Abstract
A two-compartment in vitro pharmacokinetic-pharmacodynamic model, with full computer-controlled devices, was used to accurately simulate human plasma pharmacokinetic profiles after multidose oral regimens of ciprofloxacin (750 mg every 12 h) and moxifloxacin (400 mg every 24 h) during 48 h. Pharmacodynamics of these drugs was investigated against three quinolone-susceptible strains of Stenotrophomonas maltophilia (MICs of ciprofloxacin and moxifloxacin of 0.5 to 2 and 0.0625 to 0.5 μg/ml, respectively). The first dose of ciprofloxacin and moxifloxacin reduced the bacterial count by 1 and 2 log CFU/ml, respectively, prior to a bacterial regrowth that reached the plateau value of the growth control curve at 13 to 24 h versus 24 to 36 h and persisted despite repeated administration of both drugs. The surviving bacterial cells were quinolone-resistant mutants (2 to 128 times the MIC) that exhibited cross-resistance to unrelated antibiotics. Their antibiotic resistance probably resulted from the overproduction of different multidrug resistance efflux system(s). Cmax/MIC and area under the concentration-time curve from 0 to 24 h (AUC0-24)/MIC values were at least threefold higher for moxifloxacin than for ciprofloxacin. Moreover, integral parameters of ciprofloxacin and moxifloxacin, in particular the area under the killing and regrowth curve from 0 to 48 h (AUBC0-48, 342.3 to 401.3 versus 295.2 to 378.7 h × log CFU/ml, respectively) and the area between the control growth curve and the killing and regrowth curve from 0 to 48 h (ABBC0-48, 40.4 to 101.1 versus 72.9 to 144.7 h × log CFU/ml, respectively), demonstrated a better antibacterial effect of moxifloxacin than ciprofloxacin on S. maltophilia. However, selection of resistant mutants by both fluoroquinolones, although delayed with moxifloxacin, emphasizes the need to use maximal dosages and combined therapy in the treatment of systemic S. maltophilia infections.
Over the past 15 years, Stenotrophomonas maltophilia has emerged as an important cause of nosocomial infections, mainly in severely debilitated or immunocompromised patients (13, 18, 32). This bacterial species is responsible for a wide spectrum of diseases with substantial morbidity and mortality, including respiratory tract infections, especially in patients with cystic fibrosis, bacteremia and, more rarely, wound and urinary tract infections (13, 18, 32). Therapy for these infections is problematic because of the common multidrug resistance (MDR) of the strains. Indeed, S. maltophilia is inherently resistant to most antimicrobials due to a low outer membrane permeability and/or natural MDR efflux systems, coupled with specific resistance mechanisms, such as the production of two inducible chromosomally encoded β-lactamases, L1 and L2, and an aminoglycoside acetyltransferase (13, 20, 30). The few available antibiotics that are naturally active against S. maltophilia include cotrimoxazole, some β-lactams (principally the combination ticarcillin-clavulanic acid), and fluoroquinolones (essentially ciprofloxacin) (13, 18, 32). Moreover, acquired resistances to these antibiotics are frequent (2 to 10%, 10 to 29%, and 21 to 53%, respectively) (18). Indeed, mutants exhibiting pleiotropic resistance, including to quinolones, are readily selected in vitro (1, 19, 22, 39, 40) as in vivo (19, 36, 38). In contrast to other gram-negative organisms, the primary determinant of quinolone resistance in S. maltophilia has been identified as the overproduction of efflux pumps, SmeDEF being the most clearly involved (1-3), rather than mutations in the type II topoisomerases targets (30, 35).
Newer fluoroquinolones, such as moxifloxacin, appear to be more active against S. maltophilia than the older agents of this family (28, 37). Moreover, the development of resistance has been shown to be less pronounced with moxifloxacin than with ciprofloxacin in Escherichia coli, Staphylococcus aureus, and Streptococcus pneumoniae (7, 12, 29). Therefore, this drug might be adopted to replace ciprofloxacin in the treatment of S. maltophilia infections.
We have previously developed an in vitro pharmacokinetic-pharmacodynamic (PK/PD) model that accurately simulates the variations of the antibiotic concentrations versus time in a defined body compartment and to investigate their efficiency on bacterial strains (4). The aim of the present study was to apply this model to the human serum pharmacokinetics of ciprofloxacin and moxifloxacin, administered at recommended dosing regimens, and to compare their pharmacodynamics against quinolone-susceptible strains of S. maltophilia, with emphasis on the emergence of resistant mutants.
MATERIALS AND METHODS
PK/PD model
The two-compartment PK/PD model with full computer-controlled devices has been previously described in detail elsewhere (4). The model was designed to simulate the same pharmacokinetic profile in the two compartments. Flow rates were adjusted according to reference pharmacokinetic parameters of ciprofloxacin (10, 17) and moxifloxacin (33). The target values for ciprofloxacin and moxifloxacin, respectively, were as follows: maximal concentration (Cmax), 2.94 and 2.50 μg/ml; absorption rate constant (Ka), 2.70 and 2 h−1; and elimination half-lives (t1/2β), 4.70 and 15.6 h. All experiments were performed in Mueller-Hinton (MH) broth (A.D.L., Tresses, France) at 37°C.
Antibiotics and simulated dosage regimens
Ciprofloxacin obtained as a reference powder (84.4% free base) and as a marketed infusion solution (Ciflox; 200 mg/100 ml), as well as moxifloxacin as reference powder (95.2% free base), were provided by Bayer Pharma (Puteaux, France). Human plasma pharmacokinetic profiles were simulated according to multidose oral regimens of ciprofloxacin (a route consistent with that of moxifloxacin, 750 mg every 12 h) and moxifloxacin (400 mg every 24 h) for 48 h. Ciprofloxacin infusion solution was directly used to simulate the oral administration. The simulation of the oral administration of moxifloxacin was performed with a 2-mg/ml solution.
Bacterial strains and antimicrobial susceptibility testing
The clinical isolate of S. maltophilia Sm206 was selected for the validation of the model, and two reference strains from the Collection de l'Institut Pasteur, CIP 54.90 and CIP 60.77T (ATCC 13637), were added for data confirmation. The initial strains and the surviving bacterial cells obtained during ciprofloxacin and moxifloxacin administration simulation were identified by conventional tests and by using the API 20NE system (bioMérieux, La Balme les Grottes, France). Their antibiotypes were established by the disk diffusion method (28 discriminant antibiotics) according to official guidelines (http://www.sfm.asso.fr). MICs were determined in at least three independent experiments by an agar dilution method on MH medium after a 24-h incubation at 37°C (http://www.sfm.asso.fr). Antibiotic reference powders were kindly supplied by their manufacturers (ciprofloxacin and moxifloxacin [Bayer Pharma]; norfloxacin [Merck Sharp and Dohme-Chibret]; nalidixic acid [Sanofi]; tetracycline and chloramphenicol [Aventis]; sulfamethoxazole, trimethoprim, and their cotrimoxazole combination [5:1 m/m] [Roche]; ticarcillin-clavulanic acid [Glaxo Smith Kline]; cefepime and amikacin [Bristol Myers Squibb]) or were purchased (erythromycin [Fluka]). Quality control strains included E. coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and S. aureus ATCC 25923.
Quantitation of bacterial growth
The peripheral compartment (PCp) of the model was inoculated with each bacterial strain 3 h before performing the first sampling, in order to obtain exponentially growing culture of 107 CFU/ml in a 50-ml culture volume. Since the volume of fresh antibiotic-free medium flowing into the central compartment (CCp) depended on the antibiotic absorption, distribution, and elimination half-lives, the absence of biased results due to the influence of the amount of nutrient on growth characteristics was controlled by performing growth control curves in triplicate (Sm206) or in duplicate (CIP 54.90 and CIP 60.77T) for each simulation condition.
Operating procedure and antimicrobial assays
During the validation tests with Sm206, a total of 65 (for ciprofloxacin kinetics, at 0.35, 0.4, 0.5, 0.75, 1, 1.25, 1.50, 1.75, 2, 2.50, 3, 4, 6, 8, 10, and 12 h after the beginning of antibiotic administration for each regimen interval) or 30 (for moxifloxacin kinetics, at 0.15, 0.25, 0.50, 0.75, 0.90, 1, 1.50, 2, 2.50, 3, 4, 6, 8, 12, and 24 h) 200-μl samples of each were drawn from the CCp by means of a computer-controlled fraction collector and from the PCp by manual sampling with sterile Vacutainer tubes (PolyLabo, Strasbourg, France); the number of samples was reduced for the reference strains (23 and 15 for ciprofloxacin and moxifloxacin kinetics, respectively). A 10-μl aliquot of each sample was used for antibiotic chromatographic assays consisting of isocratic reversed-phase high-pressure liquid chromatography with column switching and direct injection of broth (5, 6). A 100-μl volume from PCp samples was devoted for the quantification of bacterial growth. For bacterial counts in the absence of antibiotics, a total of 20 samples were drawn from the PCp at 0.08, 1, 2, 3, 4, 4.50, 5, 5.50, 6, 6.50, 7, 7.50, 8, 8.50, 9, 10, 12, 14, 18, 30, 36, 42, and 54 h after bacteria inoculation. Bacteria were counted by making 10-fold dilutions of the samples with a sterile 0.9% NaCl solution and then plating 100 μl on MH agar. In the case of treatment simulations, the emergence of resistant mutants was monitored by plating the same diluted samples on MH agar supplemented with 1, 4, 16, and 64 times the MIC of ciprofloxacin or moxifloxacin. Plates were read after 48 h of incubation by visual inspection. The lower detection and counting limits were 2 and 3 log CFU/ml, respectively. Colony counts were expressed as log10 CFU/ml.
Data analysis. (i) Pharmacokinetic analysis
Compartmental analysis of drug experimental concentration data was performed with the software Pharmacokin (G. Kister, J. Bres, and G. Cassanas, Prog. Abstr. 2nd Sci. Meet. Assoc. Pharmacy Faculties Pharmacologists, abstr. 13, p. 14, 1998). The goodness of fit for each concentration-time curve was evaluated both by the correlation coefficient between experimental and software-calculated data, and the objective function F (data not shown), which represents the sum of weighted squared deviations. The Cmax, the residual concentration at the end of the administration interval (Cres.), and the time to reach the peak (Tmax) were taken directly from concentration-time profiles, whereas the t1/2β, mean residence time, Ka, area under the concentration-time curve (AUC) within the different dosing intervals, apparent oral total clearance (CLtot/F), and apparent oral volume of distribution (V/F) were calculated.
(ii) Quantitation of bacterial growth and evaluation of the antibacterial effect
Doubling time, defined as time necessary for a twofold increase of the bacterial cell count, was determined by linear regression analysis of the log10 CFU/milliliter versus time during exponential phase (from 3 to 5.5 h postinoculation). In order to compare the in vitro antibacterial effect of the two antibiotics under human simulated concentration profiles, several parameters were determined: the MIC-related pharmacokinetic parameters were the inhibitory quotient (Cmax/MIC) (14) and the AUC divided by the MIC (AUC/MIC) (9, 15, 26), and the indices of bacterial killing in the presence of antibiotic were calculated as the difference between the inoculum at the beginning of the treatment and the bacterial concentration at a defined time (Δlog CFU/milliliter) (9, 24), the area under the bacterial killing and regrowth curve from 0 to 48 h (AUBC0-48), and the area between the control growth curve and the bacterial killing and regrowth curve from the zero point to 48 h (ABBC0-48) (16). The levels of resistant mutants were expressed as the percentage of the control population obtained with the same bacterial suspension sample but on antibiotic-free agar plates.
RESULTS
Antibiotic assays and pharmacokinetic data
Antibiotic assays exhibited the same reliable performances as previously described (5, 6). Fluorescence detection allowed a quantification limit of 0.078 and 0.050 μg/ml for ciprofloxacin and moxifloxacin, respectively. The standard curve was linear between 0.078 and 1.25 μg/ml for ciprofloxacin and between 0.050 and 3.2 μg/ml for moxifloxacin. Intraday and interday coefficients of variation within the linearity range varied for ciprofloxacin from 0.30 to 1.04% and 2.09 to 7.07%, respectively, and the corresponding imprecisions for moxifloxacin were ≤4.76 and ≤5.75%, respectively. Intraday and interday accuracies ranged from −2.64 to 4.04% and from −3.54 to 1.25% for ciprofloxacin, respectively, and from −2.93 to 4.50% and −1.10 to 6.00% for moxifloxacin, respectively. Mean concentration-time curves from triplicate experiments for the two compartments during Sm206 strain tests are depicted in Fig. 1A1 and B1 for ciprofloxacin and moxifloxacin, respectively. The corresponding mean pharmacokinetic parameters ± the standard deviation (SD) for PCp are shown in Table 1 for both ciprofloxacin and moxifloxacin. The coefficient of correlation between experimental data and the calculated pharmacokinetic profile was always greater than 0.99. Mean concentration curves (n = 2) for the compartments during CIP 54.90 and CIP 60.77T strains tests are shown in Fig. 2 and 3, respectively. Pharmacokinetic parameter values obtained from these curves (data not shown) were similar to those obtained in the Sm206 tests.
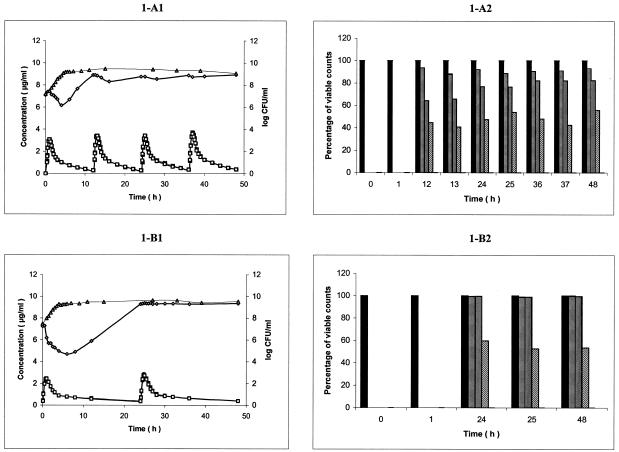
FIG. 1.
(A1 and B1) Ciprofloxacin (A1) and moxifloxacin (B1) pharmacokinetics and pharmacodynamic effect on S. maltophilia Sm206 during simulation of twice-daily oral administration of 750 mg of ciprofloxacin over 48 h and once-daily oral administration of 400 mg of moxifloxacin, respectively, in the in vitro PK/PD model (mean, n = 3). Symbols: ▪, CCp concentration-time curve; □, PCp concentration-time curve; ▵, control growth curve; ⋄, killing and regrowth curve. (A2 and B2) Emergence of Sm206 ciprofloxacin (A2)- and moxifloxacin (B2)-resistant mutants during the corresponding simulations (mean percentage of viable counts; n = 3). Bars: ▪, control; ▤, resistant to 1 times the MIC; ▥; resistant to 4 times MIC; ▧, resistant to 16 times the MIC.
TABLE 1.
Mean pharmacokinetic parameters for the PCp after simulation with twice-daily oral administration of 750 mg of ciprofloxacin and once daily oral administration of 400 mg of moxifloxacin over 48 h and the corresponding human reference data.
| Parameter | Mean value ± SD (n = 3) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Ciprofloxacin |
Moxifloxacin |
|||||||
| Dosing interval |
Human data (n = 6) | Dosing interval |
Human data (n = 12) | |||||
| 0-12 h | 12-24 h | 24-36 h | 36-48 h | 0-24 h | 24-48 h | |||
| Cmax (μg/ml) | 3.08 ± 0.43 | 3.39 ± 0.06 | 3.43 ± 0.21 | 3.60 ± 0.17 | 2.94 ± 0.42a | 2.45 ± 0.11 | 2.82 ± 0.23 | 2.5 ± 1.29c |
| Tmax (h) | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.2 ± 0.5a | 0.90 ± 0.00 | 0.90 ± 0.00 | 2 (0.5-6.0)c |
| Cres. (μg/ml) | 0.29 ± 0.02 | 0.29 ± 0.10 | 0.34 ± 0.05 | 0.37 ± 0.08 | 0.36 ± 0.02 | 0.39 ± 0.06 | ||
| Ka (h−1) | 2.72 ± 0.47 | 2.68 ± 0.44 | 2.37 ± 0.10 | 2.18 ± 0.13 | 2.70 ± 1.22b | 2.09 ± 0.80 | 1.99 ± 0.68 | 2-3c |
| t1/2β (h) | 4.59 ± 0.09 | 4.77 ± 1.03 | 5.19 ± 0.33 | 5.15 ± 0.59 | 4.70 ± 0.83a | 16.5 ± 0.5 | 16.9 ± 2.9 | 15.6 ± 1.15c |
| MRTd (h) | 6.72 ± 0.10 | 6.81 ± 0.99 | 7.57 ± 0.47 | 7.73 ± 0.83 | 5.28 ± 0.89a | 22.0 ± 0.7 | 22.5 ± 3.8 | |
| AUC (h × μg/ml) | 11.5 ± 0.6 | 12.5 ± 1.2 | 12.8 ± 0.6 | 14.2 ± 1.6 | 11.53 ± 2.21a | 17.6 ± 0.3 | 19.3 ± 0.5 | |
| CLtot/F (liter) | 56.0 ± 2.5 | 52.1 ± 8.7 | 48.9 ± 3.9 | 44.6 ± 6.9 | 45.0 ± 5.9a | 15.3 ± 0.3 | 13.7 ± 1.8 | |
| V/F (liter) | 371.1 ± 14.3 | 350.8 ± 31.3 | 365.1 ± 10.3 | 328.2 ± 11.9 | 304.0 ± 61.0a | 364.4 ± 5.1 | 324.0 ± 34.1 | |
| Correlation coefficient | 0.996 ± 0.003 | 0.998 ± 0.000 | 0.996 ± 0.000 | 0.993 ± 0.006 | 0.996 ± 0.005 | 0.997 ± 0.002 | ||
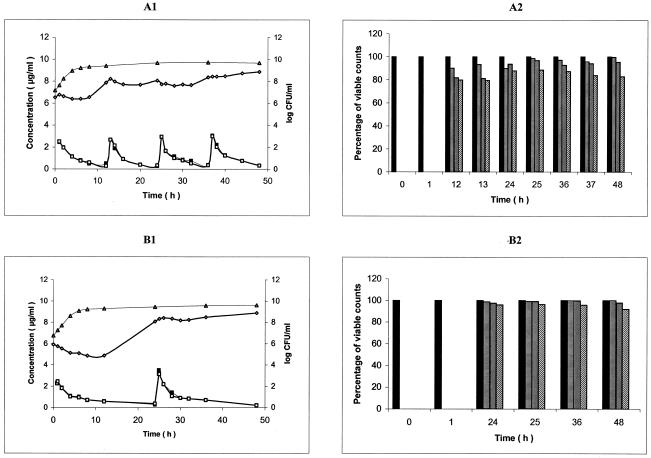
FIG. 2.
(A1 and B1) Ciprofloxacin (A1) and moxifloxacin (B1) pharmacokinetics and pharmacodynamic effect on S. maltophilia CIP 54.90, during simulation of twice daily oral administration of 750 mg of ciprofloxacin over 48 h and once daily oral administration of 400 mg of moxifloxacin, respectively, in an in vitro PK/PD model (mean, n = 2). Symbols: ▪, CCp concentration-time curve; □, PCp concentration-time curve; ▵, control growth curve; ⋄, killing and regrowth curve. (A2 and B2) Emergence of CIP 54.90 ciprofloxacin (A2)- and moxifloxacin (B2)-resistant mutants during the corresponding simulations (mean percentage of viable counts, n = 2). Bars: ▪, control; ▤, resistant to 1 times the MIC; ▥; resistant to 4 times MIC; ▧, resistant to 16 times the MIC.
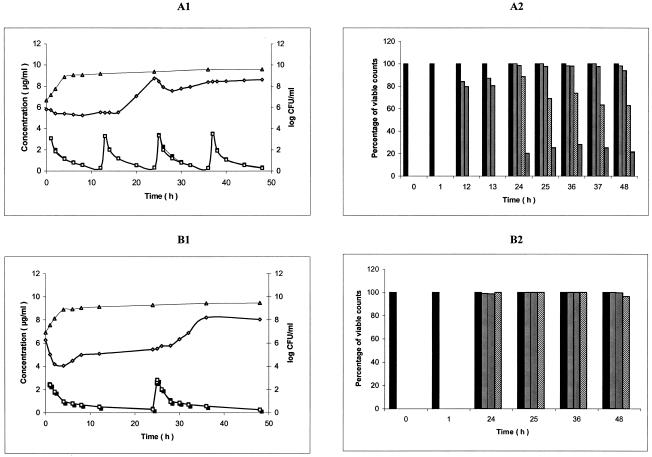
FIG. 3.
(A1 and B1) Ciprofloxacin (A1) and moxifloxacin (B1) pharmacokinetics and pharmacodynamic effect on S. maltophilia CIP 60.77T (ATCC 13637) during simulation of twice-daily oral administration of 750 mg of ciprofloxacin over 48 h and once-daily oral administration of 400 mg of moxifloxacin, respectively, in an in vitro PK/PD model (mean, n = 2). Symbols: ▪, CCp concentration-time curve; □, PCp concentration-time curve; ▵, control growing curve; ⋄, killing and regrowth curve. (A2 and B2) Emergence of CIP 60.77T ciprofloxacin (A2)- and moxifloxacin (B2)-resistant mutants during the corresponding simulations (mean percentage of viable counts, n = 2). Bars: ▪, control; ▤, resistant to 1 times the MIC; ▥; resistant to 4 times MIC; ▧, resistant to 16 times the MIC; , resistant to 64 times the MIC.
Growth control curves
Growth control curves of strain Sm206 for ciprofloxacin and moxifloxacin administration simulation (means of three curves) are shown in Fig. A1 and B1, respectively. At 3 h after inoculation of the PCp, the bacterial concentration (initial inoculum for experiments), expressed in log10 CFU/milliliter were similar for ciprofloxacin and moxifloxacin administration simulation: 7.165 ± 0.309 and 7.463 ± 0.236, respectively. Exponential growth continued for up to 4 h after the initial sampling time (t0). The doubling times during the exponential phase were 0.65 ± 0.10 h and 0.59 ± 0.06 h for ciprofloxacin and moxifloxacin administration simulation, respectively. Then, a plateau at ∼9 log10 CFU/ml was observed under the two conditions. Similar results were obtained for CIP 54.90 (Fig. 2A1 and B1) and CIP 60.77T (Fig. 3A1 and B1).
Pharmacodynamic data. (i) Killing curves
Killing curves of strain Sm206 exposed to ciprofloxacin and moxifloxacin (means of three curves) are shown in Fig. 1A1 and B1, respectively. During exposure at the initial dose of the simulation of twice-daily oral administration of 750 mg of ciprofloxacin, bacterial counts increased until the antibiotic concentration in the PCp reached the peak. Then, a 1-log decrease within 3 h was observed, followed by a rapid regrowth. Bacterial concentration reached 8.9 log CFU/ml at 12 h, just before the second dose, which induced a slight decrease of 0.6 log CFU/ml within 4 h. Regrowth occurred again until the third dose, which produced a similar effect. However, the fourth dose did not lead to any appreciable reduction in the number of viable organisms. After the first dose of moxifloxacin (400 mg per 24 h), an immediate reduction in the number of viable counts was seen, from 7.25 ± 0.05 to 4.69 ± 0.36 log CFU/ml over 6 h. Then, a relatively slow regrowth started and, at 24 h, the bacterial population was close to the plateau value of the control (9.28 ± 0.09 versus 9.48 ± 0.22 log CFU/ml). The second dose at 24 h had no appreciable influence on the growth pattern of Sm206. Similar results were obtained for CIP 54.90 (Fig. 2A1 and B1, means of two curves). Both for ciprofloxacin and moxifloxacin, the population of the more susceptible strain, CIP 60.77T, took longer to attain the plateau value (24 and 36 h, respectively) (Fig. 3A1 and B1, means of two curves).
(ii) MIC-related pharmacokinetic parameters and quantitative evaluation of in vitro antibacterial effect
Mean parameters (± the SD for Sm206) calculated from antibiotic experimental concentrations, pharmacokinetic parameters, growth control curves, and killing and regrowth curves are presented in Table 2 for both ciprofloxacin and moxifloxacin. Considering the MICs of ciprofloxacin and moxifloxacin for Sm206 (2 and 0.5 μg/ml, respectively), CIP 54.90 (1 and 0.25 μg/ml, respectively), and CIP 60.77T (ATCC 13637, 0.5 and 0.0625 μg/ml, respectively), the Cmax/MIC ratios ranged from 1.54 to 6.70 and from 4.90 to 45.1 for ciprofloxacin and moxifloxacin, respectively. The AUC/MIC ratios for a 24-h interval ranged from 12 to 50.2 h and from 35.1 to 286.7 h for ciprofloxacin and moxifloxacin, respectively. The Δlog CFU/milliliter 1 h after the beginning of the first administration at 1 h (at t1) was positive for ciprofloxacin against Sm206 and CIP 54.90 but negative for moxifloxacin against the three tested strains. This parameter showed that ciprofloxacin had an appreciable effect at 6 h, although it remained sixfold lower than that of moxifloxacin. Bacterial counts of Sm206 and CIP 54.90 overtook the initial inoculum 12 h after the first dose of ciprofloxacin, while the regrowth was still slowed down in the presence of moxifloxacin. The population of CIP 60.77T was less than the initial inoculum 12 h after administration of both ciprofloxacin and moxifloxacin. However, at 48 h after the beginning of the treatment, a similar level of regrowth was observed for ciprofloxacin and moxifloxacin, although CIP 60.77T seemed to be more susceptible to moxifloxacin. The AUBC0-48 was slightly greater in the presence of ciprofloxacin than in the presence of moxifloxacin for the three strains. The difference in effect between the two antibiotics was still more pronounced when the ABBC0-48 was measured.
TABLE 2.
MIC-related pharmacokinetic parameters and antibacterial effect indices of the simulation of oral administration of ciprofloxacin (750 mg/12 h over 48 h) and moxifloxacin (400 mg/24 h over 48 h) against a clinical isolate of S. maltophilia (Sm206) and two reference strains, CIP 54.90 and CIP 60.77T
| Parameter | Mean or mean ± SD for strain: |
|||||
|---|---|---|---|---|---|---|
| Sm206 |
CIP 54.90 |
CIP 60.77T (ATCC 13637) |
||||
| Ciprofloxacin (MIC = 2 μg/ml; n = 3) | Moxifloxacin (MIC = 0.5 μg/ml; n = 3) | Ciprofloxacin (MIC = 1 μg/ml; n = 2) | Moxifloxacin (MIC = 0.25 μg/ml; n = 2) | Ciprofloxacin (MIC = 0.5 μg/ml; n = 2) | Moxifloxacin (MIC = 0.0625 μg/ml; n = 2) | |
| Doubling time (h) | 0.65 ± 0.10 | 0.59 ± 0.06 | 0.61 | 0.57 | 0.55 | 0.55 |
| Cmax/MIC at: | ||||||
| 0.90 h | 4.90 ± 0.23 | |||||
| 1 h | 1,54 ± 0.21 | 2.48 | 9.80 | 6.20 | 38.5 | |
| 24.90 h | 5.63 ± 0.46 | 12.6 | ||||
| 25 h | 1.72 ± 0.11 | 2.96 | 6.70 | 45.1 | ||
| AUC0-24/MIC (h) | 12.0 ± 0.7 | 35.1 ± 0.6 | 22.6 | 68.8 | 49.4 | 267.2 |
| AUC24-48/MIC (h) | 13.5 ± 1.1 | 38.6 ± 0.9 | 25.2 | 77.6 | 50.2 | 286.7 |
| Δlog CFU/ml at: | ||||||
| t1 | 0.32 ± 0.09 | −1.08 ± 0.17 | 0.24 | −0.15 | −0.07 | −1.24 |
| t6 | −0.42 ± 0.30 | −2.56 ± 0.33 | −0.12 | −0.81 | −0.49 | −1.77 |
| t12 | 1.79 ± 0.22 | −1.37 ± 0.89 | 1.35 | −1.03 | −0.30 | −1.17 |
| t24 | 1.66 ± 0.12 | 2.08 ± 0.02 | 1.53 | 2.17 | 2.93 | −0.80 |
| t25 | 1.65 ± 0.19 | 2.13 ± 0.03 | 1.21 | 2.42 | 2.64 | −0.73 |
| t48 | 1.81 ± 0.13 | 2.08 ± 0.13 | 2.34 | 2.97 | 2.81 | 1.79 |
| AUBC0-48 (h × log CFU/ml) | 401.3 ± 3.7 | 378.7 ± 13.8 | 371.8 | 343.2 | 342.3 | 295.2 |
| ABBC0-48 (h × log CFU/ml) | 40.4 ± 3.7 | 72.9 ± 13.8 | 81.5 | 101.8 | 101.1 | 144.7 |
(iii) Emergence of resistant mutants
Survivors able to grow in the presence of concentrations equal or superior to the MICs for the parental strains remained quinolone resistant after repeated subcultures on antibiotic-free media, demonstrating that they were stable mutants rather than transiently induced cells. Inoculation of the surviving cells on antibiotic-containing media showed that the mutants emerged from the 12th hour for ciprofloxacin and from the 24th hour for moxifloxacin (Fig. 1 to 3). At 12 h, Sm206 and CIP 54.90 mutants tolerated 16 times the MIC of ciprofloxacin for the initial strains (45 and 80% of the population, respectively), while mutants of the more susceptible CIP 60.77T strain were only resistant to 4 times the MIC (80%). Nevertheless, CIP 60.77T led to mutants with the highest increases in MICs (64 times the MIC) from the 24th hour. In contrast, mutants derived from the three strains could be cultivated on medium containing 16 times the MIC of moxifloxacin from the 24th hour, but none could be cultivated at 64 times the MIC.
Antibiograms and MIC determinations were performed for 53 mutants selected at different sampling times and antibiotic concentrations. These mutants exhibited increases not only in resistance to quinolones but also in resistance to other structurally unrelated antibiotics. When mutants derived from the same parental strain were compared, they showed different resistance patterns, although most of them harbored the same profile, as indicated by the mode MICs (Table 3). Furthermore, substantial variations were noticed between mutants according to the parental strain. For example, the increases in the mode MICs of nalidixic acid were 2 to 4 times, 8 times, and 64 times the MIC for Sm206, CIP 54.90, and CIP 60.77T, respectively. Similarly, increases in erythromycin resistance were observed only in the Sm206 and CIP 54.90 series, while no decrease in amikacin susceptibility was recorded only for the CIP 60.77T derivatives. Finally, for a given parental strain, no significant differences were observed between resistance profiles of mutants grown under either ciprofloxacin or moxifloxacin (data not shown).
TABLE 3.
Comparative susceptibilities to antibiotics of parental strains and their MDR-derived mutantsa
| Antibiotic | Sm206 |
CIP 54.90 |
CIP 60.77T (ATCC 13637) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Parent (MIC, μg/ml) | MDR mutant (MIC×) (n = 26) |
Parent (MIC, μg/ml) | MDR mutant (MIC×) (n = 14) |
Parent (MIC, μg/ml) | MDR mutant (MIC×) (n = 13) |
||||
| Range | Mode MIC | Range | Mode MIC | Range | Mode MIC | ||||
| Ciprofloxacin | 2 | 4-64 | 16 | 1 | 16-32 | 32 | 0.5 | 16-128 | 64 |
| Moxifloxacin | 0.5 | 4-64 | 16 | 0.25 | 32-64 | 32 | 0.0625 | 8-64 | 64 |
| Nalidixic acid | 16 | 2-16 | 2-4 | 8 | 8-16 | 8 | 4 | 32-64 | 64 |
| Norfloxacin | 16 | 2-64 | 8 | 8 | 16-128 | 64 | 4 | 16-128 | 32 |
| Tetracycline | 8 | 1-8 | 2 | 4 | 4-8 | 4-8 | 1 | 1-8 | 2 |
| Chloramphenicol | 4 | 1-16 | 4 | 8 | 2-4 | 4 | 4 | 8-64 | 16-32 |
| Erythromycin | 128 | 1-4 | 4 | 256 | 2-4 | 2 | 16 | 0.5-1 | 1 |
| Trimethoprim | 8 | 1-16 | 8 | 8 | 2-4 | 4 | 16 | 8-32 | 32 |
| Sulfamethoxazole | 8 | 2-4 | 1 | 8 | 0.125-0.5 | 0.5 | 16 | 1-8 | 2 |
| Cotrimoxazole | 8 | 1-4 | 1 | 4 | 1 | 1 | 4 | 1-16 | 4 |
| Ticarcillin plus CAb | 16 | 1-2 | 1 | 16 | 0.5-1 | 1 | 8 | 0.5-2 | 1 |
| Cefepime | 16 | 1-4 | 1 | 2 | 2-4 | 2 | 8 | 0.5-4 | 2 |
| Amikacin | 64 | 0.125-4 | 0.5 | 16 | 0.25-0.5 | 0.5 | 4 | 1-4 | 1-2 |
n, Number of tested MDR mutants; MIC×, multiple of the MIC compared to that for the parental strain. The range gives the lowest and highest increases in MICs, and the mode MIC corresponds to the value obtained for the majority (at least half) of the strains.
CA, clavulanic acid at a fixed concentration of 2 μg/ml.
DISCUSSION
Cotrimoxazole has traditionally been regarded as the agent of choice for the therapy of S. maltophilia infection, with ticarcillin-clavulanate being an alternative in intolerant individuals (13, 32). Due to an increasing frequency of cotrimoxazole resistance, ciprofloxacin has eventually become the empirical therapy, but a parallel increase in quinolone resistance was noted (36). Previous in vitro studies suggest that moxifloxacin can offer a better therapeutic option (28, 37). However, they are based on MICs that are static measures and have been found to be unreliable for S. maltophilia (13). Models that interpret both pharmacokinetics and bacterial response may provide more clinically meaningful information. In the present study, a PK/PD model was used to compare the potential efficacy of ciprofloxacin and moxifloxacin against three quinolone-susceptible strains of S. maltophilia.
The model was first validated at the pharmacokinetic level. Concentration-time curves of ciprofloxacin and moxifloxacin showed a good reproducibility of sequential experiments, and pharmacokinetic parameters obtained for both CCp and PCp ranged within the confidence intervals of the means of human data (10, 17, 33). The model was also validated for the different growth conditions since similar doubling times were obtained. Finally, quinolone susceptibility of the test organisms was precisely determined (MICs of ciprofloxacin and moxifloxacin, 0.5 to 2 μg/ml and 0.0625 to 0.5 μg/ml, respectively) and found to be consistent with the literature (28, 37, 40).
Time-kill experiments indicated that ciprofloxacin reduced the bacterial counts by ca. 1 log CFU/ml prior to a rapid bacterial regrowth up to the plateau value of the growth control curve at 12 h (i.e., at the end of the first dosing interval) for Sm206 and CIP 54.90, or 24 h for CIP 60.77T. After moxifloxacin first dose, bacterial counts were reduced by ∼2 log CFU/ml, and the regrowth rate was slower than that observed with ciprofloxacin, since the plateau value was reached at the 24th hour (just before the second administration) or the 36th hour. Further doses of both agents did not reduce the bacterial counts. A ciprofloxacin Cmax/MIC greater than 10 in a dose ranging study in an in vitro model has been considered to be a strong predictor of clinical success against P. aeruginosa (27), an organism similarly saprophytic, multiresistant, and pathogenic for debilitated patients as S. maltophilia. In our study, only moxifloxacin led to upper values for the reference strains and the two MIC-related parameters were at least threefold higher for moxifloxacin than for ciprofloxacin. During simulation infection with Sm206 and CIP 54.90, the Δlog CFU/milliliter was negative only at 6 h for ciprofloxacin, whereas it remained negative until 12 h for moxifloxacin. In a recent study using an in vitro pharmacodynamic model to assess the best measure of antibacterial effect for moxifloxacin (25), the AUBC appeared to be the optimum one. In the present investigation, AUBC values were slightly smaller for moxifloxacin than for ciprofloxacin (295.2 to 378.7 versus 342.3 to 401.3 h × log CFU/ml). In another study comparing several parameters in an in vitro dynamic model with ciprofloxacin (16), better unbiased presentation of the effect was obtained with the ABBC. The values of ABBC0-48 were ∼1.5-fold higher for moxifloxacin than for ciprofloxacin (72.9 to 144.7 versus 40.4 to 101 h × log CFU/ml). Consequently, moxifloxacin compared favorably to ciprofloxacin by all antibacterial effect indices, the AUBC0-48 and ABBC0-48 values in particular.
Non-protein-supplemented broth was used for all experiments that simulated human total (free plus bound) concentrations of the antibiotics in plasma. Protein binding appears to be only a modest contributor to factors involved in the therapeutic effectiveness of fluoroquinolones (8). Indeed, fluoroquinolones exhibit a low level of protein binding, usually <50% (8, 34). Moreover, the magnitude of this parameter is similar for ciprofloxacin and moxifloxacin (20 to 40%) (8, 34). Thus, this factor is not expected to introduce a bias effect in the comparison of the intrinsic antibacterial effect of the two compounds.
Both fluoroquinolones selected for resistant mutants that expressed cross-resistance to unrelated antibiotics, as previously reported in strains overproducing MDR efflux systems (22, 39). Mutants derived from the same parental strain exhibited various antibiotic resistance patterns. Such variations, including erythromycin resistance, in particular, together with increased amikacin susceptibility, versus no changes toward these antibiotics have previously been ascribed to overproduction of different MDR efflux systems rather than to hyperexpression at different levels of a single pump (39). However, from each parental strain, mutants with a predominant resistance profile were selected, which might reflect strain-specific propensity for the overexpression of different systems; similar resistance profiles were obtained after ciprofloxacin or moxifloxacin exposure. The reduced potential of moxifloxacin to select for bacterial resistance has been assessed for organisms in which high-level quinolone resistance implies target alterations that occur at low frequencies (10−7 to 10−9) and/or multistep mutations (7, 21, 23, 25, 29). In S. maltophilia, mutants overproducing their efflux pumps are present at high frequencies (10−5 to 10−6) and exhibit clinically relevant increases in MICs (1, 22). Our results highlight the importance of achieving drug concentrations that exceed the MIC of not only the original isolate but also resistant subpopulations. Although noteworthy, the risk of emergence of resistant mutants is certainly overestimated in our PK/PD model, in which the initial inoculum is high and host defenses are absent. Moreover, moxifloxacin concentrates in particular body sites, such as the lungs (7, 11, 29).
In conclusion, the antibacterial effect indices obtained in our PK/PD model demonstrated a better efficacy of moxifloxacin than ciprofloxacin against S. maltophilia under conditions simulating human pharmacokinetics of the two compounds. Thus, when a fluoroquinolone has to be used for treating S. maltophilia infections, e.g., in the case of a fluoroquinolone-susceptible strain either resistant to cotrimoxazole and/or ticarcillin-clavulanate or in a cotrimoxazole-allergic patient, moxifloxacin is preferable to ciprofloxacin. However, due to the selection of resistant mutants by both fluoroquinolones, moxifloxacin should be administered at the maximum tolerated doses and in combination. Given the existence of cross-resistance between fluoroquinolones and trimethoprim, the combination of moxifloxacin plus ticarcillin-clavulanate might represent the most promising regimen, as much as synergy has been demonstrated between ciprofloxacin and ticarcillin-clavulanate (31).
Acknowledgments
We thank Catherine André for expert technical assistance and Pierre Arvis (Bayer France), Isabelle Maachi-Guillot (Haut Levêque Hospital Pharmacy Service), Sylvie Darmagnac (Fresenius Medical Care), Vincent Marque, and all of the Pellegrin Hospital Sterilization Service staff for their help.
This study was supported by grants from the Ministère de l'Education Nationale et de la Recherche (EA 525), Université de Bordeaux 2, Bordeaux, France.
REFERENCES
- 1.Alonso, A., and J. L. Martinez. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41:1140-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, A., and J. L. Martinez. 2000. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 44:3079-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso, A., and J. L. Martinez. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:1879-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ba, B. B., A. Bernard, A. Iliadis, C. Quentin, D. Ducint, R. Etienne, M. Fourtillan, I. Maachi-Guillot, and M.-C. Saux. 2001. New approach for accurate simulation of human pharmacokinetics in an in vitro pharmacodynamic model: application to ciprofloxacin. J. Antimicrob. Chemother. 47:223-227. [DOI] [PubMed] [Google Scholar]
- 5.Ba, B. B., D. Ducint, M. Fourtillan, and M.-C. Saux. 1998. Fully automated high-performance liquid chromatography of ciprofloxacin with direct injection of plasma and Mueller-Hinton broth for pharmacokinetic/pharmacodynamic studies. J. Chromatogr. B 714:317-324. [DOI] [PubMed] [Google Scholar]
- 6.Ba, B. B., R. Etienne, D. Ducint, C. Quentin, and M.-C. Saux. 2001. Determination of moxifloxacin in growth media by high-performance liquid chromatography. J. Chromatogr. B 754:107-112. [DOI] [PubMed] [Google Scholar]
- 7.Balfour, J. A., and H. M. Lamb. 2000. Moxifloxacin: a review of its clinical potential in the management of community-acquired respiratory tract infections. Drugs 59:115-139. [DOI] [PubMed] [Google Scholar]
- 8.Bergogne-Bérézin, E. 2002. Clinical role of protein binding of quinolones. Clin. Pharmacokinet. 41:741-750. [DOI] [PubMed] [Google Scholar]
- 9.Bowker, K. E., M. Wootton, C. A. Rogers, R. Lewis, H. A. Holt, and A. P. MacGowan. 1999. Comparison of in-vitro pharmacodynamics of once and twice daily ciprofloxacin. J. Antimicrob. Chemother. 44:661-667. [DOI] [PubMed] [Google Scholar]
- 10.Crump, B., R. Wise, and J. Dent. 1983. Pharmacokinetics and tissue penetration of ciprofloxacin. Antimicrob. Agents Chemother. 24:784-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalhoff, A. 1999. Pharmacodynamics of fluoroquinolones. J. Antimicrob. Chemother. 43(Suppl. B):51-59. [DOI] [PubMed] [Google Scholar]
- 12.Dalhoff, A., U. Petersen, and R. Endermann. 1996. In vitro activity of BAY 12-8039, a new 8-methoxyquinolone. Chemotherapy 42:410-425. [DOI] [PubMed] [Google Scholar]
- 13.Denton, M., and K. G. Kerr. 1998. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microb. Rev. 11:57-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellner, P. D., and H. C. Neu. 1981. The inhibitory quotient: a method for interpreting minimum inhibitory concentration data. JAMA 246:1575-1578. [DOI] [PubMed] [Google Scholar]
- 15.Firsov, A. A., A. A. Shevchenko, S. N. Vostrov, and S. H. Zinner. 1998. Inter- and intraquinolone predictors of antimicrobial effect in an in vitro dynamic model: new insight into a widely used concept. Antimicrob. Agents Chemother. 42:659-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firsov, A. A., S. N. Vostrov, A. A. Shevchenko, and G. Cornaglia. 1997. Parameters of bacterial killing and regrowth kinetics and antimicrobial effect examined in terms of area under the concentration-time curve relationships: action of ciprofloxacin against Escherichia coli in an in vitro dynamic model. Antimicrob. Agents Chemother. 41:1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fourtillan, J. B. 1990. Caractéristiques pharmacocinétiques de la ciprofloxacine par voies orale et intraveineuse. Med. Mal. Infect. 20:33-37. [Google Scholar]
- 18.Gales, A. C., R. N. Jones, K. R. Forward, J. Liñares, H. S. Sader, and J. Verhoef. 2001. Emerging importance of multidrug-resistant Acinetobacter species and Stenotrophomonas maltophilia as pathogens in seriously ill patients: geographic patterns, epidemiological features, and trends in the SENTRY antimicrobial surveillance program (1997-1999). Clin. Infect. Dis. 32(Suppl. 2):104-113. [DOI] [PubMed] [Google Scholar]
- 19.Garrison, M. W., D. E. Anderson, D. M. Campbell, K. C. Carroll, C. L. Malone, J. D. Anderson, R. J. Hollis, and M. A. Pfaller. 1996. Stenotrophomonas maltophilia: emergence of multidrug-resistant strains during therapy and in an in vitro pharmacodynamic chamber model. Antimicrob. Agents Chemother. 40:2859-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock, R. E. W. 1998. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis. 27(Suppl. 1):93-99. [DOI] [PubMed] [Google Scholar]
- 21.Klepser, M. E., E. J. Ernst, C. R. Petzold, P. Rhomberg, and G. V. Doern. 2001. Comparative bactericidal activities of ciprofloxacin, clinafloxacin, grepafloxacin, levofloxacin, moxifloxacin, and trovafloxacin against Streptococcus pneumoniae in a dynamic in vitro model. Antimicrob. Agents Chemother. 45:673-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecso-Bornet, M., J. Pierre, D. Sarkis-Karam, S. Lubera, and E. Bergogne-Berezin. 1992. Susceptibility of Xanthomonas maltophilia to six quinolones and study of outer membrane proteins in resistant mutants selected in vitro. Antimicrob. Agents Chemother. 36:669-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lister, P. D. 2001. Pharmacodynamics of moxifloxacin and levofloxacin against Staphylococcus aureus and Staphylococcus epidermidis in an in vitro pharmacodynamic model. Clin. Infect. Dis. 32(Suppl. 1):33-38. [DOI] [PubMed] [Google Scholar]
- 24.MacGowan, A. P., K. E. Bowker, M. Wootton, and H. A. Holt. 1999. Exploration of the in-vitro pharmacodynamic activity of moxifloxacin for Staphylococcus aureus and streptococci of Lancefield group A and G. J. Antimicrob. Chemother. 44:761-766. [DOI] [PubMed] [Google Scholar]
- 25.MacGowan, A., C. Rogers, H. A. Holt, M. Wootton, and K. Bowker. 2000. Assessment of different antibacterial effect measures used in in vitro models of infection and subsequent use in pharmacodynamic correlations for moxifloxacin. J. Antimicrob. Chemother. 46:73-78. [DOI] [PubMed] [Google Scholar]
- 26.Madaras-Kelly, K. J., B. E. Ostergaard, L. B. Hovde, and J. C. Rotschafer. 1996. Twenty-four-hour area under the concentration-time curve/MIC ratio as a generic predictor of fluoroquinolone antimicrobial effect by using three strains of Pseudomonas aeruginosa and an in vitro pharmacodynamic model. Antimicrob. Agents Chemother. 40:627-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchbanks, C. R., J. R. McKiel, D. H. Gilbert, N. J. Robillard, B. Painter, S. H. Zinner, and M. N. Dudley. 1993. Dose ranging and fractionation of intravenous ciprofloxacin against Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro model of infection. Antimicrob. Agents Chemother. 37:1756-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muñoz Bellido, J. L., F. J. Sánchez-Hernández, M. N. Gutiérrez Zufiaurre, and J. A. García-Rodríguez. 2000. In vitro activity of newer fluoroquinolones against Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 46:323-342. [DOI] [PubMed] [Google Scholar]
- 29.Nightingale, C. H. 2000. Moxifloxacin, a new antibiotic designed to treat community-acquired respiratory tract infections: a review of microbiologic and pharmacokinetic-pharmacodynamic characteristics. Pharmacotherapy 20:245-256. [DOI] [PubMed] [Google Scholar]
- 30.Poole, K. 2001. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3:255-264. [PubMed] [Google Scholar]
- 31.Poulos, C. D., S. O. Matsumura, B. M. Willey, D. E. Low, and A. McGeer. 1995. In vitro activities of antimicrobial combinations against Stenotrophomonas (Xanthomonas) maltophilia. Antimicrob. Agents Chemother. 39:2220-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robin, T., and J. M. Janda. 1996. Pseudo-, Xantho-, Stenotrophomonas maltophilia: an emerging pathogen in search of a genus. Clin. Microbiol. Newsl. 18:9-13. [Google Scholar]
- 33.Stass, H., and D. Kubitza. 1999. Pharmacokinetics and elimination of moxifloxacin after oral and intravenous administration in man. J. Antimicrob. Chemother. 43(Suppl. B):83-90. [DOI] [PubMed] [Google Scholar]
- 34.Stein, G. E. 1996. Pharmacokinetics and pharmacodynamics of newer fluoroquinolones. Clin. Infect. Dis. 23(Suppl. 1):19-24. [DOI] [PubMed] [Google Scholar]
- 35.Valdezate, S., A. Vindel, A. Echeita, F. Baquero, and R. Cantón. 2002. Topoisomerase II and IV quinolone resistance-determining regions in Stenotrophomonas maltophilia clinical isolates with different levels of quinolone susceptibility. Antimicrob. Agents Chemother. 46:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vartivarian, S., E. Anaissie, G. Bodey, H. Sprigg, and K. Rolston. 1994. A changing pattern of susceptibility of Xanthomonas maltophilia to antimicrobial agents: implications for therapy. Antimicrob. Agents Chemother. 38:624-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss, K., C. Restieri, E. De Carolis, M. Laverdière, and H. Guay. 2000. Comparative activity of new quinolones against 326 clinical isolates of Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 45:363-365. [DOI] [PubMed] [Google Scholar]
- 38.Wüst, J., R. Frei, H. Günthard, and M. Altwegg. 1995. Analysis of restriction fragment length polymorphism and ribotyping of multiresistant Stenotrophomonas maltophilia isolated from persisting lung infection in a cystic fibrosis patient. Scand. J. Infect. Dis. 27:499-502. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, L., X.-Z. Li, and K. Poole. 2000. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob. Agents Chemother. 44:287-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, L., X.-Z. Li, and K. Poole. 2001. Fluoroquinolone susceptibilities of efflux-mediated multidrug-resistant Pseudomonas aeruginosa, Stenotrophomonas maltophilia, and Burkholderia cepacia. J. Antimicrob. Chemother. 48:549-552. [DOI] [PubMed] [Google Scholar]