Abstract
Bacteriophage genomes found in a range of bacterial pathogens encode a diverse array of virulence factors ranging from superantigens or pore forming lysins to numerous exotoxins. Recent studies have uncovered an entirely new class of bacterial virulence factors, called effector proteins or effector toxins, which are encoded within phage genomes that reside among several pathovars of Escherichia coli and Salmonella enterica. These effector proteins have multiple domains resulting in proteins that can be multifunctional. The effector proteins encoded within phage genomes are translocated directly from the bacterial cytosol into their eukaryotic target cells by specialized bacterial type three secretion systems (T3SSs). In this review, we will give an overview of the different types of effector proteins encoded within phage genomes and examine their roles in bacterial pathogenesis.
Keywords: Escherichia coli, Salmonella enterica, phage encoded exotoxins and effector proteins
Introduction
Bacteriophage-encoded virulence genes can convert their bacterial host from a non-pathogenic strain to a virulent strain, or to a strain with increased virulence, by providing novel mechanisms to the bacterial cell that enable attachment, invasion and survival within their eukaryotic host. Sometimes the loss of a bacteriophage by a pathogen can render the bacterium non-pathogenic, however this is rare since many of the bacteriophages carrying bacterial virulence genes are temperate, forming stable lysogens within the host genome. Various phage-encoded exotoxins are found in many Gram-negative and Gram-positive bacteria, including Corynebacterium diphtheriae, Escherichia coli, Shigella spp, Pseudomonas aeruginosa, Vibrio cholerae, Staphylococcus aureus and Streptococcus pyogenes.1,2 These exotoxins may be cytotoxic, enterotoxic or neurotoxic, causing an array of diseases ranging from mild gastrointestinal disease to life-threatening toxemia and sepsis. A recently discovered group of virulence factors to be included in the list of phage-encoded virulence factors are effector proteins or toxins (Table 1).1-4 These are multi-domain multifunctional proteins that are secreted by specialized type three secretion systems (T3SSs) found in Gram-negative animal and plant pathogens. In this review, we begin with a brief discussion of the classical phage-encoded exotoxins including their distribution and function in bacterial pathogenesis. We will then explore the range of phage-encoded effector proteins that are described in pathovars of E. coli and Salmonella enterica.
Table 1. Phage-encoded effector proteins in E. coli and S. enterica.
| Phage |
Effector protein |
Function |
|---|---|---|
| E. coli | ||
| lambdoid |
Cif |
Cyclomodulin |
| lambdoid |
EspJ |
Transmission |
| SpLE3-like |
EspL2 |
Actin remodeling |
| lambdoid |
NleA |
Golgi localized |
| lambdoid |
NleG |
Ubiquitin ligase |
| lambdoid |
NleH |
NF-kappaB activity |
| lambdoid | EspF (TccP) | Actin remodeling |
| S. enterica | ||
|---|---|---|
|
Gifsy-1 |
GogB |
Anti-inflammatory effector |
|
Gifsy-1 |
GipA |
Invasion |
| SopEphi |
SopE |
Guanine exchange factor mimic |
| Phage-like |
SopE2 |
Guanine exchange factor mimic |
| Phage-like |
SspH2 |
Inhibits actin polymerization |
| Phage-like |
SseJ |
Deacylase |
| ST64B |
SseK3 |
Invasion |
|
Gifsy-2 |
SspH1 |
Inhibits actin polymerization |
| Gifsy-3 | SseI (GtgB) | E3 ubiquitin ligase |
Classical Phage-Encoded Exotoxins
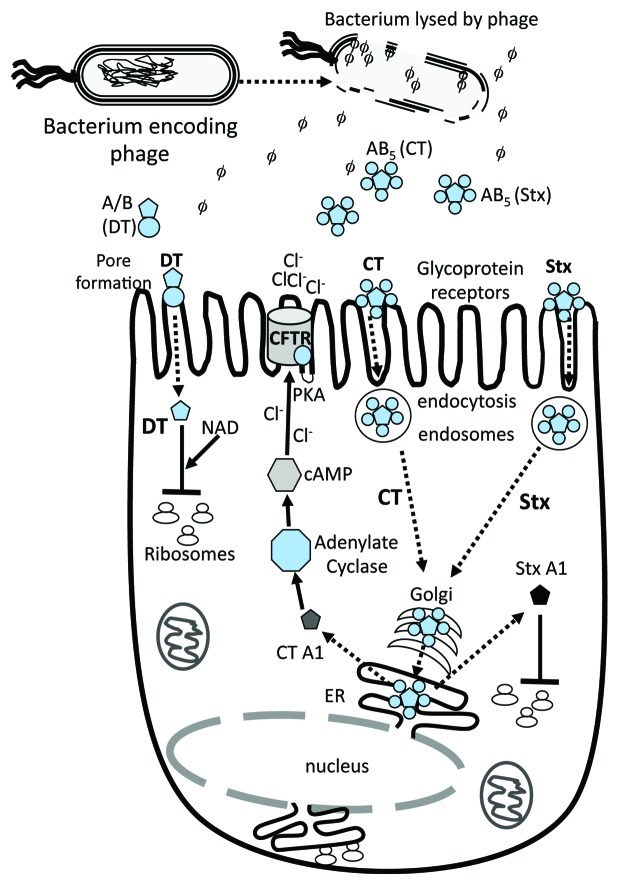
The effects of different phage-encoded exotoxins depend on the specificity of their target cell and target site within the cell. Three phage-encoded AB type exotoxins such as diphtheria toxin (DT), cholera toxin (CT) and Shiga toxin (Stx) are encoded by C. diphtheria, V. cholerae and E. coli and Shigella species, respectively. Each of them has very different cellular target sites and consequently diverse disease outcomes (Fig. 1).
Figure 1. Schematic of toxins function in eukaryotic cells. Dotted arrows indicate steps in bacterial lysis with phage/toxin release, toxin uptake by host cell and target activity. CTXphi which encodes cholera toxin (CT) does not lyse its bacterial host cell. Black solid arrows or t-bars represent activation or blocking of specified bacterial function by toxin. AB denotes a two subunit protein, subunit A is the catalytic subunit and B is the receptor binding or docking subunit on target cell. AB5 denotes a toxin that contains one A subunit and 5 B subunits. Three examples are given; the A/B diphtheria toxin (DT) and the AB5 toxins CT and shiga toxin (Stx). DT gains direct entry into target cell by pore formation. The second method of toxin uptake is by receptor mediated endocytosis, a mechanism used by CT and Stx. Only the A subunit of DT enters the cell cytosol and along with NAD, ADP-ribosylates Elongation factor 2 blocking protein synthesis. Both CT and Stx are retrograde trafficked to the Golgi apparatus. In CT, the A subunit is dissociated from B subunits in the endoplasmic recticulum (ER) and released into the cytosol where it activates adenylate cyclase increasing cAMP which in turn activates PKA resulting in the CFTR membrane Cl- channel activation. The A subunit of Stx ADP- ribosylates the 28S rRNA ribosomal subunit, thus blocking protein synthesis.
DT is an AB type toxin made up of a single A and B subunit where A is the catalytic subunit and B is the receptor binding subunit on the eukaryotic target cell. The tox gene which encodes DT, is encoded by corynebacteriophage β and is located directly adjacent to the phage attachment site, attR, suggesting that this gene was acquired by imprecise excision from a donor host genome.5 Expression of the tox gene is regulated by iron availability and is activated when iron levels are low.6,7 DT is released from the bacterial cell and attaches to a receptor on the cell surface of respiratory epithelium cells. The toxin is internalized by pore formation and transported to its site of activity, the ribosome, where it ADP-ribosylates Elongation factor 2, thereby halting protein synthesis followed by subsequent cell death (Fig. 1). Little is known about the diversity of phages that encode the tox gene but the phages that have been examined appear to be highly similar.
Cholera toxin is the main cause of secretory diarrhea, a characteristic of cholera.8 CT is an AB5 toxin, consisting of a single A subunit and five B subunits. It is encoded within the genome of a filamentous phage named CTXphi, a 7 kb single-stranded DNA phage found in serogroup O1 and O139 isolates of V. cholerae.9 The ctxAB genes are found directly adjacent to the phage attachment site, attR.9 The location of these genes at the terminal region of the phage genome and the isolation of functional CTXphi genomes lacking these genes suggests that they were a later addition to the phage genome.10 CTXphi can integrate into either of the host’s two chromosomes and only at a single site, named dif1 and dif2.11-13 In V. cholerae, CT production is regulated by the bacterial encoded regulator ToxRS and its production increases under conditions encountered in vivo such as low pH, temperature and bile salt concentration.14-16 CT is secreted from V. cholerae cells by the host Type II secretion system17 and subsequently attaches to its target intestinal epithelium cell receptor, the GM1 ganglioside.18 The B subunits are required for attachment and the toxin is taken up by endopinocytosis within a vacuole. The toxin is retrograde trafficked to the Golgi via early and late endosomes (Fig. 1). In the Golgi, the A subunit separates from the B subunits and enters the endoplasmic reticulum (ER) via coat protein I (COPI) vesicles.19 The active A1 subunit is released into the cytosol where it stimulates adenylate cyclase, an important signaling molecule that increases cyclic AMP. Increased cAMP levels activate protein kinase A (PKA) permanently, thus opening the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) chloride channel and ultimately causing chloride secretion from the cell (Fig. 1). CTXphi genomes are identical to each other in terms of gene arrangement, content and general sequence identity. The exception is the phage regulatory gene, rstR, which shows high sequence divergence.20 The bacterial host receptor for CTXphi attachment has been identified as a Type IV pilus, named the toxin co-regulated pilus (TCP).9
Shiga toxin (Stx) and the shiga-like toxin are also AB5 toxins consisting of one active A-subunit and five identical B-subunits, similar to CT. Stx and Stx-like toxins are present on diverse lambdoid phages that insert at multiple locations in the genome of Shigella species and select E. coli pathovars.21,22 Unlike the genes that encode DT and CT, the stx genes are not located adjacent to the phage attachment site but instead are present in the phage late gene region of the phage genome. The genome arrangement of the stx genes suggests that they were not picked up by imprecise prophage excision.23 Moreover, it has now been shown that Stx production is tightly linked to prophage induction and the phage regulatory circuit rather than by host regulatory systems.24,25 The Stx toxin is released from the bacterial cell upon phage lysis of the cell. The Stx B subunits bind to glycolipids on the host eukaryotic cell, specifically the cell surface receptor globotriosyl ceraminde (Gb3) (Fig. 1). Stx enters the host cell by a clathrin-dependent pathway and is retrograde trafficked directly from early/recycling endosomes to the Golgi and then to the ER via a COPI-independent pathway.26 The A1-subunit is released into the cytosol and inactivates the 60S ribosome subunit by N-glycosidic bond cleavage causing disruption of protein synthesis and cell death27 (Fig. 1). The bacterial receptor for Stx phage binding and uptake is the highly conserved YaeT protein required for insertion of proteins into the outer membrane of Gram-negative bacteria.28 Allison’s group has demonstrated that 70% of Stx phages recognize the YaeT surface molecule via conserved tail spike proteins associated with a short tailed morphology.28
Shiga Toxin Encoding E. coli Pathovars
Escherichia coli is a commensal of the gastrointestinal tract of warm-blooded animals, the most common facultative anaerobe in the human intestines. In addition, many E. coli strains are pathogenic and cause enteric diseases ranging from severe watery diarrhea, dysentery and hemorrhagic colitis to extraintestinal infections such as cystitis, septicemia and meningitis. Diversity in the number and type of toxins produced by E. coli, as well as diversity in the bacteriophages involved, has enabled E. coli to become a versatile pathogen. To date, eight different E. coli pathovars have been identified; Enterohemorrhagic E. coli (EHEC), enteropathogenic E. coli (EPEC), enteroinvasive E. coli (EIEC), entrotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC) and diffusely adherent E. coli (DAEC) and the extra-intestinal pathovars Neonatal Meningitis E. coli (NMEC) and Uropathogenic E. coli (UPEC).29 EHEC, EPEC, ETEC and EAEC are all extracellular pathogens that cause secretory diarrhea, whereas EIEC is an intracellular pathogen that causes inflammatory diarrhea. EHEC, EIEC and EAEC are associated with infection of the large intestine whereas EPEC, ETEC, DAEC and EAEC are associated with infection of the small intestine. UPEC is associated with urinary tract infection.29
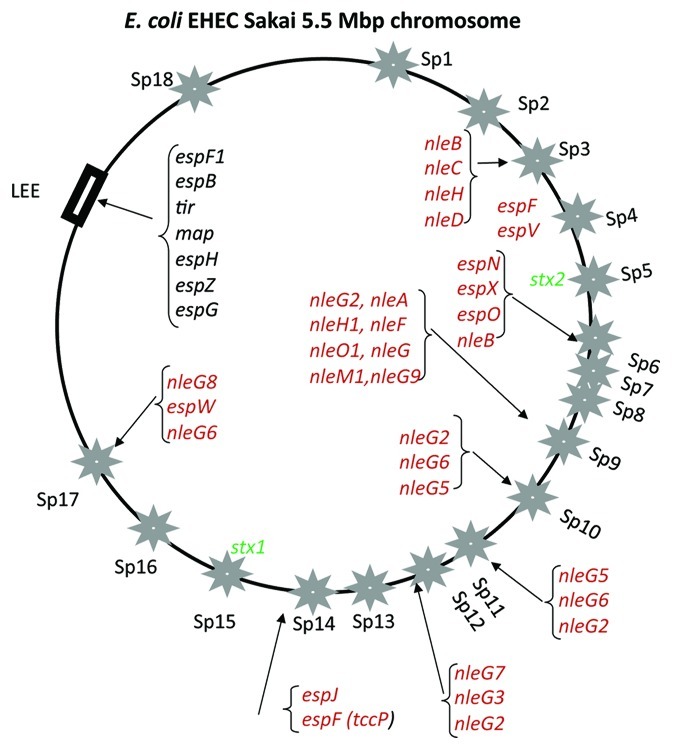
In enterohemorrhagic E. coli (EHEC) and Shiga toxin E. coli (STEC) strains responsible for hemorrhagic colitis and hemolytic uremic syndrome (HUS), the shiga-toxin (Stx) encoded by stx1 or stx2, and enterohemolysin encoded by the hly locus are present within lambdoid phage genomes.21,22,30-32 Since the first discovery of Stx in E. coli O157:H7 in 1983, which expresses the somatic (O) 157 and flagellar (H) 7 antigens, Stx has been identified in over 500 different E. coli serogroups.33 The lambdoid phage (denoted as Sp15 in Figure 2) containing the stx operon encoding stxA and stxB genes in E. coli O157:H7 Sakai and EDL strains, is integrated at the yehV gene whereas the Stx2 encoding bacteriophage (denoted as Sp5 in Figure 2) is integrated at the wrbA gene. The majority of E. coli O157:H7 strains encode Stx2 and over 75% also encode Stx1. Among these Stx1 negative strains, a truncated prophage is always present at the yehV locus suggesting that these strains all contained the Stx1 phage at one point that was subsequently lost in a subset of strains.34 In Stx1 negative strains, it was found that Stx2 was not at the yehV site, evidence that a diversity of phages are involved in Stx transfer.34 Similarly, it was shown that different STEC isolates have varying bacteriophage titers and toxin production.35 Structural analysis of bacteriophage-borne stx1, stx2 and flanking sequences in E. coli O157, STEC and Shigella dysenteriae Type 1 strains demonstrated bacteriophage genomic variation indicating that unrelated bacteriophages encode shiga toxin genes.31,32 In S. dysenteriae serotype 1 strains, Stx2 has only 56% identity to Stx1.
Figure 2. Prophage location on the E. coli O157:H7 strain Sakai genome map. Stars indicate prophages Sp1-Sp18 as named using the convention of.38 Rectangular black box indicates the position of the LEE pathogenicity island. Four letter designations indicate effector genes (red) and toxins (green) identified within prophages and at the LEE island (black).55
Some of the first studies to compare the whole genomic sequence of pathogenic E. coli strains examined O157 strains, a pathogen that emerged as a worldwide public health threat in the past three decades, and the laboratory-maintained commensal E. coli K12 strain.36-38 These ground breaking studies determined, for the first time, the source and cause of the genome size variation previously found among E. coli strains based on pulsed-field gel electrophoresis.39 These genomic studies were the first to show that E. coli strains contain a core-conserved sequence that is common to all genomes within the species as well as being interspersed with sequences unique to each strain. The presence of a core set of genes augmented with accessory genes has since been shown to be true for all free-living bacterial species. In one study, the E. coli O157 strain examined contained 1.3 million base pairs of strain-specific DNA, which included a bacteriophage containing Stx and a pathogenicity island carrying the locus of enterocyte effacement (LEE).36 Genomic comparison of two E. coli O157 strains, Sakai and EDL933, also revealed large differences in their prophage content.37,38 Strain EDL933 contained 12 prophage genomes whereas strain Sakai contained 18 prophage genomes (Fig. 2).37,38 In silico genomic analysis of the two E. coli O157 strains identified numerous virulence factors encoded by these prophages.37,38 The pathogenic potential of these two strains correlates with their bacteriophage-encoded virulence factors, demonstrating that acquisition of phages played a decisive role in the emergence of O157 as a foodborne pathogen. Whittam and colleagues demonstrated that the O157:H7 EHEC pathogen evolved in a stepwise process by the acquisition of prophages and conversion to a new serotype.40-44 The stepwise evolutionary model proposed that the highly virulent O157:H7 EHEC strains emerged from an O55:H7 EPEC ancestor by sequential acquisition of virulence factors predominantly encoded on prophages. EPEC is a major cause of infant diarrhea in the developing world and is mainly distinguished from EHEC by the absence of the stx genes. EPEC contains the LEE island and the EPEC adherence factor (EAF) plasmid. Using whole genome microarray, Whittam and colleagues examined a number of O157:H7 strains and demonstrated that 85% of the genes that were variably absent/present were phage derived, in total 463 such phage-related genes were found.41 Their analysis demonstrated the dominance of phages in the diversification of the E. coli O157:H7 genome.41
The 2011 Germany Outbreak E. coli O104:H4 Clone
A Stx2-producing E. coli O104:H4 clone emerged in the summer of 2011 in northern Germany and was responsible for one of the largest hemolytic uremic syndrome (HUS) outbreaks.45,46 The Germany O104:H4 clone caused nearly 3,000 cases of diarrhea without HUS and nearly 900 cases with HUS leading to 50 combined deaths.47 Unlike O157:H7, the O104:H4 serotype was not associated with animals prior to the outbreak and was rarely associated with humans in Germany. Using next generation sequencing technology, several groups sequenced multiple O104:H4 outbreak strains and determined that these strains carried genes present in enteroaggregative E. coli (EAEC) and EHEC pathovars.48-50 The Germany O104:H4 strains contained a plasmid carrying an aggregative adherence fimbria type (AAF/I) common to EAEC and a prophage carrying the Stx2 toxin inserted in chromosomal gene wbrA common to STEC. Phylogenetic analysis of 53 E. coli strains indicated that the O104:H4 strains from Germany formed a clade with O104:H4 strains from Africa, as well as with some EAEC strains.50 The O104:H4 strains differed from the EAEC strains due to the presence of the Stx2 prophage as well as the antibiotic resistance genes, TEM-1 and CTX-M-15.50 Similar to Stx containing prophage 933W from O157:H7, it was found that the stxAB genes in the O104:H4 prophage, were located between the antitermination Q gene and the S/R lysis genes of a lambdoid phage. In addition, there was a high degree of sequence identity between the O157:H7 and O104:H4 prophages suggesting recent horizontal gene transfer.47 Thus, emergence of this highly virulent strain is a direct consequence of phage acquisition.
Phage-Encoded Effector Proteins
Effector proteins or toxins are large proteins contained within a range of bacterial species that are translocated directly from the bacterial cytosol into the target eukaryotic cell cytosol using contact dependent bacterial secretion systems. Within their eukaryotic host cell, these effector proteins cause a variety of effects and in many cases encode overlapping functions. Effector proteins contain several distinct domains or modules that appear to have arisen through recombination reassortment events.51 Thus, the N-terminal region of one effector may show similar to the C-terminal region of another effector but lack any other homology. Most effector proteins also require a chaperone and thus have a specific chaperone-binding domain. Redundancy in functions encoded by these proteins is well documented.52 Effector proteins function by targeting a range of structures within the eukaryotic cell such as actin filaments, microtubules, mitochrondria, lysosomes, the nucleus, the inner cell membrane and tight junctions, as well as disrupting many different cell signaling pathways such as MAPK, cyclic AMP and GTPases Rho, Rac and Cdc42.52,53 Bacterial effector proteins function in many cases by molecular mimicry, hijacking eukaryotic cell function and signaling pathways for their own advantage. To date, effector proteins have only been identified on phage genomes in two species E. coli and S. enterica (Table 1).
E. coli Phage-Encoded Effector Proteins
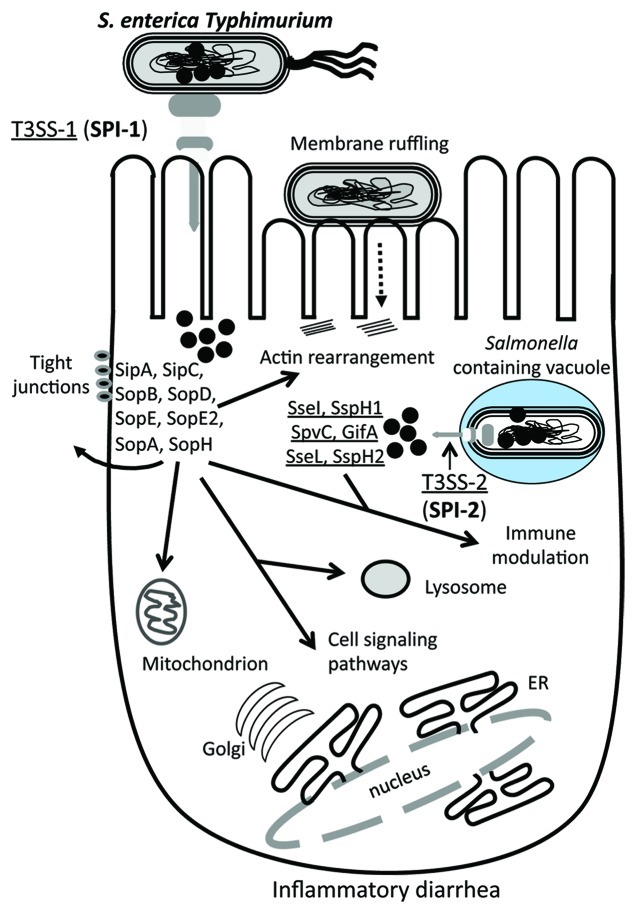
Nearly a hundred effector proteins have been identified among E. coli strains that are translocated via a common Type III secretion system (T3SS). Some of these effectors are encoded within prophages, some within pathogenicity islands and some elsewhere on the bacterial genome. As stated previously, EPEC and EHEC isolates are distinguished from each other mainly by the presence of the stx genes in EHEC strains. However, both EPEC and EHEC isolates along with ETEC isolates share the attaching and effacing (A/E) lesion phenotype. These isolates attach to the host cell surface resulting in cytoskeleton rearrangement and pedestal formation (Fig. 3). The factors involved in this host cell surface change are encoded within the LEE pathogenicity island. The canonical LEE island is a 35 kb region that encodes the Esc-Esp T3SS as well as effector proteins, their chaperones and regulators.54 T3SSs are contact dependent secretion systems found in a large number of Gram-negative species. These secretion systems are multi-component structures that span the bacterial cell wall and protrude from the cell surface that can inject into a eukaryotic host cell to translocate effector proteins directly into the eukaryotic cell cytoplasm.
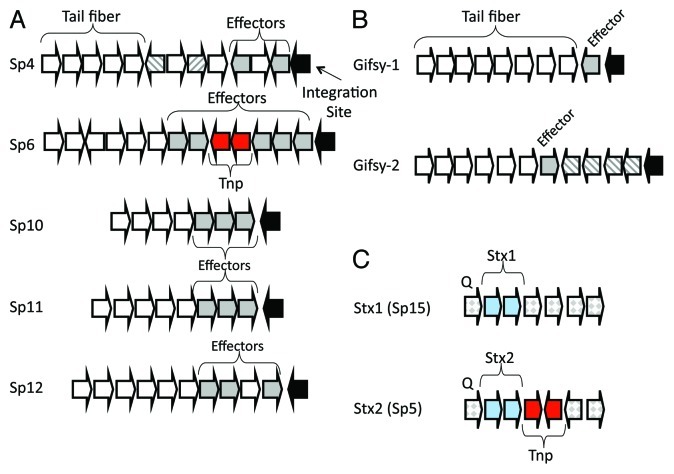
Figure 3. Location of effector genes and stx genes within prophages in E. coli and S. enterica. Partial prophage genomic maps are shown. (A) Terminal region of five lambdoid prophages from E. coli strain Sakai, (B) terminal region of S. enterica Gifsy-1 and Gifsy-2 prophages and (C) central region of the genome of Stx1 (Sp15) and Stx2 (Sp5) prophages encoding stx1AB and stx2AB genes, respectively, shown as blue arrows. Tail fiber genes are represented by white arrows and effector genes are shown as dark gray arrows. Putative additional virulence genes are shown as striped arrows. Black block arrows indicate the prophage site of genomic integration. Arrows in red represent transposase genes.
Of the effector proteins identified in E. coli, 60 have been identified within prophages in the EHEC Sakai strain, 39 of which were experimentally shown to be translocated into eukaryotic cells55 (Fig. 2). Of the 13 lambdoid prophages present within the genome, 9 of these prophages encode effector proteins55 (Fig. 2). The two shiga toxin-encoding phages Sp5 and Sp15 in strain Sakai do not encode any effector genes. Similar to many phage-encoded toxin genes, with the exception of stx genes, the majority of effector toxins in prophage genomes are found in the terminal region of the genome, after the tail fiber genes, and adjacent to the phage attachment site1,55 (Fig. 3A and C). The location of multiple effector genes at the end of phage genomes suggests acquisition by imprecise excision from a previous host genome (Fig. 3). Similar to the ctxAB genes that encode CT, the percent guanine-cytosine of the effector genes differs significant from the rest of the phage genome again suggesting that they are not true phage genes per se.55 The E. coli effectors identified were named following the esp (E. coli secreted protein) and nle (non-lee encoded effector) nomenclature (Fig. 3).
The effectors identified in the E. coli Sakai strain fell into greater than 20 protein families, the largest of which is NleG and has14 representatives in this strain.55 Savchenko and colleagues recently identified an ubiquitin E3 ligase function in the C-terminal region of NleG.56 Ubiquitination is an essential eukaryotic function that involves attachment of an ubiquitin polypeptide to a lysine residue of a target protein. The targeted protein is either destined for degradation by the host proteasome or its function is regulated. It has now been demonstrated that the NleG family function in vitro as E3 ubiquitin ligases by acting as a scaffold between E2 enzyme and the target protein in ubiquitination.56 The target protein(s) as well as the role in pathogenesis of NleG effectors have yet to be identified. The few previous studies on effectors with E3 ubiquitin ligase function suggest they are involved in suppression of the host immune response by targeting immune-related host proteins to proteasome degradation.57-59
Of the many different effector genes that have been identified within pathovars of E. coli only a handful have known host cell target sites and to date, most have unknown functions.52,60 Both Tir and the phage encoded EspF (aka TccP) effector contain mitochondrial targeted sequences (MTS) within their N-terminal regions52,60 and a recent study suggested that EspF targets and disrupts nucleolus function by a mitochondrial based mechanism (Fig. 4). EspF as well as Map have also been shown to disrupt tight junctions resulting in induction of apoptosis. The phage-encoded NleA and its homologs were shown to target the cell tight junctions (disrupting cellular barrier function) as well as mediate trafficking to the Golgi52,60 (Fig. 4).
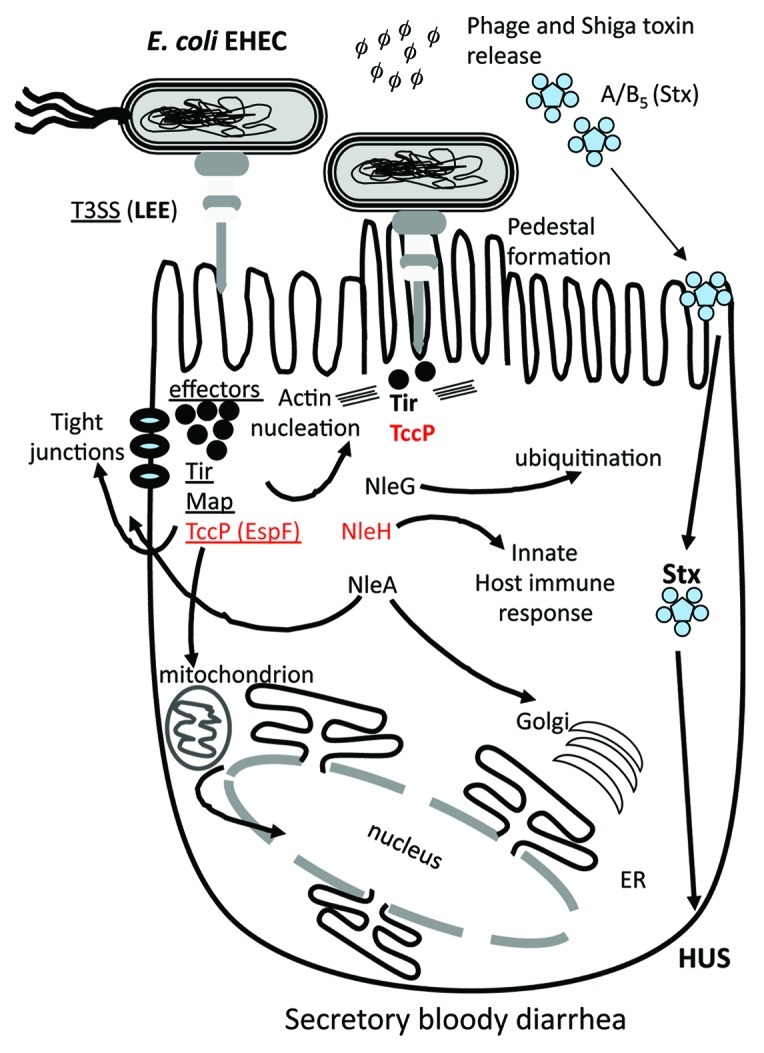
Figure 4. Schematic of the role of different effector proteins in EHEC colonization and pathogenesis. The LEE encoded T3SS is required for intimate binding and adherence of the bacterium to the eukaryotic cell surface which is mediated by the translocation of effector proteins Tir, Map and TccP (EspF). Tir and TccP are required for pedestal formation by directing actin rearrangement within the cell. The NleG family of effector proteins is the largest found in pathogenic E. coli and has been shown to be involved in protein ubiquitination. Many effector proteins are multi-functional such as TccP, which is also involved in cell tight junction disruption and interactions with the nucleus. Prophage-encoded effector genes are shown in red.
Salmonella enterica Phage-Encoded Effector Proteins
Salmonella enterica can cause acute infections, ranging from gastroenteritis (S. enterica serovar Typhimurium) to enteric fever (S. enterica serovar Typhi) depending on the serovar and strain examined.61Salmonella enterica is a facultative intracellular food-borne pathogen that causes morbidity and mortality worldwide as well as economic losses due to food recalls.62 This bacterium can colonize birds, reptiles and mammals and is subdivided into seven subspecies: I, II, IIIa, IIIb, IV, VI and VII.63-66 Infections in warm-blooded animals such as humans are generally caused by subspecies I isolates, which encompass more than 2,300 different serovars.64,66S. enterica serovar Typhimurium is one of the most common causes of food-borne infections causing inflammatory diarrhea.
The key virulence genes of S. enterica serovar Typhimurium are encoded on mobile and integrative genetic elements that were acquired by horizontal gene transfer (Fig. 5).67-71 Numerous prophages and pathogenicity islands have been identified and characterized from S. enterica Typhimurium genome sequences (Fig. 5). Five pathogenicity islands named Salmonella pathogenicity island 1 (SPI-1) to SPI-5 have been identified within the genome (Fig. 5).70,72-78 Two of these pathogenicity islands, SPI-1 and SPI-2, contain T3SSs and effector proteins and are present in all S. enterica strains. Eight different phages or phage-like region have also been characterized and shown to carry effector proteins and are variably present among strains (Fig. 5).69,70,79-81
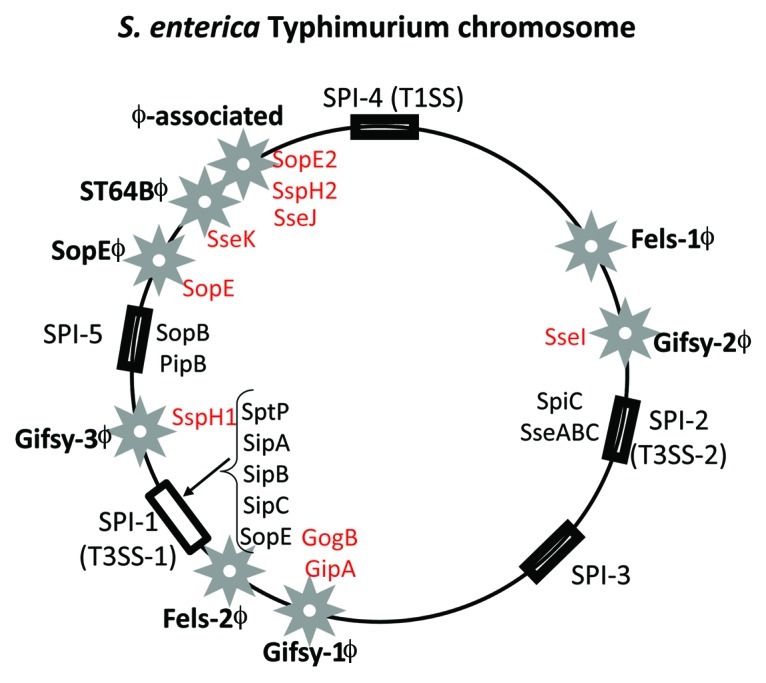
Figure 5. Prophage and pathogenicity island location on the S. enterica serovar Typhimurium genome map. Stars indicate prophages and rectangular black boxes indicate the position of Salmonella pathogenicity islands (SPIs). Four letter designations indicate effector genes identified within prophages (red) and SPIs (black).
Pathogenicity Islands (PAIs) are a group of integrative elements which encode multiple virulence factors that confer an advantage to the bacterium within its target eukaryotic host and were first described in an E. coli uropathogenic strain 536.82-85 PAIs integrate into host genome usually at tRNA sites by site-specific recombination mediated by the PAI-encoded integrase. In E. coli, PAI-encoded integrases have been shown to be unrelated to integrases present in phages indicating that phages and PAIs are diverse elements not closely related to one another.86,87 SPI-1 and SPI-2 encodes multiple virulence genes involved in invasion and enteropathogenesis including T3SSs and several effector proteins (Fig. 5).72,88,89 SPI-3 and SPI-4 are much smaller regions and appear to have a less significant role in pathogenesis. The SPI-5 region carries a number of effector proteins that are secreted by both T3SSs encoded on SPI-1 and SPI-2 (Fig. 6).78
Figure 6. Schematic of the role of different effector protein functions and targets in S. enterica. Salmonella enterica encodes two T3SSs, each within a Salmonella pathogenicity island, SPI-1 and SPI-2. Similar to EHEC effector proteins, a number of effectors are required for intimate binding to the host cell surface, which leads to the uptake into a Salmonella containing vacuole (SCV). Within the SCV, the T3SSs translocates a range of effector proteins into the cytosol that allow for intracellular survival. The S. enterica effector proteins target the tight junctions, mitochondria and lysosomes, as well as several cell signaling pathways. Prophage-encoded effectors are shown in red. See text for details.
Among S. enterica strains, approximately 30 different effector proteins have been identified, most of which are encoded within the same region that contains their cognate T3SS (SPI-1 and SPI-2).90 As was the case for E. coli, the S. enterica phage-encoded effector genes are found at the terminal region of the phage genome but generally only occur as a single gene within the phage genome (Fig. 3B).3 Serovar Typhimurium effectors secreted by T3SS-1 trigger membrane ruffling and internalization within a vacuole and are required for host cell invasion (Fig. 6). These effectors activate cytoskeleton rearrangement by causing actin nucleation, which disrupts the microvilli on the apical cell surface and aid in bacterial uptake, creating a Salmonella containing vacuole (SCV) within the host cell cytosol (Fig. 6). Salmonella then secretes effector proteins into the cytosol to modulate intracellular survival, first by preventing lysosome fusion with the SCV (Fig. 6).91 Both T3SSs are involved in translocating proteins from the SCV into the host cell cytosol, many of which are required for modulation of the host immune response.90,91 Among S. enterica strains examined nine effector proteins are found on prophage genomes, SseI within Gifsy-2ϕ, GogB and GipA within Gifsy-1 ϕ, a homology of SspH1 on Gifsy-3ϕ, SseK3 on phage ST64B, SopE within SpoEϕ, and SopE2, SspH2 and SseJ on a phage-like region.3 T3SS-1 translocated effectors SopE and SopE2 and T3SS-2 translocated effectors SseI and SspH1 are required for modulation of the host immune response.90 SopE and SopE2 are also involved in activating Rho GTPases by mimicking guanine exchange factors.90
Conclusion
Key to effector protein function is their delivery into their target cell. Bacteria encode three contact-dependent secretion systems named T3SS, T4SS and T6SS that translocate effector proteins directly from the bacterial cytosol into the eukaryotic cytosol. Interestingly the evolutionary origin of these three systems is very different as are the effector proteins translocated by each. The T3SS shows homology to the bacterial flagellar system, the T4SS shows homology to the bacterial conjugation system and the T6SS shows homology to a contractile phage tail-like structure. It was shown that this structure contains an exterior sheath wrapped around an interior tube, similar to that in phage T4. However, the structural similarities do not translate into significant primary sequence homology and it remains to be determined whether functional similarity exists.92 The T6SSs are a recent discovery and only a handful of effector proteins have been identified and one could speculate that some of their effector proteins may be phage encoded. Although effector proteins identified to date number nearly 150, the function(s) of the vast majority of these proteins remains unknown. In addition as more species and strains are sequenced the number of phage-encoded effector proteins is sure to increase and with it our knowledge of how phages encoding cargo genes interact with their host genomes and the host regulatory systems.
Acknowledgments
We thank members of our group for their enthusiasm and hard work: Laura Powell, Sai S. Kalburge, Luke Raber, Brandy Haines Menges, Jean Bernard Lubin, Serge Ongagna-Yhombi and W. Brian Whitaker. Work in the Boyd group is support by a National Science Foundation CAREER grant DEB-0844409. Literature citations have been limited in many cases to review articles due to space consideration and we apologize to those authors whose primary research is not included or cited.
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/21658
References
- 1.Boyd EF, Brüssow H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002;10:521–9. doi: 10.1016/S0966-842X(02)02459-9. [DOI] [PubMed] [Google Scholar]
- 2.Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehrbar K, Hardt WD. Bacteriophage-encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium. Infect Genet Evol. 2005;5:1–9. doi: 10.1016/j.meegid.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Boyd EF. Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions. Adv Virus Res. 2012;82:91–118. doi: 10.1016/B978-0-12-394621-8.00014-5. [In Press.] [DOI] [PubMed] [Google Scholar]
- 5.Holmes RK, Barksdale L. Genetic analysis of tox+ and tox- bacteriophages of Corynebacterium diphtheriae. J Virol. 1969;3:586–98. doi: 10.1128/jvi.3.6.586-598.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd J, Oza MN, Murphy JR. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci U S A. 1990;87:5968–72. doi: 10.1073/pnas.87.15.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao X, Boyd J, Murphy JR. Specific binding of the diphtheria tox regulatory element DtxR to the tox operator requires divalent heavy metal ions and a 9-base-pair interrupted palindromic sequence. Proc Natl Acad Sci U S A. 1992;89:5897–901. doi: 10.1073/pnas.89.13.5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De SN. Enterotoxicity of bacteria-free culture-filtrate of Vibrio cholerae. Nature. 1959;183:1533–4. doi: 10.1038/1831533a0. [DOI] [PubMed] [Google Scholar]
- 9.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–4. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 10.Boyd EF, Heilpern AJ, Waldor MK. Molecular analyses of a putative CTXphi precursor and evidence for independent acquisition of distinct CTX(ϕ)s by toxigenic Vibrio cholerae. J Bacteriol. 2000;182:5530–8. doi: 10.1128/JB.182.19.5530-5538.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Val ME, Bouvier M, Campos J, Sherratt D, Cornet F, Mazel D, et al. The single-stranded genome of phage CTX is the form used for integration into the genome of Vibrio cholerae. Mol Cell. 2005;19:559–66. doi: 10.1016/j.molcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Das B, Bischerour J, Val ME, Barre FX. Molecular keys of the tropism of integration of the cholera toxin phage. Proc Natl Acad Sci U S A. 2010;107:4377–82. doi: 10.1073/pnas.0910212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd EF. Efficiency and specificity of CTXphi chromosomal integration: dif makes all the difference. Proc Natl Acad Sci U S A. 2010;107:3951–2. doi: 10.1073/pnas.1000310107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mekalanos JJ. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–63. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 15.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A. 1987;84:2833–7. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–92. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis BM, Lawson EH, Sandkvist M, Ali A, Sozhamannan S, Waldor MK. Convergence of the secretory pathways for cholera toxin and the filamentous phage, CTXphi. Science. 2000;288:333–5. doi: 10.1126/science.288.5464.333. [DOI] [PubMed] [Google Scholar]
- 18.Holmgren J. Actions of cholera toxin and the prevention and treatment of cholera. Nature. 1981;292:413–7. doi: 10.1038/292413a0. [DOI] [PubMed] [Google Scholar]
- 19.Majoul I, Sohn K, Wieland FT, Pepperkok R, Pizza M, Hillemann J, et al. KDEL receptor (Erd2p)-mediated retrograde transport of the cholera toxin A subunit from the Golgi involves COPI, p23, and the COOH terminus of Erd2p. J Cell Biol. 1998;143:601–12. doi: 10.1083/jcb.143.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis BM, Kimsey HH, Chang W, Waldor MK. The Vibrio cholerae O139 Calcutta bacteriophage CTXphi is infectious and encodes a novel repressor. J Bacteriol. 1999;181:6779–87. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Brien AD, Marques LR, Kerry CF, Newland JW, Holmes RK. Shiga-like toxin converting phage of enterohemorrhagic Escherichia coli strain 933. Microb Pathog. 1989;6:381–90. doi: 10.1016/0882-4010(89)90080-6. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien AD, Newland JW, Miller SF, Holmes RK, Smith HW, Formal SB. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226:694–6. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 23.Huang A, Friesen J, Brunton JL. Characterization of a bacteriophage that carries the genes for production of Shiga-like toxin 1 in Escherichia coli. J Bacteriol. 1987;169:4308–12. doi: 10.1128/jb.169.9.4308-4312.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waldor MK, Friedman DI. Phage regulatory circuits and virulence gene expression. Curr Opin Microbiol. 2005;8:459–65. doi: 10.1016/j.mib.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Wagner PL, Waldor MK. Bacteriophage control of bacterial virulence. Infect Immun. 2002;70:3985–93. doi: 10.1128/IAI.70.8.3985-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wälchli S, Skånland SS, Gregers TF, Lauvrak SU, Torgersen ML, Ying M, et al. The Mitogen-activated protein kinase p38 links Shiga Toxin-dependent signaling and trafficking. Mol Biol Cell. 2008;19:95–104. doi: 10.1091/mbc.E07-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxena SK, O’Brien AD, Ackerman EJ. Shiga toxin, Shiga-like toxin II variant, and ricin are all single-site RNA N-glycosidases of 28 S RNA when microinjected into Xenopus oocytes. J Biol Chem. 1989;264:596–601. [PubMed] [Google Scholar]
- 28.Smith DL, James CE, Sergeant MJ, Yaxian Y, Saunders JR, McCarthy AJ, et al. Short-tailed stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J Bacteriol. 2007;189:7223–33. doi: 10.1128/JB.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 30.O’Brien AD, Lively TA, Chang TW, Gorbach SL. Purification of Shigella dysenteriae 1 (Shiga)-like toxin from Escherichia coli O157:H7 strain associated with haemorrhagic colitis. Lancet. 1983;2:573. doi: 10.1016/S0140-6736(83)90601-3. [DOI] [PubMed] [Google Scholar]
- 31.Recktenwald J, Schmidt H. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage phiP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect Immun. 2002;70:1896–908. doi: 10.1128/IAI.70.4.1896-1908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unkmeir A, Schmidt H. Structural analysis of phage-borne stx genes and their flanking sequences in shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun. 2000;68:4856–64. doi: 10.1128/IAI.68.9.4856-4864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonald JE, Smith DL, Fogg PC, McCarthy AJ, Allison HE. High-throughput method for rapid induction of prophages from lysogens and its application in the study of Shiga Toxin-encoding Escherichia coli strains. Appl Environ Microbiol. 2010;76:2360–5. doi: 10.1128/AEM.02923-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaikh N, Tarr PI. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J Bacteriol. 2003;185:3596–605. doi: 10.1128/JB.185.12.3596-3605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner PL, Acheson DW, Waldor MK. Isogenic lysogens of diverse shiga toxin 2-encoding bacteriophages produce markedly different amounts of shiga toxin. Infect Immun. 1999;67:6710–4. doi: 10.1128/iai.67.12.6710-6714.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–33. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 37.Ohnishi M, Kurokawa K, Hayashi T. Diversification of Escherichia coli genomes: are bacteriophages the major contributors? Trends Microbiol. 2001;9:481–5. doi: 10.1016/S0966-842X(01)02173-4. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 39.Bergthorsson U, Ochman H. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J Bacteriol. 1995;177:5784–9. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng PC, Monday SR, Lacher DW, Allison L, Siitonen A, Keys C, et al. Genetic diversity among clonal lineages within Escherichia coli O157:H7 stepwise evolutionary model. Emerg Infect Dis. 2007;13:1701–6. doi: 10.3201/eid1311.070381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wick LM, Qi W, Lacher DW, Whittam TS. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J Bacteriol. 2005;187:1783–91. doi: 10.1128/JB.187.5.1783-1791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytia-Trees EK, et al. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 2006;16:757–67. doi: 10.1101/gr.4759706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng P, Lampel KA, Karch H, Whittam TS. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–3. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 44.Whittam TS, Reid SD, Selander RK. Mutators and long-term molecular evolution of pathogenic Escherichia coli O157:H7. Emerg Infect Dis. 1998;4:615–7. doi: 10.3201/eid0404.980411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, et al. HUS Investigation Team Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365:1771–80. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 46.Denamur E. The 2011 Shiga toxin-producing Escherichia coli O104:H4 German outbreak: a lesson in genomic plasticity. Clin Microbiol Infect. 2011;17:1124–5. doi: 10.1111/j.1469-0691.2011.03620.x. [DOI] [PubMed] [Google Scholar]
- 47.Muniesa M, Hammerl JA, Hertwig S, Appel B, Brüssow H. Shiga toxin-producing Escherichia coli O104:H4: a new challenge for microbiology. Appl Environ Microbiol. 2012;78:4065–73. doi: 10.1128/AEM.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One. 2011;6:e22751. doi: 10.1371/journal.pone.0022751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho CC, Yuen KY, Lau SK, Woo PC. Rapid identification and validation of specific molecular targets for detection of Escherichia coli O104:H4 outbreak strain by use of high-throughput sequencing data from nine genomes. J Clin Microbiol. 2011;49:3714–6. doi: 10.1128/JCM.05062-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–17. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stavrinides J, Ma W, Guttman DS. Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathog. 2006;2:e104. doi: 10.1371/journal.ppat.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dean P, Kenny B. The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr Opin Microbiol. 2009;12:101–9. doi: 10.1016/j.mib.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marlovits TC, Stebbins CE. Type III secretion systems shape up as they ship out. Curr Opin Microbiol. 2010;13:47–52. doi: 10.1016/j.mib.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–8. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, Fivian A, et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A. 2006;103:14941–6. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu B, Skarina T, Yee A, Jobin MC, Dileo R, Semesi A, et al. NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases. PLoS Pathog. 2010;6:e1000960. doi: 10.1371/journal.ppat.1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singer AU, Rohde JR, Lam R, Skarina T, Kagan O, Dileo R, et al. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol. 2008;15:1293–301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Higashide WM, McCormick BA, Chen J, Zhou D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol. 2006;62:786–93. doi: 10.1111/j.1365-2958.2006.05407.x. [DOI] [PubMed] [Google Scholar]
- 59.Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev. 2011;35:1100–25. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- 61.Tsolis RM, Young GM, Solnick JV, Bäumler AJ. From bench to bedside: stealth of enteroinvasive pathogens. Nat Rev Microbiol. 2008;6:883–92. doi: 10.1038/nrmicro2012. [DOI] [PubMed] [Google Scholar]
- 62.Miller MA, Sentz J, Rabaa MA, Mintz ED. Global epidemiology of infections due to Shigella, Salmonella serotype Typhi, and enterotoxigenic Escherichia coli. Epidemiol Infect. 2008;136:433–5. doi: 10.1017/S095026880800040X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reeves MW, Evins GM, Heiba AA, Plikaytis BD, Farmer JJ., 3rd Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989;27:313–20. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Le Minor L, Popoff MY, Bockemühl J. Supplement 1989 (n. 33) to the Kauffmann-White scheme. Res Microbiol. 1990;141:1173–7. doi: 10.1016/0923-2508(90)90090-D. [DOI] [PubMed] [Google Scholar]
- 65.Boyd EF, Wang FS, Whittam TS, Selander RK. Molecular genetic relationships of the salmonellae. Appl Environ Microbiol. 1996;62:804–8. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Popoff MY, Bockemühl J, Gheesling LL. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res Microbiol. 2004;155:568–70. doi: 10.1016/j.resmic.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 67.Groisman EA, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–9. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 68.Ochman H, Bergthorsson U. Genome evolution in enteric bacteria. Curr Opin Genet Dev. 1995;5:734–8. doi: 10.1016/0959-437X(95)80005-P. [DOI] [PubMed] [Google Scholar]
- 69.Porwollik S, Boyd EF, Choy C, Cheng P, Florea L, Proctor E, et al. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J Bacteriol. 2004;186:5883–98. doi: 10.1128/JB.186.17.5883-5898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Porwollik S, Wong RM, McClelland M. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc Natl Acad Sci U S A. 2002;99:8956–61. doi: 10.1073/pnas.122153699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lan R, Reeves PR, Octavia S. Population structure, origins and evolution of major Salmonella enterica clones. Infect Genet Evol. 2009;9:996–1005. doi: 10.1016/j.meegid.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 72.Mills DM, Bajaj V, Lee CA. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–59. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 73.Hensel M, Shea JE, Gleeson C, Jones MD, Dalton E, Holden DW. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–3. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 74.Blanc-Potard AB, Solomon F, Kayser J, Groisman EA. The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol. 1999;181:998–1004. doi: 10.1128/jb.181.3.998-1004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blanc-Potard AB, Groisman EA. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–85. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blanc-Potard AB, Solomon F, Kayser J, Groisman EA. The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol. 1999;181:998–1004. doi: 10.1128/jb.181.3.998-1004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wong KK, McClelland M, Stillwell LC, Sisk EC, Thurston SJ, Saffer JD. Identification and sequence analysis of a 27-kilobase chromosomal fragment containing a Salmonella pathogenicity island located at 92 minutes on the chromosome map of Salmonella enterica serovar typhimurium LT2. Infect Immun. 1998;66:3365–71. doi: 10.1128/iai.66.7.3365-3371.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wood MW, Jones MA, Watson PR, Hedges S, Wallis TS, Galyov EE. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol Microbiol. 1998;29:883–91. doi: 10.1046/j.1365-2958.1998.00984.x. [DOI] [PubMed] [Google Scholar]
- 79.Fang FC, DeGroote MA, Foster JW, Bäumler AJ, Ochsner U, Testerman T, et al. Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc Natl Acad Sci U S A. 1999;96:7502–7. doi: 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Figueroa-Bossi N, Bossi L. Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999;33:167–76. doi: 10.1046/j.1365-2958.1999.01461.x. [DOI] [PubMed] [Google Scholar]
- 81.Figueroa-Bossi N, Uzzau S, Maloriol D, Bossi L. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol Microbiol. 2001;39:260–71. doi: 10.1046/j.1365-2958.2001.02234.x. [DOI] [PubMed] [Google Scholar]
- 82.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschäpe H, et al. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–14. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–97. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 84.Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2:376–81. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–24. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 86.Boyd EF, Almagro-Moreno S, Parent MA. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 2009;17:47–53. doi: 10.1016/j.tim.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 87.Napolitano MG, Almagro-Moreno S, Boyd EF. Dichotomy in the evolution of pathogenicity island and bacteriophage encoded integrases from pathogenic Escherichia coli strains. Infect Genet Evol. 2011;11:423–36. doi: 10.1016/j.meegid.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 88.Galán JE. Molecular and cellular bases of Salmonella entry into host cells. Curr Top Microbiol Immunol. 1996;209:43–60. doi: 10.1007/978-3-642-85216-9_3. [DOI] [PubMed] [Google Scholar]
- 89.Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1996;93:2593–7. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol. 2009;12:117–24. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ibarra JA, Steele-Mortimer O. Salmonella--the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell Microbiol. 2009;11:1579–86. doi: 10.1111/j.1462-5822.2009.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and Regulation of the Type VI Secretion System. Annu Rev Microbiol. 2012 doi: 10.1146/annurev-micro-121809-151619. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]