Abstract
Background
Recent molecular studies have revealed a highly complex bacterial assembly in the canine intestinal tract. There is mounting evidence that microbes play an important role in the pathogenesis of acute and chronic enteropathies of dogs, including idiopathic inflammatory bowel disease (IBD). The aim of this study was to characterize the bacterial microbiota in dogs with various gastrointestinal disorders.
Methodology/Principal Findings
Fecal samples from healthy dogs (n = 32), dogs with acute non-hemorrhagic diarrhea (NHD; n = 12), dogs with acute hemorrhagic diarrhea (AHD; n = 13), and dogs with active (n = 9) and therapeutically controlled idiopathic IBD (n = 10) were analyzed by 454-pyrosequencing of the 16S rRNA gene and qPCR assays. Dogs with acute diarrhea, especially those with AHD, had the most profound alterations in their microbiome, as significant separations were observed on PCoA plots of unweighted Unifrac distances. Dogs with AHD had significant decreases in Blautia, Ruminococcaceae including Faecalibacterium, and Turicibacter spp., and significant increases in genus Sutterella and Clostridium perfringens when compared to healthy dogs. No significant separation on PCoA plots was observed for the dogs with IBD. Faecalibacterium spp. and Fusobacteria were, however, decreased in the dogs with clinically active IBD, but increased during time periods of clinically insignificant IBD, as defined by a clinical IBD activity index (CIBDAI).
Conclusions
Results of this study revealed a bacterial dysbiosis in fecal samples of dogs with various GI disorders. The observed changes in the microbiome differed between acute and chronic disease states. The bacterial groups that were commonly decreased during diarrhea are considered to be important short-chain fatty acid producers and may be important for canine intestinal health. Future studies should correlate these observed phylogenetic differences with functional changes in the intestinal microbiome of dogs with defined disease phenotypes.
Introduction
Recent molecular-phylogenetic studies have revealed a complex assembly of bacteria in the mammalian gastrointestinal (GI) tract [1]–[3]. Intestinal microbes play a crucial role in the maintenance of host health. They act as a defending barrier against transient pathogens, support the host in digestion and energy harvest from the diet, stimulate the immune system, and provide nutritional support for enterocytes [4].
The intestinal microbiota has also been implicated in the pathogenesis of various canine GI disorders, either associated with the presence of specific pathogens (e.g., enterotoxigenic C. perfringens, Salmonella, viruses, and parasites) in acute episodes of diarrhea [5], [6], or a non-specific dysbiosis such as that described in dogs with idiopathic inflammatory bowel disease [7]–[10]. Canine idiopathic IBD is one of the most common causes of chronic GI disease in dogs and encompasses a group of chronic enteropathies of unknown cause, which are characterized by infiltration of the intestinal mucosa with inflammatory cells [11]. Although histopathologic changes may be found in any segment of the GI tract, the small intestine is typically the most frequently affected segment. The diagnosis of idiopathic IBD is made after known causes of GI inflammation have been ruled out, the animal has not shown a favorable response to a dietary and antibiotic therapeutic trial, and typically requires immunosuppressive or anti-inflammatory therapy [11].
Molecular-phylogenetic studies have revealed a bacterial and/or fungal dysbiosis in the duodenum of dogs with idiopathic IBD. Most commonly, a decrease in the proportions of Clostridiales and an increase in Proteobacteria is observed [7], [9], [10], [12]. Only few molecular studies have described the fecal microbiota of dogs with acute and chronic GI disorders. One study, using fluorescent in situ hybridization (FISH) probes, found Bacteroides counts to be significantly increased in Beagle dogs with chronic diarrhea [13]. In contrast, using 454-pyrosequencing of the cpn60 gene, significantly decreased proportions of Bacteroidetes were observed in dogs with unspecified diarrhea [14]. Using terminal restriction fragment length polymorphism (T-RFLP) analysis and quantitative PCR (qPCR), an increased abundance of Clostridium perfringens, Enterococcus faecalis, and E. faecium was observed in dogs during diarrheic episodes [15]. While these studies suggest a dysbiosis present in fecal samples of dogs with diarrhea, additional studies using high-throughput sequencing technologies in dogs with well-defined acute and chronic disease phenotypes are needed to further characterize changes in the fecal microbiome. In addition, comparison of fecal findings in dogs with IBD with those previously observed in duodenal biopsies is of interest [7], as collection of fecal samples is more practical. Furthermore, it is unclear if the pattern of dysbiosis observed in dogs with IBD is specific for this disorder, or if similar patterns are present in acute GI diseases.
This study compared the fecal microbiome of healthy dogs, dogs with acute non-hemorrhagic diarrhea (NHD), dogs with acute hemorrhagic diarrhea (AHD), and dogs with active and therapeutically controlled clinically insignificant IBD. The results indicate differences in the fecal microbiome among the dogs with various GI diseases. Dogs with acute diarrhea had the most pronounced changes, with several bacterial groups altered when compared to healthy dogs. Only Faecalibacterium spp. and Fusobacteria were decreased in dogs with clinically active IBD, but increased during time periods of clinically insignificant IBD.
Materials and Methods
Ethics Statement
The collection and analysis of fecal samples was approved by the institutional Clinical Research Review Committee of the College of Veterinary Medicine, Texas A&M University (CRRC#09-06).
Animals and Sample Collection
Fecal samples from a total of 76 dogs were analyzed. These dogs were either healthy (n = 32), or had signs of either acute non-hemorrhagic diarrhea (NHD; n = 12), acute hemorrhagic diarrhea (AHD, n = 13), active inflammatory bowel disease (A-IBD; n = 9), or therapeutically controlled clinically insignificant IBD (S-IBD; n = 10), as scored by a published canine clinical IBD activity index (CIBDAI) [16]. Left-over naturally-passed feces collected for routine fecal examination were frozen within a few hours of collection at either −20°C or −80°C, and were stored frozen until processing of samples for DNA extraction. The summary of the baseline characteristics for each animal group is listed in Table 1, and detailed descriptions of each enrolled dog are listed in supplementary Tables S1–S3.
Table 1. Summary of basic characteristics and alpha diversity measures.
| Healthy | NHD | AHD | A_IBD | S_IBD | p-value | |
| Age (years; median, range) | 4.6, 0.3–15.0 | 5.3, 0.5–15.0 | 5.0, 2.0–16.0 | 3.5, 0.6–7.6 | 5.7, 3.7–8.7 | 0.496 |
| Weight (lbs; median, range) | 47.0, 5.8–81.5 | 47.1, 5.5–75.0 | 19.8, 4.0–68.3 | 55.0, 9.0–130.0 | 56.1, 18.5–91.7 | 0.574 |
| CIBDAI (median, range) | N/A | N/A | N/A | 7, 5–9 | 1.5, 1–2 | <0.001 |
| gender (female/male) | 14/18 | 8/4 | 7/6 | 3/5 | 3/7 | 0.449 |
| Country | Sweden n = 8; USA n = 24 | USA | Germany | Sweden | Sweden | n/a |
| OTU97 (mean ± SD) | 242±93 | 188±95 | 175±57 | 163±91 | 119±85 | 0.111 |
| Shannon index (mean ± SD) | 3.3±0.8 | 2.4±1.4 | 2.6±0.9 | 2.3±1.1 | 1.9±1.0 | 0.104 |
| Chao1 (mean ± SD) | 504±181 | 390±232 | 327±126 | 357±212 | 251±203 | 0.053 |
IBD = inflammatory bowel disease.
CIBDAI = canine IBD disease activity index.
NHD = acute non-hemorrhagic diarrhea, AHD = acute hemorrhagic diarrhea, A_IBD = active IBD, S_IBD = clinically insignificant IBD.
Healthy controls
Fecal samples from a total of 32 pet dogs were analyzed by 454-pyrosequencing and quantitative PCR assays (qPCR). All dogs were privately owned and lived in diverse home environments, were on a variety of commercial diets, and none of the dogs had a history of gastrointestinal signs or administration of antibiotics for at least the past 3 months (Table S1). Eight healthy dogs lived in Sweden, while the remaining 24 healthy dogs lived in Texas, USA.
Dogs with acute non-hemorrhagic diarrhea (NHD)
Fecal samples from a total of 12 pet dogs that presented to a first-opinion practice (Austin, TX) with acute, uncomplicated, non-hemorrhagic diarrhea were evaluated (duration of diarrhea <3 days). Of those, 7 samples were analyzed by 454-pyrosequencing, while all 12 samples were analyzed by qPCR assays. None of the dogs had a previous history of GI signs or had received antibiotics within the previous 3 months (Table S2). Diagnostic evaluation included complete blood count (CBC), serum chemistry profiles, and partial fecal analysis for enteric pathogens by fecal flotation and fecal cytology. Clostridium perfringens enterotoxin and C. difficile toxin A/B were analyzed using commercially available ELISA kits (C. perfringens Enterotoxin Test™ and C. difficile Tox A/B II™, TechLab, Blacksburg, VA). Based on review of the medical records, all dogs in this group recovered after non-specific symptomatic therapy (e.g., fluid supplementation, gastric acid blockers) within a few days.
Dogs with acute hemorrhagic diarrhea (AHD)
Fecal samples were analyzed from a total of 13 pet dogs that presented to the Clinic of Small Animal Medicine, LMU University of Munich, Germany, with acute hemorrhagic diarrhea (duration of <3 days). None of the dogs had a previous history of GI signs or had received antibiotics within the previous 3 months (Table S2). Diagnostic evaluation included CBC, serum chemistry profiles, and partial fecal analysis for enteric pathogens (C. perfringens enterotoxin ELISA, C. difficile toxin A/B ELISA, and fecal culture).
Dogs with idiopathic IBD
Fecal samples were analyzed from pet dogs that had been presented to the Helsingborg Referral Animal Hospital, Helsingborg, Sweden with signs of chronic GI disease. Dogs underwent clinical evaluation by a veterinary internist (LT). Diagnostic tests that were performed included a CBC, serum chemistry profiles, fecal flotation, serum concentrations of cobalamin and folate, and depending on the clinical signs, serum concentrations of trypsin-like immunoreactivity (cTLI) and pancreatic lipase-immunoreactivity (cPLI). During the months of diagnostic work-ups, dogs underwent various forms of antibiotic and/or dietary trials. All dogs failed the trials and subsequently underwent endoscopy with collection of intestinal biopsies. All dogs then received and responded to immunosuppressive therapy, leading to a diagnosis of idiopathic IBD (Table S3).
The disease activity of these dogs was scored using the published clinical canine IBD activity index (CIBDAI) [16]. The CIBDAI is based on 6 criteria, each scored on a scale from 0–3: attitude/activity, appetite, vomiting, stool consistency, stool frequency, and weight loss. The total composite score is determined to be clinically insignificant (score 0–3), mild (score 4–5), moderate (score 6–8), or severe (score 9 or greater). We analyzed a total of 19 dogs (Table S3). Of those 19 dogs, 9 were newly diagnosed with active IBD (A_IBD) as judged by their CIBDAI score (median 7, range 5–9), and fecal samples collected at the time of diagnosis were analyzed (5 samples were analyzed by pyrosequencing; all 9 samples were analyzed by qPCR). The other 10 dogs had been on medical treatment (Table S3) for their idiopathic IBD for several months to years (therapeutically controlled stable IBD; S_IBD) and had clinically insignificant or no signs of IBD as scored by the CIBDAI (median 1.5, range 1–2) at the time of sample collection. From the latter group of dogs all 10 samples were analyzed by pyrosequencing and qPCR assays. None of these 19 dogs received antibiotics for at least 2 months before sample collection.
In addition, paired samples were obtained from 8 dogs, representing time points when, based on CIBDAI scoring, the dogs showed either a clinically significant CIBDAI (median 5, range 4–9) or a clinically insignificant CIBDAI (median 1.5, range 1–3). The time period between the collections of repeated samples ranged from 2–8 months (median 5.5 months). These paired samples were analyzed separately by qPCR assays, as these follow up samples were obtained after the pyrosequencing analysis had been completed.
DNA Extraction
An aliquot of 100 mg (wet weight) of each fecal sample was extracted by a bead-beating method using a commercial DNA extraction kit (ZR Fecal DNA Kit™, Zymo Research Corporation) following the manufacturer’s instructions. The bead beating step was performed on a homogenizer (FastPrep-24, MP Biomedicals) for 60 s at a speed of 4 m/s.
454-pyrosequencing
Bacterial tag-encoded FLX-titanium amplicon pyrosequencing (bTEFAP) based on the V1–V3 region (E. coli position 27–519) of the 16 S rRNA gene was performed on 67 of the 77 samples as described previously with primers forward 28F: GAGTTTGATCNTGGCTCAG and reverse 519R: GTNTTACNGCGGCKGCTG [3], [17]. Raw sequence data were screened, trimmed, denoised, and filtered with the QIIME pipeline version 1.4.0 (http://qiime.sourceforge.net) [18] with the following settings: minimum read length of 300 bp; no ambiguous base calls; no homopolymeric runs longer than 8 bp; average quality value>q25 within a sliding window of 50 bp. Chimeras were excluded using the software B2C2 (http://www.researchandtesting.com/B2C2.html) [7], [19]. Operational taxonomic units (OTUs) were defined as sequences with at least 97% similarity using QIIME. For classification of sequences on a genus level the naïve Bayesian classifier within the Ribosomal Database Project (RDP, v10.28) was used. The confidence threshold in RDP was set to 80% [7].
Quantitative PCR (qPCR)
For validation of pyrosequencing results and/or to evaluate bacterial groups that are typically present at very low abundance or typically not detected in sequence data based on the authors’ experience from previous studies (i.e., Bifidobacterium spp.; Faecalibacterium spp.) [3], [20], [21], qPCR assays for selected bacterial groups were performed: total bacteria, Bacteroidetes, Fusobacteria, Blautia, Ruminococcaceae, Faecalibacterium spp., Turicibacter spp., Bifidobacterium spp., and Clostridium perfringens. PCR was also used to detect the genes encoding C. perfringens enterotoxin (cpe) and C. difficile toxin B (tcd B). Real-time PCR conditions were performed as described previously [22]. The oligonucleotide sequences of primers and probes, and respective annealing temperatures are summarized in Table 2. The qPCR data was expressed as log amount of DNA (fg) for each particular bacterial group per 10 ng of isolated total DNA.
Table 2. Oligonucleotides primers/probes used in this study.
| qPCR primers/probe | Sequence (5′-3′) | Target | Annealing (°C) | Reference |
| CFB555f | CCGGAWTYATTGGGTTTAAAGGG | Bacteroidetes | 60 | [38] |
| CFB968r | GGTAAGGTTCCTCGCGTA | |||
| BifF | TCGCGTCYGGTGTGAAAG | Bifidobacterium | 60 | [39] |
| BifR | CCACATCCAGCRTCCAC | |||
| FaecaF | GAAGGCGGCCTACTGGGCAC | Faecalibacterium | 60 | [22] |
| FaecaR | GTGCAGGCGAGTTGCAGCCT | |||
| RumiF | ACTGAGAGGTTGAACGGCCA | Family Ruminococcaceae | 59 | [22] |
| RumiR | CCTTTACACCCAGTAAWTCCGGA | |||
| CPerf165F | CGCATAACGTTGAAAGATGG | |||
| CPerf269R | CCTTGGTAGGCCGTTACCC | C. perfringens 16S | 58 | [40] |
| CPerf187F (probe) | TCATCATTCAACCAAAGGAGCAATCC | |||
| TM-cpe-F | AACTATAGGAGAACAAAATACAATAG | |||
| TM-cpe-R | TGCATAAACCTTATAATATACATATTC | C. perfringens enterotoxin | 55 | [41] |
| TM-cpe-pr | TCTGTATCTACAACTGCTGGTCCA | |||
| tcdB-F | GGTATTACCTAATGCTCCAAATAG | |||
| tcdB-R | TTTGTGCCATCATTTTCTAAGC | C. difficile toxin B gene | 58 | [42] |
| tcdB-P (probe) | ACCTGGTGTCCATCCTGTTTCCCA | |||
| Fuso-F | KGGGCTCAACMCMGTATTGCGT | Fusobacterium | 51 | This study |
| Fuso-R | TCGCGTTAGCTTGGGCGCTG | |||
| Blaut-F | TCTGATGTGAAAGGCTGGGGCTTA | Blautia spp. | 56 | This study |
| Blaut-R | GGCTTAGCCACCCGACACCTA | |||
| 341-F | CCTACGGGAGGCAGCAGT | Universal Bacteria | 59 | [43] |
| 518-R | ATTACCGCGGCTGCTGG | |||
| TuriciF | CAGACGGGGACAACGATTGGA | Turicibacter | 63 | This study |
| TuricR | TACGCATCGTCGCCTTGGTA |
Statistical Analysis
To account for unequal sequencing depth across samples, and to avoid exclusion of samples with lower number of sequence reads, the subsequent analysis was performed on a randomly selected subset of 2,000 sequences per sample. Differences in microbial communities between disease groups were investigated using the phylogeny-based unweighted UniFrac distance metric, and PCoA plots and rarefaction curves were plotted using QIIME. To determine differences in microbiota composition between the animal groups, the analysis of similarities (ANOSIM) function in the statistical software package PRIMER 6 (PRIMER-E Ltd., Lutton, UK) was used on the unweighted UniFrac distance matrixes. To visualize the relative abundance of bacterial families for individual fecal samples, heat maps were generated in NCSS 2007 (NCSS, Kaysville, Utah).
For all datasets, assumption of normality was tested using the D’Agostino and Pearson normality test (Prism 5.00, GraphPad Software Inc.). Only bacterial taxa that were present in at least 50% of dogs (either healthy or diseased) were included in the analysis. Because most datasets did not meet the assumptions of normal distribution, the differences in the proportions of bacterial taxa (defined as percentage of total sequences) or qPCR results between healthy and disease groups were determined using non-parametric Kruskal-Wallis tests (Prism v5.00, GraphPad Software Inc.). The resulting p-values of the Kruskal-Wallis tests were corrected for multiple comparisons on each phylogenetic level using the Benjamini & Hochberg’s False Discovery Rate, and a p<0.05 was considered statistically significant [23]. For those bacterial groups that were still significant after p-value adjustment, a Dunns’ post-test was used to determine which disease categories were significantly different. A Fisher’s exact test was used to determine the proportions of dogs that harbored specific bacterial taxa or toxins.
Paired samples were available for 8 dogs with IBD, representing samples at time of clinically active and therapeutically controlled IBD (i.e., clinically insignificant IBD), respectively. The qPCR assays for these paired samples were performed as batch in the same assay run, and the obtained data for the paired time points were compared using a Wilcoxon signed rank test.
Linear Discriminant Analysis (LDA) was used to achieve dimensionality reduction and thereby to identify combinations of bacterial groups that would discriminate between healthy dogs and all diseased dogs (independent of disease phenotype) [24]. Using the OTUs as features for the classification, the single, two, or three feature LDA classifiers were constructed and ranked based on their error estimates [25], [26]. More detailed descriptions of LDA classification are provided as supplementary Method S1.
Results
Animals and Disease Characteristics
No significant differences in age, gender, or body weight were found among the various animal groups (Table 1).
Sequence Analysis
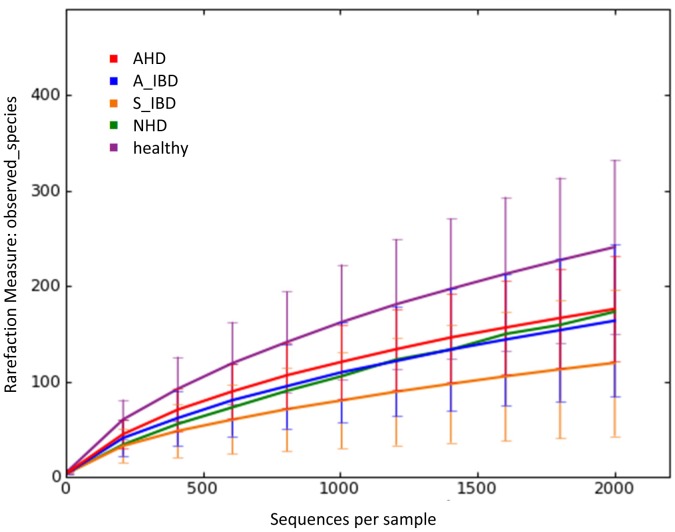
The 454- pyrosequencing pipeline yielded 189,138 quality sequences for the 67 samples analyzed. Across all disease groups, sequences were classified into nine bacterial phyla (Table 3). Figure 1 illustrates the rarefaction curves for all disease groups. No significant differences in the number of OTUs, the Shannon index, and the Chao1 metric were observed (Table 1).
Table 3. Relative percentages of the most abundant bacterial groups at the various phylogenetic levels (phylum, class, order, family, genus) based on pyrosequencing.
| Medians % (min-max%)* | ||||||
| Healthy | NHD | AHD | Active IBD | Controlled IBD | Kruskal-WallisP-value** | |
| Firmicutes | 96.6 (81–100) | 95.6 (83–100) | 56 (13–100) | 98.7 (95–100) | 98.6 (24–100) | 0.0985 |
| Clostridia | 78.1 (21–97) | 86.8 (46–99) | 55.6 (12–99) | 45.5 (1–94) | 47 1–91) | 1 |
| Clostridiales | 78.1 (21–97) | 86.8 (46–99) | 55.6 (12–99) | 45.5 (1–94) | 47 (1–91) | 1 |
| Clostridiaceae | 36.2 (6–84) | 81.5 (32–99) | 46.4 (7–99) | 26.4 (1–82) | 18.2 (1–75) | 0.302 |
| Clostridium | 33.7a (5–84) | 81.5b (32–99) | 44.0a,b (6–99) | 13.7a (0–82) | 14.2a (0–82) | 0.03 |
| Ruminococcaceae | 16.0a (0–46) | 4.7b (0–21) | 0.8b (0–18) | 5.6a,b (0–54) | 7.9a,b (0–53) | 0.004 |
| Faecalibacterium | 0.1 (0–16) | 0 (0–5) | 0 (0–3) | 0 (0–0) | 0.3 (0–3) | 1 |
| Ruminococcus | 15.4a (0–46) | 4.7b (0–16) | 0.7b (0–18) | 5.6a,b (0–54) | 6.8a,b (0–53) | 0.008 |
| Lachnospiraceae | 0.4 (0–2) | 0.1 (0–3) | 0 (0–1) | 0.3 (0–1) | 0.3 (0–3) | 0.114 |
| Blautia | 9.9a (0–28) | 0.2b (0–17) | 0.2b (0–4) | 5.9a,b (0–9) | 3.6b (0–16) | 0.002 |
| Roseburia | 0.2 (0–1) | 0.1 (0–3) | 0 (0–1) | 0.1 (0–0) | 0.1 (0–1) | 0.642 |
| Coprococcus | 0 (0–1) | 0 (0–0) | 0 (0–0) | 0.1 (0–0) | 0.1 (0–1) | 1 |
| Veillonellaceae | 0 (0–4) | 0 (0–0) | 0 (0–1) | 0 (0–0) | 0 (0–0) | 1 |
| Eubacteriaceae | 0.2 (0–2) | 0.1 (0–13) | 0.1 (0–17) | 0.2 (0–0) | 0.5 (0–4) | 1 |
| Eubacterium | 0.8 (0–27) | 0.1 (0–13) | 0.1 (0–17) | 0.3 (0–1) | 1 (0–5) | 0.564 |
| Erysipelotrichi | 7.8a (0–45) | 0.7a,b (0–9) | 0.1b (0–2) | 0.8a,b (0–99) | 0.8b (0–8) | 0.0009 |
| Erysipelotrichales | 7.8a (0–45) | 0.7a,b (0–9) | 0.1b (0–2) | 0.8a,b (0–99) | 0.8b (0–8) | 0.0009 |
| Erysipelotrichaceae | 7.8a (0–45) | 0.7a,b (0–9) | 0.1b (0–2) | 0.8a,b (0–99) | 0.8b (0–8) | 0.0009 |
| Turicibacter | 0.5 (0–39) | 0.1 (0–4) | 0 (0–0) | 0 (0–0) | 0.1 (0–1) | 0.138 |
| Allobaculum | 0.3 (0–14) | 0.4 (0–8) | 0 (0–1) | 0 (0–3) | 0 (0–2) | 1 |
| Bacilli | 0 (0–15) | 0.2 (0–16) | 0 (0–3) | 0 (0–1) | 0 (0–1) | 0.2169 |
| Lactobacillales | 0.2 (0–74) | 1.7 (0–29) | 0.4 (0–5) | 18.2 (0–60) | 7.9 (0–98) | 1 |
| Streptococcaceae | 0.1 (0–74) | 0.3 (0–19) | 0.1 (0–4) | 6 (0–60) | 2.7 (0–95) | 1 |
| Streptococcus | 0 (0–74) | 0.3 (0–19) | 0.1 (0–4) | 3.4 (0–60) | 2.5 (0–95) | 1 |
| Lactobacillaceae | 0 (0–61) | 0 (0–2) | 0 (0–0) | 0 (0–11) | 0.2 (0–98) | 0.86 |
| Enterococcaceae | 0 (0–3) | 0 (0–2) | 0 (0–3) | 0 (0–1) | 0 (0–1) | 1 |
| Proteobacteria | 0.30a (0–3) | 1.3a,b (0–16) | 4.3b (0–17) | 0.1a (0–1) | 0.1a (0–46) | 0.016 |
| Betaproteobacteria | 0.0a (0–0) | 0.0a,b (0–3) | 2.1b (0–14) | 0.0a,b (0–0) | 0.0a (0–8) | 0.0099 |
| Sutterella | 0.0a (0–0) | 0.0a (0–0) | 1.6b (0–14) | 0.0a (0–0) | 0.0a (0–1) | 0.008 |
| Gammaproteobacteria | 0 (0–3) | 1 (0–16) | 0.6 (0–15) | 0 (0–0) | 0 (0–29) | 0.0648 |
| Enterobacteriales | 0 (0–3) | 0.2 (0–16) | 0.1 (0–13) | 0 (0–0) | 0 (0–19) | 1 |
| Enterobacteriaceae | 0 (0–0) | 0.2 (0–16) | 0.1 (0–13) | 0 (0–0) | 0 (0–19) | 1 |
| Alphaproteobacteria | 0.1 (0–0.9) | 0.1 (0–1.2) | 0 (0–0.2) | 0.1 (0–0.3) | 0 (0–2) | 0.063 |
| Rickettsiales | 0.1a (0–0.9) | 0.0a,b (0–0.2) | 0.0b (0–0.2) | 0.1a,b (0–0.3) | 0.0b (0–0.4) | 0.0072 |
| Anaplasmataceae | 0.1a (0–0.9) | 0.0a,b (0–0.2) | 0.0b (0–0.2) | 0.1a,b (0–0.3) | 0.0b (0–0.4) | 0.016 |
| Anaplasma | 0.1a (0–0.9) | 0.0a,b (0–0.2) | 0.0b (0–0.2) | 0.1a,b (0–0.3) | 0.0b (0–0.4) | 0.016 |
| Bacteroidetes | 0 (0–18) | 0 (0–3) | 0.5 (0–17) | 0 (0–0) | 0 (0–12) | 0.0685 |
| Bacteroidia | 0 (0–18) | 0 (0–0) | 0.5 (0–17) | 0 (0–0) | 0 (0–11) | 0.1683 |
| Bacteroidales | 0 (0–18) | 0 (0–0) | 0.5 (0–17) | 0 (0–0) | 0 (0–11) | 0.1683 |
| Bacteroidaceae | 0 (0–4) | 0 (0–0) | 0.5 (0–10) | 0 (0–0) | 0 (0–3) | 0.524 |
| Bacteroides | 0 (0–3) | 0 (0–0) | 0.5 (0–10) | 0 (0–0) | 0 (0–3) | 0.696 |
| Actinobacteria | 1.8a (0–13) | 1.4a.b (0–6) | 0.2b (0–3) | 0.8a,b (0–5) | 1.0a,b (0–15) | 0.019 |
| Actinobacteria (class) | 1.8a (0–13) | 1.4a.b (0–6) | 0.2b (0–3) | 0.8a,b (0–5) | 1.0a,b (0–15) | 0.0342 |
| Coriobacteriales | 1.8a (0–13) | 1.0a,b (0–6) | 0.1b (0–2) | 0.8a,b (0–5) | 0.7a,b (0–15) | 0.0162 |
| Coriobacteriaceae | 1.8a (0–13) | 1.0a,b (0–6) | 0.1b (0–2) | 0.8a,b (0–5) | 0.7a,b (0–15) | 0.036 |
| Collinsella | 1.5a (0–13) | 1.0a,b (0–4) | 0.0b (0–2) | 0.7a,b (0–5) | 0.5a,b (0–13) | 0.018 |
| Fusobacteria | 0.1 (0–4) | 0 (0–2) | 23.5 (0–75) | 0 (0–0) | 0 (0–17) | 0.0865 |
| Fusobacteriales | 0 (0–4) | 0 (0–2) | 23.4 (0–0) | 0 (0–0) | 0 (0–14) | 0.1647 |
| Fusobacteriaceae | 0 (0–4) | 0 (0–2) | 23.4 (0–0) | 0 (0–0) | 0 (0–14) | 0.366 |
| Fusobacterium | 0 (0–4) | 0 (0–2) | 23.2 (0–0) | 0 (0–0) | 0 (0–6) | 0.5 |
Taxa present in at least 50% of dogs (either healthy or diseased) included in analysis.
p-values adjusted based on the Benjamini and Hochberg False discovery rate.
Medians not sharing a common superscript are significantly different (p<0.05 based on a Dunn’s multiple comparisons test).
NHD = acute hemorrhagic diarrhea; AHD = acute hemorrhagic diarrhea; IBD = inflammatory bowel disease.
Figure 1. Rarefaction analysis of 16 S rRNA gene sequences obtained from canine fecal samples.
Lines represent the average of each group, while the error bars represent the standard deviations. The analysis was performed on a randomly selected subset of 2,000 sequences per sample. A_IBD = active IBD; S_IBD = therapeutically controlled IBD; NHD = acute non-hemorrhagic diarrhea; AHD = acute hemorrhagic diarrhea.
Microbial Communities in Controls and in Dogs with Acute and Chronic Gastrointestinal Diseases
No significant differences were observed in total bacterial abundance among the groups based on qPCR analysis (Table 4; p = 0.09). Significant differences in microbial communities were, however, observed among the various groups. According to the linear discriminant analysis (LDA; Method S1), the triple combination of Blautia spp., Faecalibacterium spp., and Turicibacter spp. had the highest discriminatory power when healthy dogs were compared to all the dogs with gastrointestinal disease (independent of disease phenotype).
Table 4. Summary statistics for qPCR results.
| Medians (min-max)* log DNA | ||||||
| Healthy | NHD | AHD | Active IBD | Controlled IBD | Kruskal-WallisP-value | |
| Bacteroidetes | 6.4a (0.0–12.1) | 4.5b (0.0–6.1) | 6.7a (4.7–9.9) | 7.3a (0.0–9.3) | 5.3a,b (0.0–7.5) | 0.0009 |
| Bifidobacterium | 2.9 (0.0–7.3) | 3.1 (0.0–6.2) | 0 (0.0–3.9) | 2.2 (0.0–6.6) | 3.8 (0.0–7.5) | 0.0959 |
| Blautia | 9.7a (8.2–10.7) | 6.3b (5.7–10.2) | 8.2b (6.9–10.2) | 9.2a,b (7.3–9.9) | 9.5a,b (5.9–9.9) | 0.0003 |
| C. perfringens | 2.0a (0.0–6.1) | 4.0a,b (0.0–7.4) | 6.2b (0.6–7.6) | 3.0a,b (0.0–6.7) | 2.4a,b (0.6–5.7) | 0.0002 |
| Faecalibacterium | 5.8a (4.1–7.9) | 0.0b (0.0–7.7) | 4.7b (0.0–7.8) | 4.2b (0.0–6.3) | 5.5a,b (0.0–7.3) | 0.0002 |
| Fusobacteria | 7.3a,b (5.5–8.8) | 6.9a,c (3.9–8.6) | 8.2b (6.0–10.3) | 6.4c (4.7–6.7) | 7.1a,b,c (3.0–7.9) | 0.0014 |
| Ruminococcaceae | 7.6a (2.4–8.9) | 5.7b (2.7–7.1) | 5.6b (0.0–7.5) | 7.3a,b (0.0–8.6) | 7.9a (6.5–8.6) | <0.0001 |
| Turicibacter | 2.9a (0.0–7.7) | 0.0b (0.0–4.8) | 0.0b (0.0–0.0) | 1.5a,b (0.0–6.6) | 3.8a (0.0–5.9) | 0.0003 |
| Universal | 12.0 (10.9–13.2) | 10.8 (8.3–12.7) | 11.6 (10.7–12.4) | 12.0 (9.7–12.2) | 12.3 (8.2–12.8) | 0.0935 |
Medians not sharing a common superscript are significantly different (p<0.05 based on a Dunn’s multiple comparisons test).
NHD = acute non-hemorrhagic diarrhea; AHD = acute hemorrhagic diarrhea; IBD = inflammatory bowel disease
PCoA plots (Fig. 2) were constructed to compare the individual groups of dogs, and showed notable differences between healthy dogs and dogs with acute GI disorders (ANOSIM; NHD vs. healthy dogs, p = 0.003; AHD vs. healthy dogs, p = 0.001). Furthermore, both acute disease groups differed significantly from each other (Fig. 2) (ANOSIM; NHD vs. AHD, p = 0.004).
Figure 2. Principal Coordinates Analysis (PCoA) of unweighted UniFrac distances of 16 S rRNA genes.
(A) Analysis for healthy dogs (blue), dogs with acute non-hemorrhagic diarrhea (NHD; green), and dogs with acute hemorrhagic diarrhea (AHD; red). These results indicate that fecal microbial communities differ in dogs with acute forms of diarrhea compared to healthy control dogs. Statistical analysis revealed a significant separation between samples obtained from NHD and AHD (ANOSIM; p = 0.004) and both groups were also significantly different from the healthy dogs (ANOSIM; NHD vs. healthy dogs, p = 0.003; AHD vs. healthy dogs, p = 0.001). (B) Analysis for healthy dogs (blue), dogs with active IBD (red), and dogs with therapeutically controlled IBD (green). In contrast to the dogs with acute diarrhea, fecal communities in dogs with chronic forms of diarrhea (active idiopathic IBD) were not significantly different from healthy dogs in this study.
In contrast, neither the A_IBD nor the S_IBD group were significantly different from the healthy group or different from each other based on PCoA plots (ANOSIM; A_IBD vs. S_IBD, p = 0.91; A_IBD vs. controls, p = 0.75; S_IBD vs. controls, p = 0.07). However, sequence analysis and qPCR results revealed that Faecalibacterium spp. and Fusobacteria were significantly lower in dogs with clinically active IBD when compared to healthy dogs (see below).
Dogs with acute hemorrhagic diarrhea (AHD)
Based on PCoA plots, dogs with AHD had profound microbiome changes. Several bacterial groups were altered in dogs with AHD compared to the healthy dogs, but also to dogs with other forms of GI disease (Table 3). Increases in proportions were observed for the genus Sutterella (class β-Proteobacteria) and Clostridium perfringens-like sequences (Table 3, Fig. 3). The phylum Fusobacteria was also increased in dogs with AHD (Fig. 3), but this difference did not reach significant differences when p-values were adjusted for multiple comparisons (p = 0.08). Decreases in proportions were observed for Actinobacteria (i.e., Coriobacteriaceaea) and several members within the Firmicutes, most notably Ruminococcaceae, Blautia spp. (Lachnospiraceae), and Turicibacter spp. (Erysipelotrichaceae) (Table 3, Fig. 3).
Figure 3. Results of sequence analysis for selected bacterial groups.
H = healthy, NHD = acute non-hemorrhagic diarrhea, AHD = acute hemorrhagic diarrhea, A_IBD = active IBD, S_IBD = therapeutically controlled, clinically insignificant IBD. Columns not sharing a common superscript are significantly different (P<0.05).
Results of qPCR assays confirmed significant decreases in Blautia, Ruminococcaceae including Faecalibacterium, and Turicibacter spp. (Fig. 4). Clostridium perfringens was significantly increased only in the dogs with AHD when compared to healthy dogs.
Figure 4. Results of quantitative PCR assays for selected bacterial groups.
H = healthy, NHD = acute non-hemorrhagic diarrhea, AHD = acute hemorrhagic diarrhea, A_IBD = active IBD, S_IBD = therapeutically controlled, clinically insignificant IBD. Columns not sharing a common superscript are significantly different (P<0.05).
Dogs with acute non-hemorrhagic diarrhea (NHD)
PCoA plots also revealed shifts in the fecal microbiome of dogs with NHD (ANOSIM; NHD vs. healthy dogs, p = 0.003). While several bacterial taxa showed similar trends as observed for dogs with AHD, only few groups reached significance (Table 3). Sequence analysis revealed decreased proportions of Blautia spp. and Ruminococcaceae. Analysis by qPCR confirmed these decreases and also revealed decreases in Turicibacter spp., Faecalibacterium spp., and Bacteroidetes (Table 4).
Dogs with idiopathic IBD
PCoA plots did not indicate a significant separation between dogs with idiopathic IBD and control dogs (Fig. 2.). However, trends were observed for decreases in proportions of Faecalibacterium spp. and Turicibacter spp. (p = 0.06) in active IBD when compared to the healthy dogs, and the proportions of these groups tended to be similar to control dogs in the samples of dogs with controlled IBD. One reason for lack of significance in the sequencing results was most likely due to the low percentage of sequencing tags for Faecalibacterium, with medians of 0.1% in healthy dogs and 0.0% in diseased animals. However, Faecalibacterium was detectable in 23/32 healthy dogs, 6/10 dogs with controlled IBD, but only 1/5 with active IBD (p = 0.04 vs. healthy dogs). Results of qPCR analysis for Faecalibacterium spp. confirmed the trend observed for the pyrosequencing results and showed a significant decrease in dogs with active IBD, but no significant difference between the healthy dogs and dogs with clinically insignificant IBD. Furthermore, when paired samples were analyzed from dogs (n = 8) at time periods of active and clinically insignificant IBD, the abundance of Faecalibacterium was significantly higher in samples collected during time periods of clinically insignificant IBD (Fig. 5, p = 0.0313).
Figure 5. Faecalibacterium spp. and the phylum Fusobacteria in active and non-active IBD.
Using qPCR, paired fecal samples were analyzed from dogs (n = 8) at time periods of active and clinically insignificant IBD as scored by a clinical IBD disease activity index (CIBDAI). The time period between the collections of repeated samples ranged from 2–8 months (median 5.5 months). None of the other bacterial groups evaluated by qPCR, including total bacteria, revealed significant differences between the paired time periods.
Based on qPCR analysis, also Fusobacteria were significantly decreased in dogs with active IBD compared to the healthy dogs. Fusobacteria were also significantly increased in samples collected during time periods of clinically insignificant IBD compared to those during active disease when paired samples were analyzed (Fig. 5, p = 0.0148).
None of the other bacterial groups evaluated by qPCR, including total bacteria, revealed significant differences between healthy dogs and those with active IBD, or between time periods of active disease vs. time periods of clinically insignificant IBD.
Discussion
This study evaluated the fecal microbiome of healthy dogs and dogs with acute and chronic GI disorders. Significant differences were observed in microbiome composition among the various disease groups. Dogs with acute diarrhea showed the most profound alterations in their microbiome. Faecalibacterium spp. and the phylum Fusobacteria were decreased in active IBD, but not significantly different during time periods of clinically insignificant IBD. Rarefaction curves (Fig. 1.) and alpha diversity measures (Table 1) suggested a trend for lower species richness and microbial diversity in the diseased groups. However, statistical differences (e.g., p = 0.053 for the Chao1) were not identified, most likely due to the large inter-animal variation and the relative small number of animals analyzed.
Various pathogens, but also other causes such as hypersensitivities, have been associated with causing acute diarrhea [27], [28]. In this study only a partial evaluation for bacterial and parasitic enteropathogens was conducted. Potential pathogens identified were E. coli, Isospora, Giardia/Cryptosporidium, enterotoxigenic C. perfringens, and toxigenic C. difficile (Table S2), but no clear evidence for an association between specific pathogens and acute diarrhea was identified. Patients were, therefore, classified based on clinical signs (i.e., AHD and NHD) rather than the presence of specific pathogens. The results of this study are in general agreement with previous molecular studies examining the fecal microbiota of dogs with diarrhea. In one study, dogs with acute episodes of diarrhea had an increased abundance of C. perfringens and Enterococcus spp., and a decrease in Bacteroides spp. [15]. Decreased proportions of Bacteroidetes were also observed in 9 dogs with unspecified diarrhea when compared to 9 healthy dogs [14]. Similarly, this study observed significant decreases in Bacteroidetes in the dogs with acute non-hemorrhagic diarrhea, but interestingly not in dogs with AHD. We also observed significant increases in Clostridium spp. in dogs with NHD and increases in C. perfringens in dogs with AHD (Fig. 4). Additionally, dogs with AHD showed increases in Fusobacteria and the genus Sutterella (family Alcaligenaceae). We also observed that dogs with acute diarrhea displayed significant decreases in prominent members of the intestinal microbiota, such as Erysipelotrichaceae (i.e., genus Turicibacter), Ruminococcaceae (i.e., Ruminocococcus, Faecalibacterium) and Lachnospiraceae (i.e., Blautia) (Fig. 6). Some of these groups are believed to be important producers of short-chain fatty acids (SCFA), which are important for intestinal health. For example, butyrate protects against colitis by inducing apoptosis in cells with DNA damage, while acetate beneficially modulates intestinal permeability [29], [30]. The members of the intestinal microbiota produce various other metabolites (e.g., indole) that have direct immunomodulatory properties [30], [31]. Future studies are warranted to evaluate microbiome function (e.g., microbial derived metabolites) in dogs with acute diarrhea.
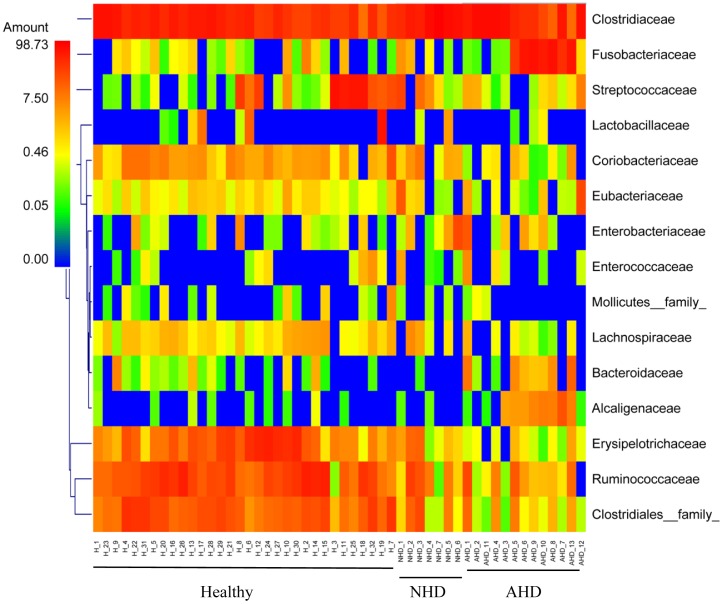
Figure 6. Heatmap illustrating the relative abundance of predominant bacterial families in fecal samples of healthy dogs and dogs with acute diarrhea based on 454-pyrosequencing.
Samples from healthy dogs (H), dogs with acute non-hemorrhagic diarrhea (NHD), and dogs with acute hemorrhagic diarrhea (AHD) are shown. The heatmap represents the relative percentage of each family within each sample.
Idiopathic IBD is a common chronic GI disorder of dogs. As in humans, an interplay between the intestinal microbiota together with an underlying genetic susceptibility of the host and dietary and environmental factors, are implicated in the development of disease [7], [32]. While recent studies have reported a dysbiosis in duodenal samples of dogs with IBD, limited data are available describing the fecal microbiota in these dogs [7]–[10]. In this study, Faecalibacterium spp. and Fusobacteria were significantly decreased in dogs with active IBD. Furthermore, when paired fecal samples were analyzed, the abundances of Faecalibacterium spp. and Fusobacteria were significantly decreased in samples collected during episodes of clinically active disease vs. periods of clinically insignificant IBD (Fig. 5). In contrast to previous findings in duodenal biopsies of dogs with IBD [7], we did not observe significant differences in members of Proteobacteria between healthy dogs and dogs with IBD. Proteobacteria were only significantly higher in dogs with acute diarrhea.
Faecalibacterium spp. were found decreased in dogs with acute diarrhea and active IBD. Faecalibacterium prausnitzii has garnered attention as it is often observed to be decreased in humans with IBD [33]. Furthermore it has been shown to secrete anti-inflammatory peptides in in-vitro studies [34]. Recent studies suggest that Faecalibacterium spp. are prominent members of the canine gut microbiota, as FISH analysis of fecal samples of healthy dogs estimated the abundance of the Faecalibacterium–Subdoligranulum group as a median 16% of total bacterial counts [21]. It has been suggested that dogs may harbor Faecalibacterium spp. other than F. prausnitzii, as an initial phylogenetic assessment of a near-full-length 16S rRNA gene clone obtained from the canine colon clustered distinct from a human strain of F. prausnitzii (AJ270469) [35]. Therefore, the phylogenetic classification of Faecalibacterium spp and their functional properties in the canine intestine deserve further research.
As a limitation to this study, we evaluated only a small number of animals in the various disease groups. We initially evaluated only a subset of animals by 454-pyrosequencing, as these samples were initially available at the time of sequence analysis. Therefore, due to the small sample size we may have missed some bacterial groups that are altered in certain groups of diseased dogs. Our study population was also not homogenous, as some of the dogs lived in different countries. Geographical differences in fecal microbiota have not been well examined in dogs. In this study we evaluated samples from control dogs and diseased dogs from Sweden, and also control and diseased dogs (NHD) from Texas. We did not have matching controls from Germany, the site where samples from dogs with hemorrhagic diarrhea were collected, as these were part of an unrelated study. For initial evaluation for potential differences in the canine fecal microbiome based on country and other variables, we performed clustering based on the Unifrac distance metric on all healthy dogs in this study and did not observe clustering based on country of origin (USA vs. Sweden), weight or gender (data not shown). We also performed a separate Unifrac analysis on all dogs from Sweden (healthy vs. active IBD vs. controlled IBD), and similar to the analysis that contained all dogs regardless of country of origin, we did not observe clustering between healthy dogs and dogs with IBD, but sequence and qPCR analysis revealed similar results as observed when all dogs were included in the analysis. These results would also suggest that dog microbiota could potentially be classified into specific enterotypes [36], [37]. In humans, enterotypes are characterized as compositional categories of organisms, most notably Bacteroides, Prevotella, or Ruminococcus, respectively, which are typically independent of nationality, gender, age, or short-term dietary interventions [36], [37]. Definition of specific microbial community enterotypes may open up new therapeutic approaches to companion animal health, by designing or prescribing appropriate diets for specific disease phenotypes.
In conclusion, we observed differences in the fecal microbiome composition between dogs with acute and chronic diarrhea compared to healthy dogs. These changes were more profound in dogs with acute diarrhea, and were not identified in dogs with therapeutically controlled IBD. These results form a roadmap for additional studies focused on a more defined population of diseased dogs.
Supporting Information
Control dogs enrolled into the study.
(PDF)
Dogs with acute hemorrhagic diarrhea (AHD) and acute non-hemorrhagic diarrhea (NHD).
(PDF)
Dogs with active IBD and with therapeutically controlled IBD.
(PDF)
Linear discriminant analysis.
(PDF)
Funding Statement
These authors have no support or funding to report.
References
- 1. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, et al. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ritchie LE, Steiner JM, Suchodolski JS (2008) Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol Ecol 66: 590–598. [DOI] [PubMed] [Google Scholar]
- 3. Handl S, Dowd SE, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS (2011) Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol 76: 301–310. [DOI] [PubMed] [Google Scholar]
- 4. Suchodolski JS (2011) Companion animals symposium: Microbes and gastrointestinal health of dogs and cats. J Anim Sci 89: 1520–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marks SL, Rankin SC, Byrne BA, Weese JS (2011) Enteropathogenic bacteria in dogs and cats: diagnosis, epidemiology, treatment, and control. J Vet Intern Med 25: 1195–1208. [DOI] [PubMed] [Google Scholar]
- 6.Kempf C, Schulz B, Strauch C, Sauter-Louis C, Tuyen U, et al.. (2010) Virusnachweis in Kotproben und klinische sowie labordiagnostische Befunde von Hunden mit akutem haemorrhagischem Durchfall. Tieraerztliche Praxis 38 79–86. [PubMed]
- 7. Suchodolski JS, Dowd SE, Wilke V, Steiner JM, Jergens AE (2012) 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. Plos ONE 7: e39333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allenspach K, House A, Smith K, McNeill FM, Hendricks A, et al. (2010) Evaluation of mucosal bacteria and histopathology, clinical disease activity and expression of Toll-like receptors in German shepherd dogs with chronic enteropathies. Vet Microbiol 146: 326–335. [DOI] [PubMed] [Google Scholar]
- 9. Suchodolski JS, Xenoulis PG, Paddock CG, Steiner JM, Jergens AE (2010) Molecular analysis of the bacterial microbiota in duodenal biopsies from dogs with idiopathic inflammatory bowel disease. Vet Microbiol 142: 394–400. [DOI] [PubMed] [Google Scholar]
- 10. Xenoulis PG, Palculict B, Allenspach K, Steiner JM, Van House AM, et al. (2008) Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol Ecol 66: 579–589. [DOI] [PubMed] [Google Scholar]
- 11. Simpson KW, Jergens AE (2011) Pitfalls and progress in the diagnosis and management of canine inflammatory bowel disease. Vet Clin North Am Small Anim Pract 41: 381–398. [DOI] [PubMed] [Google Scholar]
- 12. Suchodolski JS, Morris EK, Allenspach K, Jergens AE, Harmoinen JA, et al. (2008) Prevalence and identification of fungal DNA in the small intestine of healthy dogs and dogs with chronic enteropathies. Vet Microbiol 132: 379–388. [DOI] [PubMed] [Google Scholar]
- 13. Jia J, Frantz N, Khoo C, Gibson GR, Rastall RA, et al. (2010) Investigation of the faecal microbiota associated with canine chronic diarrhoea. FEMS Microbiol Ecol 71: 304–312. [DOI] [PubMed] [Google Scholar]
- 14. Chaban B, Links MG, Hill JE (2012) A Molecular Enrichment Strategy Based on cpn60 for Detection of Epsilon-Proteobacteria in the Dog Fecal Microbiome. Microb Ecol 63: 348–357. [DOI] [PubMed] [Google Scholar]
- 15. Bell JA, Kopper JJ, Turnbull JA, Barbu NI, Murphy AJ, et al. (2008) Ecological characterization of the colonic microbiota of normal and diarrheic dogs. Interdiscip Perspect Infect Dis 2008: 149694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jergens AE, Schreiner CA, Frank DE, Niyo Y, Ahrens FE, et al. (2003) A scoring index for disease activity in canine inflammatory bowel disease. J Vet Intern Med 17: 291–297. [DOI] [PubMed] [Google Scholar]
- 17. Suchodolski JS, Dowd SE, Westermarck E, Steiner JM, Wolcott RD, et al. (2009) The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiol 9: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gontcharova V, Youn E, Wolcott RD, Hollister EB, Gentry TJ, et al. (2010) Black Box Chimera Check (B2C2): a Windows-Based Software for Batch Depletion of Chimeras from Bacterial 16S rRNA Gene Datasets. Open Microbiol J 4: 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ritchie LE, Burke KF, Garcia-Mazcorro JF, Steiner JM, Suchodolski JS (2010) Characterization of fecal microbiota in cats using universal 16S rRNA gene and group-specific primers for Lactobacillus and Bifidobacterium spp. Vet Microbiol 144: 140–146. [DOI] [PubMed] [Google Scholar]
- 21. Garcia-Mazcorro JD, Dowd SE, Poulsen J, Steiner JM, Suchodolski JS (2012) Abundance and short-term temporal variability of fecal microbiota in healthy dogs. Microbiology Open 1: 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia-Mazcorro JF, Suchodolski JS, Jones KR, Clark-Price SC, Dowd SE, et al. (2012) Effect of the proton pump inhibitor omeprazole on the gastrointestinal bacterial microbiota of healthy dogs. FEMS Microbiol Ecol 80: 624–636. [DOI] [PubMed] [Google Scholar]
- 23. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57: 289–300. [Google Scholar]
- 24. Kachroo P, Ivanov I, Davidson LA, Chowdhary BP, Lupton JR, et al. (2011) Classification of diet-modulated gene signatures at the colon cancer initiation and progression stages. Dig Dis Sci 56: 2595–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chapkin RS, Zhao C, Ivanov I, Davidson LA, Goldsby JS, et al. (2010) Noninvasive stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells. Am J Physiol Gastrointest Liver Physiol 298: G582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sima C, Braga-Neto UM, Dougherty ER (2011) High-dimensional bolstered error estimation. Bioinformatics 27: 3056–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Unterer S, Strohmeyer K, Kruse BD, Sauter-Louis C, Hartmann K (2011) Treatment of aseptic dogs with hemorrhagic gastroenteritis with amoxicillin/clavulanic acid: a prospective blinded study. J Vet Intern Med 25: 973–979. [DOI] [PubMed] [Google Scholar]
- 28. Weese JS (2011) Bacterial enteritis in dogs and cats: diagnosis, therapy, and zoonotic potential. Vet Clin North Am Small Anim Pract 41: 287–309. [DOI] [PubMed] [Google Scholar]
- 29. Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ (2002) The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217: 133–139. [DOI] [PubMed] [Google Scholar]
- 30. Fukuda S, Toh H, Taylor TD, Ohno H, Hattori M (2012) Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes 3: 449–454. [DOI] [PubMed] [Google Scholar]
- 31. Bansal T, Alaniz RC, Wood TK, Jayaraman A (2010) The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A 107: 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Packey CD, Sartor RB (2009) Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis 22: 292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, et al. (2009) Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 15: 1183–1189. [DOI] [PubMed] [Google Scholar]
- 34. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, et al. (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suchodolski JS, Camacho J, Steiner JM (2008) Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol Ecol 66: 567–578. [DOI] [PubMed] [Google Scholar]
- 36. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, et al. (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, et al. (2011) Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muhling M, Woolven-Allen J, Murrell JC, Joint I (2008) Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J 2: 379–392. [DOI] [PubMed] [Google Scholar]
- 39. Malinen E, Rinttila T, Kajander K, Matto J, Kassinen A, et al. (2005) Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol 100: 373–382. [DOI] [PubMed] [Google Scholar]
- 40. Wise MG, Siragusa GR (2005) Quantitative detection of Clostridium perfringens in the broiler fowl gastrointestinal tract by real-time PCR. Appl Environ Microbiol 71: 3911–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gurjar AA, Hegde NV, Love BC, Jayarao BM (2008) Real-time multiplex PCR assay for rapid detection and toxintyping of Clostridium perfringens toxin producing strains in feces of dairy cattle. Mol Cell Probes 22: 90–95. [DOI] [PubMed] [Google Scholar]
- 42. Houser BA, Hattel AL, Jayarao BM (2010) Real-time multiplex polymerase chain reaction assay for rapid detection of Clostridium difficile toxin-encoding strains. Foodborne Pathog Dis 7: 719–726. [DOI] [PubMed] [Google Scholar]
- 43. Lubbs DC, Vester BM, Fastinger ND, Swanson KS (2009) Dietary protein concentration affects intestinal microbiota of adult cats: a study using DGGE and qPCR to evaluate differences in microbial populations in the feline gastrointestinal tract. J Anim Physiol Anim Nutr (Berl) 93: 113–121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Control dogs enrolled into the study.
(PDF)
Dogs with acute hemorrhagic diarrhea (AHD) and acute non-hemorrhagic diarrhea (NHD).
(PDF)
Dogs with active IBD and with therapeutically controlled IBD.
(PDF)
Linear discriminant analysis.
(PDF)