaturase regulation and lipid metabolism. The mechanisms whereby this strain acts directly on the worm lipid homeostasis remain unknown; notwithstanding, a possible correlation between these results and the increased lifespan of worms fed on CNCM I-3690 cannot be ruled out.
Although further in vivo corroboration is required, these results suggest that some L. rhamnosus CNCM I-3690 ligands could act direct or indirectly on the DAF-16/FOXO and/or SKN-1 pathways through the lumen of C. elegans intestine, by a mechanism as yet unknown. One may hypothesize that some bacterial ligands could trigger the DAF-16 transcription factor by its de-phosphorylation, either by the upstream AKT kinase cascade or by another mechanism that allows DAF-16 to translocate to the nucleus [62]. L. rhamnosus CNCM I-3690 has a strong anti-inflammatory profile in co-culture with intestinal epithelial cell-lines, in vitro and this was confirmed in a TNBS-induced colitis model in mice.
Interestingly, similar effects have been observed in other L. rhamnosus strains. In this respect, L. rhamnosus GR-1 suppresses TNFα signaling in macrophages through a G-CSF-mediated inhibition of JNK [63]. Furthermore, recent findings have shown that Lactobacillus rhamnosus GG generates Extracellular Related Kinase (ERK)-mediated cellular ROS through formyl peptide receptors and modulation of MAP kinase phosphatase redox status [64], [65].
These results correlate the antioxidant effects of L. rhamnosus CNCM I-3690 in C. elegans with the strong anti-inflammatory profile of this strain when co-cultured with human epithelial and/or Dendritic Cells and, likewise, its ability to reduce inflammation in a murine model of colitis. New in vivo assays are currently underway to shed light on the pathways involved in the anti-inflammatory effect in mice.
In summary, this study shows that C. elegans is a promising pre-clinical model for screening potential probiotic strains in a faster and simpler way than other more classical approaches. This simple model enables us to study potential probiotic effects based on simple read-outs, such as oxidative stress, and then correlate these effects with more complex models in vitro or in vivo. Further studies must be performed to better characterize the potential probiotic effects identified here.
Supporting Information
C. elegans BA17 can be fed with Bifidobacteria , Lactobacilli and Streptococci strains. Survival of C. elegans BA 17 strain fed for 3 days with Bifidobacteria, Lactobacilli and Streptococci strains in liquid medium was followed, after larvae 1st stage synchronization with E. coli OP 50 and without any oxidative stress. Protection by E. coli OP50 is fixed at 100%.
(TIF)
Genera and species specifications for all the bacterial strains used in this work. Genera, species and subspecies are specified for each bacterial strain used in this study, with their corresponding code names.
(DOCX)
DAF-16, DAF-2 and SKN-1 play an essential role in C. elegans longevity resulting from feeding with Lactobacillus rhamnosus CNCM I-3690. See corresponding data in Figure 2. *NS: No significant differences between control conditions (NGM+E. coli OP50) and treatment conditions (NGM + CNCM I-3690). Statistical analysis was performed with GraphPad prism 4 using Log Rank Test.
(DOCX)
LAB and bifidobacteria growth curves. Growth curves of representative strains for each genus, Streptococcus CNCM I-2778, Lactobacillus CNCM I-3064 and Bifidobacterium Bal7, and detailed protocols are presented.
(DOCX)
List of the 1278 differentially expressed genes in C. elegans fed with L. rhamnosus CNCM I-3690 versus E. coli OP50. C. elegans N2 wild-type strain was fed with L. rhamnosus strain CNCM I-3690 and E. coli strain OP50 during 3 days. Transcriptome profiling was performed comparing worms fed with the two bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 18 genes differentially expressed in C. elegans fed with L. rhamnosus CNCM I-3690 versus L. rhamnosus CNCM I-4317 and in C. elegans fed with L. rhamnosus CNCM I-3690 versus E. coli OP50. C. elegans N2 wild-type strain was fed with the three strains L. rhamnosus CNCM I-3690, L. rhamnosus CNCM I-4317 and E. coli OP50 during 3 days. Transcriptome profiling was performed comparing worms fed with the three bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 7 differentially expressed genes not shared between C. elegans fed with L. rhamnosus CNCM I-3690 versus L. rhamnosus CNCM I-4317 and in C. elegans fed with L. rhamnosus CNCM I-3690 versus E. coli OP50. C. elegans N2 wild-type strain was fed with the three strains L. rhamnosus CNCM I-3690, L. rhamnosus CNCM I-4317 and E. coli OP50 during 3 days. Transcriptome profiling was performed comparing worms fed with the three bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 4 differentially expressed genes C. elegans fed for 10 days with L. rhamnosus CNCM I-3690 versus L. rhamnosus CNCM I-4317. C. elegans N2 wild-type strain was fed with both L. rhamnosus strains CNCM I-3690 and CNCM I-4317 during 10 days. Transcriptome profiling was performed comparing worms fed with the two bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 133 differentially expressed genes C. elegans fed for 10 days with L. rhamnosus CNCM I-3690 versus E. coli OP50. C. elegans N2 wild-type strain was fed with L. rhamnosus CNCM I-3690 and E. coli OP50 strains during 10 days. Transcriptome profiling was performed comparing worms fed with the two bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 196 differentially expressed genes C. elegans fed for 10 days with L. rhamnosus CNCM I-4317 versus E. coli OP50. C. elegans N2 wild-type strain was fed with L. rhamnosus CNCM I-4317 and E. coli OP50 strains during 10 days. Transcriptome profiling was performed comparing worms fed with the two bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
Confirmation of microarray data by RT-qPCR for arf-1.1 and gst-22 genes. Expression ratios for arf-1.1 and gst-22 genes were obtained by RT-qPCR. All experiments were repeated at least three times (biological replicates) and were internally controlled (technical replicate). Expression changes were obtained by calculating the relative expression levels using the 2−ΔΔCT method. See Materials and Methods for details.
(DOCX)
Acknowledgments
We thank Johan Van Hylckama Vlieg, Jean-Michel Faurie, Jean-Michel Antoine and Bruno Pot for critical reading of the manuscript and helpful discussions.
Funding Statement
Funders of this work were: ANII (Agencia Nacional de Investigacion e Innovacion, URUGUAY): PE_ALI_1_1702 and Danone Research. ANII had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Danone Research had no role in study design, data collection and analysis, but checked the manuscript before publication.
References
- 1. Sanz A, Stefanatos RK (2008) The mitochondrial free radical theory of aging: a critical view. Curr Aging Sci 1: 10–21. [DOI] [PubMed] [Google Scholar]
- 2. Harman D (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11: 298–300. [DOI] [PubMed] [Google Scholar]
- 3. Harman D (1972) The biologic clock: the mitochondria? J Am Geriatr Soc 20: 145–147. [DOI] [PubMed] [Google Scholar]
- 4. Sohal RS, Weindruch R (1996) Oxidative stress, caloric restriction, and aging. Science 273: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, et al.. (2007) Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol 39: 44–84. S1357-2725(06)00219-6 [pii];10.1016/j.biocel.2006.07.001 [doi]. [DOI] [PubMed]
- 6.Sohal RS (2002) Oxidative stress hypothesis of aging. Free Radic Biol Med 33: 573–574. S0891584902008857 [pii]. [DOI] [PubMed]
- 7.Sohal RS (2002) Role of oxidative stress and protein oxidation in the aging process. Free Radic Biol Med 33: 37–44. S0891584902008560 [pii]. [DOI] [PubMed]
- 8. Pravda J (2005) Radical induction theory of ulcerative colitis. World J Gastroenterol 11: 2371–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rezaie A, Parker RD, Abdollahi M (2007) Oxidative stress and pathogenesis of inflammatory bowel disease: an epiphenomenon or the cause? Dig Dis Sci 52: 2015–2021. 10.1007/s10620-006-9622-2 [doi]. [DOI] [PubMed]
- 10.Hekimi S, Lapointe J, Wen Y (2011) Taking a “good” look at free radicals in the aging process. Trends Cell Biol 21: 569–576. S0962-8924(11)00134-6 [pii];10.1016/j.tcb.2011.06.008 [doi]. [DOI] [PMC free article] [PubMed]
- 11.Ristow M, Schmeisser S (2011) Extending life span by increasing oxidative stress. Free Radic Biol Med 51: 327–336. S0891-5849(11)00312-1 [pii];10.1016/j.freeradbiomed.2011.05.010 [doi]. [DOI] [PubMed]
- 12.Tomas-Barberan FA, Cienfuegos-Jovellanos E, Marin A, Muguerza B, Gil-Izquierdo A, et al.. (2007) A new process to develop a cocoa powder with higher flavonoid monomer content and enhanced bioavailability in healthy humans. J Agric Food Chem 55: 3926–3935. 10.1021/jf070121j [doi]. [DOI] [PubMed]
- 13.Butalla AC, Crane TE, Patil B, Wertheim BC, Thompson P, et al.. (2012) Effects of a carrot juice intervention on plasma carotenoids, oxidative stress, and inflammation in overweight breast cancer survivors. Nutr Cancer 64: 331–341. 10.1080/01635581.2012.650779 [doi]. [DOI] [PubMed]
- 14.Ellinger S, Muller N, Stehle P, Ulrich-Merzenich G (2011) Consumption of green tea or green tea products: is there an evidence for antioxidant effects from controlled interventional studies? Phytomedicine 18: 903–915. S0944-7113(11)00191-7 [pii];10.1016/j.phymed.2011.06.006 [doi]. [DOI] [PubMed]
- 15.Frankel EN (2011) Nutritional and biological properties of extra virgin olive oil. J Agric Food Chem 59: 785–792. 10.1021/jf103813t [doi]. [DOI] [PubMed]
- 16.Rijkers GT, Bengmark S, Enck P, Haller D, Herz U, et al.. (2010) Guidance for substantiating the evidence for beneficial effects of probiotics: current status and recommendations for future research. J Nutr 140: 671S-676S. jn.109.113779 [pii];10.3945/jn.109.113779 [doi]. [DOI] [PubMed]
- 17.Lin MY, Yen CL (1999) Inhibition of lipid peroxidation by Lactobacillus acidophilus and Bifidobacterium longum. J Agric Food Chem 47: 3661–3664. jf981235l [pii]. [DOI] [PubMed]
- 18.Lin MY, Yen CL (1999) Antioxidative ability of lactic acid bacteria. J Agric Food Chem 47: 1460–1466. jf981149l [pii]. [DOI] [PubMed]
- 19. Lin MY, Chang FJ (2000) Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Dig Dis Sci 45: 1617–1622. [DOI] [PubMed] [Google Scholar]
- 20.Sun J, Hu XL, Le GW, Shi YH (2010) Lactobacilli prevent hydroxy radical production and inhibit Escherichia coli and Enterococcus growth in system mimicking colon fermentation. Lett Appl Microbiol 50: 264–269. LAM2786 [pii];10.1111/j.1472-765X.2009.02786.x [doi]. [DOI] [PubMed]
- 21.Han W, Mercenier A, Ait-Belgnaoui A, Pavan S, Lamine F, et al.. (2006) Improvement of an experimental colitis in rats by lactic acid bacteria producing superoxide dismutase. Inflamm Bowel Dis 12: 1044–1052. 10.1097/01.mib.0000235101.09231.9e [doi];00054725-200611000-00004 [pii]. [DOI] [PubMed]
- 22.LeBlanc JG, del Carmen S, Miyoshi A, Azevedo V, Sesma F, et al.. (2011) Use of superoxide dismutase and catalase producing lactic acid bacteria in TNBS induced Crohn's disease in mice. J Biotechnol 151: 287–293. S0168-1656(10)02041-9 [pii];10.1016/j.jbiotec.2010.11.008 [doi]. [DOI] [PubMed]
- 23. Richmond J (2000) FRAME annual lecture. The three Rs: a journey or a destination? Altern Lab Anim 28: 761–773. [DOI] [PubMed] [Google Scholar]
- 24. Richmond J (2002) Refinement, reduction, and replacement of animal use for regulatory testing: future improvements and implementation within the regulatory framework. ILAR J 43 Suppl: S63–S68 [DOI] [PubMed] [Google Scholar]
- 25.Kaletsky R, Murphy CT (2010) The role of insulin/IGF-like signaling in C. elegans longevity and aging. Dis Model Mech 3: 415–419. dmm.001040 [pii];10.1242/dmm.001040 [doi]. [DOI] [PubMed]
- 26.Zhou KI, Pincus Z, Slack FJ (2011) Longevity and stress in Caenorhabditis elegans. Aging (Albany NY ) 3: 733–753. 100367 [pii]. [DOI] [PMC free article] [PubMed]
- 27.Giordano-Santini R, Dupuy D (2011) Selectable genetic markers for nematode transgenesis. Cell Mol Life Sci 68: 1917–1927. 10.1007/s00018-011-0670-1 [doi]. [DOI] [PMC free article] [PubMed]
- 28. Larsen PL (1993) Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc Natl Acad Sci U S A 90: 8905–8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikeda T, Yasui C, Hoshino K, Arikawa K, Nishikawa Y (2007) Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against salmonella enterica serovar enteritidis. Appl Environ Microbiol 73: 6404–6409. AEM.00704-07 [pii];10.1128/AEM.00704-07 [doi]. [DOI] [PMC free article] [PubMed]
- 30.Zhou YM, Shao L, Li JA, Han LZ, Cai WJ, et al.. (2011) An efficient and novel screening model for assessing the bioactivity of extracts against multidrug-resistant Pseudomonas aeruginosa using Caenorhabditis elegans. Biosci Biotechnol Biochem 75: 1746–1751. JST.JSTAGE/bbb/110290 [pii]. [DOI] [PubMed]
- 31.Kenyon C (2011) The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Philos Trans R Soc Lond B Biol Sci 366: 9–16. 366/1561/9 [pii];10.1098/rstb.2010.0276 [doi]. [DOI] [PMC free article] [PubMed]
- 32.Amrit FR, May RC (2010) Younger for longer: insulin signalling, immunity and ageing. Curr Aging Sci 3: 166–176. BSP/CAS/E-Pub/000010 [pii]. [DOI] [PubMed]
- 33.Finkel T (2011) Signal transduction by reactive oxygen species. J Cell Biol 194: 7–15. jcb.201102095 [pii];10.1083/jcb.201102095 [doi]. [DOI] [PMC free article] [PubMed]
- 34.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, et al.. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. 10.1093/biostatistics/4.2.249 [doi];4/2/249 [pii]. [DOI] [PubMed]
- 35.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I (2001) Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125: 279–284. S0166-4328(01)00297-2 [pii]. [DOI] [PubMed]
- 36.Montaner D, Dopazo J (2010) Multidimensional gene set analysis of genomic data. PLoS One 5: e10348. 10.1371/journal.pone.0010348 [doi]. [DOI] [PMC free article] [PubMed]
- 37.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al.. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. gb-2004-5-10-r80 [pii];10.1186/gb-2004-5-10-r80 [doi]. [DOI] [PMC free article] [PubMed]
- 38.Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315. 10.1093/bioinformatics/btg405 [doi];20/3/307 [pii]. [DOI] [PubMed]
- 39.Smyth GK, Michaud J, Scott HS (2005) Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21: 2067–2075. bti270 [pii];10.1093/bioinformatics/bti270 [doi]. [DOI] [PubMed]
- 40.Tien MT, Girardin SE, Regnault B, Le BL, Dillies MA, et al.. (2006) Anti-inflammatory effect of Lactobacillus casei on Shigella-infected human intestinal epithelial cells. J Immunol 176: 1228–1237. 176/2/1228 [pii]. [DOI] [PubMed]
- 41. Dauer M, Obermaier B, Herten J, Haerle C, Pohl K, et al. (2003) Mature dendritic cells derived from human monocytes within 48 hours: a novel strategy for dendritic cell differentiation from blood precursors. J Immunol 170: 4069–4076. [DOI] [PubMed] [Google Scholar]
- 42.Foligne B, Nutten S, Steidler L, Dennin V, Goudercourt D, et al.. (2006) Recommendations for improved use of the murine TNBS-induced colitis model in evaluating anti-inflammatory properties of lactic acid bacteria: technical and microbiological aspects. Dig Dis Sci 51: 390–400. 10.1007/s10620-006-3143-x [doi]. [DOI] [PubMed]
- 43.Wallace JL, MacNaughton WK, Morris GP, Beck PL (1989) Inhibition of leukotriene synthesis markedly accelerates healing in a rat model of inflammatory bowel disease. Gastroenterology 96: 29–36. S0016508589000120 [pii]. [DOI] [PubMed]
- 44. Ameho CK, Adjei AA, Harrison EK, Takeshita K, Morioka T, et al. (1997) Prophylactic effect of dietary glutamine supplementation on interleukin 8 and tumour necrosis factor alpha production in trinitrobenzene sulphonic acid induced colitis. Gut 41: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashrafi K (2007) Obesity and the regulation of fat metabolism. WormBook 1–20. 10.1895/wormbook.1.130.1 [doi]. [DOI] [PMC free article] [PubMed]
- 46.Salminen A, Ojala J, Huuskonen J, Kauppinen A, Suuronen T, et al.. (2008) Interaction of aging-associated signaling cascades: inhibition of NF-kappaB signaling by longevity factors FoxOs and SIRT1. Cell Mol Life Sci 65: 1049–1058. 10.1007/s00018-008-7461-3 [doi]. [DOI] [PMC free article] [PubMed]
- 47.Salminen A, Kauppinen A, Suuronen T, Kaarniranta K (2008) SIRT1 longevity factor suppresses NF-kappaB -driven immune responses: regulation of aging via NF-kappaB acetylation? Bioessays 30: 939–942. 10.1002/bies.20799 [doi]. [DOI] [PubMed]
- 48.Fasseas MK, Fasseas C, Mountzouris KC, Syntichaki P (2012) Effects of Lactobacillus salivarius, Lactobacillus reuteri, and Pediococcus acidilactici on the nematode Caenorhabditis elegans include possible antitumor activity. Appl Microbiol Biotechnol. 10.1007/s00253-012-4357-9 [doi]. [DOI] [PubMed]
- 49.Lee J, Yun HS, Cho KW, Oh S, Kim SH, et al.. (2011) Evaluation of probiotic characteristics of newly isolated Lactobacillus spp.: immune modulation and longevity. Int J Food Microbiol 148: 80–86. S0168-1605(11)00267-4 [pii];10.1016/j.ijfoodmicro.2011.05.003 [doi]. [DOI] [PubMed]
- 50.Wang C, Wang J, Gong J, Yu H, Pacan JC, et al.. (2011) Use of Caenorhabditis elegans for preselecting Lactobacillus isolates to control Salmonella Typhimurium. J Food Prot 74: 86–93. 10.4315/0362-028X.JFP-10-155 [doi]. [DOI] [PubMed]
- 51. C. elegans Sequencing Consortium (1998) Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- 52.Silverman GA, Luke CJ, Bhatia SR, Long OS, Vetica AC, et al.. (2009) Modeling molecular and cellular aspects of human disease using the nematode Caenorhabditis elegans. Pediatr Res 65: 10–18. 10.1203/PDR.0b013e31819009b0 [doi]. [DOI] [PMC free article] [PubMed]
- 53.Shaye DD, Greenwald I (2011) OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One 6: e20085. 10.1371/journal.pone.0020085 [doi];PONE-D-11-05121 [pii]. [DOI] [PMC free article] [PubMed]
- 54.Chen L, Wang Z, Ghosh-Roy A, Hubert T, et al.. (2011) Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron 71: 1043–1057. S0896-6273(11)00606-4 [pii];10.1016/j.neuron.2011.07.009 [doi]. [DOI] [PMC free article] [PubMed]
- 55.Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, et al.. (2007) Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog 3: e18. 06-PLPA-RA-0392R3 [pii];10.1371/journal.ppat.0030018 [doi]. [DOI] [PMC free article] [PubMed]
- 56.Ewbank JJ, Zugasti O (2011) C. elegans: model host and tool for antimicrobial drug discovery. Dis Model Mech 4: 300–304. dmm.006684 [pii];10.1242/dmm.006684 [doi]. [DOI] [PMC free article] [PubMed]
- 57.Glavis-Bloom J, Muhammed M, Mylonakis E (2012) Of model hosts and man: using Caenorhabditis elegans, Drosophila melanogaster and Galleria mellonella as model hosts for infectious disease research. Adv Exp Med Biol 710: 11–17. 10.1007/978-1-4419-5638-5_2 [doi]. [DOI] [PubMed]
- 58.Van Gilst MR, Hadjivassiliou H, Yamamoto KR (2005) A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc Natl Acad Sci U S A 102: 13496–13501. 0506234102 [pii];10.1073/pnas.0506234102 [doi]. [DOI] [PMC free article] [PubMed]
- 59.Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR (2005) Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol 3: e53. 10.1371/journal.pbio.0030053 [doi]. [DOI] [PMC free article] [PubMed]
- 60.Magner DB, Antebi A (2008) Caenorhabditis elegans nuclear receptors: insights into life traits. Trends Endocrinol Metab 19: 153–160. S1043-2760(08)00058-1 [pii];10.1016/j.tem.2008.02.005 [doi]. [DOI] [PMC free article] [PubMed]
- 61.Horikawa M, Sakamoto K (2010) Polyunsaturated fatty acids are involved in regulatory mechanism of fatty acid homeostasis via daf-2/insulin signaling in Caenorhabditis elegans. Mol Cell Endocrinol 323: 183–192. S0303-7207(10)00137-1 [pii];10.1016/j.mce.2010.03.004 [doi]. [DOI] [PubMed]
- 62.Cohen E, Dillin A (2008) The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat Rev Neurosci 9: 759–767. nrn2474 [pii];10.1038/nrn2474 [doi]. [DOI] [PMC free article] [PubMed]
- 63.Kim SO, Sheikh HI, Ha SD, Martins A, Reid G (2006) G-CSF-mediated inhibition of JNK is a key mechanism for Lactobacillus rhamnosus-induced suppression of TNF production in macrophages. Cell Microbiol 8: 1958–1971. CMI763 [pii];10.1111/j.1462-5822.2006.00763.x [doi]. [DOI] [PubMed]
- 64.Wentworth CC, Jones RM, Kwon YM, Nusrat A, Neish AS (2010) Commensal-epithelial signaling mediated via formyl peptide receptors. Am J Pathol 177: 2782–2790. S0002-9440(10)62907-0 [pii];10.2353/ajpath.2010.100529 [doi]. [DOI] [PMC free article] [PubMed]
- 65.Wentworth CC, Alam A, Jones RM, Nusrat A, Neish AS (2011) Enteric commensal bacteria induce extracellular signal-regulated kinase pathway signaling via formyl peptide receptor-dependent redox modulation of dual specific phosphatase 3. J Biol Chem 286: 38448–38455. M111.268938 [pii];10.1074/jbc.M111.268938 [doi]. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C. elegans BA17 can be fed with Bifidobacteria , Lactobacilli and Streptococci strains. Survival of C. elegans BA 17 strain fed for 3 days with Bifidobacteria, Lactobacilli and Streptococci strains in liquid medium was followed, after larvae 1st stage synchronization with E. coli OP 50 and without any oxidative stress. Protection by E. coli OP50 is fixed at 100%.
(TIF)
Genera and species specifications for all the bacterial strains used in this work. Genera, species and subspecies are specified for each bacterial strain used in this study, with their corresponding code names.
(DOCX)
DAF-16, DAF-2 and SKN-1 play an essential role in C. elegans longevity resulting from feeding with Lactobacillus rhamnosus CNCM I-3690. See corresponding data in Figure 2. *NS: No significant differences between control conditions (NGM+E. coli OP50) and treatment conditions (NGM + CNCM I-3690). Statistical analysis was performed with GraphPad prism 4 using Log Rank Test.
(DOCX)
LAB and bifidobacteria growth curves. Growth curves of representative strains for each genus, Streptococcus CNCM I-2778, Lactobacillus CNCM I-3064 and Bifidobacterium Bal7, and detailed protocols are presented.
(DOCX)
List of the 1278 differentially expressed genes in C. elegans fed with L. rhamnosus CNCM I-3690 versus E. coli OP50. C. elegans N2 wild-type strain was fed with L. rhamnosus strain CNCM I-3690 and E. coli strain OP50 during 3 days. Transcriptome profiling was performed comparing worms fed with the two bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 18 genes differentially expressed in C. elegans fed with L. rhamnosus CNCM I-3690 versus L. rhamnosus CNCM I-4317 and in C. elegans fed with L. rhamnosus CNCM I-3690 versus E. coli OP50. C. elegans N2 wild-type strain was fed with the three strains L. rhamnosus CNCM I-3690, L. rhamnosus CNCM I-4317 and E. coli OP50 during 3 days. Transcriptome profiling was performed comparing worms fed with the three bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 7 differentially expressed genes not shared between C. elegans fed with L. rhamnosus CNCM I-3690 versus L. rhamnosus CNCM I-4317 and in C. elegans fed with L. rhamnosus CNCM I-3690 versus E. coli OP50. C. elegans N2 wild-type strain was fed with the three strains L. rhamnosus CNCM I-3690, L. rhamnosus CNCM I-4317 and E. coli OP50 during 3 days. Transcriptome profiling was performed comparing worms fed with the three bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 4 differentially expressed genes C. elegans fed for 10 days with L. rhamnosus CNCM I-3690 versus L. rhamnosus CNCM I-4317. C. elegans N2 wild-type strain was fed with both L. rhamnosus strains CNCM I-3690 and CNCM I-4317 during 10 days. Transcriptome profiling was performed comparing worms fed with the two bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 133 differentially expressed genes C. elegans fed for 10 days with L. rhamnosus CNCM I-3690 versus E. coli OP50. C. elegans N2 wild-type strain was fed with L. rhamnosus CNCM I-3690 and E. coli OP50 strains during 10 days. Transcriptome profiling was performed comparing worms fed with the two bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
List of the 196 differentially expressed genes C. elegans fed for 10 days with L. rhamnosus CNCM I-4317 versus E. coli OP50. C. elegans N2 wild-type strain was fed with L. rhamnosus CNCM I-4317 and E. coli OP50 strains during 10 days. Transcriptome profiling was performed comparing worms fed with the two bacterial strains. RNA samples were hybridized (in triplicate) onto C. elegans Genome Array (Affymetrix), for details see Materials. Genes were considered differentially expressed when the multiple testing adjusted P-value<0,01.
(XLS)
Confirmation of microarray data by RT-qPCR for arf-1.1 and gst-22 genes. Expression ratios for arf-1.1 and gst-22 genes were obtained by RT-qPCR. All experiments were repeated at least three times (biological replicates) and were internally controlled (technical replicate). Expression changes were obtained by calculating the relative expression levels using the 2−ΔΔCT method. See Materials and Methods for details.
(DOCX)