Abstract
While cervical cancer screening relies on cervical cytology and high-risk HPV detection, histologic diagnosis, and specifically lesion grade, is the main parameter that drives clinical management of screen-positive women. Morphologically diagnosed squamous intraepithelial lesions (SIL/CIN) regress spontaneously in more than half of the cases, but identifying those likely to persist and progress is not currently possible based upon morphology. Lack of major capsid protein L1 expression has been suggested as a feature in progressive lesions, while expression of the minor capsid protein L2 has not been extensively evaluated. The goal of this study is to evaluate immunohistochemical expression of L1 and L2 in SILs in correlation with lesion grade.
One hundred fifty cervical specimens with SILs were selected based on HPV 16 or HPV 18 detection by Q-PCR. These included 89 low-grade SILs (LSIL/CIN1) and 123 high-grade SILs (75 HSIL/CIN2 and 48 HSIL/CIN3). More than one lesion/grade was identified in 53 specimens. The presence and grade of SIL was determined by a panel of pathologists. Capsid protein expression was assessed by immunohistochemistry using MAB 837 for L1 and RG-1 for L2. Lesions of different grades in the same specimen were scored separately.
Expression of capsid proteins was detected in 34/89 (40%) LSIL/CIN1, 5/75 (6%) HSIL/CIN2 and none of 48 HSIL/CIN3. L1 and L2 were co-expressed in the same area of the lesion in 22 cases. In addition, L1 alone was expressed in 6 lesions and L2 alone in 11 lesions. Among the cases with multiple lesion grades in the same specimen, none with HSIL/CIN 3 expressed capsid proteins in any portion/grade of the lesion.
HPV capsid proteins are expressed almost exclusively in LSIL/CIN1 and rarely in HSIL/CIN 2. Additional studies are warranted to examine lack of L1 and L2 expression in LSIL/CIN1 as a predictor of persistence or progression to HSIL/CIN 3, the precursor of cervical cancer.
Keywords: cervical cancer precursors, biomarkers, HPV, capsid proteins, immunohistochemistry
Introduction
Although the incidence of invasive cervical cancer in the United States is decreasing, the rates of viral infection leading to development of carcinoma precursors (squamous intraepithelial lesions, SILs) are extremely high. Management of SILs remains an important health care problem despite recent advances, including the development of effective prophylactic HPV vaccines. While cervical cancer screening relies on cervical cytology and high-risk HPV detection, the histologic diagnosis, and specifically lesion grade, is the main parameter that drives clinical management of screen-positive women. Histologically diagnosed SILs (traditionally labeled as cervical intraepithelial neoplasia [CIN]) regress spontaneously in more than half of the cases, but morphology alone is an insufficient predictor of lesion behavior. (1–3) Development of markers that can predict the lesion regression versus persistence/progression would be extremely useful for clinical management.
It has been suggested that the expression and distribution of viral gene products can serve as potential markers for distinction of regressive versus progressive lesions. (4,5,6) These studies indicate that the inability of virus to complete its productive cycle is associated with increased lesion grade.
Productive viral infection is characterized by genome amplification and expression of late viral genes responsible for virion assembly, notably the capsid proteins (L1 and L2). This late gene expression is restricted to terminally-differentiated, superficial squamous epithelial cells. Major capsid protein L1 constitutes the primary structural element of viral capsid. It also represents a major target for the cell-mediated immune response. L2 is a minor component of the capsid, thought to aid in the assembly and the packaging of viral DNA within the virions.(7) Aberrant squamous cell differentiation that is considered a feature of a high-grade SIL is associated with a failure to express the capsid proteins and thus to complete the life cycle. Absence of L1 expression was a feature of progressive lesions in several studies using cytologic specimens. (8,9) Expression of L2 protein in clinical lesions has not been extensively evaluated, although it has been suggested that L2 is also expressed predominantly in LSIL/CIN, consistent with productive infection in these low-grade lesions.(10) The goal of this study is to evaluate immunohistochemical expression of L1 and L2 in SILs in correlation with lesion grade.
Material and Methods
Case selection
The study was approved by the Johns Hopkins University School of Medicine Institutional Review Board. Three hundred forty-two cervical specimens including cervical biopsies and excisional specimens (loop electrosurgical excision procedure [LEEP] and cone biopsy specimens) were retrieved from the files of The Johns Hopkins Hospital. Of these, 150 cervical tissue specimens containing 212 morphologically distinct SILs were selected for analysis based on HPV 16 or HPV 18 DNA detection by Q-PCR (see below). The study was limited to HPV 16 and HPV 18 positive cases to match the specificity of the antibodies used for immunohistochemical analysis of L1 and L2 expression. The 212 SILs included 89 low-grade squamous intraepithelial lesions (LSIL/CIN 1) and 123 high-grade squamous intraepithelial lesions (HSIL), with the latter including 75 CIN 2 and 48 CIN 3 (53 cases had more than one grade of lesion in the same specimen). Of these 150 samples, 112 contained HPV16 DNA and 32 contained HPV18 DNA; both HPV types were detected in 6 specimens (adjacent SILs of different grades present in the same specimen were not separately analyzed for HPV type). The presence and the grade of SIL were confirmed by thorough histologic review by two observers (AY and BMR).
Human Papillomavirus (HPV) DNA Detection
DNA was isolated from two 10-μm paraffin-embedded tissue sections after octane deparaffinization and digestion with buffer containing proteinase K. A 2–5 μl aliquot of purified DNA was tested for the presence and number of copies of HPV16 DNA and HPV18 DNA as previously described using real-time TaqMan PCR methods. (11) All viral quantities were normalized to total human cell equivalents tested by quantitating the ERV-3 human endogenous retrovirus gene from each sample. (12)
Immunohistochemical analysis
For immunohistochemical analysis 4–5μm -thick tissue sections were deparaffinized in xylene and rehydrated. Epitope retrieval was performed by placing the slides in Citrate Buffer Solution (pH 6.0, ZYMED laboratories, Life Technologies, Carlsbad, CA, USA) at 95°C for 20 minutes. Endogenous peroxidase activity was blocked with Peroxidase Blocking Solution (DAKO North America, Inc., Carpinteria, CA), then the slides were incubated with Protein Block (Biogenex, Fremont, CA, USA) for 30 min. Primary antibody incubation was performed for 1 hour at room temperature with MAB 837 clone 1H8 (1:1000 dilution; final antibody concentration – 1μg/ml; (Chemicon/Millipore, Billerica, MA, USA) for L1 and for 1.5 hours with RG-1 (1:500; final antibody concentration – 3.1 μg/ml) for L2. (13) These antibodies recognize the respective capsid proteins of HPV 16 and 18; MAB 837 also recognizes HPV 1, 6, 11, and 31; the specificity of RG-1 for other HPV types is less well characterized. Staining was detected with anti-mouse HRP Polymer (DAKO) for 30 minutes at room temperature. Staining was repeated in triplicate in all positive cases, all cases with discordant L1/L2 expression, and in a random 10% of negative cases; all results were consistently reproduced. In cases with more than one grade of SIL present in the same slide, immunostaining in each distinct grade was analyzed separately. Any nuclear staining was considered a positive result, regardless of the quantity of cells having expression. For descriptive purposes, the extent of the staining was also semi-quantitatively assessed: positive - ≥10 positive nuclei, focal positive - >2 but <10 positive nuclei, or limited positive - 1 or 2 positive nuclei. Expression of either L1 or L2 or both was considered as evidence of capsid protein expression for statistical analysis. In the cases with multiple lesions/grades present, the expression was recorded for each grade separately and reported as such. For the purposes of data presentation the following categories were distinguished: LSIL alone – the only grade present in the specimen; LSIL with HSIL/CIN 2 – coexisting LSIL and HSIL/CIN 2 within the same specimen, but the expression is assessed in LSIL only; LSIL with HSIL/CIN 3 – coexisting LSIL and HSIL/CIN 3 within the same specimen, but the expression is assessed in LSIL only; LSIL total – all LSILs alone or associated with a higher grade SIL. Similar categories were distinguished for HSIL/CIN 2 and HSIL/CIN 3 (HSIL/CIN 2 alone; HSIL/CIN 2 with LSIL; etc.)
Statistical analysis
The number and percentage of SIL cases expressing any L1 and/or L2 capsid protein are presented by each grade of lesion present in the specimen. If more than one morphologically distinct SIL was present in a specimen, the data are further stratified by co-occurring lesion grades. Chi-squared test or Fisher’s exact test statistics, when the number in any category was 5 or less, were calculated to compare the proportion of cases expressing any capsid protein between two mutually exclusive groups based on SIL grade: LSIL/CIN 1, HSIL/CIN2, and HSIL/CIN3. Cochran-Armitage test was used to determine if there was a significant trend in the proportion of cases expressing capsid protein across the ordered categories LSIL/CIN1 alone, HSIL/CIN2 alone, and HSIL/CIN3 alone. The level of significance was p<0.05 and no results were corrected for multiple comparisons. All analyses were conducted using SAS version 9.2 (SAS, Cary, NC).
Results
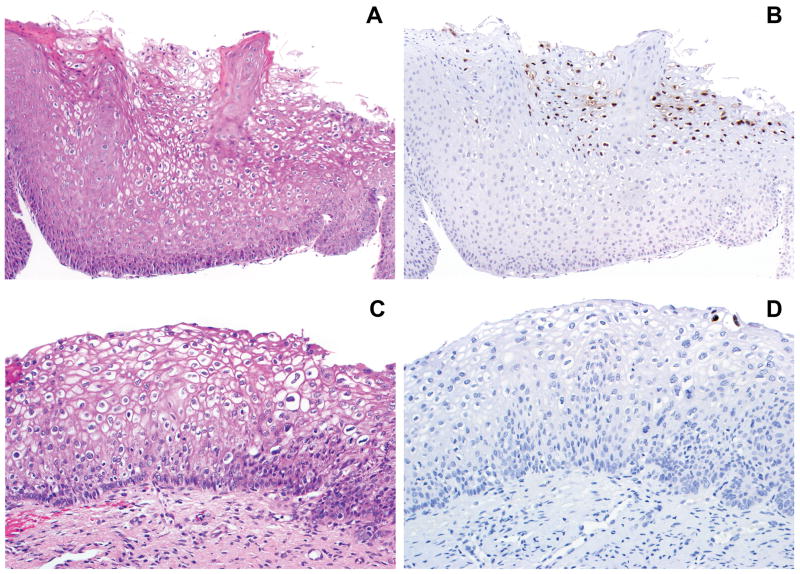
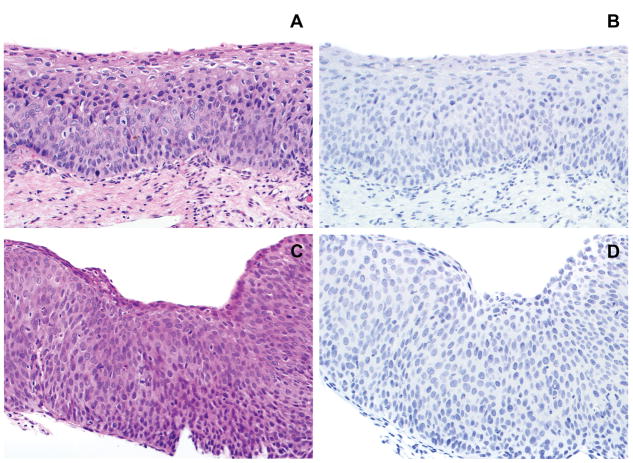
Expression of either L1 or L2 or both was observed in 39/212 lesions (18%) overall. These included 34 LSIL/CIN 1 and 5 HSIL/CIN 2. Co-expression of both L1 and L2 in the same area of the lesion was observed in 22 lesions, including 18 LSIL/CIN 1 and 4 HSIL/CIN 2 (Figure 1). L1 alone was expressed in 6 lesions, all of which were LSIL/CIN 1; L2 alone was expressed in 11 lesions, including 10 LSIL/CIN 1 and 1 HSIL/CIN 2. Neither L1 nor L2 was expressed in any of the 48 HSIL/CIN 3 lesions (Figure 2). The positive nuclei were always situated in the upper epithelial layers of the lesional squamous epithelium (Figure 1). Extent of staining was positive (≥10 cells) in 21 lesions, focal positive in 13 lesions, and limited positive in 5 lesions (Figure 1). The latter included 2 LSIL/CIN 1 and 3 HSIL/CIN 2. The immunohistochemical staining results in SILs stratified by grade are summarized in table 1.
Figure 1.
A, C. Low grade squamous intraepithelial lesion (LSIL/CIN 1). B. Extensive L2 expression in the superficial epithelial layers; L1 showed similar findings (not shown). D. Limited L1 expression (two positive nuclei) in the most superficial epithelial layer; the distribution of L2 expression was identical (not shown).
Figure 2.
A. High grade squamous intraepithelial lesion (HSIL/CIN 2). B. Absent expression of L1; L2 showed similar findings (not shown). C. High grade squamous intraepithelial lesion (HSIL/CIN 3). D. The lesion lacks expression of L1; L2 expression was absent as well (not shown).
Table 1.
Expression of HPV capsid proteins (L1 and L2) in squamous intraepithelial lesions
| Squamous intraepithelial lesion grade | Number of Lesions/Grade (n) | Number of Lesions Expressing Capsid Proteins (n/%)* |
|---|---|---|
| LSIL/CIN 1 - total | 89 | 34 (38%) |
| LSIL/CIN 1 alone | 49 | 24 (49%) |
| LSIL/CIN 1 with HSIL/CIN 2 | 29 | 10 (34%) |
| LSIL/CIN 1 with HSIL/CIN 3 | 2 | 0 (0%) |
| LSIL/CIN 1 with HSIL/CIN 2 and HSIL/CIN 3 | 9 | 0 (0%) |
| HSIL CIN 2 and CIN 3 - total | 123 | 5 (4%) |
| HSIL/CIN 2 - total | 75 | 5 (7%) |
| HSIL/CIN 2 alone | 24 | 3 (13%) |
| HSIL/CIN 2 with LSIL/CIN 1 | 29 | 2 (7%) |
| HSIL/CIN 2 with HSIL/CIN 3 | 13 | 0 (0%) |
| HSIL/CIN 2 with LSIL/CIN 1 and HSIL/CIN 3 | 9 | 0 (0%) |
| HSIL/CIN 3 - total | 48 | 0 (0%) |
| HSIL/CIN 3 alone | 25 | 0 (0%) |
| HSIL/CIN 3 with LSIL/CIN 1 | 2 | 0 (0%) |
| HSIL/CIN 3 with LSIL/CIN 1 and HSIL/CIN 2 | 9 | 0 (0%) |
| HSIL/CIN 3 with HSIL/CIN 2 | 13 | 0 (0%) |
The number of positive cases is reported for the first lesion grade in each category.
The difference in capsid protein expression in LSIL/CIN 1 alone (49%) versus HSIL/CIN 2 alone (13%) or any HSIL [CIN2 and CIN 3] (5%) was statistically significant (p=0.0039 and p<0.0001, respectively). There were no differences in expression between LSIL/CIN 1 alone (49%) and LSIL/CIN 1 with HSIL/CIN 2 (34%) groups (p=0.2). However, expression in LSIL/CIN 1 with or without HSIL/CIN 2 (43%) was different from expression in LSIL/CIN1 with adjacent HSIL/CIN 3 (0%) (p=0.006). Within the HSIL/CIN 2 group, there were no differences in expression between HSIL/CIN 2 alone (13%) and HSIL/CIN 2 with LSIL/CIN 1 (7%) groups (p=0.108). However, expression in HSIL/CIN 2 with or without LSIL/CIN 1 (9%) was different from expression in HSIL/CIN 2 with adjacent HSIL/CIN 3 (0%) (p=0.008). The expression of capsid proteins decreased with increase in lesion grade (LSIL/CIN 1 alone vs. HSIL/CIN 2 alone vs. HSIL/CIN 3 alone - ptrend < 0.0001).
Discussion
The need for an immunohistochemical marker of HPV-related precancer that is likely to progress is indisputable. The majority of the biomarkers used in pathologic diagnosis of squamous intraepithelial lesions (SILs), including p16, PCNA, MCM, ProEx C, reflect the presence of a high-risk HPV-related lesion, but are not specific for identification of progressive disease with potential to develop into an invasive carcinoma. It has been suggested that patterns of viral protein expression may be used to differentiate between self-limited productive viral infection and a true precancer. (6)
Immunohistochemical detection of viral protein expression has been described in prior studies in cervical specimens. (14–16) Kurman et al. reported immunohistochemical expression of PV (papillomavirus) common (structural) proteins in 43% of mild dysplasia, 15% of moderate dysplasia, and only in a rare case of severe dysplasia. (16) In one study, immunohistochemical expression of L1 and L2 in two cases of condyloma and a CIN 2 lesion was identified in nuclei restricted to the upper epithelial layers. (14)
A number of more recent studies have evaluated the expression of major capsid protein L1 in cytologic specimens. It has been shown that 30% to 75% of LSILs and 33% to 40% of HSILs express L1 in cytology specimens. (8,9,17–21) Some of these studies evaluated the prognostic utility of L1 immunocytochemistry on a Pap sample in predicting lesion behavior. Most of the studies that relied on “cytologic regression” with variable length of follow-up (up to 2 years) associated the presence of L1expression with clinical “regression”. However, lesion regression is difficult to reliably establish in the clinical setting because histologic evaluation of the entire cervix is not possible in most cases and all other measures of outcome (Pap cytology or colposcopy with cervical biopsies) have imperfect sensitivity for detecting significant lesions. In addition, the diagnostic biopsy itself in some cases may be curative and result in removal of the entire lesion.
Our data are consistent with previously published studies using histologic specimens that demonstrated L1 expression in 30–65% of LSIL/CIN1. (22–25) A lower proportion of the positive cases in histology compared to cytology specimens is likely related to the fact that in the majority of lesions the expression of capsid proteins is restricted to the most superficial layers of the epithelium. Thus, the keratinocytes in the superficial layers are most readily detached and harvested during sampling for a Pap test, but might also be inadvertently removed from the epithelial surface during colposcopy (mucous clearing etc.) and tissue handling during pathologic processing. A larger number of positive cases in cytology specimens compared to matched histologic preparations was previously described.(8,26,27) Some studies evaluating the correlation of L1 and p16 expression in LSIL/CIN 1 have observed that lesions lacking L1 and demonstrating diffuse p16 expression are more likely to persist and progress.(25) Diffuse p16 expression in these cases is most likely related to the presence of high-risk HPV within these lesions, most commonly HPV 16 and 18; these types are associated with lower regression rates compared to LSIL/CIN 1 harboring other HPV types,(28) thus, the prognostic value of p16 expression is likely not independent of the HPV type implicated in the lesion development.
In our study, a significant fraction of LSIL/CIN 1 cases were L1 negative, but the value of L1 expression alone as a predictor of behavior in tissue specimens is unclear. The inability to detect L1 in these LSIL/CIN 1 cases may reflect inadequate sampling, given the extremely focal staining in some cases noted in this and other studies.(22) Using cytology Pap samples likely provide a more representative sample of superficial lesional cells for analysis of L1 expression.
We have previously reported expression of L2 in 89% of LSIL/CIN 1 lesions and none of HSIL/CIN 2 and HSIL/CIN 3 in a study with a smaller number of cases.(10) This data and our current data are different from what has been previously reported in another prior study that observed L2 expression in a significant proportion of moderate and severe dysplasias;(29) this could be due to differences in histologic grading.
The discordant expression of L1 and L2 in the same lesion that was observed in 17 cases (43% of positive cases) in our study is surprising. This may be related to infection with multiple HPV types, with discordant expression related to antibody specificity in the setting of an HPV type other than 16 or 18. Thus, detection of both L1 and L2 proteins might not have added value if a more broadly HPV type-specific antibody for either L1 or L2 had been used.
All cases of HSIL/CIN3, either alone or with adjacent LSIL/CIN 1 or HSIL/CIN2, lacked= expression of capsid proteins. Statistically, expression of capsid proteins was negatively associated with the grade of the lesion (ptrend < 0.0001).
The absence of capsid protein expression observed in this study differs from data reported by Galgano and colleagues that recorded L1 expression in 16.5% of HSIL/CIN 3. (30) These discordant findings could be related to fact that the current study was restricted to HPV16 and HPV18-related lesions, while the reported expression of L1 in HSIL/CIN 3 in the Galgano study was based on SILs irrespective of HPV type. Reported L1 staining in HSIL/CIN 3 in some other studies may also be related to the known world wide difference in histologic grading of SILs.
Recent studies evaluating methylation of HPV genes, specifically L1, found high levels of methylation of this gene associated with high-grade cytology. (31,32) Consistent intense methylation of L1 gene was shown in HPV 18-related cervical carcinomas.(33) These findings may provide further insight into the mechanisms of loss of capsid protein expression in higher grade SILs.
In our study, 62% of all LSIL/CIN1 lacked expression of both L1 and L2. The significance of this finding is not clear. Lack of detected expression in some of the negative cases could be a reflection of focality of staining and sampling. In other cases, it could represent a false negative result related to the presence of HPV types other than those that are covered by the antibodies used. Nonetheless, further investigation of the viral capsid protein expression as a marker of progression/persistence in LSILs in correlation with methylation analysis of the viral genome is warranted.
Eleven specimens contained discrete foci of both LSIL/CIN 1 and HSIL/CIN 3 (with or without HSIL/CIN 2). No expression of either L1 or L2 was observed in HSIL/CIN 3 foci. Interestingly, in these cases with multiple lesions/grades, the adjacent lower-grade components (LSIL/CIN 1 and HSIL/CIN 2) of these cases also lacked expression. The absence of capsid proteins expression in LSIL associated HSIL/CIN 3 may be related to molecular alterations that are associated with lesion progression but occur before the morphologic changes take place (e.g. integration, gene methylation, etc.). However, the exact mechanism responsible for the loss of L1 and L2 expression in these lesions is not known. It is also possible that these different grades of SIL represent independent lesions due to multiple HPV types that are not reactive with the antibodies used in this study. However, the number of specimens containing adjacent lesions of different grades is too small for drawing meaningful conclusions. Nonetheless, the hypothesis suggested by this observation, namely, that the presence of L1 and L2 expression might be used as a marker of absence of adjacent HSIL/CIN 3 in the same specimen, would be extremely useful clinically and warrants further examination.
Footnotes
Disclosure/Conflict of Interest: This study was supported by NIH/NCI/R21CA150033. Dr. Yemelyanova is supported by SPORE in Cervical Cancer carrier development award NIH/NCI/P50 CA098252. The authors declare no conflict of interest.
References
- 1.Castle PE, Schiffman M, Wheeler CM, Solomon D. Evidence for frequent regression of cervical intraepithelial neoplasia-grade 2. Obstet Gynecol. 2009;113:18–25. doi: 10.1097/AOG.0b013e31818f5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho GY, Bierman R, Beardsley L, Chang CJ, Burk RD. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338:423–428. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- 3.Moscicki AB, Shiboski S, Hills NK, et al. Regression of low-grade squamous intra-epithelial lesions in young women. Lancet. 2004;364:1678–1683. doi: 10.1016/S0140-6736(04)17354-6. [DOI] [PubMed] [Google Scholar]
- 4.Doorbar J, Cubie H. Molecular basis for advances in cervical screening. Mol Diagn. 2005;9:129–142. doi: 10.1007/BF03260081. [DOI] [PubMed] [Google Scholar]
- 5.Middleton K, Peh W, Southern S, et al. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J Virol. 2003;77:10186–10201. doi: 10.1128/JVI.77.19.10186-10201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doorbar J. Papillomavirus life cycle organization and biomarker selection. Dis Markers. 2007;23:297–313. doi: 10.1155/2007/613150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck CB, Cheng N, Thompson CD, et al. Arrangement of L2 within the papillomavirus capsid. J Virol. 2008;82:5190–5197. doi: 10.1128/JVI.02726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H, Lee KJ, Jung CK, et al. Expression of HPV L1 capsid protein in cervical specimens with HPV infection. Diagn Cytopathol. 2008;36:864–867. doi: 10.1002/dc.20922. [DOI] [PubMed] [Google Scholar]
- 9.Melsheimer P, Kaul S, Dobeck S, Bastert G. Immunocytochemical detection of HPV high-risk type L1 capsid proteins in LSIL and HSIL as compared with detection of HPV L1 DNA. Acta Cytol. 2003;47:124–128. doi: 10.1159/000326491. [DOI] [PubMed] [Google Scholar]
- 10.Lin Z, Yemelyanova AV, Gambhira R, et al. Expression pattern and subcellular localization of human papillomavirus minor capsid protein L2. Am J Pathol. 2009;174:136–143. doi: 10.2353/ajpath.2009.080588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravitt PE, Peyton C, Wheeler C, Apple R, Higuchi R, Shah KV. Reproducibility of HPV 16 and HPV 18 viral load quantitation using TaqMan real-time PCR assays. J Virol Methods. 2003;112:23–33. doi: 10.1016/s0166-0934(03)00186-1. [DOI] [PubMed] [Google Scholar]
- 12.Yuan CC, Miley W, Waters D. A quantification of human cells using an ERV-3 real time PCR assay. J Virol Methods. 2001;91:109–117. doi: 10.1016/s0166-0934(00)00244-5. [DOI] [PubMed] [Google Scholar]
- 13.Gambhira R, Karanam B, Jagu S, et al. A protective and broadly cross-neutralizing epitope of human papillomavirus L2. J Virol. 2007;81:13927–13931. doi: 10.1128/JVI.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firzlaff JM, Kiviat NB, Beckmann AM, Jenison SA, Galloway DA. Detection of human papillomavirus capsid antigens in various squamous epithelial lesions using antibodies directed against the L1 and L2 open reading frames. Virology. 1988;164:467–477. doi: 10.1016/0042-6822(88)90561-2. [DOI] [PubMed] [Google Scholar]
- 15.Kurman RJ, Shah KH, Lancaster WD, Jenson AB. Immunoperoxidase localization of papillomavirus antigens in cervical dysplasia and vulvar condylomas. Am J Obstet Gynecol. 1981;140:931–935. doi: 10.1016/0002-9378(81)90087-9. [DOI] [PubMed] [Google Scholar]
- 16.Kurman RJ, Jenson AB, Lancaster WD. Papillomavirus infection of the cervix. II. Relationship to intraepithelial neoplasia based on the presence of specific viral structural proteins. Am J Surg Pathol. 1983;7:39–52. [PubMed] [Google Scholar]
- 17.Griesser H, Sander H, Hilfrich R, Moser B, Schenck U. Correlation of immunochemical detection of HPV L1 capsid protein in pap smears with regression of high-risk HPV positive mild/moderate dysplasia. Anal Quant Cytol Histol. 2004;26:241–245. [PubMed] [Google Scholar]
- 18.Griesser H, Sander H, Walczak C, Hilfrich RA. HPV vaccine protein L1 predicts disease outcome of high-risk HPV+ early squamous dysplastic lesions. Am J Clin Pathol. 2009;132:840–845. doi: 10.1309/AJCPCU0HBFFFGDTV. [DOI] [PubMed] [Google Scholar]
- 19.Xiao W, Bian M, Ma L, et al. Immunochemical analysis of human papillomavirus L1 capsid protein in liquid-based cytology samples from cervical lesions. Acta Cytol. 2010;54:661–667. doi: 10.1159/000325229. [DOI] [PubMed] [Google Scholar]
- 20.Yu L, Wang L, Zhong J, Chen S. Diagnostic value of p16INK4A, Ki-67, and human papillomavirus L1 capsid protein immunochemical staining on cell blocks from residual liquid-based gynecologic cytology specimens. Cancer Cytopathol. 2010;118:47–55. doi: 10.1002/cncy.20061. [DOI] [PubMed] [Google Scholar]
- 21.Rauber D, Mehlhorn G, Fasching PA, Beckmann MW, Ackermann S. Prognostic significance of the detection of human papilloma virus L1 protein in smears of mild to moderate cervical intraepithelial lesions. Eur J Obstet Gynecol Reprod Biol. 2008;140:258–262. doi: 10.1016/j.ejogrb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Choi YS, Kang WD, Kim SM, et al. Human papillomavirus L1 capsid protein and human papillomavirus type 16 as prognostic markers in cervical intraepithelial neoplasia 1. Int J Gynecol Cancer. 2010;20:288–293. doi: 10.1111/IGC.0b013e3181cd184c. [DOI] [PubMed] [Google Scholar]
- 23.Hilfrich R, Hariri J. Prognostic relevance of human papillomavirus L1 capsid protein detection within mild and moderate dysplastic lesions of the cervix uteri in combination with p16 biomarker. Anal Quant Cytol Histol. 2008;30:78–82. [PubMed] [Google Scholar]
- 24.Hoshikawa S, Sano T, Yoshida T, Ito H, Oyama T, Fukuda T. Immunohistological analysis of HPV L1 capsid protein and p16 protein in low-grade dysplastic lesions of the uterine cervix. Pathol Res Pract. 2010;206:816–820. doi: 10.1016/j.prp.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Negri G, Bellisano G, Zannoni GF, et al. p16 ink4a and HPV L1 immunohistochemistry is helpful for estimating the behavior of low-grade dysplastic lesions of the cervix uteri. Am J Surg Pathol. 2008;32:1715–1720. doi: 10.1097/PAS.0b013e3181709fbf. [DOI] [PubMed] [Google Scholar]
- 26.Huang MZ, Li HB, Nie XM, Wu XY, Jiang XM. An analysis on the combination expression of HPV L1 capsid protein and p16INK4a in cervical lesions. Diagn Cytopathol. 2010;38:573–578. doi: 10.1002/dc.21258. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida T, Sano T, Kanuma T, et al. Immunochemical analysis of HPV L1 capsid protein and p16 protein in liquid-based cytology samples from uterine cervical lesions. Cancer. 2008;114:83–88. doi: 10.1002/cncr.23366. [DOI] [PubMed] [Google Scholar]
- 28.Trimble CL, Piantadosi S, Gravitt P, et al. Spontaneous regression of high-grade cervical dysplasia: effects of human papillomavirus type and HLA phenotype. Clin Cancer Res. 2005;11:4717–4723. doi: 10.1158/1078-0432.CCR-04-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auvinen E, Kujari H, Arstila P, Hukkanen V. Expression of the L2 and E7 genes of the human papillomavirus type 16 in female genital dysplasias. Am J Pathol. 1992;141:1217–1224. [PMC free article] [PubMed] [Google Scholar]
- 30.Galgano MT, Castle PE, Atkins KA, Brix WK, Nassau SR, Stoler MH. Using biomarkers as objective standards in the diagnosis of cervical biopsies. Am J Surg Pathol. 2010;34:1077–1087. doi: 10.1097/PAS.0b013e3181e8b2c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandsma JL, Sun Y, Lizardi PM, et al. Distinct human papillomavirus type 16 methylomes in cervical cells at different stages of premalignancy. Virology. 2009;389:100–107. doi: 10.1016/j.virol.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun C, Reimers LL, Burk RD. Methylation of HPV16 genome CpG sites is associated with cervix precancer and cancer. Gynecol Oncol. 2011;121:59–63. doi: 10.1016/j.ygyno.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turan T, Kalantari M, Calleja-Macias IE, et al. Methylation of the human papillomavirus-18 L1 gene: a biomarker of neoplastic progression? Virology. 2006;349:175–183. doi: 10.1016/j.virol.2005.12.033. [DOI] [PubMed] [Google Scholar]