Abstract
The outcome of infection in the host snail Biomphalaria glabrata with the digenean parasite Schistosoma mansoni is determined by the initial molecular interplay occurring between them. The mechanisms by which schistosomes evade snail immune recognition to ensure survival are not fully understood, but one possibility is that the snail internal defence system is manipulated by the schistosome enabling the parasite to establish infection. This study provides novel insights into the nature of schistosome resistance and susceptibility in B. glabrata at the transcriptomic level by simultaneously comparing gene expression in haemocytes from parasite-exposed and control groups of both schistosome-resistant and schistosome-susceptible strains, 2 h post exposure to S. mansoni miracidia, using an novel 5K cDNA microarray. Differences in gene expression, including those for immune/stress response, signal transduction and matrix/adhesion genes were identified between the two snail strains and tests for asymmetric distributions of gene function also identified immune-related gene expression in resistant snails, but not in susceptible. Gene set enrichment analysis revealed that genes involved in mitochondrial electron transport, ubiquinone biosynthesis and electron carrier activity were consistently up-regulated in resistant snails but down-regulated in susceptible. This supports the hypothesis that schistosome-resistant snails recognize schistosomes and mount an appropriate defence response, while in schistosome-susceptible snails the parasite suppresses this defence response, early in infection.
Introduction
The tropical freshwater snail Biomphalaria glabrata is an intermediate host for several digenean trematode parasitic worms, including Schistosoma mansoni, the causative agent of human intestinal schistosomiasis. Human schistosomiasis is the most widespread trematode infection affecting around 200 million people, leading to a chronic debilitating disease and up to 200,000 deaths per year, across 75 developing countries [1]. Because of its medical importance, the B. glabrata/S. mansoni system has also emerged as a model for studies into multicellular host-parasite co-evolution, driven by reciprocal evolution of host resistance and parasite infectivity and/or virulence [2], [3]. The initial interactions between snail and invading schistosome are considered to define their respective future reproduction and survival; the parasite transforming from a short-lived free-living form in freshwater to a longer-term asexual parasitic stage in the snail hosts. If the snail cannot suppress and eliminate the invading schistosome quickly it risks parasitic castration (reviewed in [4]) followed by early death [5], [6]. The initial molecular interplay between snails and schistosomes is complex and there exists an urgent need to determine the principal pathways controlling this response, since identifying those factors involved in the intricate balance between the snail internal defence system (IDS) and trematode infectivity mechanisms that determine the success or failure of an infection (reviewed in [7]–[9]) may provide insight into approaches to disrupt the parasitic infection in the snail and break transmission. Furthermore, by understanding the basis of compatibility and the mechanisms underlying snail susceptibility to schistosome infection, the levels of compatibility in field situations can be assessed, leading to enhanced understanding of transmission dynamics which could ultimately inform control strategies.
Susceptibility of B. glabrata to S. mansoni is a heritable trait [10], with both snail and parasite genes influencing the outcome of infection [11]. In incompatible interactions, the schistosome fails to recognize, penetrate or develop within the snail, or may be destroyed by the IDS; such killing is mediated by haemocytes, ‘macrophage-like’ defence cells, encapsulating and eliminating non-compatible parasites [12]. The “schistosome-resistant” phenotype is defined as individuals or strain refractory to infection by a normally compatible schistosome strain. To establish an infection in a compatible strain, the schistosome larva must prevent the snail from detecting and/or eliminating it. Two hypotheses are that either the parasite remains undetected by the host and therefore no defence response is mounted [13], [14], or that the parasite is able to interfere with or suppress the host response to enable it to establish an infection [15]–[17]. Haemocyte-derived molecules thought to be key regarding snail defence to schistosomes include a diverse family of secreted lectins called fibrinogen-related proteins (FREPs), co-determinants of resistance as shown by RNAi knockdown [12], [18]–[20] that form complexes with schistosome mucins [21]–[23]; and lysosomal enzymes, and reactive oxygen/nitrogen intermediates [8], [23]–[25] which facilitate killing of the parasite. A cytosolic copper/zinc superoxide dismutase (SOD1) has also been associated with the schistosome-resistant phenotype [26], [27]. Moreover, the snail host oxidant response to schistosome infection has been investigated from the perspective of molecular co-evolution, through evaluation of reciprocal anti-oxidant responses of S. mansoni [28]; parasite anti-oxidant capacities appear to match closely host haemocyte oxidant responses in sympatric B. glabrata/S. mansoni combinations, highlighting the importance of oxidant production by resistant phenotype haemocytes [28]. Migration and recognition/adhesion of haemocytes to transforming miracidia/developing sporocysts are also likely important determinants of the resistance response. Integrin-like cell surface receptors [29] are known to regulate haemocyte adhesion and motility [30]–[32] and a tandem-repeat galectin has been found to bind haemocytes and the tegument of S. mansoni sporocysts [33] making it a candidate anti-schistosome pattern recognition receptor. Interestingly, S. mansoni excretory-secretory products (ESPs), released by transforming miracidia, have been shown to suppress extracellular signal-regulated kinase (ERK) signalling in haemocytes of susceptible, but not resistant B. glabrata [34] demonstrating that in compatible hosts, schistosomes can interfere with pathways that regulate snail haemocyte defence responses such as nitric oxide production [25]. Finally, knock-down of the recently-characterized B. glabrata cytokine Macrophage Migration Inhibitory Factor (BgMIF) was shown to reduce encapsulation of S. mansoni sporocysts in vitro and increase mother sporocyst survival in vivo [35]. Hence, the complex nature of the snail haemocyte defence response to schistosomes, outlined above, makes analysis of global gene expression a vital component of research aimed at elucidating the array of underlying mechanisms of snail-schistosome compatibility.
Both a cDNA microarray [36] and a specific stress/immune gene selected oligo array [20], [37], [38] have been employed previously for gene expression analysis in B. glabrata. This widely used method of global gene expression analysis involves hybridizing reverse-transcribed cDNA to the array to indicate relative gene expression for each arrayed gene or EST; this approach does not rely on prior knowledge of candidate genes or mechanisms in advance [39]. We previously developed a 2K cDNA microarray for B. glabrata [36], using randomly sequenced ORESTES-derived expressed sequence tags (ESTs) of various snail tissues including haemocytes [40], together with sequences derived from earlier differential gene expression analyses associated with schistosome resistance in snails using differential display (DD) [41]–[43] and suppression subtractive hybridization (SSH) [44]. We have therefore included many genes on the array which may be involved in snail parasite interactions, without making a priori assumptions of their functions based on homology to genes characterised in other organisms. Previous experiments using this microarray to compare gene expression in vivo in haemocytes from schistosome-exposed B. glabrata strains exhibiting resistant or susceptible phenotypes identified more than 90 strain-specific differentially expressed genes [36]. Unlike techniques such as qPCR, the strength of microarray analysis is that it provides a global view of changes in gene expression through simultaneous comparison of large numbers of genes, indicating cellular pathways and processes involved in response to parasite challenge. The 2K microarray was significantly expanded for use in this study, with the addition of more than 3K ESTs.
With the aim of identifying snail strain differences in early haemocyte responses to schistosomes, we furthered past approaches towards defining genes and pathways involved in host defence responses by the first use of gene set enrichment analysis (GSEA) [45] and FatiScan [46] in snail-schistosome interaction studies, using the most comprehensive B. glabrata cDNA microarray to date. A key question was to determine if strain specific responses to parasite exposure were at an early stage post exposure and to assess if the parasite was influencing (suppressing) the normal snail defence response in the susceptible strain. During invasion of the snail host, molecules are released from the miracidial penetration glands and within 1 hour the ciliated epidermal plates covering the miracidium are released allowing the parasite to transform into a post-miracidium that lacks a protective surface [47]; other ESPs are also released from the schistosome during such early post-embryonic development and ESPs can modulate kinase signalling [34] and protein expression [48] in B. glabrata haemocytes in vitro within a similar time frame. Thus, 2 hours post-exposure was selected as an appropriate time frame for our investigation to compare haemocyte gene expression changes in both snail strains in the context of schistosome invasion and early schistosome development in the host. The transcriptomic responses of these schistosome-resistant and -susceptible phenotypes to S. mansoni provide novel insights into the nature of early stage interactions that are likely to define trematode resistance and susceptibility.
Methods
Microarray Construction
In addition to the 2053 ORESTES, SSH and DD clones printed on the 2K B. glabrata cDNA microarray [36], a further 3174 were available from ORESTES libraries [14], [40], and a haemocyte cDNA library [49]. Selected clones, supplied spotted onto Whatman FTATM cards, were prepared by eluting the DNA from small discs punched from the card using Multiscreen PCR filter plates (Millipore, Billerica, USA); the resultant DNA was used for 100 µl PCRs containing 1×NH4 reaction buffer (Bioline, London, UK), 2.5 mM MgCl2, 0.2 mM dNTP, 0.2 µM each M13 forward and reverse primers and 0.025 U/µl PCR Taq polymerase (Bioline). Cycling conditions were: 94°C for 2 min, then 35 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 90 s, then a single cycle of 10 min at 72°C. We specifically included several genes implicated in the defence response of resistant snails, including FREP2 [50]; Cu/Zn superoxide dismutase (SOD) [26], [27] and Mn SOD [51]. The array also contained antioxidant genes such as thioredoxin peroxidase, peroxiredoxin 6, peroxinectin, peroxidasin and dual oxidase I and genes for cell signalling proteins including nuclear factor-kappa B (NF-κB), focal adhesion kinase (FAK) and I-kappa-B kinase complex associated protein (IKAP). PCR products were amplified for 7 specific B. glabrata genes (chosen as the sequences were available on GenBank, but not already represented in the ESTs from SSH, ORESTES or cDNA library), using primers designed from their sequences (Table 1). For all clones, 2 µg PCR product was transferred to 384-well plates (Molecular Devices (Genetix), New Milton, Hampshire, UK) using a Microlab Star robotic work-station (Hamilton Robotics, Birmingham, UK). A total of 5234 cDNA clones (50–200 ng/µl) were selected for printing. Controls were also included: yeast tRNA (250 ng/µl); B. glabrata genomic DNA from snail strains (NHM laboratory code) NHM3017 (derived from BS90 [52]), NHM1742, NHM3036 (derived from BB02 [53]) (200 ng/µl) and genomic DNA from Biomphalaria tenogophila, Bi straminea, Bi pfeifferi, Bi alexandrina, Bulinus globosus and Bu truncatus; pGem (purified vector with no insert) (75 ng/µl), two specific genes, (ribosomal 18s and cytochrome oxidase I) amplified from S. mansoni and blanks containing spotting buffer only. 15 µl aliquots were transferred to a further 384-well plate and 5 µl 4× spotting buffer (600 mM sodium phosphate; 0.04% SDS) added. The clones were printed (in duplicate) in 16 sub-arrays (4 columns×4 rows), with 26×26 clones in each sub-array on aminopropyl silane coated glass slides (Corning® GAPS™ II), at the Microarray Facility, Department of Pathology, University of Cambridge, UK; this array is the second generation B. glabrata microarray. Microarrays were processed by baking for 2 h at 80°C and UV cross-linking at 600 mJ. (GenePix Array List (GAL) file: NHM-ABDN B.glabrata 5K v1 ArrayExpress Archive accession: Array A-MEXP-1401). Differential expression of haemocyte genes using this B. glabrata cDNA microarray technology has previously been confirmed by quantitative qPCR [36] and the array was found to deliver robust results in our hands.
Table 1. Biomphalaria glabrata gene-specific primers used to amplify specific gene fragments included on the microarray.
| Code | Acc No | Gene | Primer sequence | |
| BgB | AB210096 | Dermatopontin 1 | F | GGTTATGCCAATGACTTCGGAC |
| R | GATTGACTTGCTCGCTCACG | |||
| BgG | AF179902 | Integrin interactor protein | F | CCTTGGGAATGTCATTGCTTG |
| R | GACCATTCCACCCTGATTGC | |||
| BgI | AF302260 | Serine protease B | F | CTAAGATACGGTGCTGGCTCG |
| R | GCGTAGACACCTGGTCTGCC | |||
| BgK | AY026258 | Thioredoxin peroxidase | F | CACTCACCTTGCATGGACTAATG |
| R | CAAGCGCAGTGTCTCATCAAC | |||
| BgP | AY678119 | Type 2 cystatin | F | CAAAATTGTCCACGCCACATC |
| R | GATGGTGTTCCCTGTAGTTGGG | |||
| BgQ | DQ087398 | guanine nucleotide-binding protein Rho | F | GGCAGCAATACGTAAGAAGCTTG |
| R | GCTGTGTCCCATAAGGCTAGTTC | |||
| BgSOD | AY505496 | Cu/Zn superoxide dismutase | F | GGTGATGATGGTGTTGCTGA |
| R | GATACCAATGACACCACAAGCTAA |
F-forward facing primer. R- Reverse facing primer.
Snail Material and Parasite Exposure
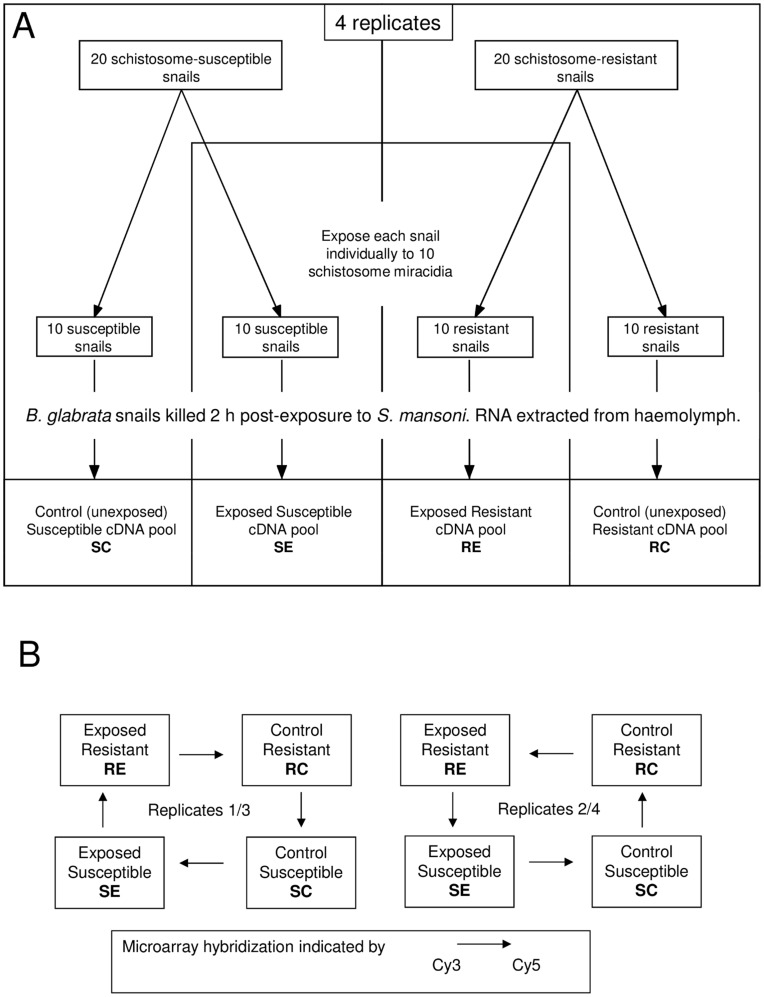
Four replicate experiments were performed (Fig. 1A). For each replicate, 20 adult B. glabrata from the susceptible strain (NHM1742) and 20 from the resistant strain (NHM3017) were maintained overnight in autoclaved snail water containing 100 µg/ml ampicillin. 10 snails from each strain were exposed individually to 10 S. mansoni miracidia (Belo Horizonte strain), while 10 were kept in identical conditions, but not exposed to miracidia. Snails were killed swiftly by decapitation 2 h post-exposure, and the exuded haemolymph collected. Haemolymph was pooled for each sample group (resistant exposed (RE), resistant control (RC), susceptible exposed (SE), susceptible control (SC)) and haemocytes pelleted by centrifuging at 10,000×g for 20 min at 4°C. The lymph was then removed and the haemocyte pellet frozen in liquid nitrogen and stored at –80°C.
Figure 1. Experimental design for the simultaneous comparison of haemocyte gene expression in Biomphalaria glabrata strains upon exposure to Schistosoma mansoni.
A. Resistant (R) and susceptible (S) strains of B. glabrata were exposed to S. mansoni (E) or kept unexposed as controls (C) B. Microarray hybridizations: 16 array hybridizations in double loop design with dye swaps were performed.
Microarray Hybridization
Total RNA was extracted from pooled haemocytes, using the SV RNA extraction kit (Promega UK Ltd, Southampton, UK) according to the manufacturer’s protocol. This kit includes DNAse treatment to eliminate genomic DNA contamination. cDNA was synthesized from 100 ng total RNA using the SMART PCR [54] cDNA synthesis kit (BD Biosciences, Oxford. UK) according to the manufacturer’s instructions and labelled with both Cy3 and Cy5 in 2 separate reactions using the BioPrime DNA labeling system (Invitrogen, Paisley, UK). 16 microarray hybridizations were carried out as described previously [36] using a loop design with dye swaps (Fig. 1B). The loop design [55] allowed direct comparison of results from resistant and susceptible snails and control and exposed snails, by comparing, directly on the arrays: i) control and parasite-exposed snails of both the resistant snail line and of the susceptible snail line (4 replicates, 2 with Cy dyes in one orientation and 2 swapped over) and ii) resistant and susceptible snail lines both for control snails and parasite-exposed snails (again with 4 replicates, 2 labelled in one orientation, 2 in the other).
Microarray Scanning and Analysis
Microarray slides were scanned sequentially for each Cy dye, at 10 µm resolution using an Axon GenePix 4100A scanner (Molecular Devices (UK) Ltd, Wokingham, UK). Photo multiplier tube values were adjusted to give an average intensity ratio between channels of approximately 1. Spot finding and intensity analysis was carried out using GenePix Pro 5.0. Data from these microarray experiments have been deposited with ArrayExpress: Experiment E-MEXP-1882. 16 GenePix output files were analysed using LIMMA (Linear models for microarray data, Bioconductor [56]). Print-tip loess normalization was used for within-array normalization [57] including log-transformation of the gene intensities. Moderated t-statistics were employed to assess change significance and a moderated f-statistic was used to test whether all contrasts were zero simultaneously, that is, whether there was no difference between strains before or after exposure or whether a gene showed an overall effect [58]. Within array duplicates were averaged and showed a good correlation of 0.84. One array displayed weak hybridization and was thus removed from the analysis. LIMMA was also used to define and test for certain contrasts, e.g. the difference of the fold change susceptible/resistant strains between the exposed and control groups (p-values were adjusted for multiple testing using the false Benjamini-Hochberg method [59], which controls the false discovery rate (FDR)).
Functional Analysis of Genes
Cluster analysis was performed in SeqTools (http://www.seqtools.dk/) using BlastN score values. Basic Local Alignment Search Tool (BLAST) searches, GO (gene ontology) and KEGG (Kyoto Encyclopaedia of Genes and Genomes) annotation and Interpro scans were performed using Blast2GO [60]. An annotation file was generated for the B. glabrata microarray (File S1).
Gene Set Enrichment Analysis
Gene set analysis for the data was performed using the Bioconductor function geneSetTest, which is available within the LIMMA library. The analysis was restricted to those GO terms that had at least 5 genes corresponding to them on the array (333 sets/terms overall). For each of the 5 comparisons (REvSE, RCvSC, REvRC, SEvSC and RE/SEvRC/SC) we tested whether the calculated p-values were more significant for the Gene Set/GO-term in question than for a random selection of genes to generate a Gene Set p-value for each combination of GO-Term and each comparison, and an average p-value across all the genes for each comparison. P-values were adjusted for multiple testing using the Bonferroni method of correction.
FatiScan
FatiScan ([46], [61], Babelomics: http://babelomics.bioinfo.cipf.es) was employed to identify significant asymmetrical distributions of biological labels (such as GO terms) associated with the ranked genes (based on fold change) for each comparison (REvRC, SEvSC, RCvSC and REvSE), using custom B. glabrata microarray annotations generated by Blast2Go.
Results
Annotation of Array Genes
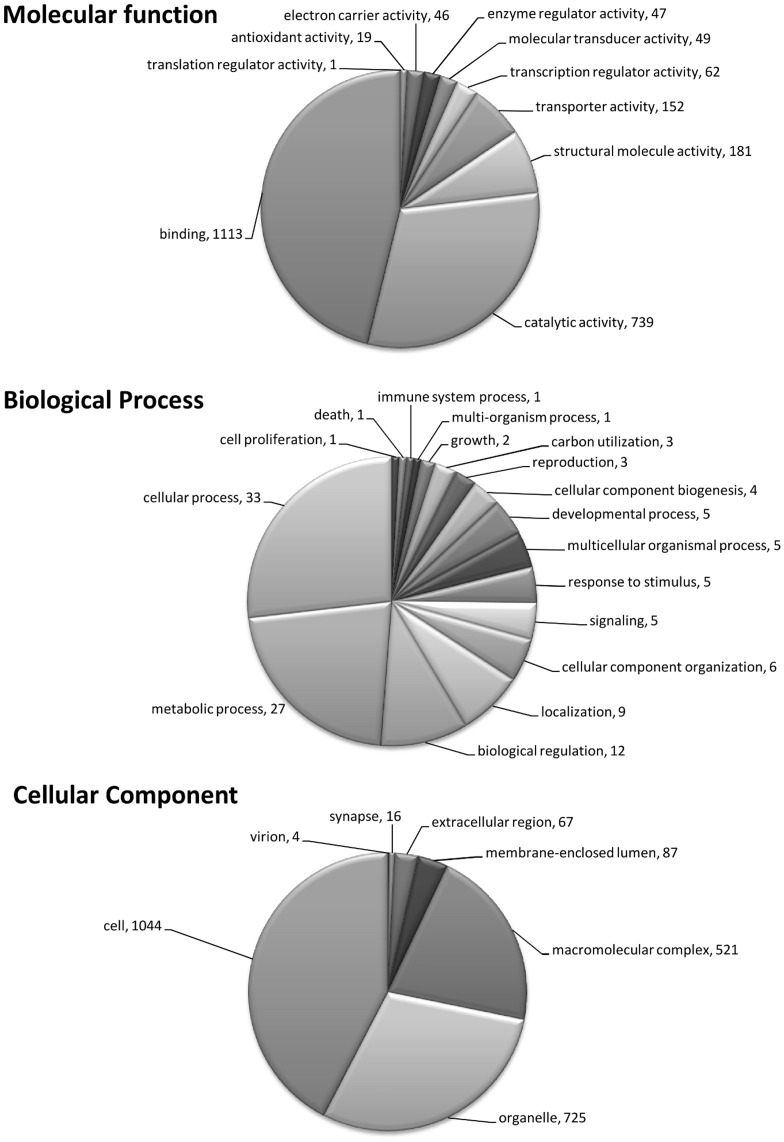
Genes with significant homology to previously characterized genes have GO terms assigned to them on the basis of that homology (Fig. 2). This produced a reference set of annotations associated with the sequences of the genes spotted on the 5K B. glabrata microarray (File S1).
Figure 2. Graphical representation of annotation for version 2 of the B. glabrata microarray.
The pie charts illustrate the number of genes in each GO assignment for molecular function, biological process and cellular component, for those genes with known functions. Genes that were not assigned are not represented here and each individual sequence may have more than one assignment.
Haemocyte Genes Differentially Expressed in Response to Parasite Exposure
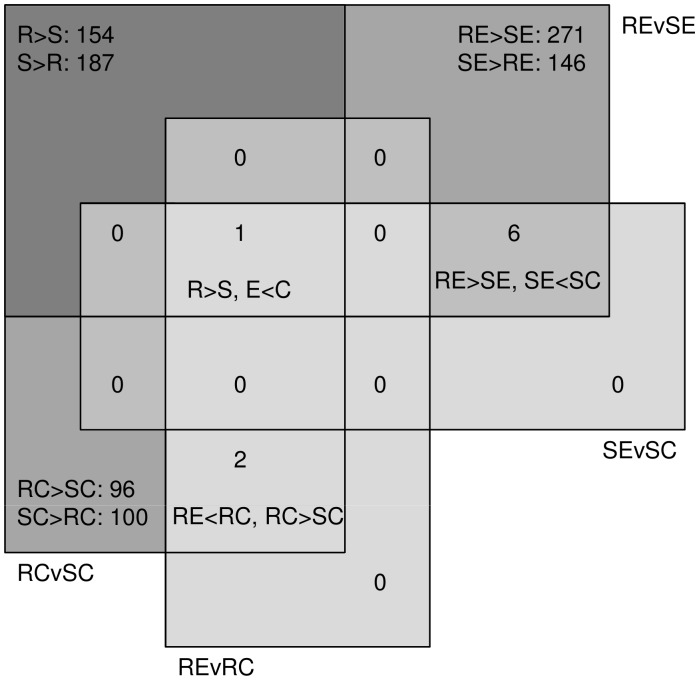
mRNA from haemocytes of both control and schistosome-exposed resistant and susceptible snails was compared using the 5K B. glabrata microarray (these data are available at ArrayExpress Archive accession: Array A-MEXP-1401). From the analysis we identified genes that were differentially expressed between control and parasite exposed snails in both strains and between resistant and susceptible strains both for control and parasite exposed snails. The numbers of identified, and the classes in which they demonstrated differential expression, are summarized in the Venn diagram (Fig. 3). Firstly, genes expressed in haemocytes sampled 2 h post exposure to S. mansoni miracidia, were compared to unexposed controls to investigate the initial response of each snail strain to the parasite. Analysis of differential gene expression from the microarray identified 9 genes demonstrating a significant difference (p<0.01) in intensity between the compared samples in each category, with some genes located in more than one category (Fig. 3; see also Table 2). One gene (CV548474: unknown) was identified as having significantly higher expression in resistant snails before and after parasite exposure (R>S), and this same gene was also found to be down-regulated in both strains after exposure (E<C). Two genes (EW997021 and EW997112, both unknown, Table 2) were identified which showed significantly higher expression in the resistant control snails (RC>SC) and were down-regulated in the resistant snails after exposure (RE<RC). Additionally, 6 genes were shown to be significantly down-regulated in susceptible snails after infection (SE<SC), which were expressed less in susceptible exposed compared with resistant exposed snails (RE>SE); these included 3 unknown genes, 2 myosin II heavy chain genes and 1 alpha actin gene (Table 2).
Figure 3. Significant differentially expressed B. glabrata haemocyte genes identified from microarray comparisons.
The Venn diagram shows the number of identified significantly differentially expressed genes in each category. Some genes were identified which were differentially expressed in more than one comparison and hence lie in the overlapping regions of the diagram. Key to symbols: R, resistant B. glabrata; S, susceptible B. glabrata; E, S. mansoni exposed snails; C, control snails. > greater than, < less than.
Table 2. Genes identified as differentially expressed upon exposure to S. mansoni in resistant and susceptible B. glabrata strains.
| Acc No | Name | Organism | Blast match Acc No | E value | fc(RE/SE) | fc(RC/SC) | fc(RE/RC) | fc(SE/SC) |
| EW997021 | Unknown | 2.67 | 3.81 | −5.89 | −4.12 | |||
| EW997112 | Unknown | 2.21 | 2.78 | −4.32 | −3.43 | |||
| CV548474 | Unknown | 3.10 | 3.19 | −5.58 | −5.43 | |||
| EW997519 | Unknown | 1.74 | 1.27 | −1.94 | −2.66 | |||
| EW997424 | Unknown | 3.86 | 1.41 | 1.15 | −2.37 | |||
| EW997386 | myosin II | Nasonia vitripennis | XP_001607303 | 5.45E−35 | 3.02 | 1.16 | 1.18 | −2.21 |
| EW997539 | Unknown | 2.49 | −1.04 | 1.13 | −2.29 | |||
| EW997462 | myosin II | Placopecten magellanicus | 2EC6-A | 3.65E−75 | 2.43 | −1.01 | 1.09 | −2.24 |
| EW997421 | alpha 2 actin | Dicentrarchus labrax | ACN66629 | 1.74E−28 | 2.22 | −1.02 | 1.09 | −2.07 |
fc – fold change; fc figures in bold indicate a significant difference. Resistant control (RC); susceptible control (SC); resistant exposed (RE); susceptible exposed (SE).
Differences between Schistosome-resistant and Schistosome-susceptible Strains
Secondly, comparison of haemocytes from schistosome-resistant and schistosome-susceptible snails (Fig. 3) revealed large numbers of genes to be differentially expressed between strains, before (196, comprising 96 resistant-specific, and 100 susceptible-specific transcripts) and after exposure (417, comprising 146 in susceptible and 271 in resistant snails). Additionally, 341 genes were differentially expressed regardless of schistosome infection (Fig. 3); without exception, all 187 susceptible-specific genes remained such either before or after exposure, as did all 154 resistant-specific genes (data not shown).
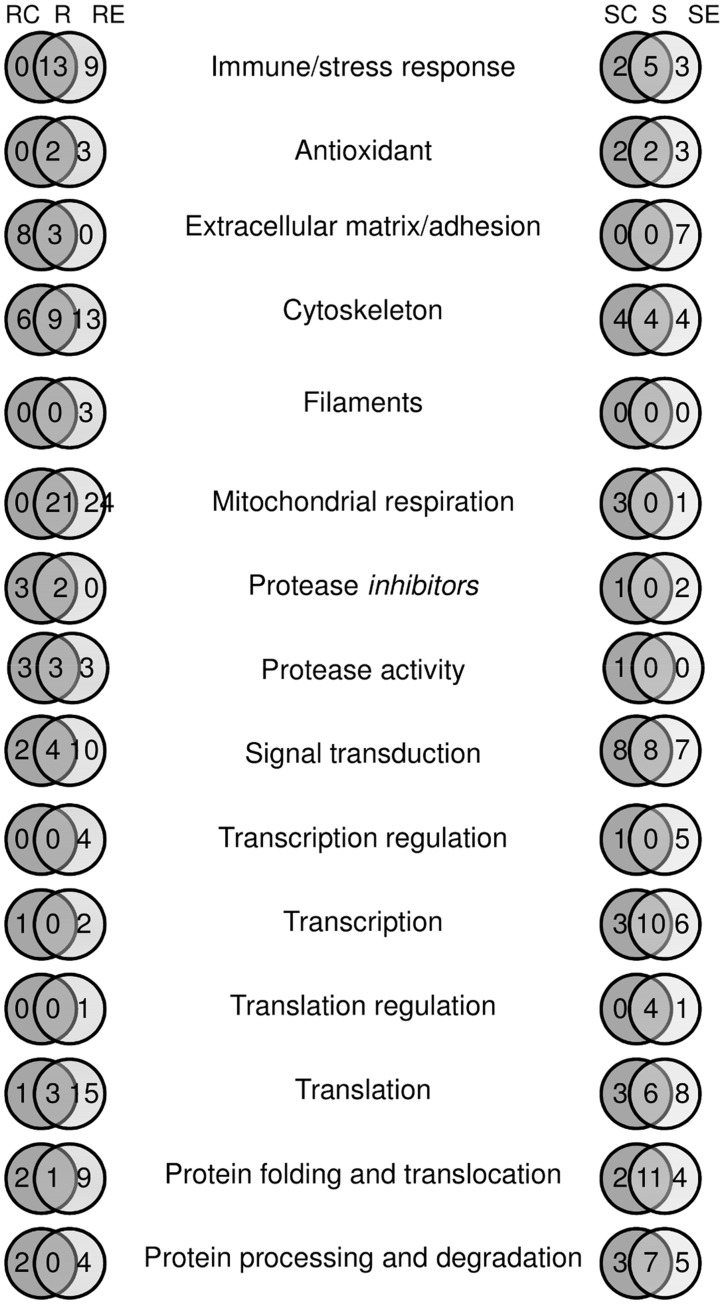
Genes found differently expressed between schistosome-resistant and schistosome-susceptible snails might be highly relevant for the interaction of those snails with S. mansoni. Differential levels of constitutive gene expression in haemocytes of the different strains before exposure may be responsible for the speedy response and elimination of S. mansoni in the resistant strain upon infection. In addition, differences between strains after exposure might give insight into the mechanism(s) of parasite elimination in resistant snails, since these are mounting a defence response; conversely, dissimilarities might indicate parasite interference with gene expression in susceptible snails. Therefore strain-specific differences should not be dismissed, although it is important to remember that other resistant and susceptible snail strains may show different responses. Gene homologues were identified for these differentially expressed transcripts and all were assigned GO annotations based on these homologies. The genes with identified GO categories were then classified into functional groups accordingly (File S2). The main functional groups represented cluster genes for known immune/stress response proteins, extracellular matrix/adhesion components, cytoskeletal proteins, mitochondrial respiratory chain proteins, signalling proteins, transcription and translation proteins and proteins that facilitate protein folding and degradation (Fig. 4). Immune/stress response genes with greater expression in resistant snail haemocytes included peptidoglycan recognition protein 1, FREPs 1 and 2, gram-negative binding protein, allograft inflammatory factor 1, heat shock protein (HSP) 40, ferritin, and glutathione-s-transferases (GSTs), all of which were differentially expressed irrespective of exposure, while FREPs 3 and 12, HSPs 70 and 90 showed greater expression post infection (File S2). Interestingly, HSP 60 was expressed to a greater extent in haemocytes of unexposed susceptible snails, with genes for the antimicrobial peptides hydramacin and neuromacin differentially expressed both before and after infection. Extracellular matrix/adhesion genes such as matrilin, dermatopontin 2, VWA domain-containing proteins and fibrillin were differentially expressed in resistant snail haemocytes with EGF-like domain containing protein, agrin, and a tandem repeat galectin showing lower expression after schistosome exposure. Unlike susceptible snails, a large number (45) of genes involved in mitochondrial respiration showed greater expression in haemocytes of resistant snails, with 24 of these showing greater expression post infection (Fig. 4). These genes included cytochrome b, cytochrome C oxidase subunits I-III, NADH dehydrogenase subunits 1, 3, 4 and 5, and ATPase subunits (File S2). In terms of signal transduction, differences were more balanced between the two strains with 16 genes showing greater expression in resistant snail haemocytes across all exposure regimes, as opposed to 23 genes in the susceptible (Fig. 4). Notable genes in the resistant phenotype included the protein tyrosine kinase src and protein kinase D, with 14-3-3 protein, G-protein coupled receptor kinase 2, twitchin, titin and nuclear factor κB (NFκB) inhibitor differentially expressed only following infection (File S2). In susceptible snail haemocytes, genes for rho GTPase activating protein, IKAP, and phosphoglycerate kinase were differentially expressed, with a dual specificity kinase and transforming growth factor β (TGFβ) receptor 1 involved only after infection (File S2). In terms of genes for cytoskeletal proteins, there was a preponderance of molecules involved in actin-related processes in haemocytes of the resistant strain compared to tubulin-related molecules in the susceptible strain (File S2); this situation persisted post-infection. Finally, encompassing all GO categories, genes that displayed the largest differences in expression (3.5-fold or greater) between snails included FREP2, Gram-negative bacteria binding protein, GSTσ, cytochrome b, NADH dehrogenase subunits 1 and 4, elastase 2, cystatin b, and endo 1,4 β glucanase in resistant snail haemoctyes, and polyprotein, endonuclease G, endonuclease mitochondrial precursor, and ATP-synthase-like protein, in susceptible snail haemocytes. HSP 90, and type 2 cystatin and fibropellin differed to such a degree only after infection for resistant and susceptible snail haemocytes, respectively (File S2).
Figure 4. B. glabrata genes in different GO categories found to be differentially expressed.
The number of differentially expressed genes in haemocytes from resistant (R) and susceptible (S) strains, before (C, control), both before and after (R/ S), and after S. mansoni exposure (E, exposed).
Gene Set Enrichment Analysis
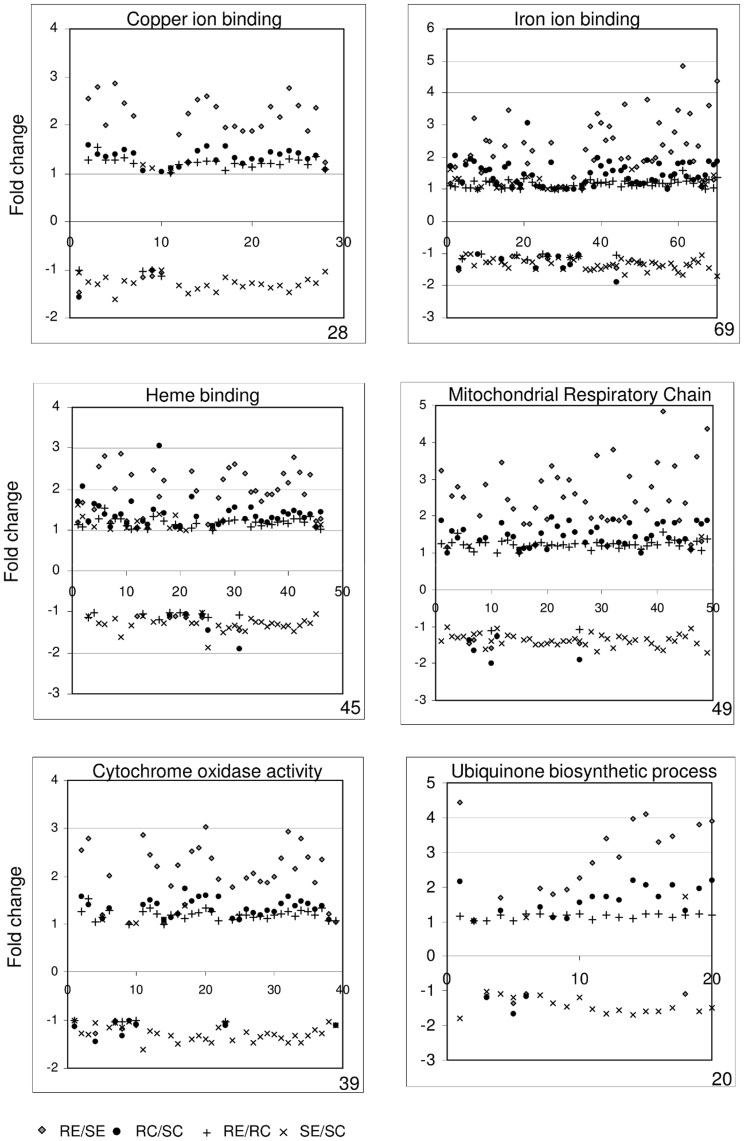
Gene set enrichment analysis (GSEA) [45], that identifies significant changes in expression of gene groups based on their function, rather than single genes, was employed for REvSE, RCvSC, RCvRE, SCvSC. We identified groups of GO terms (represented in bold in Table 3), all of which had higher expression levels in exposed resistant snails compared to unexposed (up-regulated on exposure), higher in resistant control snails compared to susceptible (strain-specific), higher in resistant exposed than susceptible exposed, and less in exposed susceptible snails than controls, (down-regulated in susceptible on exposure). Not every gene in each GO category followed the same trend, but the GSEA tests whether a significant number (more than would be expected by chance) are differently expressed in a particular category. For example, Fig. 5 illustrates 6 examples of GO categories in which the associated genes in general show positive fold changes (>+1) for REvSE, RCvSC and REvRC, and negative fold changes (>−1) for SEvSC. This indicates that even between control and unexposed snails, expression in R was greater than S, and that a significant number of the associated genes are up-regulated post-exposure in the schistosome-resistant snails, but down-regulated in the schistosome-susceptible snails; there is also, therefore, an even larger difference in expression of these genes when comparing RE with SE, with resistant being greater than susceptible. The gene groups identified as following these trends are primarily involved in mitochondrial respiratory process and ubiquinone biosynthetic processes, both indicative of increased metabolic activity consistent with mounting a defence response.
Table 3. Gene Set Enrichment Analysis (GSEA).
| Upregulation (AvB A>B) | |||||||
| GO ID | Gene Ontology | Type of GO | No.genes | REvSE | RCvSC | REvRC | SEvSC |
| GO:0009060 | aerobic respiration | biological_process | 21 | 2.78E−12 | 4.31E−09 | 1.88E E−09 | 1 |
| GO:0005507 | copper ion binding | molecular_function | 28 | 2.47E E−11 | 3.33E E−08 | 9.73E E−10 | 1 |
| GO:0004129 | cytochrome-c oxidase activity | molecular_function | 39 | 3.91E E−12 | 4.96E E−07 | 7.38E E−11 | 1 |
| GO:0009055 | electron carrier activity | molecular_function | 47 | 1.85E E−06 | 4.19E E−08 | 2.85E E−05 | 0.996695 |
| GO:0006118 | electron transport | biological_process | 106 | 3.97E E−12 | 8.96E E−11 | 8.43E E−12 | 1 |
| GO:0020037 | heme binding | molecular_function | 45 | 1.56E E−13 | 1.30E E−10 | 8.37E E−07 | 0.99999 |
| GO:0016021 | integral to membrane | cellular_component | 126 | 2.74E E−10 | 7.52E E−11 | 9.89E E−12 | 1 |
| GO:0005506 | iron ion binding | molecular_function | 69 | 0 | 2.15E E−14 | 3.33E E−12 | 1 |
| GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | biological_process | 39 | 3.91E E−12 | 4.96E E−07 | 7.38E E−11 | 1 |
| GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | biological_process | 23 | 3.43E E−05 | 0.001246 | 4.49E E−06 | 0.999995 |
| GO:0005746 | mitochondrial respiratory chain | cellular_component | 49 | 0 | 1.16E E−12 | 0 | 1 |
| GO:0008137 | NADH dehydrogenase (ubiquinone) activity | molecular_function | 19 | 9.68E E−08 | 1.41E E−05 | 2.55E E−06 | 1 |
| GO:0015992 | proton transport | biological_process | 73 | 1.15E E−12 | 8.49E E−07 | 8.75E E−12 | 1 |
| GO:0045277 | respiratory chain complex IV | cellular_component | 36 | 2.46E E−12 | 2.73E E−07 | 5.24E E−11 | 1 |
| GO:0022904 | respiratory electron transport chain | biological_process | 7 | 6.79E E−05 | 0.000196 | 0.000403 | 0.995547 |
| GO:0006814 | sodium ion transport | biological_process | 24 | 3.16E E−07 | 5.69E E−05 | 0.000109 | 0.999999 |
| GO:0006810 | transport | biological_process | 58 | 0.0003 | 7.62E E−06 | 3.29E E−05 | 0.953241 |
| GO:0006744 | ubiquinone biosynthetic process | biological_process | 20 | 3.49E E−07 | 7.20E E−06 | 1.02E E−06 | 0.999997 |
| Downregulation (AvB A<B) | |||||||
| GO ID | Gene Ontology | Type of GO | No.genes | REvSE | RCvSC | REvRC | SEvSC |
| GO:0009060 | aerobic respiration | biological_process | 21 | 1 | 1 | 1 | 3.72E E−07 |
| GO:0005507 | copper ion binding | molecular_function | 28 | 1 | 1 | 1 | 5.21E E−09 |
| GO:0005524 | ATP binding | molecular_function | 154 | 0.66052 | 0.04391 | 0.334357 | 2.19E E−06 |
| GO:0004129 | cytochrome-c oxidase activity | molecular_function | 39 | 1 | 1 | 1 | 9.28E E−10 |
| GO:0006118 | electron transport | biological_process | 106 | 1 | 1 | 1 | 3.10E E−09 |
| GO:0020037 | heme binding | molecular_function | 45 | 1 | 1 | 0.999999 | 9.86E E−06 |
| GO:0016021 | integral to membrane | cellular_component | 126 | 1 | 1 | 1 | 2.11E E−07 |
| GO:0005506 | iron ion binding | molecular_function | 69 | 1 | 1 | 1 | 1.40E E−09 |
| GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen | biological_process | 39 | 1 | 1 | 1 | 9.28E E−10 |
| GO:0006120 | mitochondrial electron transport, NADH to ubiquinone | biological_process | 23 | 0.99997 | 0.998755 | 0.999996 | 4.82E E−06 |
| GO:0005746 | mitochondrial respiratory chain | cellular_component | 49 | 1 | 1 | 1 | 0 |
| GO:0005739 | mitochondrion | cellular_component | 48 | 0.93988 | 0.800927 | 0.817296 | 8.18E E−06 |
| GO:0008137 | NADH dehydrogenase (ubiquinone) activity | molecular_function | 19 | 1 | 0.999986 | 0.999997 | 2.42E E−07 |
| GO:0015992 | proton transport | biological_process | 73 | 1 | 0.999999 | 1 | 5.85E E−14 |
| GO:0045277 | respiratory chain complex IV | cellular_component | 36 | 1 | 1 | 1 | 9.42E E−11 |
| GO:0006814 | sodium ion transport | biological_process | 24 | 1 | 0.999943 | 0.999891 | 8.06E E−07 |
| GO:0006744 | ubiquinone biosynthetic process | biological_process | 20 | 1 | 0.999993 | 0.999999 | 2.96E E−06 |
| GO:0005840 | ribosome | cellular_component | 85 | 0.03859 | 1.20E E−05 | 0.723301 | 0.000821 |
| GO:0006457 | protein folding | biological_process | 21 | 0.00041 | 3.17E E−06 | 0.55687 | 0.00087 |
| GO:0006094 | gluconeogenesis | biological_process | 22 | 0.01078 | 3.09E E−05 | 0.163101 | 0.002104 |
| GO:0005634 | nucleus | cellular_component | 93 | 0.00057 | 7.58E E−08 | 0.533302 | 0.00296 |
| GO:0006096 | glycolysis | biological_process | 20 | 0.00633 | 2.13E E−05 | 0.220296 | 0.003285 |
| GO:0003735 | structural constituent of ribosome | molecular_function | 105 | 0.05398 | 6.79E E−06 | 0.970198 | 0.021822 |
| GO:0006412 | translation | biological_process | 98 | 0.04483 | 6.10E E−05 | 0.88094 | 0.028665 |
| GO:0042254 | ribosome biogenesis | biological_process | 108 | 0.04124 | 2.39E E−06 | 0.989179 | 0.03393 |
| GO:0005615 | extracellular space | cellular_component | 26 | 0.00024 | 0.000126 | 0.019181 | 0.044914 |
| GO:0005515 | protein binding | molecular_function | 240 | 0.00035 | 3.62E E−06 | 0.836396 | 0.33465 |
| GO:0003743 | translation initiation factor activity | molecular_function | 9 | 5.79E E−05 | 8.26E E−06 | 0.278141 | 0.050926 |
| GO:0006446 | regulation of translational initiation | biological_process | 8 | 9.10E E−05 | 1.99E E−05 | 0.249216 | 0.074297 |
| GO:0051082 | unfolded protein binding | molecular_function | 27 | 0.00013 | 2.34E E−05 | 0.878669 | 0.135328 |
| GO:0003924 | GTPase activity | molecular_function | 67 | 9.99E E−05 | 9.11E E−05 | 0.262811 | 0.315466 |
| GO:0007018 | microtubule-based movement | biological_process | 47 | 9.39E E−06 | 3.30E E−05 | 0.422782 | 0.744616 |
| GO:0005874 | microtubule | cellular_component | 59 | 6.63E E−06 | 1.04E E−05 | 0.440339 | 0.745536 |
| GO:0003723 | RNA binding | molecular_function | 55 | 1.00E E−05 | 1.02E E−05 | 0.730866 | 0.809142 |
| GO:0051258 | protein polymerization | biological_process | 32 | 3.37E E−06 | 0.000186 | 0.848843 | 0.988085 |
The gene ontologies (GO) listed were significantly different (adjusted p <0.01) in at least one of the 4 comparisons. Significant p values (Bonferri adjusted) are shown in bold. GOs that were found to be up-regulated in resistant snails and down-regulated in susceptible are also shown in bold.
Figure 5. Gene set enrichment analysis (GSEA).
The collated fold changes for each comparison (RE/SE, RC/SC, RE/RC, SE/SC) is shown for the genes in six selected GO categories. The order of the genes along the x-axis is arbitrary.
FatiScan Analysis to Identify Trends in Gene Expression
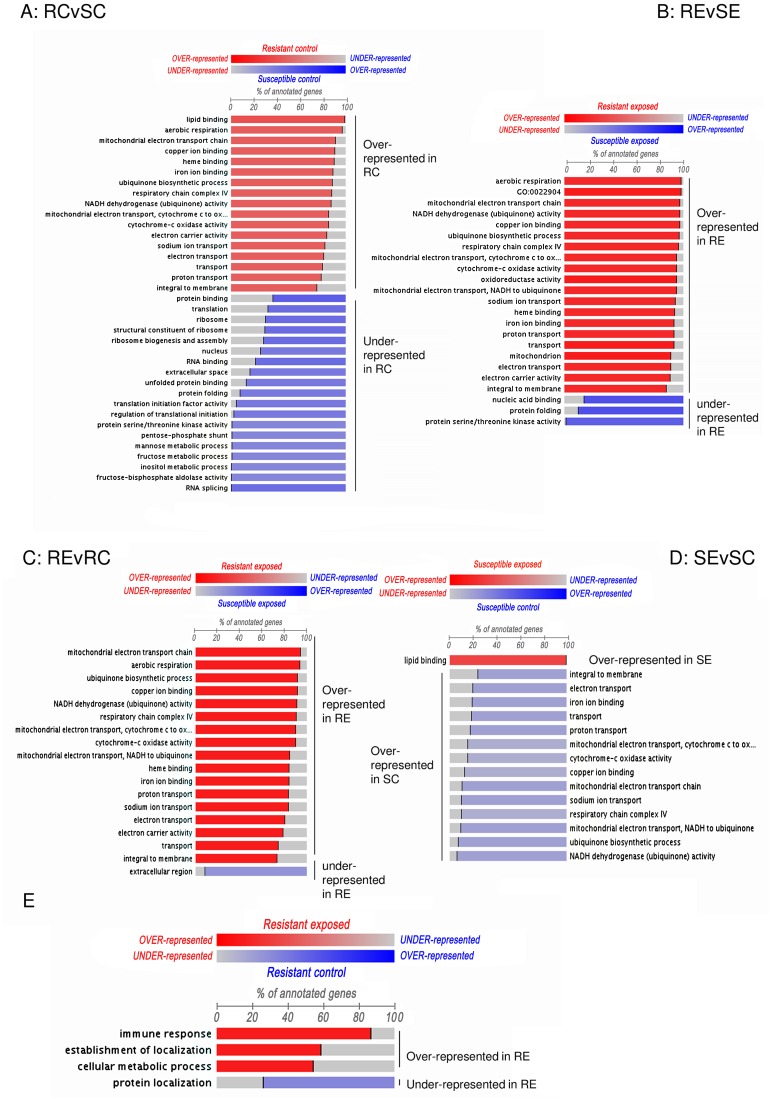
FatiScan analysis [46] was used to detect asymmetrical distribution of GO categories from the fold change ranked list for each of the 4 haemocyte comparisons (RCvSC, REvSE, REvRC, and SEvSC). Distinct differences were identified between the snail strains both before and after infection, and the types of genes found to differ between haemocytes confirm the GSEA results (Fig. 6 A–B). Most striking, however, were the differences in response to the schistosome shown by the snail strains. Haemocytes of resistant snails exposed to S. mansoni showed an over-representation of haemocyte genes involved in mitochondrial respiratory processes and ubiquinone biosynthetic processes (Fig. 6C), demonstrating these are up-regulated in response to the parasite (as well as being already elevated in control resistant snails compared to susceptible), while in haemocytes of the susceptible snails these same genes were under-represented (Fig. 6D) indicating that their expression was suppressed by the parasite. This is also consistent with the findings of the GSEA analysis. FatiScan analysis of level 3 GO annotations also identified that the resistant exposed snails switched on immune response genes upon exposure to S. mansoni (Fig. 6E), again showing an active response to infection.
Figure 6. FatiScan analysis illustrating gene expression fold change ranked lists from B. glabrata microarray comparisons using custom array annotation.
A. Resistant control (RC) compared to susceptible control (SC). B. Resistant exposed (RE) compared to susceptible exposed (SE). C. Resistant exposed (RE) to resistant control (RC). D. Susceptible exposed (SE) compared to susceptible control (SC). E. RE compared to RC using level 3 GO annotations. Significantly differentially represented GO categories are listed for each comparison. Bar leading from the left, with GO category label on the left indicates over representation in the upper category (shown above each diagram), label on the right means under represented. Bar leading from the right and label on the right indicates over-representation in the lower category and label on the left under-represented.
Discussion
The genes differentially expressed between haemocytes of the schistosome-susceptible and schistosome-resistant B. glabrata strains offer great insight into the complex molecular processes that are involved in the defence response to the parasite. Fifty-nine of the 98 genes that we identified in our initial investigation of strain differences using the considerably smaller (2K versus 5K) previous array platform [36] have been confirmed, and added to, by the current experiment. Again, we observed differential expression (elevated in resistant snails) of genes involved in energy metabolism, and transcription and translation indicating a general increase in cellular activity, consistent with generating the necessary components for mounting a defence response. Perhaps of greater interest is the finding of a considerably different response in susceptible and resistant snail strains as early as 2 hr post exposure to S. mansoni.
Stress response genes were identified in the set of genes that were present in the resistant snails both before and after exposure, as well as in the exposed resistant snails. FREPs, a unique family of molluscan calcium-dependent lectins, are known to be up-regulated following parasite infection and to bind to parasite surfaces [9], likely through interaction with parasite mucins [21], [22]. Knockdown of FREP3 in B. glabrata resistant to E. paraensei by RNA interference (RNAi) resulted in a phenotype switch, whereby 31% of RNAi-treated snails became susceptible to the parasite [11]. Suppression of FREP3 in B. glabrata resistant to S. mansoni has also been shown to increase susceptibility to this schistosome with 20% of snails becoming infected [20]. Thus FREP3 seems to play some role in defence against S. mansoni concordant with the view that the primary function of fibrinogen-domain containing proteins in invertebrates is in protection against infection, rather than coagulation [62]. Here, we have demonstrated that FREP3 and FREP12 expression was greater in haemocytes of resistant snails compared to susceptible snails post miracidial exposure, whereas FREPS 1 and 2 were differentially expressed irrespective of exposure. Thus it seems possible that in the B. glabrata/S. mansoni infection model, FREP3 might represent a molecule vital to the maintenance of the resistance phenotype and that high levels of FREP expression in general might facilitate early parasite recognition.
Expression of a HSP 70 gene was also greater in resistant snail haemocytes after schistosome infection confirming of our previous findings [36], [42], but this contrasts with the findings of Ittiprasert et al (2009), who demonstrated up-regulation of HSP70 in susceptible juvenile snails but not resistant [63]. Temperature stresses have also recently been suggested to affect susceptibility of snails to schistosome infection in conjunction with changes to expression levels of HSP transcripts [64]. Interestingly Zahoor et al (2010) demonstrated that S. mansoni ESPs, derived from larvae transforming from miracidia to mother sporocysts, reduced the quantity of HSP 70 protein in haemocytes of both snail strains 1 h after exposure to ESPs and that HSP 70 protein levels were also lower 35 days after infection [48]. Given that this molecule has important intracellular chaperone and extracellular immunomodulatory capacities [65], it would be valuable to elucidate the temporal dynamics of schistosome infection on HSP 70 gene and protein expression in detail together with that for the three other differentially expressed HSP genes, in order to fully understand the role they may play in snail responses to schistosome infection.
Differential expression between haemocytes of resistant and susceptible control snails, of matrillin, dermatopontin and other transcripts involved in cell adhesion may also have a significant bearing on host-parasite interactions. Bouchut et al (2006) investigated gene expression of several cell adhesion genes in B. glabrata strains resistant or susceptible to E. caproni and found dermatopontin 2 and matrillin to be differentially expressed [66]; however, in their snail strains matrillin was over expressed in snails susceptible to a different parasite, E. caproni, and they hypothesized that susceptible snails may possess a more potent haemolymph coagulation system preventing or slowing haemocyte migration [66]. Here we identified six of the genes investigated by these authors as being significantly present in resistant control snails and three (two the same) that were different between haemocytes of resistant snails and susceptible snails both before and after exposure. Although different expression patterns were found in this study, it may emphasize the importance of these genes in snail-parasite interactions, although their roles are not currently clear. That fewer differentially expressed genes were found after exposure may suggest that the susceptible snails up-regulated these genes, whereas the resistant snails already expressed these genes, even in control snails. A gene for tandem repeat galectin showed greater expression in susceptible snail haemocytes after S. mansoni infection. This finding is curious given that these molecules are expressed on the surface of ∼60% of B. glabrata haemocytes, that they bind the S. mansoni sporocyst tegument via interaction with surface-exposed LacNAc sugars, and that susceptible snail haemocytes do not encapsulate S. mansoni miracidia/sporocysts [33]. However, if developing larvae release sufficient LacNAc into the haemolymph during transformation, binding to the haemocyte receptors could serve to dampen recognition of and/or responses to the parasite either directly, or through interference of haemocyte signalling mechanisms as demonstrated for other sugar molecules [67], [68]. Cell surface receptors such as integrins bind to extracellular matrix components and facilitate cell migration through tissues towards invading pathogens. These events are communicated intracellularly and cell movements, including encapsulation and phagocytosis, are then facilitated by actin and a variety of actin binding proteins. In this context it is notable that resistant snail haemocytes displayed a preponderance of genes involved in actin-related processes either in the presence or absence of schistosome infection. This implies that these haemocytes might display enhanced phagocytic and migratory capacity when compared to their susceptible snail counterparts benefiting the anti-parasite response.
The GSEA and FatiScan analyses also highlight that genes involved in mitochondrial respiration and ubiquinone degradation were already active in haemocytes of the resistant snails prior to schistosome exposure and that in these snails they were further activated, while in the susceptible snails they were suppressed, demonstrating a significant difference in the response of resistant and susceptible snails to schistosome exposure. Genes required for copper, iron and heme-binding were also differentially expressed, although they may function in energy production too; for example, haemoglobins that possess a high oxygen affinity are present as blood respiratory proteins in B. glabrata [69]. Hanelt et al (2008) also found indications of up-regulation of heme and metal ion-binding in response to bacterial and S. mansoni challenge (12hr post-exposure) [14]. Ferritin, also identified in resistant snails in earlier gene expression studies [44] and recently identified in an RNA-seq based approach to identify immune responses following bacterial or yeast challenges in B. glabrata [70] stores iron in a non-toxic form, enabling deposition of iron in a safe form and transport to areas where iron is required. Ferrous iron (Fe2+) is toxic to cells as it acts as a catalyst in the formation of the hydroxyl free radical (OH•) from hydrogen peroxide (H2O2). Given the importance of these reactive oxygen species to killing of S. mansoni sporocysts [8], [28], [71], greater ferritin expression could be relevant in terms of its capacity to affect the cellular balance of H2O2 and OH• possibly influencing the outcome of infection. In the current study, resistant snail haemocytes were found to also differentially express GSTs that would serve as antioxidants to prevent cellular damage to the haemocytes. In the context of energy production, genes involved in respiratory chain and ATP production have been shown to differ between two oyster species that vary in their response to the parasite Perkinsus marinus [72]. The above suggests that the resistant snails were already expressing many of the genes required for defence responses prior to exposure.
This study provides the first evidence from global gene expression analyses that not only is there is a fundamental difference in the defence physiology between the snail strains used here before infection, but that the resistant snails actively respond to the schistosome, while the susceptible snails react in an opposing fashion by suppressing expression of the types of genes which are activated in a responding (resistant) snail, consistent with the notion that the schistosome is producing molecules that interfere with the snail’s defence response [68]. Hanington et al (2010) found that an initial (0.5–2 day post infection) up-regulation of immune/stress response genes in susceptible B. glabrata was followed by a stronger down-regulation later during infection with S. mansoni, that was in contrast to E. paraensei exposed snails which showed down-regulation from 0.5 day post-infection [38]. They concluded that both parasites were able to interfere with host defense responses, but that E. paraensei was able to do this more rapidly and robustly than S. mansoni. Our results also suggest interference by the parasite, but in contrast to Hanington et al (2010) [38], we show that this phenomenon is occurring only 2 hours after exposure. Such early inactivation is coincident with early post-embryonic development of the parasitic sporocyst larval stage, a crucial phase when the schistosome lays down a new tegument and is perhaps more exposed to the host immune system while the snail is exposed to ESPs and ciliated plates released from schistosomes during their development [73].
In conclusion, this microarray experiment, by determining the expression of a large number of genes simultaneously, many more than can be investigated by qPCR, has enabled the construction of a framework of processes involved in haemocyte responses during the first phase of schistosome infection. The resistant snails, even before infection, express many different genes compared with susceptible snails and in many respects seem to be primed and ready to respond to schistosome attack. The resistant snails also demonstrated activation of defence processes, while the susceptible snails displayed inactivation. Pinpointing individual genes significantly affected by parasite exposure may have been made more difficult by biological variation both in schistosome penetration time (after addition of miracidia to snail water) and in individual snail responses since to obtain sufficient material it was necessary to pool haemocytes. Alternatively, the actual gene expression changes at this early stage after schistosome exposure, may be subtle and therefore difficult to detect resulting in a skewed outcome whereby strain-specific differences in gene expression outweighed parasite-induced changes. Nevertheless, by integrating GSEA and FatiScan analysis, the outcomes detailed in this paper have enabled a holistic view of changes in gene expression as a consequence of phenotype and exposure regime. Statistical analysis of clustered gene expression within any particular category has provided enhanced confidence in the relative importance of changes that might result in altered cellular physiology. In this way, this study provides a new way of assessing the complex biology of snail-schistosome interactions giving insight that will help future studies to identify mechanisms of compatibility in this fascinating host-parasite system.
Supporting Information
Annotation file for the genes represented on the 5 K B. glabrata microarray.
(TXT)
Genes identified as significantly differentially expressed between resistant and susceptible B. glabrata snails, before exposure (C-control), after exposure to S. mansoni (E-exposed) and both before and after (not affected by exposure). Genes with no known homologues are not shown. fc- fold change, figure in grey = no significant difference. *Genes previously identified as being significantly different between schistosome-resistant and -susceptible strains of B. glabrata [36]
(DOC)
Acknowledgments
We would like to thank Jayne King and Mike Anderson, NHM, for snail and parasite culture, and Julia Llewellyn-Hughes and Steve Llewellyn-Hughes for using the Microlab Star robotic work station (Hamilton) to pick the clones. Microarrays were printed at the Department of Pathology, Cambridge University, by Anthony Brown.
Funding Statement
This work was carried out with funding from the Wellcome Trust (068589/Z/02/Z). CMA and BH acknowledge support from the United States National Institutes of Health (NIH), USA R01AI052363 (CMA) and COBRE 1P20RR018754 IdEA, NCRR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chitsulo L, Loverde R, Engels D, Barakat R, Colley D, et al. (2004) Schistosomiasis. Nature Reviews Microbiology 2: 12–13. [DOI] [PubMed] [Google Scholar]
- 2. Webster JP, Davies CM (2001) Coevolution and compatibility in the snail-schistosome system. Parasitology 123: S41–S56. [DOI] [PubMed] [Google Scholar]
- 3. El-Ansary A, Al-Daihan S (2006) Important aspects of Biomphalaria snail-schistosome interactions as targets for antischistosome drug. Medical Science Monitor 12: RA282–RA292. [PubMed] [Google Scholar]
- 4. Lafferty KD, Kuris AM (2009) Parasitic castration: the evolution and ecology of body snatchers. Trends in Parasitology 25: 564–572 doi:10.1016/j.pt.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 5. Webster JP, Woolhouse MEJ (1999) Cost of resistance: relationship between reduced fertility and increased resistance in a snail-schistosome host-parasite system. Proc R Soc Lond Ser B-Biol Sci 266: 391–396. [Google Scholar]
- 6. Lewis FA, Stirewalt M, Souza C, Gazzinelli G (1986) Large scale laboratory maintenance of Schistosoma mansoni with observations on 3 schistosome snail host combinations. J Parasitol 72: 813–829 doi:10.2307/3281829. [PubMed] [Google Scholar]
- 7. Lockyer AE, Jones CS, Noble LR, Rollinson D (2004) Trematodes and snails: an intimate association. Canadian Journal of Zoology 82: 251–269. [Google Scholar]
- 8. Bayne CJ (2009) Successful parasitism of vector snail Biomphalaria glabrata by the human blood fluke (trematode) Schistosoma mansoni: A 2009 assessment. Molecular and Biochemical Parasitology 165: 8–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loker ES (2011) Gastropod immunobiology. Adv Exp Med Biol 708: 17–43 doi:10.1007/978-1-4419-8059-5_2. [DOI] [PubMed] [Google Scholar]
- 10. Newton WL (1953) The inheritance of susceptibility to infection with Schistosoma mansoni in Australorbis glabratus . Experimental Parasitology 2: 242–257. [Google Scholar]
- 11. Theron A, Coustau C (2005) Are Biomphalaria snails resistant to Schistosoma mansoni? Journal of Helminthology 79: 187–191. [DOI] [PubMed] [Google Scholar]
- 12.Adema CM, Loker ES (1997) Specificity and immunobiology of larval digenean snail associations. Advances in Trematode Biology: 230 – 253.
- 13. Yoshino TP, Bayne CJ (1983) Mimicry of snail host antigens by miracidia and primary sporocysts of Schistosoma mansoni . Parasite Immunol 5: 317–328. [DOI] [PubMed] [Google Scholar]
- 14. Hanelt B, Lun CM, Adema CM (2008) Comparative ORESTES-sampling of transcriptomes of immune-challenged Biomphalaria glabrata snails. Journal of Invertebrate Pathology 99: 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boswell CA, Bayne CJ (1985) Schistosoma mansoni - lectin-dependent cyto-toxicity of hemocytes from susceptible host snails, Biomphalaria glabrata . Exp Parasitol 60: 133–138. [DOI] [PubMed] [Google Scholar]
- 16. Lie K, Heyneman D, Jeong K (1976) Studies on resistance in snails. 7. Evidence of interference with defense reaction in Biomphalaria glabrata by trematode larvae. J Parasitol 62: 608–615 doi:10.2307/3279428. [PubMed] [Google Scholar]
- 17. Loker ES, Cimino DF, Hertel LA (1992) Excretory-secretory products of Echinostoma paraensei sporocysts mediate interference with Biomphalaria glabrata hemocyte functions. J Parasitol 78: 104–115. [PubMed] [Google Scholar]
- 18. Zhang SM, Adema CM, Kepler TB, Loker ES (2004) Diversification of Ig superfamily genes in an invertebrate. Science 305: 251–254. [DOI] [PubMed] [Google Scholar]
- 19. Hanington PC, Forys MA, Dragoo JW, Zhang S-M, Adema CM, et al. (2010) Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proceedings of the National Academy of Sciences of the United States of America 107: 21087–21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanington PC, Forys MA, Loker ES (2012) A somatically diversified defense factor, FREP3, is a determinant of snail resistance to schistosome infection. Plos Neglect Trop Dis 6. doi:10.1371/journal.pntd.0001591. [DOI] [PMC free article] [PubMed]
- 21.Mone Y, Gourbal B, Duval D, Du Pasquier L, Kieffer-Jaquinod S, et al.. (2010) //000282271300016. [DOI] [PMC free article] [PubMed]
- 22. Mitta G, Adema CM, Gourbal B, Loker ES, Theron A (2012) Compatibility polymorphism in snail/schistosome interactions: From field to theory to molecular mechanisms. Dev Comp Immunol 37: 1–8 doi:10.1016/j.dci.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bayne CJ, Hahn UK, Bender RC (2001) Mechanisms of molluscan host resistance and of parasite strategies for survival. Parasitology 123: S159–167. [DOI] [PubMed] [Google Scholar]
- 24. Humphries JE, Yoshino TP (2008) Regulation of hydrogen peroxide release in circulating hemocytes of the planorbid snail Biomphalaria glabrata . Developmental and Comparative Immunology 32: 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zahoor Z, Davies A, Kirk R, Rollinson D, Walker A (2009) Nitric oxide production by Biomphalaria glabrata haemocytes: effects of Schistosoma mansoni; ESPs and regulation through the extracellular signal-regulated kinase pathway. Parasites & Vectors 2: 1–10 doi:10.1186/1756-3305-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goodall CP, Bender RC, Brooks JK, Bayne CJ (2006) Biomphalaria glabrata cytosolic copper/zinc superoxide dismutase (SOD1) gene: Association of SOD1 alleles with resistance/susceptibility to Schistosoma mansoni . Molecular and Biochemical Parasitology 147: 207–210. [DOI] [PubMed] [Google Scholar]
- 27. Bender RC, Goodall CP, Blouin MS, Bayne CJ (2007) Variation in expression of Biomphalaria glabrata SOD1: A potential controlling factor in susceptibility/resistance to Schistosoma mansoni . Developmental & Comparative Immunology 31: 874–878. [DOI] [PubMed] [Google Scholar]
- 28. Mone Y, Ribou A-C, Cosseau C, Duval D, Theron A, et al. (2011) An example of molecular co-evolution: Reactive oxygen species (ROS) and ROS scavenger levels in Schistosoma mansoni/Biomphalaria glabrata interactions. International Journal for Parasitology 41: 721–730. [DOI] [PubMed] [Google Scholar]
- 29. Davids BJ, Wu XJ, Yoshino TP (1999) Cloning of a beta integrin subunit cDNA from an embryonic cell line derived from the freshwater mollusc, Biomphalaria glabrata . Gene 228: 213–223. [DOI] [PubMed] [Google Scholar]
- 30. Davids BJ, Yoshino TP (1998) Integrin-like RGD-dependent binding mechanism involved in the spreading response of circulating molluscan phagocytes. Dev Comp Immunol 22: 39–53. [DOI] [PubMed] [Google Scholar]
- 31. Plows LD, Cook RT, Davies AJ, Walker AJ (2006) Integrin engagement modulates the phosphorylation of focal adhesion kinase, phagocytosis, and cell spreading in molluscan defence cells. Biochimica Et Biophysica Acta-Molecular Cell Research 1763: 779–786. [DOI] [PubMed] [Google Scholar]
- 32. Walker AJ, Lacchini AH, Sealey KL, Mackintosh D, Davies AJ (2010) Spreading by snail (Lymnaea stagnalis) defence cells is regulated through integrated PKC, FAK and Src signalling. Cell and Tissue Research 341: 131–145. [DOI] [PubMed] [Google Scholar]
- 33. Yoshino TP, Dinguirard N, Kunert J, Hokke CH (2008) Molecular and functional characterization of a tandem-repeat galectin from the freshwater snail Biomphalaria glabrata, intermediate host of the human blood fluke Schistosoma mansoni . Gene 411: 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zahoor Z, Davies AJ, Kirk RS, Rollinson D, Walker AJ (2008) Disruption of ERK signalling in Biomphalaria glabrata defence cells by Schistosoma mansoni: Implications for parasite survival in the snail host. Developmental and Comparative Immunology 32: 1561–1571. [DOI] [PubMed] [Google Scholar]
- 35.Baeza Garcia A, Pierce RJ, Gourbal B, Werkmeister E, Colinet D, et al.. (2010) //20886098. [DOI] [PMC free article] [PubMed]
- 36. Lockyer AE, Spinks J, Kane RA, Hoffmann KF, Fitzpatrick JM, et al. (2008) Biomphalaria glabrata transcriptome: cDNA microarray profiling identifies resistant- and susceptible-specific gene expression in haemocytes from snail strains exposed to Schistosoma mansoni . BMC Genomics 9: 634 doi:10.1186/1471-2164-9-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adema CM, Hanington PC, Lun C-M, Rosenberg GH, Aragon AD, et al. (2010) Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes). Mol Immunol 47: 849–860 doi:10.1016/j.molimm.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hanington PC, Lun C-M, Adema CM, Loker ES (2010) Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei . International Journal for Parasitology 40: 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lockhart DJ, Winzeler EA (2000) Genomics, gene expression and DNA arrays. Nature 405: 827–836. [DOI] [PubMed] [Google Scholar]
- 40. Lockyer AE, Spinks JN, Walker AJ, Kane RA, Noble LR, et al. (2007) Biomphalaria glabrata transcriptome: identification of cell-signalling, transcriptional control and immune-related genes from open reading frame expressed sequence tags (ORESTES). Dev Comp Immunol 31: 763–782 doi:10.1016/j.dci.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lockyer AE, Jones CS, Noble LR, Rollinson D (2000) Use of differential display to detect changes in gene expression in the intermediate snail host Biomphalaria glabrata upon infection with Schistosoma mansoni . Parasitology 120: 399–407. [DOI] [PubMed] [Google Scholar]
- 42. Lockyer AE, Noble LR, Rollinson D, Jones CS (2004) Schistosoma mansoni: resistant specific infection-induced gene expression in Biomphalaria glabrata identified by fluorescent-based differential display. Exp Parasitol 107: 97–104 doi:10.1016/j.exppara.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 43. Jones CS, Lockyer AE, Rollinson D, Noble LR (2001) Molecular approaches in the study of Biomphalaria glabrata-Schistosoma mansoni interactions: linkage analysis and gene expression profiling. Parasitology 123 Suppl: S181–196 [DOI] [PubMed] [Google Scholar]
- 44. Lockyer AE, Spinks J, Noble LR, Rollinson D, Jones CS (2007) Identification of genes involved in interactions between Biomphalaria glabrata and Schistosoma mansoni by suppression subtractive hybridization. Mol Biochem Parasitol 151: 18–27 doi:10.1016/j.molbiopara.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, et al. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Shahrour F, Arbiza L, Dopazo H, Huerta-Cepas J, Minguez P, et al.. (2007) //000245804600001. [DOI] [PMC free article] [PubMed]
- 47. Pan SCT (1996) Schistosoma mansoni: The ultrastructure of larval morphogenesis in Biomphalaria glabrata and of associated host-parasite interactions. Jpn J Med Sci Biol 49: 129–149. [DOI] [PubMed] [Google Scholar]
- 48. Zahoor Z, Davies AJ, Kirk RS, Rollinson D, Walker AJ (2010) Larval excretory-secretory products from the parasite Schistosoma mansoni modulate HSP70 protein expression in defence cells of its snail host, Biomphalaria glabrata . Cell Stress & Chaperones 15: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mitta G, Galinier R, Tisseyre P, Allienne JF, Girerd-Chambaz Y, et al. (2005) Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Developmental and Comparative Immunology 29: 393–407. [DOI] [PubMed] [Google Scholar]
- 50. Hertel LA, Adema CM, Loker ES (2005) Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Echinostoma paraensei . Developmental and Comparative Immunology 29: 295–303. [DOI] [PubMed] [Google Scholar]
- 51. Jung Y, Nowak TS, Zhang SM, Hertel LA, Loker ES, et al. (2005) Manganese superoxide dismutase from Biomphalaria glabrata . Journal of Invertebrate Pathology 90: 59–63. [DOI] [PubMed] [Google Scholar]
- 52. Paraense WL, Correa LR (1963) Variation in susceptibility of populations of Austrolorbis glabratus to a strain of Schistosoma mansoni . Revista do Instituto Medicina Tropica Sao Paulo 5: 15–22. [PubMed] [Google Scholar]
- 53.Adema CM, Luo MZ, Hanelt B, Hertel LA, Marshall JJ, et al. (2006) A bacterial artificial chromosome library for Biomphalaria glabrata, intermediate snail host of Schistosoma mansoni. Mem Inst Oswaldo Cruz 101: 167 – 177. [DOI] [PubMed]
- 54. Petalidis L, Bhattacharyya S, Morris GA, Collins VP, Freeman TC, et al. (2003) Global amplification of mRNA by template-switching PCR: linearity and application to microarray analysis. Nucl Acids Res 31: e142–e142 doi:10.1093/nar/gng142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kerr MK, Churchill GA (2001) Experimental design for gene expression microarrays. Biostat 2: 183–201. [DOI] [PubMed] [Google Scholar]
- 56.Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer. 397–420.
- 57. Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31: 265–273. [DOI] [PubMed] [Google Scholar]
- 58.Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 3: Article 3. [DOI] [PubMed]
- 59. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 57: 289–300. [Google Scholar]
- 60. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 61. Medina I, Carbonell J, Pulido L, Madeira SC, Goetz S, et al. (2010) Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Research 38: W210–W213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hanington PC, Zhang S-M (2011) The primary role of fibrinogen-related proteins in invertebrates is defense, not coagulation. J Innate Immun 3: 17–27 doi:10.1159/000321882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ittiprasert W, Nene R, Miller A, Raghavan N, Lewis F, et al. (2009) Schistosoma mansoni infection of juvenile Biomphalaria glabrata induces a differential stress response between resistant and susceptible snails. Exp Parasitol 123: 203–211 doi:10.1016/j.exppara.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ittiprasert W, Knight M (2012) Reversing the resistance phenotype of the Biomphalaria glabrata snail host Schistosoma mansoni infection by temperature modulation. PLoS Pathog 8. doi:10.1371/journal.ppat.1002677. [DOI] [PMC free article] [PubMed]
- 65.Van Eden W, Wick G, Albani S, Cohen I (2007) //000251158200020. [DOI] [PubMed]
- 66. Bouchut A, Roger E, Coustau C, Gourbal B, Mitta G (2006) Compatibility in the Biomphalaria glabrata/Echinostoma caproni model: Potential involvement of adhesion genes. International Journal for Parasitology 36: 175–184. [DOI] [PubMed] [Google Scholar]
- 67. Plows LD, Cook RT, Davies AJ, Walker AJ (2005) Carbohydrates that mimic schistosome surface coat components affect ERK and PKC signalling in Lymnaea stagnalis haemocytes. International Journal for Parasitology 35: 293–302. [DOI] [PubMed] [Google Scholar]
- 68. Walker AJ (2006) Do trematode parasites disrupt defence-cell signalling in their snail hosts? Trends in Parasitology 22: 154–159. [DOI] [PubMed] [Google Scholar]
- 69. Lieb B, Dimitrova K, Kang HS, Braun S, Gebauer W, et al. (2006) Red blood with blue-blood ancestry: Intriguing structure of a snail hemoglobin. Proceedings of the National Academy of Sciences of the United States of America 103: 12011–12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deleury E, Dubreuil G, Elangovan N, Wajnberg E, Reichhart J-M, et al.. (2012) Specific versus non-specific immune responses in an invertebrate species evidenced by a comparative de novo sequencing study. PLoS One 7. doi:10.1371/journal.pone.0032512. [DOI] [PMC free article] [PubMed]
- 71. Mourao M, Dinguirard N, Franco GR, Yoshino TP (2009) Role of the endogenous antioxidant system in the protection of Schistosoma mansoni primary sporocysts against exogenous oxidative stress. PLoS neglected tropical diseases 3: e550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tanguy A, Guo XM, Ford SE (2004) Discovery of genes expressed in response to Perkinsus marinus challenge in Eastern (Crassostrea virginica) and Pacific (C. gigas) oysters. Gene 338: 121–131. [DOI] [PubMed] [Google Scholar]
- 73. Walker A (2011) Insights into the functional biology of schistosomes. Parasites & Vectors 4: 1–6 doi:10.1186/1756-3305-4-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotation file for the genes represented on the 5 K B. glabrata microarray.
(TXT)
Genes identified as significantly differentially expressed between resistant and susceptible B. glabrata snails, before exposure (C-control), after exposure to S. mansoni (E-exposed) and both before and after (not affected by exposure). Genes with no known homologues are not shown. fc- fold change, figure in grey = no significant difference. *Genes previously identified as being significantly different between schistosome-resistant and -susceptible strains of B. glabrata [36]
(DOC)