Abstract
Mass spectrometry imaging (MSI) has emerged as an important tool in the last decade and it is beginning to show potential to provide new information in many fields owing to its unique ability to acquire molecularly specific images and to provide multiplexed information, without the need for labeling or staining. In MSI, the chemical identity of molecules present on a surface is investigated as a function of spatial distribution. In addition to now standard methods involving MSI in vacuum, recently developed ambient ionization techniques allow MSI to be performed under atmospheric pressure on untreated samples outside the mass spectrometer. Here we review recent developments and applications of MSI emphasizing the ambient ionization techniques of desorption electrospray ionization (DESI), laser ablation electrospray ionization (LAESI), probe electrospray ionization (PESI), desorption atmospheric pressure photoionization (DAPPI), femtosecond laser desorption ionization (fs-LDI), laser electrospray mass spectrometry (LEMS), infrared laser ablation metastable-induced chemical ionization (IR-LAMICI), liquid microjunction surface sampling probe mass spectrometry (LMJ-SSP MS), nanospray desorption electrospray ionization (nano-DESI), and plasma sources such as the low temperature plasma (LTP) probe and laser ablation coupled to flowing atmospheric-pressure afterglow (LA-FAPA). Included are discussions of some of the features of ambient MSI including the ability to implement chemical reactions with the goal of providing high abundance ions characteristic of specific compounds of interest and the use of tandem mass spectrometry to either map the distribution of targeted molecules with high specificity or to provide additional MS information in the structural identification of compounds. We also describe the role of bioinformatics in acquiring and interpreting the chemical and spatial information obtained through MSI, especially in biological applications for tissue diagnostic purposes. Finally, we discuss the challenges in ambient MSI and include perspectives on the future of the field.
Keywords: mass spectrometry, imaging, ambient ionization, ionization techniques, review
I. INTRODUCTION
Mass spectrometry imaging (MSI) refers to procedures in which both chemical information and spatial information are acquired for later processing and visualization as chemical images. The experiment can be used to record 2D or 3D images. MSI has emerged as a promising technique in many areas, including the biomedical sciences, material sciences and forensics, owing to its high chemical specificity and relative ease of application. (Chughtai & Heeren, 2010; McDonnell & Heeren, 2007; Pacholski & Winograd, 1999; Vickerman, 2011; Vidova et al., 2009)
Ambient ionization MSI is a member of the MS imaging family, in which ionization occurs at atmospheric pressure with the samples being analyzed in their native state. In ambient ionization (i) the surface is sampled with minimal or no preparation, (ii) ionization occurs externally to the mass spectrometer and (iii) ions, not the entire sample, are introduced into the mass spectrometer. (Cooks et al., 2006) More than thirty ambient ionization methods have been developed in the last eight years (Alberici et al., 2010; Chen et al., 2009; Harris et al., 2011; Huang et al., 2010; Huang et al., 2011; Ifa et al., 2010; Van Berkel et al., 2008; Venter et al., 2008; Weston, 2010) some of which are used in MSI including desorption electrospray ionization (DESI) (Ifa et al., 2007), laser ablation electrospray ionization (LAESI) (Nemes & Vertes, 2007; Nemes et al., 2008), desorption atmospheric pressure photoionization (DAPPI) (Pol et al., 2009), femtosecond laser desorption ionization (fs-LDI) (Coello et al., 2010) also known as laser electrospray ionization mass spectrometry (LEMS) when fs-LDI is combined with ESI (Judge et al., 2010), probe electrospray ionization (PESI) (Chen et al., 2009), infrared laser ablation metastable-induced chemical ionization (IR-LAMICI)(Galhena et al., 2010), liquid microjunction surface sampling probe mass spectrometry (LMJ-SSP MS)(Kertesz et al., 2005), nanospray desorption electrospray ionization (nano-DESI) (Laskin et al., 2012), and plasma sources such as low temperature plasma (LTP) (Liu et al., 2010) and laser ablation coupled to flowing atmospheric-pressure afterglow (LA-FAPA) (Shelley et al., 2008) have reported applications for MSI. Note that the liquid junction methods are not strictly ambient ionization procedures but are included because of their close relationship.
Other ambient ionization techniques, for instance, easy ambient sonic-spray ionization (EASI) and electrospray laser desorption ionization (ELDI), have recently been demonstrated to have capabilities in direct analysis of inks for document examination (Cheng et al., 2010; Lalli et al., 2010) and in characterizing organic compounds separated on thin-layer chromatography plates (Haddad et al., 2008; Lin et al., 2007). Although no MS images have been presented, the potential of EASI and ELDI for MSI is implied. Transmission mode DESI (TM-DESI) (Chipuk & Brodbelt, 2008) could also be used for imaging after blotting tissues onto the grid. Note that some atmospheric pressure imaging methods such as matrix-assisted laser desorption electrospray ionization (MALDESI) (Sampson et al., 2009) and atmospheric pressure matrix-assisted laser desorption/ionization (AP-MALDI) (Li et al., 2007) are also closely related to the ambient ionization MSI experiments discussed here. They are not considered as ambient ionization imaging techniques in this review as they require careful and specific sample preparation involving pre-application of matrix to the sample.
The most widely used of the vacuum MSI methods are matrix assisted laser desorption (MALDI) and secondary ion mass spectrometry (SIMS) (Chughtai & Heeren, 2010; McDonnell & Heeren, 2007; Pacholski & Winograd, 1999; Vickerman, 2011; Vidova, et al., 2009; Watrous et al., 2011). MALDI allows imaging of large molecules such as proteins with spatial resolution that can reach down into the tens of microns scale. SIMS has the advantage of very high spatial resolution (~100 nm) (Carado et al., 2008), and is most successful for targeted inorganic compounds or biomolecules with relatively low molecular weights. Extensive (albeit variable) fragmentation is associated with ionization, even with C60 and large solvent cluster beams. The nanostructure-initiator mass spectrometry (NIMS) technique is matrix-free, has reduced fragmentation as compared to SIMS, and allows direct characterization of peptide microarrays, direct mass analysis of single cells and tissue imaging. (Northen et al., 2007) but does require specific nanostructured surfaces or ‘clathrates’ to release and ionize the molecules adsorbed on the surface. NIMS and other LDI methods are traditionally vacuum-based and have seen application in imaging.
Ambient ionization imaging is often used to analyze compounds with molecular weights less than 2000 Da at a spatial resolution ranging from tens to a few hundred microns. (Chen, et al., 2009; Ifa, et al., 2007; Kertesz & Van Berkel, 2008; Nemes & Vertes, 2007; Shelley, et al., 2008) There are a number of inter-related parameters that can be used to characterize and compare the performance of various MSI methods. They will be weighted differently depending on the problem in hand. They include speed of analysis, sample preparation required, spatial resolution, spectral resolution, sensitivity, quantitative accuracy and precision, compatibility with exact mass and tandem mass spectrometry and with the ease of performing supplementary chemical reactions as a means of increasing specificity. Simplistically, spatial resolution is often used as the single defining characteristic of performance and at its current stage of development, this is a weak feature of most ambient MSI methods. However, reports of single cell resolution have started to appear and further improvements in spatial resolution can be foreseen. For example, a study using fs-LDI has demonstrated spatial resolution of 10 μm for imaging of onion epidermis cells under ambient conditions (Coello, et al., 2010). Metabolic analysis of single cells and small cell populations of Allium cepa and Narcissus pseudonarcissus bulb epidermis, as well as single eggs of Lytechinus pictus (~100μm in diameter) (Shrestha & Vertes, 2009) and clusters of 3–4 achlorophyllous cells in citrus leaves (Citrus aurantium) (~30μm in diameter) (Shrestha et al., 2011) have been reported by LAESI.
Advantageously, ambient ionization imaging does not require sample preparation, which can be problematic and lead to spatial dislocation or chemical modifications of endogenous compounds. An advantage is that MSI under ambient conditions is easy to implement. In addition, ionization by the spray-based ambient ionization methods such as DESI is soft, with an average internal energy deposition of ~ 2eV in typical cases, which is similar to the internal energy of electrospray ionization (ESI). (Nefliu et al., 2008) Hence DESI and other ambient imaging methods yield minimal fragmentation (Wu et al., 2010), and they are ideal for imaging in the low mass region, without interference from added matrix(cf. MALDI) or fragmentions of large molecules (cf. SIMS). The success with low mass molecules in DESI should not obscure the fact that the method has been applied to large biomolecules.(Ferguson et al., 2011) Identification of analytes if needed can be achieved by using tandem mass spectrometry (MS/MS) or accurate mass measurements. Quantification of particular analytes in their native matrix is difficult in all imaging methods, can only be done with appropriately matched standards and these can be difficult to identify for many biological samples. Some of the other features (spectral resolution, detection limits, and compatibility with exact mass measurements and with MS/MS) are properties that are more strongly controlled by the mass analyzer than by the type of ionization source chosen. The ability to implement chemical reactions in the course of the desorption and ionization steps is a potentially useful feature of MSI techniques. By implementing rapid in situ chemical reactions occurring at the spot being analyzed concurrently with acquisition of mass spectra, the sensitivity and selectivity of target molecule detection can be improved. (Nyadong et al., 2007; Shrestha et al., 2010; Wu et al., 2009) This is similar in concept, but not in method of implementation, to reactive matrix MALDI. (Mugo & Bottaro, 2007)
In this review we summarize the principles and major applications of the following ambient MSI techniques: DESI, LAESI, PESI, DAPPI, fs-LDI, LEMS, IR-LAMICI, LMJ-SSP MS, nano-DESI, LESA, LTP, LA-FAPA. The use of some of these ambient techniques for in vivo local analysis and in situ molecular tissue imaging was recently reviewed. (Nemes & Vertes, 2012) Note that these methods can be organized in different ways based on the primary sampling desorption mechanism; direct ionization, direct desorption ionization, or multiple steps desorption ionization; resolution; etc. (Harris, et al., 2011; Huang, et al., 2011) Challenges and limitations as well as the perspectives for further development of these ambient methods are also discussed.
II. GENERAL ASPECTS
The workflow in MSI normally proceeds through three steps: (i) sample preparation, (ii) data acquisition, and (iii) data analysis. Sample preparation is minimal in ambient ionization compared to traditional MSI. In some cases such as imaging of questionable documents or studies of natural products in plant material, the samples are ready for analysis without any sample preparation whatsoever. For the study of animal tissues, sectioning and drying are still necessary. However, the sample is not introduced into the vacuum environment nor are chemicals added to the sample (matrices or tagging agents) required.
Data acquisition encompasses the steps of desorption/ionization, mass analysis and ion detection. (Figure 1) Desorption/ionization includes both sampling and ionization, either accomplished in a single integrated process or in two discrete steps implemented with different agents. In some methods, the desorption and the ionization agents are clearly separated, as is the case with LAESI, DAPPI, LEMS or LMJ-SSP. For example, laser ablation is a separate process from electrospray ionization in LAESI. However, for other methods such as DESI and LTP, desorption and ionization are fully integrated and a single agent is employed. Data acquisition in MSI can be performed using two distinct approaches, as regards the method of interrogating the sample. The first and by far the more frequently used, is the microprobe mode. In this mode, the whole surface of the sample is divided conceptually into small areas (pixels) and these areas are scanned individually and sequentially in time (Figure 1). The ionization/desorption agent is set to provide the desired spatial resolution. The data from a single pixel is normally represented by a single mass spectrum or an average of two or more mass spectra acquired within of the specific spot or by continuously rastering the surface with the ionizing agent. The alternative to the microprobe method is the microscope mode, where the use of astigmatic projection of ions generated at the surface to a position-sensitive (two-dimensional) detector allows the simultaneous imaging of a larger area. This approach greatly improves the speed of the analysis. However, this method puts strict requirements on the ion optics and this type of experiment is still limited to the vacuum environment. (Heeren et al., 2009; McDonnell & Heeren, 2007) In the common microprobe mode, the use of an automated moving stage is required in order to assure the reproducibility of the scan velocity to accurately reproduce the geometry of the system. Note that the desorption/ionization agent(s) used will dictate the lateral and the depth resolution of the images that are finally acquired. The spatial resolution depends on the spot size of the ionizing agent but it can also be limited by other factors including operating conditions (substrate moving speed, step size, analyte carryover between pixels, etc.). Laser based techniques can achieve higher resolutions compared to spray based desorption ionization methods. There is an important trade-off between the time required for data acquisition and the achievable spatial resolution, with time increasing quadratically as the resolution is increased (see conclusions and perspectives, section IV). A good rule is to use the minimum resolution necessary to address the problem being investigated.
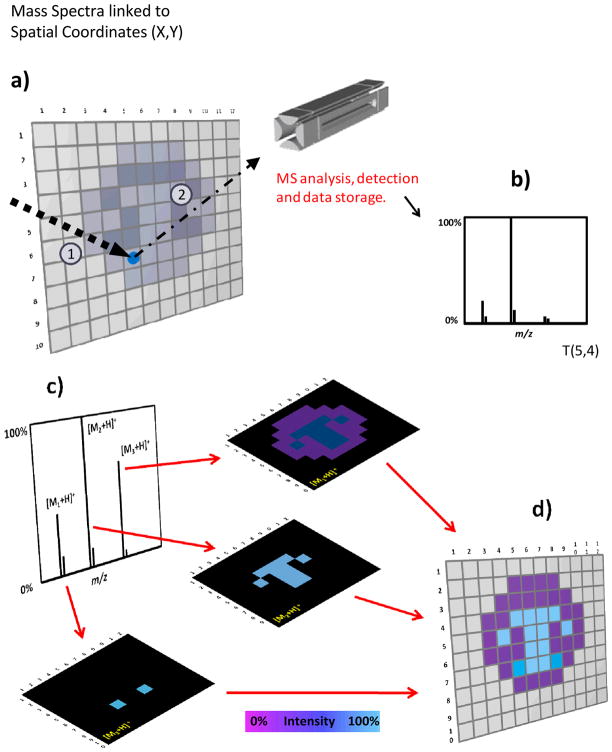
Figure 1.
MSI imaging concepts and methods (a) In a typical MSI experiment the total area is subdivided (conceptually) into pixels that are individually inspected. (b) For each pixel a single mass spectrum or the average of several mass spectra is collected and stored together with its spatial coordinates. (c) After the entire surface is scanned, an average mass spectrum can be created. The distribution of specific ions can be visualized by the creation of chemical images where the color scale (false color) represents the normalized intensity of particular ions. Each pixel from the image is associated with the original mass spectrum/mass spectra acquired at the specific point. The numbers 1 and 2 on panel A, represent the steps desorption and ionization process. (d) The aim of imaging is to display the distribution of chemicals across a surface.
Having addressed issues of sampling in two dimensions we can now turn to the subject of sampling depth. Sampling depths, which varies considerably with the different ambient ionization MS techniques, is defined as the depth from which material is removed and transmitted to the mass spectrometer during a single imaging experiment. In spray based methods like DESI, the sampling depth depends on the nature of the sample. In many cases, physical damage to the sample is minimal or, for tissue sections restricted to a few microns. For example, there is no physical damage to subtending cells of algal surfaces during DESI imaging. (Lane et al., 2009) For tissue sections, the sampling depth can easily reach 12 μm using conventional solvent systems such as MeOH/H2O (1:1). Recent experiments show sampling depths of > 50 μm for tissue imaging by DESI-MS when solvent systems such as dimethylformamide:acetonitrile (DMF:ACN,1:1) are used. Notably, these DMF based solvent systems and others, which were denoted as morphologically friendly or histologically compatible, impart no gross physical damage to the tissue section after DESI-MS imaging. (Eberlin et al., 2011) In contrast, the ablation crater in some LAESI experiments is estimated to be 30–40 μm in depth depending on sample and laser fluence. (Nemes et al., 2009) In SIMS the primary ion beam interacts exclusively with the topmost monolayers of the surface making the method much more surface sensitive than the ambient methods. Although the sample removal rate of DESI is not dissimilar to that of SIMS (Green et al., 2009), analytes are removed selectively and this does not allowing depth profiling for three-dimensional imaging, a capability that can be achieved with SIMS (Wucher et al., 2007) and LAESI (Nemes, et al., 2009). Details about LAESI fundamentals are presented in section IIIB.
After the sample has been ionized the ions either solvated in microdroplets or free gas-phase ions are directed into the mass spectrometer and mass analyzed. Several types of analyzers have been employed for ambient ionization MSI including linear triple quadrupoles (QqQ), quadrupole ion-traps (Q-Trap), OrbiTraps, Time of Flight (ToF), and Fourier Transform ion cyclotron resonance MS (FTMS). Mass analysis is followed by the data processing step, the third stage in the sequence of steps leading to creation of MS images. The recorded mass spectra are converted into a 2D image file, which can then be opened and visualized by imaging programs (e.g. the freeware BioMap or Image J or commercially available software, FlexImaging). Two-dimensional or three-dimensional ion images are displayed using a false color intensity scale with the relative ion intensity reflected by the intensity of the color. Appropriate contrast in the color bar and overlay can also be used to improve the visualization of the ion images. These ion images can be used to represent intensity distributions of ions of a single m/z ratio or they can be more complex representations, including ion populations that embody a particular chemical, biological or bioinformatics parameter e.g. a principal component derived from a multivariate statistical analysis. Processing of the imaging data, for instance background subtraction or normalization, can be done on the mass spectra prior to image creation or directly on the chemical images. The analyzer employed will dictate the mass resolution represented in the final data. When using a high mass resolution mass spectrometer for imaging, the data file size of the image can easily reach gigabytes (GB) in contrast to a few megabytes (MB) stored in low resolution experiments of the same sized sample. The data file size of the image is related to the number of pixels, the mass range used, and the mass resolution chosen. Processing and visualization of large data-size ion images require computers with large memories and relatively high speed. By carefully setting up these parameters, the data file size of the generated image can be made to fall in a range suitable for processing and storage, but the development of imaging software and hardware is an important ongoing task which is necessary to allow the full potential of high mass resolution imaging to be reached.
Three-dimensional (3D) models can be constructed from a set of 2D ion images and they provide comprehensive information on the distribution of molecules in the total object volume and facilitate correlation between molecular composition and spatial distribution to be obtained. Depth profiling and serial sectioning are the two main approaches that have been described to record the information required to create 3D MS-based images. Depth profiling has been employed to create 3D models by ablation techniques such as SIMS (Wucher, et al., 2007) and LAESI (Nemes, et al., 2009) which completely remove material while analyzing. The depth resolution of SIMS can reach <100 nm, (Wucher, et al., 2007) and the reported depth resolution of the ambient ionization method LAESI is about 30μm (Nemes, et al., 2009) in the imaging mode. DESI (Eberlin et al., 2010) and MALDI (Sinha et al., 2008) are techniques that use serial sectioning followed by analysis of each section for 3D ion image construction.
III. AMBIENT IONIZATION IMAGING TECHNIQUES
A. Desorption Electrospray Ionization (DESI)
1. Principles
In DESI, charged primary droplets generated by ESI of a solvent impact the sample-bearing surface to desorb the analyte in secondary droplets. The secondary microdroplets are generated in the open air and sucked into the inlet of the mass spectrometer (Figure 2a). To date, DESI is perhaps the most thoroughly explored of the ambient ionization methods. Desorption and ionization steps in DESI are not clearly separated, which is different from some other cases. The main DESI mechanism has been summarily described as “droplet pick-up”. (Cooks, et al., 2006) This process does not occur in a single scattering event but for most analytes and surfaces, it appears to involve initial wetting of the surface to dissolve analyte, then splashing of this liquid film upon the arrival of subsequent primary droplets with formation of secondary microdroplets. Using a phase doppler particle analyzer, it was determined that the average velocity of the primary droplets is about 120 m/s and their average diameters are about 5 μm under typical DESI experimental conditions. (Venter et al., 2006) Simulations of the DESI process show the release of dozens of microdroplets upon impact of a single high velocity primary droplet on the thin liquid film. (Costa & Cooks, 2008) The secondary droplets are generated by breakup of a thin jet of solution and the size of the measurable secondary droplets is in the range of 0.8 to 3.3 μm. Secondary droplet velocities cover a large range, depending on the size of the primary droplet, the take-off angle, and the distance from the impact point. (Venter, et al., 2006) The effects of the nebulizing gas are clearly discernible is the absence of any overt coulombic effect. The performance and characteristics of DESI were found to be dependent on the choice of the solvent (Tawiah et al., 2010), substrate (Ifa et al., 2008; Kauppila et al., 2006), and geometry (Chipuk et al., 2010; Kertesz & Van Berkel, 2008; Venter & Cooks, 2007). Experiments on solvent composition showed that different chemical information can be obtained by changing the solvent system. For example, it has been shown that a mixture of DMF with water or methanol enhances the signal of low molecular weight compounds when compared to mixtures of DMF with ACN, or other mixtures containing water, such as MeOH:H2O normally used on DESI imaging experiments. (Eberlin, et al., 2011). More importantly, such non-aqueous solvent systems are “morphologically friendly”, extracting lipids from tissue sections without causing gross changes in morphology on the microscope scale. Electrochemical processes are inherent in DESI but do not usually make a significant contribution to the mass spectra except in the case of a few particularly easily oxidized or reduced compounds. The substrate used in DESI acts as a DC capacitor, and electrochemical redox reactions can be observed at its surface. (Volny et al., 2008) Besides inherent redox electrochemical reactions, discharge-induced oxidation occurs for specific configurations in DESI.(Pasilis et al., 2008) Such oxidative modifications during ionization generally are avoided to prevent complications in spectral interpretation, but they can be advantageous when used as a deliberate in-situ derivatization method. (Wu et al., 2010)
Figure 2.
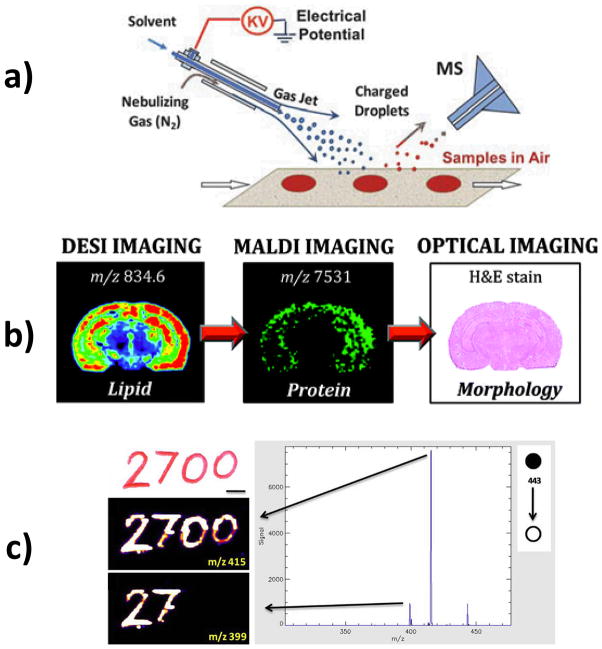
DESI imaging concepts and methods (a) DESI schematic; (b) Sequential analysis of the same coronal section of rat brain by different MS and chemical staining techniques (c) Optical image of (top) handwritten text on paper (. Chemical images of (middle) rhodamine 6G monitored by its fragment m/z 415 (neutral loss of ethylene), (bottom) by its fragment m/z 399 (neutral loss of CO2). Panels a, b, and c are reproduced with permissions from (Ifa, et al., 2010), (Eberlin, et al., 2011), and (Ifa, et al., 2009), respectively.
Optimization of the DESI sprayer is important for obtaining a high quality MS image. First, the solvent system needs to be optimized to obtain an adequate signal level, depending on the sample analyzed and the target compounds (Tawiah, et al., 2010; Wu, et al., 2009). Second, it is necessary to obtain an appropriately small spray spot by adjusting the distance between the spray tip, the substrate, and the inlet of the mass spectrometer. The size and shape of the spray spot is also affected by the solvent being used, the flow rate, the DESI source tip dimensions and the nebulizing gas pressure. (Ifa, et al., 2007; Wiseman et al., 2008) As indicated in studies of DESI sampling, there are three distinct regions in the sampling spot, with most desorption taking place in the inner region. (Green, et al., 2009; Pasilis et al., 2007) It is important to have a well-defined spot on the surface to minimize re-deposition and mixing of the sample during imaging. Third, the spatial resolution and sensitivity need to be balanced. The typical spatial resolution in DESI is ~200 μm, but this value can be reduced to 40μm under particular operating conditions (Kertesz & Van Berkel, 2008) even for biological tissue. (Campbell et al., 2012) However, it should be kept in mind that increasing the spatial resolution will lead to a decrease of signal intensity that is quadratic with spot diameter, and hence affect the quality of ion images and the time needed to record them. Fourth, the spray angle also plays an important role. For example, a typical spray angle of 52° (from the surface plane) provides higher signal intensity, whereas when the sprayer is perpendicular to the substrate (spray angle: 90°) the signal intensity decreases due to decreased secondary droplet transfer efficiency. This perpendicular spray, also called geometry independent DESI, requires an enclosure and a coaxial return tube to optimize sensitivity (Venter & Cooks, 2007) although it has an inherently higher spatial resolution.
Biological tissues can be physically damaged on a macroscopic scale during DESI imaging when conventional solvent systems such as MeOH:H2O and ACN:H2O are used. (Eberlin, et al., 2011) Depletion of tissue samples has been shown to be necessary to sustain stable and abundant signals and high quality images of small molecules in tissues with high protein content such as lens tissues. (Ellis et al., 2010) Because of the damage to the surface tissue, the sample should be rastered in a specific direction with respect to the DESI spray to avoid resampling of splashed material. (Ifa, et al., 2007; Kertesz & Van Berkel, 2008) Nevertheless, recently developed solvent systems such as ACN:DMF and EtOH:DMF (1:1) have been shown to provide abundant signal and high quality DESI images for lipids while preserving tissue morphology and protein content of biological tissues. These non-destructive, histologically compatible solvent systems allow analysis in sequence using immunohistochemistry and MALDI imaging, to be performed on the same tissue section after DESI-MS imaging (Figure 2b) (Eberlin et al., 2011).
Several different types of mass analyzers including triple quadrupoles (Kertesz et al., 2008), FT-ICR (Pol, et al., 2009), and Orbitraps (Manicke et al., 2009) have been used for DESI imaging. When using ion-trap instruments for imaging, the automatic gain control (AGC) is turned off to ensure that the scan time is the same for each pixel. Besides imaging in the full mass scan mode, MS/MS can be utilized to identify particular compounds or to map the distribution of the target molecule(s) with high specificity. (Wiseman et al., 2008) Tandem MSI is useful to provide accurate spatial distributions of particular compounds especially when there is high chemical noise during conventional MSI. Tandem MS is also important as a means of revealing the different distributions of two compounds with the same nominal mass, although the signal requirement is high and more time is required if multiple peaks need to be mapped to determine the spatial distributions of isobaric contributors. One demonstration of this application is in ink analysis. (Ifa et al., 2009) As shown in Figure 2c, the red pen used to create the characters “27” contained a mixture of the pigments rhodamine B and rhodamine 6G. The second pen, used to create the characters “00”, contained only rhodamine B. Although both pigments give ions having the same mass-to-charge ratio (m/z 443), DESI-MS/MS can easily distinguish them by their different fragmentation patterns.
The 2D imaging capabilities of DESI were developed using semi-automatic lab-built moving stages and rapidly applied to analysis of lipids from rat brain (Ifa, et al., 2007; Wiseman et al., 2006) and ink spots separated on TLC plates (Van Berkel & Kertesz, 2006). DESI imaging is readily implemented using high mass resolution mass spectrometers. Exact mass measurements on a high mass resolution mass spectrometer allow more confident assignments of endogenous compounds, and provide distributions for isobaric species, which could not be distinguished by mass spectrometers with unit mass resolution in the full-scan MS mode. Imaging using high resolution mass spectrometers solves this problem but only incompletely since isomers still cannot be distinguished. Nevertheless the improved resolution of the instrument resolves many isobaric species and increases the amount of chemical information obtained from complex samples although at a cost in terms of signal intensity and/or time. (Hu et al., 2005; Manicke, et al., 2009)
2. Applications
a. Biological Tissue Imaging
Lipids play a vital role in a variety of biological processes. Various classes of lipids have characteristic distributions in different tissues, cell membranes, and organelles. DESI imaging of lipids (Eberlin et al., 2011) has been applied to different types of tissues: dog bladder (Dill et al., 2009); rat brain (Wiseman, et al., 2006) and spinal cord (Girod et al., 2010); human prostate (Eberlin et al., 2010), human atherosclerotic plaques (Manicke et al., 2009), human lens (Ellis, et al., 2010), human kidney (Dill et al., 2010), human bladder (Dill et al., 2011) and human brain (Eberlin et al., 2010); porcine and rabbit adrenal gland (Wu, et al., 2010). In a study of human atherosclerotic plaque tissues using DESI imaging, it was found that there are regions with low lipid content in the arterial walls and there are two types of lipid-rich regions with distinctive lipid profiles. (Manicke et al., 2009) These lipid-rich regions likely correspond to areas where lipoprotein particles accumulate as atherosclerotic plaques and the differences in lipid profile are likely related to the stability of the plaque. In addition, a study has shown that lipid distributions in human lens tissue can also be mapped by DESI. (Ellis, et al., 2010) A potentially important application of lipid imaging by DESI is tumor diagnostics because lipid profiles in biological specimens sometimes serve as biomarkers for disease (see below).
Lipids can often be observed easily in DESI, especially members of those lipid classes with high ionization efficiency. However particular lipids can suppress the signals of other lipids and other endogenous compounds. It has been reported for MALDI that lipids need to be removed to achieve optimal signals for peptides in the low mass range. (Seeley & Caprioli, 2008) In DESI this is not necessary but less concentrated lipids can be suppressed and overlapped by lipids of higher concentration and ionization efficiencies in DESI. (Paglia et al.) Of course lipids are not the only endogenous compounds in tissues that can be imaged by DESI, as shown in the following sections, but ion suppression does exist and is obvious for endogenous compounds that fall in the same mass range of lipids. One of the solutions to this problem is to enhance the sensitivity of the suppressed analytes by adduct formation or by switching the polarity of ion detection. (Manicke et al., 2008) The addition of reagents to the solvent is also a good strategy to improve the sensitivity of particular compounds. (See reactive DESI, item e in this section)
DESI-MSI is being developed to differentiate tissue samples as tumor or normal (or less commonly, to identify the type and grade of the tumor) based either on a single marker compound or, more commonly, on the basis of several fatty acid or glycerophospholipid (GP) species. Thin, 15μm, sections from flash frozen tissue samples can be imaged in both the positive and negative ion modes and compared to sections stained with hematoxylin and eosin (H&E) for diagnosis. Although chemical identification of the species is not necessary for a correlation to histopathology data it can be achieved using collision induced dissociation (CID) tandem MS experiments when of interest. DESI-MSI can be used to distinguish between cancerous and adjacent non-cancerous dog bladder tissue in a small set of samples. Five different GPs and three fatty acids in the negative ion mode and four GPs in the positive ion mode showed increased absolute and relative intensities in the cancerous tissue; in addition there was one sphingolipid that showed increased intensity in the normal tissue (Dill, et al., 2009). This study was expanded to human bladder cancer and adjacent normal tissue from 20 different patients (Dill, et al., 2011). DESI imaging allowed the classification of bladder tissue as cancerous or normal based on increased absolute and relative intensities of particular fatty acids and GPs. As seen in Figure 3a, a clear distinction between cancerous and non-cancerous tissue is evident in the selected ion images as well as in the principal component analysis (PCA) derived images. PCA was employed to derive images that give a simple, unsupervised, and general way to visually inspect the data for identification of tumor and normal tissues. (Dill, et al., 2009) When the research was extended to prostate cancer tissue analysis, distinction between cancer and normal tissue was difficult when based on the fatty acid and GP profiles of the tissue sections. Fortunately, in this case cholesterol sulfate serves to make the distinction and was discovered to be present in the prostate cancer and high grade precancerous lesions in 68 tissue samples from 34 patients. (Eberlin, et al., 2010) DESI-MS imaging can be successfully used to image multiple types of tissue and provide results that can be correctly correlated to a diagnosis of cancer either on the basis of the entire lipid profile or on just a few specific marker compounds.
Figure 3.
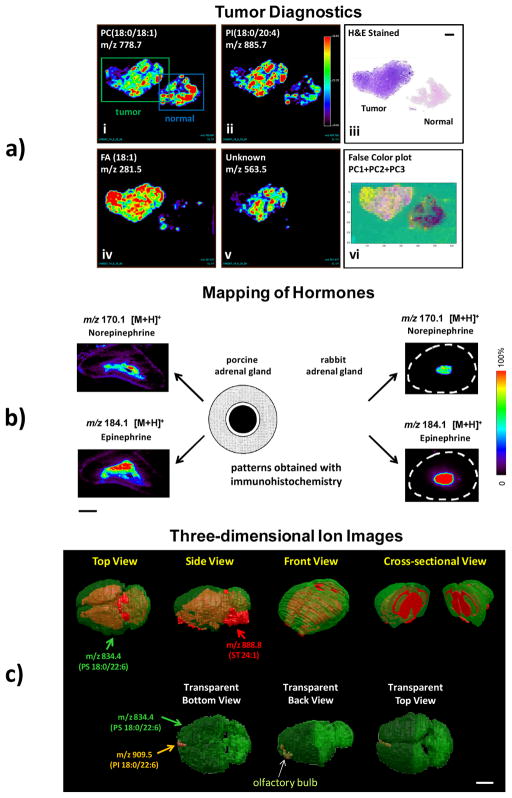
DESI-MS images of various types of tissue (a) Negative ion mode tissue imaging of human bladder tissues including areas of cancer and adjacent normal tissue (i) Ion image of m/z 788.7, PS(18:0:18:1) (ii) Ion image of m/z 885.7, PI(18:0/20:4) (iii) H&E stained tissue sections of the tumor and normal tissues. (iv) Ion image of m/z 281.5, FA(18:1) (v) Ion image of m/z 563.5, FA dimer (vi) PCA-developed images, false color plot of PC1, PC2, and PC3. (b) Two types of distribution patterns of epinephrine and norepinephrine cells in the adrenal medullae of porcine and rabbit adrenal gland, examined using immunohistochemistry. Dotted (gray) area shows adrenal cortex, and white and black areas show epinephrine (E) producing cells and norepinephrine (NE) producing cells regions in the adrenal medulla. The distributions of epinephrine and norepinephrine in the adrenal gland of the porcine and rabbit obtained with DESI imaging are compared with the patterns obtained with immunohistochemistry. The white dashed circle represents the border of rabbit adrenal gland. The distribution patterns of A and NA cells obtained from using immunohistochemistry are adapted from (Suzuki & Kachi, 1996). (c) 3D models of the mouse brain showing the distribution of PS 18:0/22:6 (in green) and ST 24:1 (in red). Length scale bars correspond to 2 mm. Panels b and c are reproduced with permissions from (Wu, et al., 2010) and (Eberlin, et al., 2010) respectively.
More recent studies have shown that DESI imaging can be used for classification of gliomas including oligodendroglioma, astrocytoma and oligoastrocytoma, of different histologic grades and tumor cell concentrations. Classification models built from lipidomic data obtained from 36 human samples showed high recognition capability with more than 97% accuracy under crossvalidation. (Eberlin, et al., 2010; Eberlin et al., 2012) When tested against a validation set for glioma subtype, grade, and tumor cell concentration, 79% agreement with histopathology was achieved, which is remarkable when considering the complexity of nervous tissue and glioma tumors.
Besides lipids, imaging of other small hormone molecules is also an important task but a difficult one in the two main vacuum-based MSI methods, SIMS and MALDI. Interpretation and assignments of small molecules in SIMS imaging is complicated by extensive fragmentation of larger molecules, although the extent of molecular ions generation has been enhanced recently by using cluster ion sources. (Touboul et al., 2004) Similarly in MALDI imaging, the presence of a matrix decreases the sensitivity to small molecules due to overlap with matrix-derived ions. Being both matrix-free and giving soft ionization, DESI allows sensitive and straightforward imaging of small molecules. It has been demonstrated that small hormones such as epinephrine and norepinephrine, antioxidants like ascorbic acid, along with different types of lipids in adrenal gland, can be successfully imaged in either the positive or negative ion mode of DESI. (Wu, et al., 2010) As shown in Figure 3b, the spatial distributions of norepinephrine and epinephrine are easily distinguished in adult porcine and rabbit adrenal glands. In porcine adrenal gland, norepinephrine is most abundant in the central area close to the vein, while epinephrine shows a complementary distribution. The epinephrine signal is greater at the top edge of the adrenal medulla. The results agree with the distribution patterns of epinephrine-storing (E) cells and norepinephrine-storing (N) cells obtained from immunohistochemistry studies (Figure 3b). DESI imaging provides more details on catecholamine distribution as compared to immunohistochemical studies owing to the molecularly specific and sensitive detection of catecholamines using DESI-MS. Another finding is that catecholamines are present not only in the porcine medulla, but also in the porcine cortex, albeit with much lower intensity. This information was not revealed in the immunohistochemical study, likely due to its lower sensitivity. Similarly, the distributions of norepinephrine and epinephrine in rabbit adrenal gland are consistent with data obtained from immunohistochemical studies (Coupland & Hopwood, 1966; Suzuki & Kachi, 1996), which demonstrate that the majority of adrenal chromaffin cells are epinephrine-storing in rabbit. A low concentration of epinephrine, but no norepinephrine, was observed in rabbit adrenal cortex. This information was not revealed from the immunohistochemical study.
Molecular imaging of small pharmaceutical drug molecules and their metabolites directly from tissue sections is one of the other important applications of DESI imaging. In one study the distributions of clozapine and its major metabolites were imaged in brain, lung, kidney, and testis tissues after rats were orally dosed at 50mg/kg and sacrificed at different time points.(Wiseman, et al., 2008) Significantly, the data agreed well with those of a parallel LC-MS study which involved time consuming extraction steps. In another study, DESI-MS/MS imaging and whole-body autoradiography (WBA) were used to image whole-body thin tissue sections of mice dosed intravenously with propranolol or radioactive [3H]propranolol.(Kertesz, et al., 2008)
The construction of 3D ion images is a recent development in ambient mass spectrometry imaging. Comprehensive information on the 3D distribution of different molecules allows direct correlation between the chemical distribution of these molecules and the structure of the tissue. (Seeley & Caprioli, 2012) DESI-MSI has been applied to construct a three-dimensional image or a model of a mouse brain. (Eberlin et al., 2010) Figure 3c shows the simplified 3D image formed by overlaying distribution of the lipids PS (assumed to be the isomer 18:0/22:6) and ST 24:1. In this case the lipids have complementary distributions and together they cover the majority of the brain, making the overlaid 3D model a representation of the entire brain. The distribution of the lipid PS 18:0/22:6 allows a full visualization of the gray-matter region while the lipid ST 24:1 represents the white-matter region of the brain. Any other lipid observed in the mass spectra can be selected to be mapped in 3D. Figure 3c, for example, shows the overlaid distribution of lipids PS 18:0/22:6, ST 24:1 and PI 18:0/22:6. The latter is exclusively distributed in a small region in the frontal part of the brain, associated with the granule cells from the glomerular layer of the olfactory bulb. As is the case for any imaging technique, a trade-off between image quality and time has to be considered in the construction of the 3D models as must the effects of approximations such as the number of tissue sections (z-axis) on resolution and quality of information.
b. Natural Products Imaging
Imaging of natural products using DESI has been demonstrated in a pioneering study by Lane et al. to provide a functional and ecological understanding of particular natural products in tropical seaweed C. serratus. (Lane, et al., 2009) Bromophycolides were found exclusively in association with heterogeneously distributed surface patches of C. serratus at concentrations sufficient for fungal inhibition. It was proposed that C. serratus secretes the antifungal bromophycolides into damaged areas as a defense against microbial infections. This study by Lane et al. is also notable for the fact that the analytes are not highly polar and are identified by their chloride adducts in the intact biological material.
Imprint techniques for indirect imaging of natural products have also been described in the literature. In this approach chemical species present on soft or irregular surfaces are initially transferred to flat hard surfaces prior to ambient MS analysis. For instance, cellulose ester filter membranes were used for imaging of bacterial metabolic exchange (Watrous et al., 2010) while porous polytetrafluoroethylene (PTFE) (Li et al., 2011; Muller et al., 2011; Thunig et al., 2011) and ordinary printer paper (Ifa et al., 2011) were used for indirect plant tissue imaging. This capability of DESI imaging, along with other MSI techniques, can be used for the studies of chemically-mediated biological processes in plant and localized ecological interactions.
c. 2D TLC separation and DESI imaging
Thin-layer chromatography (TLC) is a robust, rapid and inexpensive technique for low resolution separation of organic compounds. Direct coupling of TLC separation with DESI-MS analysis has been reported to be useful in the analysis of different classes of compounds including dyes (Van Berkel et al., 2005), alkaloids (Van Berkel et al., 2007) and peptides (Pasilis et al., 2008). With DESI working in the imaging mode, it is possible to determine the spatial distribution of peptides resulting from 2D separations of protein tryptic digests on high performance thin-layer chromatography (HPTLC) plates without derivatization, peptide ions were identified from data-dependent tandem mass spectra (product ion spectra) using protein database search software according to traditional bottom-up peptide MS/MS analysis protocols (Pasilis, et al., 2008). Investigation of extracts of Salvia divinorum leaves for salvinorin A was also performed by DESI imaging of TLC plates. (Kennedy & Wiseman, 2010)
Separation by 2D-TLC coupled with DESI imaging has also been applied to the analysis of complex lipid mixtures. (Paglia, et al., 2010) The polar lipids are identified by the position of the TLC spot which identifies the lipid class and then the identification is completed using the mass spectrum with confirmation of the identification of individual lipids coming from the MS/MS product ion spectra. This TLC-DESI combination allows identification of a much larger number of lipids than DESI analysis alone, due to ion-suppression and peak congestion in DESI performed using a unit-resolution mass spectrometer.
d. Forensic Applications
The forensic application of DESI imaging to document ink analysis (Ifa et al., 2007) and the imaging of latent fingerprints with DESI has been demonstrated. (Ifa et al., 2008) DESI is capable of producing an image based on any particular ion (drug of abuse, explosives, agrochemicals, etc.) that is present in sufficient quantities to be detected by recording data at different positions within a latent fingerprint. Endogenous compounds such as the fatty acids and lipids known to be present on the skin can also be detected and identified. Latent chemical prints which provide information on the identity of the individual and also information on his/her recent activities, have been successfully imaged from a variety of surfaces, including glass, paper, and plastic. It has also been demonstrated that overlapped prints can be distinguished. Insights may also be provided regarding an individual’s diet and medication from such fingerprints. DESI-MSI was also used for the investigation of counterfeit pharmaceuticals.(Nyadong et al., 2009) Forensic applications of ambient ionization mass spectrometry were recently reviewed. (Ifa, et al., 2009)
e. Reactive DESI Imaging
The ambient ionization methods, especially the spray methods like DESI, involve conditions (temperature, pressure, solvent system, etc.) that are remarkably similar to those encountered in ordinary solution-phase chemical reactions. Besides adjusting the solvent system to optimize desorption, it is sometimes desirable to add reagents to the spray solvent to derivatize the analyte and so enhance specificity and sensitivity. Examples have demonstrated sensitivity increases (often an order of magnitude or more) in the detection of cholesterol, (Wu, et al., 2009) steroids, (Huang et al., 2007) cyclic acetals, (Augusti et al., 2006), cis-diols(Chen et al., 2006) and esters (Song & Cooks, 2007) using different reactions. With reactive DESI in an imaging mode, endogenous compounds, which cannot be observed in conventional DESI, could be imaged. For example, naturally occurring cholesterol in rat brain tissue (~13 mg/g) is easily imaged using reactive DESI with betaine aldehyde as the reagent. The experiment is not successful in detecting cholesterol using conventional DESI with MeOH/H2O (1:1) as solvent.(Wu, et al., 2009) Another example is the enhancement of sensitivity in imaging of malondialdehyde, an effective marker to assess lipid peroxidation processes, by in situ reaction with dinitrophenylhydrazine. (Girod, et al., 2010) Similarly, it is possible to improve the sensitivity of detecting trace biologically important molecules, such as olefins by silver cationization during DESI (Jackson et al., 2010) as was done earlier in SIMS. (Hand et al., 1989; Nicola et al., 1996) The success of these derivatization experiments on the millisecond time scale used for examination of individual pixels in DESI imaging is a consequence of the rate acceleration of chemical reaction in the micro droplets (Girod, 2010). This accelerated droplet chemistry is an intrinsically interesting aspect of DESI.
f. Multivariate Statistical Analysis
One of the intrinsic features of MSI is the large amount of chemical and spatial information contained within each image acquisition. Although single ions can be manually selected and their distribution visually inspected, the total information contained in MSI experiment can be explored and interpreted in a more comprehensive and efficient way by the use of multivariate techniques. The unsupervised technique PCA has been applied in different studies to DESI-MS imaging data in order to evaluate the total information from the mass spectra from different types of tumor and normal tissue samples. For human and canine bladder cancer (Dill, et al., 2009; Dill, et al., 2011), PCA was applied to the spectra acquired from the training set of samples to generate a set of principal components (PCs). All spectra were projected onto the system of coordinates formed by the PCs, and the scores were recorded. The PCs were used to produce a synthetic image for each tissue in the validation set. These PCA images used color-coded pixels to score the underlying spectrum on the coordinate represented by the PC, where the image color indicates an approximate strength of prediction based on the PC. PCA information from all of the ions summarized in a single image was found to perform better in terms of agreement with pathological diagnosis than simple visualization of individual ions. More recently, an interactive hyperspectral approach and PCA was used to interpret the chemical and spatial information obtained from DESI-MS imaging of human cancer and normal tissues.(Pirro et al., 2012) The multivariate strategy facilitated distinction between tumor and normal tissue by automatically connecting regions of PCA score space to pixels of the 2D physical object and further correlating the information with pathological evaluation of the samples.
Supervised techniques, such as partial least squares-discriminant analysis (PLS-DA), have also been successfully used to discriminate tumor from normal tissue based on DESI-MS imaging data, specifically for kidney and bladder cancers.(Dill, et al., 2010; Dill, et al., 2011) For bladder cancer, when the predictive model generated by PLS-DA was applied to a validation set of samples, a misclassification rate of 12% was achieved. For kidney cancer, PLS-DA successfully distinguished tumor from normal tissue for both types of renal cell carcinoma, papillary and clear cell, with misclassification rates of 14.3% and 7.8%, respectively. Besides PCA and PLS-DA, algorithms such as support vector machines (SVM) have also been successfully used to generate classification models for type, grade and tumor cell concentration of human gliomas based on DESI-MS negative ion mode lipid data.(Eberlin, et al., 2012)
B. Laser Ablation Electrospray Ionization (LAESI)
1. Principles
LAESI is a hybrid ambient ionization method based on laser desorption of the analyte followed by its ionization in the course of collisions of the ablated material with charged droplets produced by ESI.(Nemes & Vertes, 2007) The samples are normally mounted on microscope slides, positioned 10–30 mm below the spray axis and 3–5 mm in front of the emitter tip, and ablated at a 90° incident angle using an Er:YAG laser (Figure 4a). A focused mid-infrared laser beam of 2940 nm wavelength excites OH vibrations of water molecules in the sample, the water acting in effect as an intrinsic matrix. The diameters of ablated craters are typically between 200–400 μm and they are 40–50 μm in depth. (Nemes & Vertes, 2007) LAESI, ELDI and LEMS are similar since all techniques rely on ESI post-ionization of neutrals from a laser plume. However, the use of nanosecond mid-IR and UV lasers makes both LAESI and ELDI resonant processes, in contrast to LEMS that uses femtosecond near-IR laser to produce non-resonant laser ablation of the sample (see next section). Calibration curves from 1nM to 10 mM showed good linearity with correlation coefficients of at least 0.995 for pure samples of verapamil and reserpine. The limits of detection for these compounds were 8 and 25 fmol, respectively.(Nemes & Vertes, 2007)
Figure 4.
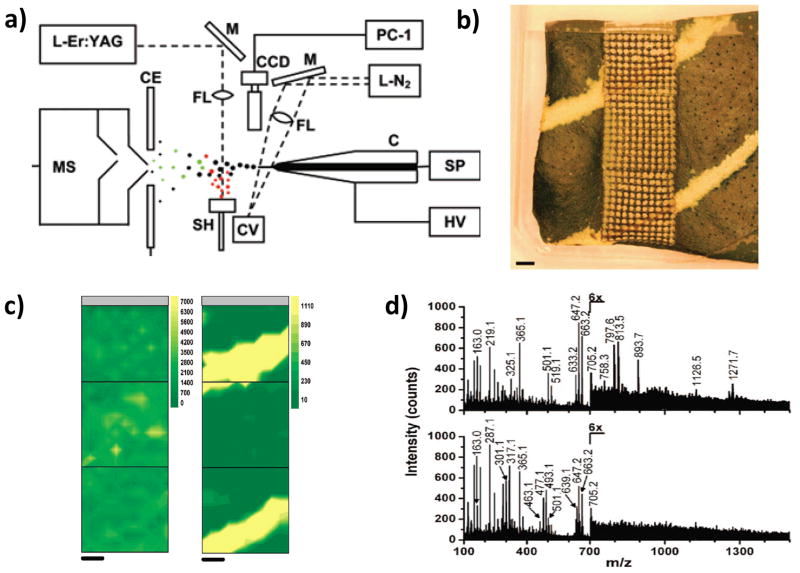
LAESI MS imaging (a) Schematic diagram of the LAESI instrumentation. (C, capillary; SP, syringe pump; HV, high-voltage power supply; L-N2, nitrogen laser; M, mirrors; FL, focusing lenses; CV, cuvette; CCD, CCD camera with short-distance microscope; CE, counter electrode; OSC, digital oscilloscope; SH, sample holder; L-Er:YAG, Er:YAG laser; MS, mass spectrometer; PC-1 to PC-3, personal computers). (b) Variegated A. squarrosa leaf probed with LAESI-MS while rastering the surface with a focused infrared laser beam over three adjacent 4 mm by 4 mm areas. The live tissue surface was ablated from a circular area (350-μm diameter) with a 400-μm step size. (c) Contour plots show the distribution of ions detected at m/z 663 and m/z 493, respectively. (d) Mass spectra obtained at different regions of the leaf (top: green sectors, bottom: yellow sectors). Length scale bars correspond to 1 mm. Panel a is reproduced from (Nemes & Vertes, 2007) with permission. Panels b–d are reproduced from (Nemes, et al., 2008) with permissions.
2. Applications
The application of LAESI to MSI was introduced by Vertes and coworkers. in 2008.(Nemes, et al., 2008) Mass spectrometric analysis of Aphelandra squarrosa leaf tissue was performed using a TOF mass spectrometer. Leaf surface of area 4 × 12 mm2 was mapped by using 400 μm × 400 μm pixels (Figure 4b). The plant data showed many singly and doubly-charged ions. The distributions of kaempferol (diacyl coumaryl-rhamnoside), m/z 663, present in both green and yellow sectors of the leaf (Figure 4c left); methoxykaempferol glucoronide, m/z 493, present only in yellow sectors (Figure 4c right); and the unassigned ion at m/z 813, more abundant in the green sectors, were in good agreement with the optical image of the variegation pattern.(Nemes, et al., 2008) More than 40 primary and secondary metabolites were detected and 36 of them were identified and assigned chemical structures (Figure 4d). The creation of 3D images based on deep profiling analysis was explored in a follow-up study where metabolites in the leaves were imaged.(Nemes, et al., 2009) Laser pulses at 0.2 Hz repetition rate were used to ablate the samples as before; the average output energy of a laser pulse was 0.1 mJ ±15% and 1.2 mJ ±10% for analysis and ablation, respectively. This translates into a fluence of ~0.1 and ~1.2 J/cm2 at the spot. Six-step depth profile acquisitions for each point on a 22 × 26 grid across a 10.5 × 12.5 mm2 area and on a 24 × 16 grid across a 11.5 × 7.5 mm2 area were performed for Spathiphyllum lynise and Aphelandra squarrosa, respectively. Several metabolites were observed and their identities assessed by tandem mass spectrometry. (Nemes, et al., 2009)
Mass spectrometric analysis of rat brain sections were also investigated. (Nemes et al., 2010) Some changes in the original set-up were implemented in order to avoid dehydratation of the sample in the ambient environment and the possibility of molecular migration upon tissue melting. A Peltier cooling stage with optimized heat transport kept the brain sections frozen during analysis and a dry nitrogen environment prevented moisture condensation on the sample surface. The lateral resolution used for these experiments was 200 μm. High-resolution MS was employed to deconvolute the distributions of different species with the same nominal mass. Pearson product-moment cross-correlation coefficients were employed to classify and correlate the distribution of the observed molecules in the rat brain. (Nemes, et al., 2010) LAESI was also used for in vitro analysis of metabolites of Torpedo californica electric organ (Sripadi et al., 2009) and for the analysis of single cells using sharped optical fiber tips.(Shrestha & Vertes, 2009) Cells with different age or pigmentation showed different metabolic profiles. The construction of molecular images using single cells as single pixels has been suggested for mapping biological tissues.(Shrestha & Vertes, 2009)
LAESI was also employed for in situ cell-by-cell imaging of plant tissues. Imaging of single epidermal cells in onion (Allium cepa) and clusters of 3–4 achlorophyllous cells in citrus leaves (Citrus aurantium) was successful with a lateral resolution of 30 μm (spot areas). Multivariate statistical analysis on LAESI-MS data was performed using the method of orthogonal projections to latent structures discriminant analysis (OPLS-DA) in order to identify the metabolites most responsible for the variance between cell populations.(Shrestha, et al., 2011)
C. Atmospheric Pressure Femtosecond Laser Desorption Ionization (fs-LDI) and Laser Electrospray Mass Spectrometry (LEMS)
1. Principles
Atmospheric pressure femtosecond laser mass spectrometry (AP fs-LDI IMS), a recently reported atmospheric pressure imaging MS technique (Coello, et al., 2010), also abbreviated as LEMS (laser electrospray mass spectrometry) when combined with ESI (Judge, et al., 2010) is unique in having some of the advantages of laser based ionization methods but for also having the possibility of being applied to in-vivo chemical imaging and other complex problems. In concept, the technique is also related to recently developed near infrared (NIR) femtosecond laser MALDI (Wichmann et al., 2009), however no matrices are needed in the AP fs-LDI experiment due to the very high peak power density of the laser.
Sample ablation and analyte ionization is achieved in AP fs-LDI MSI by using focused NIR femtosecond laser pulses without the addition of any laser absorbing matrix, either native or external. As with other ambient ionization techniques, sample preparation and handling are significantly simpler than in traditional techniques. The experiments of Coello et al. were performed using a custom-made AP femtosecond laser ion source, containing a motorized xy stage which holds the sample approximately 5 mm from the sample cone of a time-of-flight mass spectrometer. Fentosecond laser pulses (~50 fs, 1 kHz, centered at 800 nm, ~1014 W/cm2) are focused on the sample using a 5x objective, with a typical spot diameter of approximately 20 μm. Focusing of the laser at the target position is necessary to assure optimal and reproducible results. In the current set-up, the sample is normally positioned between a sample cone and a repeller electrode such that the potential difference assists in directing the ion plume toward the mass spectrometer inlet (Figure 5a).
Figure 5.
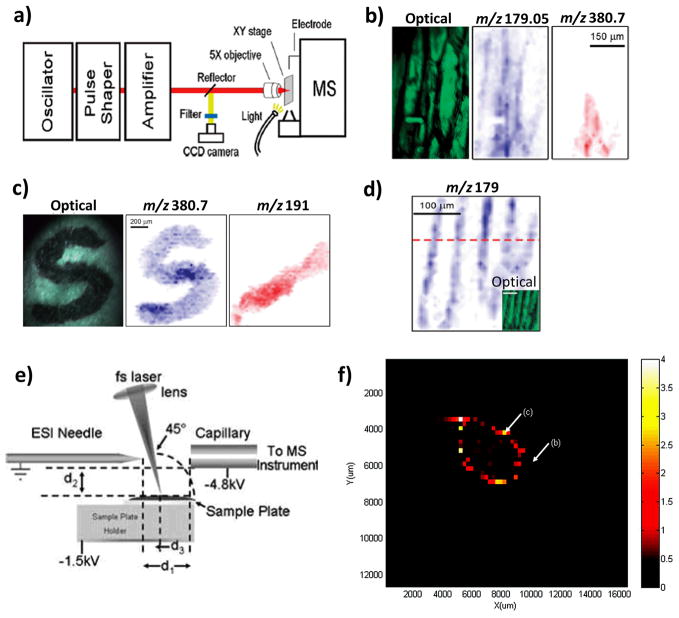
(a) Schematic of the atmospheric pressure femtosecond laser desorption ionization imaging mass spectrometer (AP fs-LDI IMS). (b) Optical image of onion epidermis cells and ion images showing the distribution of deprotonated glucose (m/z 179.05) and triiodide (m/z 380.7). (c) Optical image of a “S” character dye pattern recorded under ambient conditions. Negative ion images of the distribution of triiodide (m/z 380.7), and citrate (m/z 191.1), are also shown. (d) Negative ion image of the distribution of m/z 179 generated by probing the onion epidermis tissue at a spatial resolution of 10 μm. The inset shows the corresponding optical image. (e) Schematic of the atmospheric pressure femtosecond laser electrospray ionization mass spectrometry (LEMS) setup. (f) Positive ion LEMS image of the distribution of oxycodone (m/z 316) on a metal slide. Panels a–d are reproduced from (Coello, et al., 2010)with permissions. Panels e and f are reproduced with permissions from (Brady, et al., 2009) and (Judge, et al., 2010), respectively.
NIR femtosecond laser pulses are characterized by having very high peak power densities (~1014 W/cm2) which allows direct nonresonant desorption and ionization of analytes (Gamaly et al., 2002). For this reason, it is not necessary to add a laser absorbing matrix to assist in desorption and ionization, greatly simplifying sample preparation. At the same time, the absence of an absorbing matrix causes analytes in the sample to absorb laser radiation directly. For example, glucose arising from the fragmentation of cellulose has been observed during the analysis of onion epidermis. However, other experiments have shown that the intact molecular ion of low molecular weight compounds (<400 Da) in solid standard samples is typically present in the mass spectrum. So far, the m/z range of analysis offered by the technique is very limited (up to m/z 400) and experimental results indicate that the ion yield of AP fs-LDI decreases with increasing m/z. This effect can be explained by the inefficient ablation of heavier fragments or inefficient transport of ions from the sample surface into the mass spectrometer using the present source configuration. However, the method is still in a stage of rapid evolution and improvement.
One of the main advantages of the technique is that it offers the highest lateral resolution ever reported for an ambient ionization technique. The lateral resolution of 10 μm reported for onion epidermis is a significant improvement over what has been previously obtained in ambient imaging analysis such as DESI, PESI, LAESI (~30–50 μm) and a step forward towards chemical visualization of individual cells in ambient MSI. Another advantage of the short-pulse technique is that the damage resulting from the laser analysis is superficial and most plant and animal tissues samples could survive the analysis, opening the possibility of applications to in vivo chemical imaging. In the early studies done so far a limit of detection (LOD) for citric acid was found to ~500 fmol (Coello, et al., 2010) a value which is close to the limits of detection of few fmol reported for other ambient techniques (Nemes & Vertes, 2007).
The ablated material can also be captured by charged droplets and this event followed the ES ionization (Figure 5e). (Brady et al., 2009) The current LEMS setup uses acidified methanol:water (v/v 0.1% acetic acid) as electrospray solvent delivered at 2μL/min. Trace amounts of Claritin (750pmol), Oxycodone(650pmol) and Atenolol (430pmol) were successfully detected on different surfaces such as steel, glass and wood. (Judge, et al., 2010)
2. Applications
A few applications demonstrating analytical capabilities have been reported for this new technique. The optical image and negative ion mode image of deprotonated glucose, m/z 179.06, and triiodide, m/z 380.64 with which the onion tissue was partially stained are shown in Figure 5b,. Negative ion images of m/z 380.6 (triiodide) and m/z 191.09 (citric acid) show excellent agreement with the optical image of the “S” character and the diagonal mark of citric acid (Figure 5c). Onion epidermis tissue was analyzed at a lateral resolution of 15 μm under atmospheric pressure conditions in the negative ions mode. Finally, the technique was applied to image onion epidermis cells with lateral resolution of 10 μm (Figure 5d), seen in the negative ion image of deprotonated glucose ions at m/z 179.06. This application proves the capabilities of the technique to perform chemical imaging at a cellular level at atmospheric pressure. LEMS has been applied to map the distribution of oxycodone in a metal plate with a lateral resolution of 250μm. (Figure 5f)(Judge, et al., 2010)
D. Infrared Laser Ablation Metastable-Induced Chemical Ionization (IR-LAMICI)
1. Principles
IR-LAMICI, Figure 6a, is a hybrid plasma- and laser-based ambient ionization source that combines IR laser ablation with metastable-induced chemical ionization, the latter experiment being similar to DART.(Galhena, et al., 2010)
Figure 6.
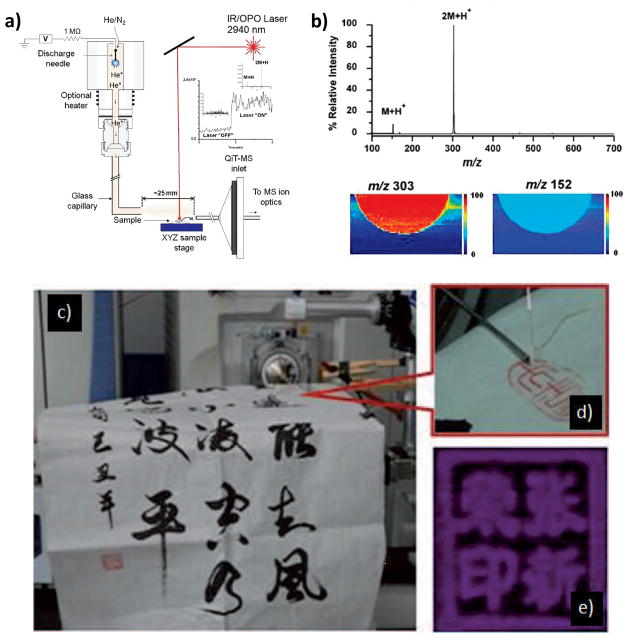
IR-LAMICI and LTP MS imaging experiments. (a) Schematic diagram of the IR-LAMICI. (b) IR-LAMICI MS analysis of a Tylenol tablet, with the insets showing false-color scale IR-LAMICI MS images of the distribution of acetaminophen monomer (m/z 152) and dimer (m/z 303) ions on the tablet. (c) Analysis of calligraphy patterns using LTP-IMS. (d) Expanded view of the LTP probe. (e) Chemical image of the inkpad of seals on rice paper recorded using a LTP probe. Panels a and b are reproduced from (Galhena, et al., 2010) with permissions. Panels c–e are reproduced from (Liu, et al., 2010) with permissions.
In IR-LAMICI a Nd:YAG laser operated at 2940 nm is pulsed orthogonally from above the sample for laser ablation with a spot size of approximately 300 μm in diameter.(Galhena, et al., 2010) A portion of the ablated material from the sample surface is directed toward the MS inlet by the flow of the metastable plume. It is within this metastable plume that the gas-phase chemical ionization of the analyte occurs, generating protonated or deprotonated species which are observed in the positive and negative ion modes. (Galhena, et al., 2010) It was observed that both the energy of the laser beam and the position of the inlet capillary of the MS greatly impact the ion intensity observed. (Galhena, et al., 2010)
In experiments conducted on red macroalga Callophycus serratus, which has also been imaged by DESI(Lane et al., 2009), considerable fragmentation was observed, suggesting that a larger amount of internal energy is deposited in IR-LAMICI than in DESI. (Galhena, et al., 2010) Imaging experiments conducted on a Tylenol tablet showed that the signal obtained from the protonated dimer was larger than that of the protonated monomer. This preference for the formation of dimers can be explained by the large concentration of neutrals generated by laser ablation, favoring self-dimerization in ion/molecule reactions, and by the use of an ambient temperature metastable flux which coupled with collisional cooling promotes clustering. (Galhena, et al., 2010) In the imaging experiment it is thought that 15–30 pg of acetaminophen, removed from the tablet in a 10 μm laser depletion layer, gives a detectable signal. While exact resolution figures were not reported, an advantage of IR-LAMICI is that resolution is determined only by the size of the laser pulse and not by the size of the metastable gas jet. (Galhena, et al., 2010)
2. Applications
IR-LAMICI has been used successfully to directly analyze pharmaceutical tablets, including Tylenol and a counterfeit artesunate antimalarial tablet. It has also been used to analyze tissue samples of the red macroalga Callophycus serratus in the negative ion mode, detecting a secondary metabolite involved in the surface-mediated chemical defense system of the algae. (Galhena, et al., 2010) Successful imaging of a Tylenol tablet, area 14 by 7 mm, took approximately 40 minutes to image at a spatial resolution of 300 μm. Figure 6b. (Galhena, et al., 2010) IR-LAMICI will further expand the range of ionization techniques available for small molecule imaging in the ambient environment.
E. Low Temperature Plasma (LTP) and Laser Ablation coupled to Flowing Atmospheric-Pressure Afterglow (LA-FAPA)
1. Principles
A number of closely related plasma-based ion sources have been reported including the plasma assisted desorption ionization (PADI) source, the dielectric barrier discharge ionization (DBDI), LTP and LA-FAPA. The LTP source is a localized ambient ionization technique developed from DBDI.(Na et al., 2007) The most effective of these ion sources employ dielectric barrier discharges produced at atmospheric pressure with a dielectric layer between two electrodes supplied with an alternating voltage. A stable low-temperature plasma with a large density of high energy electrons is formed. In the initial DBDI setup, a hollow stainless steel needle was used as the discharge electrode. A discharge gas (such as He, Ar or N2) flowed through the needle and a copper sheet was used as the counter electrode. A glass slide which operates as both the discharge barrier and sample holder was inserted between the electrodes and an alternating voltage of 3.5–4.5kV with a frequency of 20.3 kHz was used to generate a stable plasma between the tip of the needle and the glass slide. This configuration was used to desorb and ionize amino acids with good reproducibility (RDS<6%) and sensitivity (LOD< 4pmol). (Na, et al., 2007) A low temperature plasma probe of unique configuration was developed to simplify the sampling and imaging of surfaces.(Harper et al., 2008) This configuration is different from DBDI because the counter electrodes are placed within the probe without the requirement that the sample be placed between the electrodes. It allows the analysis of any type of object including bulk aqueous solutions and it has been recently applied to the evaluation of drugs of abuse in biofluids (Jackson et al., 2010), screening of agrochemicals in foodstuffs (Wiley et al., 2010), and trace detection of melamine in foods and other complex mixtures (Huang et al., 2009). An improved LTP probe was used to imaging artwork as described below. This probe consists of two aluminum foil electrodes and a fused capillary.(Liu, et al., 2010)
LA-FAPA is a hybrid laser and plasma-based ambient ionization method. The sample is first ablated by a focused UV laser beam and the resulting ablated material is transferred by a nitrogen stream to the flowing afterglow of a helium atmospheric pressure glow discharge ionization source. (Shelley, et al., 2008). The FAPA ionization source uses a glow discharge operated in helium in the flowing afterglow mode to generate excited atoms and ions at atmospheric pressure; the discharge is contained in a Teflon enclosure.(Andrade et al., 2008) Since LA-FAPA requires laser ablation and ionization chambers and it does not occur in the open environment, it is not strictly ambient, but is closely related.
In typical LA-FAPA experiments, a Nd:YAG laser employing 266nm radiation is focused on the sample surface at spot sizes between 10 and 300 μm. The resulting vapor/aerosol generated by the ablation process is carried in a stream of nitrogen through a teflon tube where it is mixed in a second chamber with the afterglow from the discharge.(Shelley, et al., 2008) Both the spatial resolution and the tendency to thermolysis are improved by the use of the laser rather than the plasma for desorption. The use of laser ablation also allows depth profiling using successive laser pulses, and a sampling depth of approximately 36 μm has been achieved.(Shelley, et al., 2008) Two-dimensional imaging can be achieved under typical conditions employing a 200 μm laser spot at 80 μm/s with a line spacing of 150 μm, the resulting horizontal resolution is 63 μm and the vertical resolution is 178 μm.(Shelley, et al., 2008) However, when a smaller laser spot of 20 μm is used with scanning at 5 μm/s, the spatial resolution is better than 20 μm.(Shelley, et al., 2008) Imaging with LA-FAPA is limited to small molecules, less than 1000 Da, but by using a plasma source a broader range of compounds can be ionized but with the decreased performance noted above.(Shelley, et al., 2008).
2. Applications
The LTP probe was recently used to image Chinese paintings and calligraphy. Figure 6c–e (Liu, et al., 2010). Since the technique does not require spray solvents or the introduction of matrices, these sources of damage, modification or contamination of the sample are avoided. Initial experiments were performed on rice paper printed with black ink and inkpads examined at different scan velocities (100–400um/s). After analysis, the samples were submitted to scanning electron microscopy (SEM) and no damage was observed on the surface. The spatial resolution, determined by a pattern of lines printed on glossy paper, was around 250 μm. The inside diameter of inner capillary is the most import parameter to control the length and diameter of the plasma plume and therefore the spatial resolution.
Imaging experiments were performed to discriminate between genuine and counterfeit seals. An area of 38x20 mm2 was scanned with a pixel size of 150x150 μm2. The chemical images of all seals were visualized by selecting a specific ion present in all samples (m/z 83). Discrimination between the genuine and counterfeit seals was achieved by mapping the ion of m/z 116 present only in genuine seals. Based on the results and the simple implementation of the technique, the authors anticipate that LTP can contribute to the identification, conservation and restoration of valuable artwork.(Liu, et al., 2010)
LA-FAPA has been used to perform chemical imaging of small molecules from solid samples. In the first imaging studies a solution of ink doped with caffeine was used to print the Indiana University logo (5.6 mm wide) which was imaged using LA-FAPA in a total time of less than 30 minutes, with 63 μm (x-driection) and 178 μm (y-direction) resolution.(Shelley, et al., 2008) In the same study the applicability of LA-FAPA for imaging tissue samples was also investigated. In the first example, lidocaine was spotted onto a slice of wet turkey tissue and imaged successfully with the chemical image matching the white-light image. In the second example, celery veins into which caffeine had been introduced were imaged using LA-FAPA. The celery was immersed in a solution of blue dye and caffeine and subsequently sliced and imaged successfully to identify the vein.(Shelley, et al., 2008) Currently the technique is limited in mass range to compounds that give ions of less than m/z 1000. Although the application of LA-FAPA to tissue imaging is at a preliminary stage, it shows promise as an additional resource to the field of ambient MSI.
F. Desorption Atmospheric Pressure Photoionization (DAPPI)
1. Principles
DAPPI was introduced by Haapala et al. in 2007 (Haapala et al., 2007) and like DAPCI it broadens the range of ambient MS analytes to include neutrals and nonpolar compounds. DAPCI uses an atmospheric plasma to create reagent ions that are directed at the sample surface for chemical desorption while DAPPI experiment uses a heated nebulizer microchip to deliver a hot mist of vaporized solvent such as toluene or acetone for sample desorption and, as ionization agent, a photoionization lamp (10eV). A heated jet of mixed nebulizer gas and spray solvent vapor is directed at the sample and promotes thermal desorption of the analytes (Figure 7a). Surface temperatures up to 350°C have been reported. The analytes then undergo the classic gas-phase mechanism of photoionization. The initial photoionization products are converted via ion/molecule reactions to [M+H]+, [M−H]−, M+•, M−•, [M−H+O]− or [M-2H+2O]− ions, depending on the analyte, spray solvent and ionization mode employed.(Luosujarvi et al., 2008) For instance, acetone and hexane were identified as good spray solvents for polar compounds such as testosterone and verapamil since they produce proton-donating reactant ions and therefore generate [M+H]+ ion species. The use of anisole and toluene as spray solvents generated M+• ions and these reagent ions are highly appropriate for the analysis of nonpolar compounds such as anthracene and tetracyclone. An ion trap mass spectrometer was used for the experiments. The thermal conductivity of the surface is an important parameter. Materials with low thermal conductivity such as polymethylmetacrylate can be locally heated to higher temperatures improving desorption of the analytes in contrast to high thermal conductivity materials such as aluminum or silicon.(Luosujarvi, et al., 2008) Limits of detection for anthracene, testosterone, MDMA (3,4-methylenedioxymethamphetamine) and verapamil were 670, 83, 56 and 56 fmol, respectively.(Haapala, et al., 2007) DAPPI has also been successfully employed for detection and analysis of illicit drugs. (Luosujarvi et al., 2009)
Figure 7.
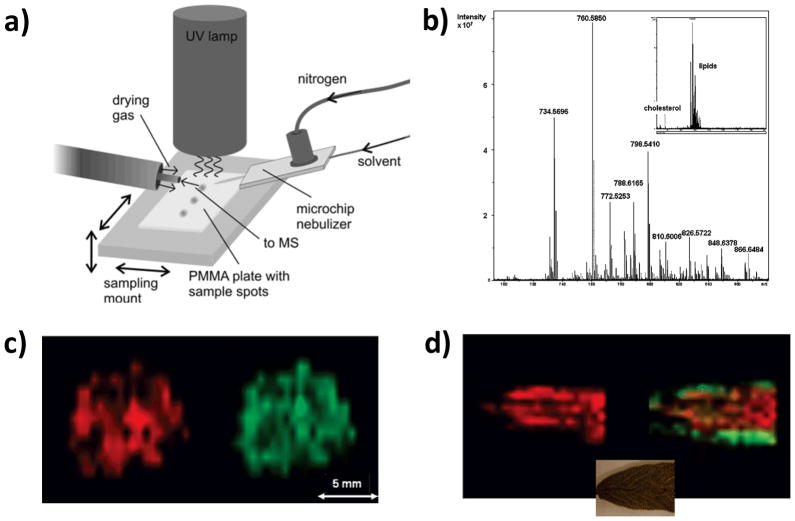
DAPPI and DESI imaging data and methods (a) Schematic diagram of the DAPPI setup. (b) DESI-MS spectrum of mouse brain tissue section obtained from a single point using an average of three scans acquired on an high-resolution mass spectrometer. Detail of the m/z 700–900 region with dominant phospholipid peaks. Inset: Overall spectrum of the whole m/z 250–1800 region. (c) Two-dimensional distributions of selected lipid-originating ions in mouse brain tissue section, m/z 760.5850 (red) and m/z 810.6006 (green). (d) Distribution of an ion at m/z 430.3810 (red) on sage leaf and combined distributions of peaks at m/z 301.2166 (green) and m/z 315.0863 (red). Whereas m/z 301 is located more on the side of the leaf, m/z 315 is centered. The inset shows the part of the leaf that was imaged. Panels a is reproduced from (Haapala, et al., 2007) with permission. Panels b–d are reproduced from (Pol, et al., 2009) with permissions.
2. Applications
The application of DAPPI to MSI was first demonstrated by Pol et al. (Pól, et al., 2009) These authors presented a lab-built modular platform for DESI and DAPPI coupling these ion sources to a commercial high-resolution Fourier transform ion cyclotron resonance mass spectrometer (FTICR-MS). After custom software modifications, the platform could be controlled via the regular user interface and the data analyzed using standard MALDI imaging software. The DAPPI set-up used was essentially the same as that proposed by Haapala et al.(Haapala, et al., 2007) Proof-of-concept images were acquired at high mass resolution from brain tissues, where dominant signals due to phospholipids could be seen (Figures 7b and c), and from dehydrated leaf of Savia (sage) (Figure 7d). Due to the heated nebulizer microchip and the glass surface used in the experiments, the maximum spatial resolution was limited to approximately 1 mm. Chemical species such as tocopherol (seen as the radical cation, m/z 430), carnosol ( as the protonated molecule, m/z 331), and methyl carnosic acid (m/z 301) were mapped by DAPPI-MS on sage leaf. For brain tissues, the cholesterol fragment at nominal mass 369 was practically the only peak in the spectrum of DAPPI when acetone was used as a dopant. (Pol, et al., 2009). One disadvantage of this technique is the possibility of thermal degradation of the sample.
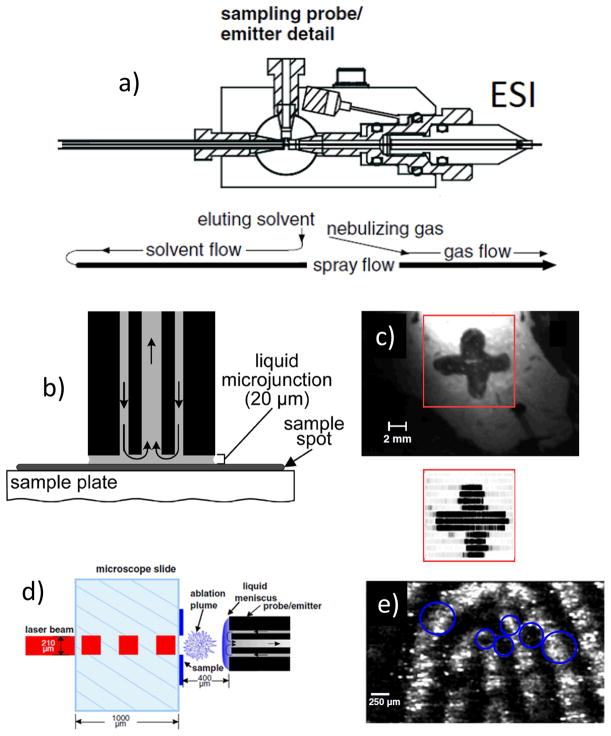
G. Probe Electrospray Ionization (PESI)
1. Principles
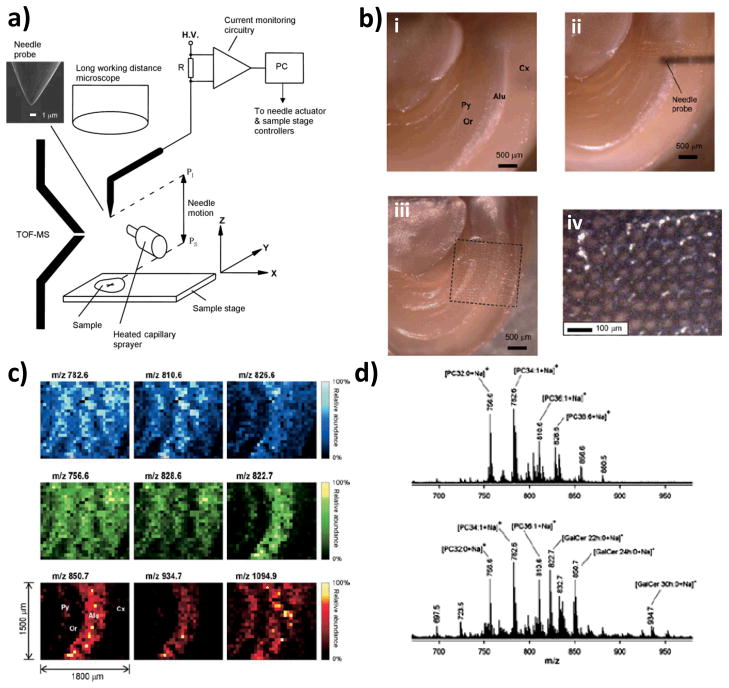
Probe electrospray ionization (PESI) is performed in the ambient environment and it requires taking multiple individual samples with a solid needle. This method, initially applied to the analysis of proteins and peptides in liquid samples (Hiraoka et al., 2007), is unique in that an electrospray is generated directly from a sharp solid needle containing a thin film of sample on which high voltage is applied. The needle used in PESI has two functions, one being as the sampling probe and the other as the solid ESI emitter. Therefore, the hallmark of this method as applied to mass spectrometry imaging arises from the fact that the sampling occurs under ambient conditions and the sampling area is dependent on the needle probe dimension, yielding lateral resolution of ~100 μm or less.
In imaging PESI experiments, sampling occurs when the needle probe, attached to the automated PESI source, is stuck into the surface of a solid sample present on a surface below the probe position. (Chen, et al., 2009). Penetration of the needle into the sample transfers a small amount of biological material from the sample. The needle is withdrawn then moved to a position close to the ion-sampling orifice of the mass spectrometer, where a high voltage is applied and electrospray is initiated (Figure 8a). The high electric field generated on the tip of the sharp needle is indispensable for ionization and desorption to occur (Chen et al., 2008). In fact, electrospray only occurs once a sufficient electric field is established, allowing mass spectra of the molecules in the biological material present in the probe needle to be obtained. It is important to note that in the imaging configuration of the PESI source, an auxiliary sprayer is used to supply uncharged solvent molecules to the probe tip when the needle is at the ionization position. This auxiliary spray is necessary because the amount of biological fluid that is picked up by the needle is so small that it can dry out very quickly preventing electrospray from even being initiated. The water arising from the heated capillary sprayer, which is aligned perpendicularly to the needle probe, condenses on the needle tip forming water droplets, and these droplets successfully re-wet the sample adhering to the needle probe so facilitating the electrospray process. Figure 8a shows a schematic diagram of the PESI-MSI system.
Figure 8.
PESI-MS imaging (a) Simplified schematic of imaging PESI-MS system consisting of a PESI ion source, an auxiliary heated capillary sprayer, and a three-axis precision sample stage. PI = ionization position and PS = sampling position. b) Microscopic images of the mouse brain section taken (i) before the measurement, (ii) during the raster scan of the imaging PESI-MS, and (iii) after the completion of the measurement and (iv) close-up inspection of the holes made by the solid needle. (c) PESI ion images of the mouse brain section for different ions. (d) Positive ion mode PESI mass spectra obtained from two sampling spots from a rat brain. All panels are reproduced from (Chen, et al., 2009) with permissions.
The x, y and z-axes of the sample stage are driven by stepper motors and the motion of the probe needle, sample stage and mass spectrum acquisition are controlled by a computer program. This automated process allows reliable mass spectrometric images to be recorded, in which each mass spectrum corresponds to the ions originating from a particular spot. The sampling and ionization cycles are repeated until the entire area of interest within the sample has been analyzed. Because sampling is performed consecutively by purely physical means, it is expected that sample carrying-over between adjacent spots might occur in PESI imaging. This is avoided, however, by addition of solvent from the heated capillary sprayer to the needle tip effectively cleaning the needle. Another important parameter to be considered in PESI imaging is the sampling depth of the needle probe. Depending on the depth of the needle penetration and how flat the sample is, the amount of sample picked up by the needle could vary during raster scanning, affecting reproducibility. Therefore, a constant sample depth, normally set at 100μm, has to be maintained in the PESI imaging experiments. The depth of penetration also influences the lateral resolution, which is determined by the diameter of the needle probe and the spacing in between two adjacent holes. Resolution of 60μm has been reported for PESI imaging of mouse brain even though higher resolution is possible by decreasing the size of the needle probe. In a recent report, resolution of 50μm was demonstrated with a heated probe using printed patters of ink on paper as test materials. This technique, termed thermal desorption proximal probe with secondary ionization (TD/SI-MS) (Ovchinnikova et al., 2011), is related to PESI considering that both techniques require an initial contact between the probe and the material on the surface to produce images. However, TD/SI-MS promotes thermal desorption of the analytes in a similar way to DAPPI (see section F) in contrast to PESI in which the probe is used as an ESI emitter (Figure 8a).
2. Applications
Proof-of-concept experiments using PESI imaging (Chen, et al., 2009) included studies of the distribution of different phospholipids in a rat brain section (Figure 8b). This study used PESI imaging at a lateral resolution of 60μm and a penetration depth of 100μm. Ion images (30 × 25 pixels) showing the distribution of different ions observed in the grey and white matter of the brain are shown in Figure 8c.
The main ions observed in the positive ion mode are sodiated and potassiated phosphatidylcholines (PC) and galactoceramides (GalCer), Figure 8d. Studies of depth profiling by PESI-MSI were also performed on porcine retina on which the results showed that the distribution of PC’s vary with depth.
While this method is advantageous in terms of the lateral resolution it provides with a better defined analysis size, it should be noted that cross-talk of ion signal between pixels and contaminations of compounds could occur. Furthermore, the unknown amount of sample that is probed by the solid needle could compromise accurate quantitative and even qualitative analysis. An encouraging recent development is the observation that different types of compounds are released or electrosprayed from the needle as a function of time, in the order of their surface activity. (Mandal et al., 2011) For example, for human breast cancer tissue, proteins such as α and β chains of hemoglobin were observed first in the mass spectrum as the dominant ions, while lipids were observed at a much later stage, just before the liquid droplet on the needle was depleted. These results demonstrate that PESI has the advantage of the suppression effect with analytes being detected separately in the order of their surface activity values.
H. Liquid Microjunction Surface-Sampling (LMJ-SSP), Nanospray Desorption Electrospray Ionization (nano-DESI) and Liquid Extraction Surface Analysis (LESA)
1. Principles
The liquid microjunction sampling technique LMJ-SSP uses a system of coaxial tubes in which a solvent flow captures the analytes at one extremity and allows their ionization by ESI at the other extremity (Figure 9a). The system can be used by creating a direct liquid microjunction between the probe and the sample surface (Figure 9b) (Van Berkel et al., 2002) or else in a non-contact mode. In this latter case, transmission geometry laser ablation is used to desorb the sample into an ablation plume which is captured by the liquid meniscus of the probe (Figure 9d). (Ovchinnikova et al., 2011) A methanol/water solvent is normally used and the flow rate (10–20 μL/min) adjusted depending on the system details. Other examples of solvent probe methods include the scanning mass spectrometry (SMS) probe (Kottke et al., 2010), nano-DESI (Roach et al., 2010), and LESA (Eikel et al., 2011). Nano-DESI uses minute amounts of solvent confined between two capillaries comprising the nano-DESI probe and the solid analyte for controlled desorption of molecules present on the substrate followed by ionization through self-aspirating nanospray. (Laskin, et al., 2012) In LESA, a pipette tip is robotically moved close to the surface of interest and dispenses a small volume of approximately 1 μL of extraction solvent (MeOH:H2O 80:20 v/v 0.1% formic acid) forming a liquid junction that extracts the material from the surface. After the extraction, the solvent is aspirated back and the tip placed in an ESI chip to generate a nano-electrospray. (Eikel, et al., 2011). Note that LMS-SSP, nano-DESI, and LESA are quite similar to each other since all techniques employ liquid extraction or solubilization of the analytes followed by classic ESI or nanoESI.
Figure 9.
(a) Schematic of LMJ-SSP/ESI. (b) Liquid microjunction formed between the probe and the surface of the sample plate. (c) Rat liver section with reserpine (+ sign), chemical image shown in the inset. (d) Laser ablation and capture of the ablated material by the liquid meniscus of the probe. (e) Chemical image of a fingerprint blotted on glass with ink. The blue circles show the regions of interest with an apparent resolution of 100μm. Panels a and c are reproduced from (Van Berkel, et al., 2008), panel d and e are reproduced from (Ovchinnikova, et al., 2011), and panel b is reproduced from (Van Berkel et al., 2009) with permission.
2. Applications
The application of LMJ-SSP probe in conjunction with ESI was successfully demonstrated for MSI. In an initial experiment, an ink stamp (37 mm × 10mm) containing rhodamine B was mapped on paper. Thirteen lanes were scanned with a distance of 1 mm between the lanes. The eluting solvent consisted of a mixture of methanol/0.1% acetic acid 1/1 (v/v), delivered at a flow rate of 15μL/min. The distribution of rhodamine B was monitored in the SRM mode by the transition m/z 443 -> 399. (Kertesz, et al., 2005) In subsequent work, a cross was drawn onto a rat liver section (10 mm × 10 mm) with a solution of reserpine (5 pmol total). The area was imaged using a mixture of methanol/water 1/1 (v/v) as eluting solvent at a flow rate of 20 μL/min. Sixteen lines were scanned with 0.67 mm spacing. The distribution of reserpine was monitored by the transition m/z 609 -> 195 during this experiment. (Figure 9c) (Van Berkel et al., 2008) In both experiments, good correlations between the optical and chemical images were observed.
The ability to create chemical images using the non-contact LMJ-SSP was demonstrated with a hand-written inked text and a fingerprint blotted onto a glass slide as test substrates. A 10 Hz, 337 nm nitrogen laser with 11ns pulse width was used to ablate the material in transition geometry. The text area (4 mm × 9 mm) was imaged with spatial resolution of 100–150 μm based on the smallest distinguishable features. The fingerprint sample area (2 mm × 4 mm) was imaged at an apparent spatial resolution of 100 μm (Figure 9e). Both experiments used a solvent mixture of 80/20/0.1 (v/v/v) MeOH/H2O/FA delivered at a flow rate of 20 μL/min. Good correlations between the optical and chemical images were observed. The imaging resolution was limited by the laser ablation area (~210 μm) and although it can be reduced to ~20 μm or less since laser ablation spots of this size had been reported in other transmission geometry laser ablation/mass spectrometry experiments,(Richards et al., 2011) the increased resolution found in the non-contact mode(100–150 μm) compared to the direct contact (0.67–1mm) mode represents an exciting advance for LMJ-SSP MSI. (Ovchinnikova, et al., 2011)
Nano-DESI has been applied to the imaging of rat brain and human papillary renal cell carcinoma tissue sections. (Laskin, et al., 2012) The positive ion mode spectrum obtained for rat brain tissue was dominated by alkali adducts of phosphatidylcholines (PC) and also an abundant peak corresponding to [M- H2O + H]+ ion of cholesterol (m/z 369.3) with high signal-to-noise ratio (S/N ~100). In the case of human papillary renal cell carcinoma, the positive ion mode spectrum was also dominated by the peak corresponding to water loss from cholesterol (m/z 369.3) but contained a broad distribution of peaks in the m/z 530–1000 range and m/z 1200–1400 range, which were identified as the sodium and potassium adducts of cholesteryl esters by MS/MS experiments and high-resolution data. For example, the peak at m/z 673.5 was assigned as the [M + Na]+ ion of cholesteryl oleate based on MS/MS fragmentation. Remarkably, the spatial resolution reported for these experiments was reported to be of less than 12 μm. LESA was applied to map the distribution of terfanadine and fenofexadine in mouse brains of whole body cryo-sections. The spatial resolution was 1mm and the distribution in good agreement with LC-MS/MS analysis of the whole tissues. (Eikel, et al., 2011)
IV. CONCLUSIONS AND PERSPECTIVES
Mass spectrometric imaging is an important and growing area of chemical and biochemical analysis. When performed under ambient conditions the spatial distributions of a wide variety of chemicals (inks, drugs of abuse, pharmaceutical molecules, or biological molecules) in various objects (fingerprints, documents, tissue sections, etc.) become readily accessible. The ambient nature and gentleness of these techniques means that the native sample can be easily examined for information on surface chemical distributions. These advantages make is certain that chemical and biochemical mass spectrometry imaging under ambient conditions is likely to receive much increased attention in the future. Such a development will be accelerated by progress in the following main areas.
Better understanding of ion suppression and matrix effects intrinsic to ambient analysis. The presence and amounts of certain classes of compounds such as lipids or salts can interfere with the ionization of other analytes. This ion suppression phenomenon is well known and is inherent to almost all ionization techniques, ambient or not. However, because a great advantage of ambient ionization techniques is minimal or no sample preparation, it is expected that ambient methods would be more susceptible to ion suppression/enhancement when compared to those techniques that require extensive sample preparation. For instance, wash steps are normally used to remove lipids and salts for protein analysis by MALDI.(Seeley & Caprioli, 2008) The number of experiments that clearly show effects of ion suppression/enhancement and the best way to correct for them in ambient MSI is still limited and represents challenges for the future development of the techniques.
Improvement in experimental setup and ambient MSI source robustness for increased analysis reproducibility. Accurate chemical images require every pixel in the sample to be interrogated under the same conditions during imaging. Ambient ionization methods include different configurations and operating parameters as presented in the previous sections of this review, such as laser power, gas and solvent flow rates, distances and angles between the MS inlet and the surface, within others. To avoid limitations to reproducibility, it is important that these parameters are kept constant during the MSI experiment. One requirement to maximize accuracy of images created by MSI is the flatness of the sample. If the surface is uneven, the change in the geometry of system during the scanning process can result in the misinterpretation of the ion intensities in specific areas.(Kertesz & Van Berkel, 2008) Thus, for in situ analysis of irregular surfaces such as intact biological organs, ambient methods should be used for profiling (collection of information at specific points on the surface) rather than imaging (collection of information over an entire area). The use of compounds homogeneously distributed on the surface could be used as internal standards (IS) and the data reported as the IS to the analyte ratios.(Solon et al., 2010; Stoeckli et al., 2007)
Molecular identification through tandem mass spectrometry and high resolution mass spectrometry. Assignments of endogenous compounds in complex samples like biological tissues are complicated, especially for isomers and isobaric ions. Such identification while not necessary when authentic samples with characteristic spectra are available as reference methods is nevertheless often of great interest. Some classes of compounds such as the phospholipids occur naturally as particularly complex mixtures of closely related compounds. With the popularization of high resolution mass spectrometers, MSI will be more frequently implemented on high mass resolution instruments to minimize chemical noise, identify endogenous compounds, and differentiate isobaric ions. Tandem MSI is a largely untapped resource, with capabilities for imaging with greater molecular specificity than is possible based on ion mass alone. Different MS/MS scan modes (Schwartz et al., 1990) can be used to image for whole classes of compounds based on characteristic neutral losses or fragment ion formation.
Development of improved imaging software and bioinformatics tools for data reduction and manipulation. MS images are extremely rich in information. Ion images of relatively large tissue sections (1×1 cm or larger) obtained with high spatial resolution and high mass resolution, yield data files of a size that can barely be handled and processed with current software. Reduction and compression of data files is desired for the storage of MS images. Validated bioinformatics tools are needed for tissue diagnostics to avoid empirical judgments on classification. Libraries of imaging spectral data that are authenticated against non-MS methods are required. In addition to validity, the bioinformatics and experimental protocols needed to establish disease states requires that speed of acquiring and evaluating MSI data be maximized. This is an additional demanding requirement that has barely been considered. (Balog et al., 2010)
Further enhancement of spatial resolution via improvements in the ion source. For example, at present most DESI imaging is done at 180–220 μm spatial resolution. Under well-controlled experimental conditions, 40 μm spatial resolution can be achieved. (Kertesz & Van Berkel, 2008). Single cell analysis has started to be reported by LAESI. (Shrestha & Vertes, 2009; Shrestha, et al., 2011). However, note that the time required to obtain an image is also a factor that will determine the applicability of imaging methods. The imaging time depends on the analysis time per pixel (related to the speed of the mass spectrometer scan, the frequency of the laser, etc.) and the number of pixels (related to the dimensions of the sample and the spatial resolution). Note that in many problems, exemplified by clinical tumor screening, speed can be more important than spatial resolution allowing low resolution or spot sampling to be used. On the other hand, a high spatial resolution is critical in fundamental studies of cellular activities, and -not surprisingly this represents an area of still limited activity in ambient ionization.
Improvement of sensitivity and specificity of molecular identification. Chemical identification in MSI has been achieved using mass measurements almost exclusively. Ambient ionization is performed under conditions in which chemical reactions can also occur and the deliberate use of reagents and conditions during desorption/ionization that favors such reactions adds a source of independent information on molecular structure. More chemical reactions with high reaction efficiency and well-established molecular specificity are necessary to improve detection and distribution of trace amounts of biologically important molecules, such as low concentrations of endogenous hormones (testosterone, androstenedione, estradiol, etc.). In some cases, reactive versions of ambient ionization are needed to mitigate the effects of low ionization efficiency. In other cases, the chemical reactions could be used to distinguish species not (or not easily) distinguished by mass measurement. By using conventional DESI (positive ion and negative ion mode) as well as reactive DESI, one can obtain more complete MS images by including more biological molecules as analytes. Besides reactive DESI imaging, optimization of the polarity of the spray solvent, or addition of surfactants into the solvent can enhance the desorption efficiency and hence improve sensitivity for imaging. This could be especially valuable for ambient imaging of proteins from biological samples, which has so far not been achieved by any of the ambient MSI techniques. Reactive LAESI was recently introduced with in-plume reactions with lithium sulfate being used to enhance structure-specific fragmentation of lipid ions. This configuration can provide additional structural information and increase the sensitivity of the method without contaminating the biological sample with the chemical reactant.(Shrestha, et al., 2010)
Miniature mass spectrometers capable of imaging will contribute to in situ applications of MSI, especially in such applications as crime scene investigations and in situ diagnostics of diseased tissues. The rapid development of ambient mass spectrometry likely will accelerate interest in miniature mass spectrometers in turn.
Quantitation is also an important emerging topic that needs more attention. Similar to in-vacuum IMS, ambient ionization techniques require the use of internal standards to minimize the effect of ion suppression. For example, clozapine was mapped and quantified by DESI in the lateral ventricles of rat brains using loxapine as an internal standard. The amounts of clozapine found were in good agreement with a cross-validation study performed by LC-MS/MS. (Vismeh et al., 2012) The selection and application of internal standards into the samples still represent real challenges to quantitative analysis.
Finally, the combination of complementary imaging techniques is always more likely to give a more complete “picture” of the sample being analyzed. For example, histological staining protocols used in light microscopy have been developed which are compatible with MALDI imaging.(Chaurand et al., 2004) Other techniques such as magnetic resonance (Sinha, et al., 2008), atomic force microscopy (AFM) (Wucher, et al., 2007) and electron microscopy (Altelaar et al., 2005) have also been combined with SIMS or MALDI. DESI imaging can be coupled to the complementary SIMS and MALDI methods (Eberlin, et al., 2011), and also be used in conjunction with non-MS imaging techniques. Fourier-transform infrared imaging has been used together with DESI imaging for counterfeit drug analysis. (Ricci et al., 2007) UV illumination and visualization of TLC plates is often used to compare with DESI imaging results. The combination of MSI with conventional microscopy has also been realized.(Harada et al., 2009) Whole-body autoradiography (WBA) and light microscopy analysis of stained tissue were also combined and compared with DESI imaging for drug distribution mapping (Kertesz, et al., 2008) and tumor diagnostics (Dill, et al., 2009).
Acknowledgments
This work was supported by US National Institutes of Health (NIH grant 1R21EB009459-01), the Office of Naval Research (N00014-05-1-0405), York University Internal Research Funds (YUIR) and the Natural Sciences and Engineering Research Council of Canada (NSERC).
ABREVIATIONS
- AP-MALDI
Atmospheric pressure MALDI
- CID
Collisionaly induced dissociation
- DAPPI
Desorption atmospheric pressure photo-ionization
- DART
Direct analysis in real time
- DBDI
Dielectric barrier discharge ionization
- DESI
Desorption electrospray ionization
- EASI
Easy ambient sonic-spray ionization
- ELDI
Electrospray-assisted laser desorption ionization
- ESI
Electrospray ionization
- FA
Fatty acid
- fs-LDI
femtosecond laser desorption ionization
- FTMS
Fourier transform mass spectrometry
- GalCer
Galactosylceramide
- GP
Glycerophospholipid
- H&E
hematoxylin and eosin
- HPLC
High-performance liquid chromatography
- IR-LAMICI
Infrared laser ablation metastable-induced chemical ionization
- LAESI
Laser ablation electrospray ionization
- LA-FAPA
Laser ablation flowing atmospheric pressure afterglow
- LEMS
Laser electrospray mass spectrometry
- LOD
Limit of detection
- LMJ-SSP
Liquid microjunction surface sampling probe
- LTP
Low temperature plasma
- MALDESI
Matrix-assisted laser desorption electrospray ionization
- MALDI
Matrix-assisted laser desorption ionization
- MS
Mass spectrometry
- MS/MS
Tandem mass spectrometry
- MSI
Mass spectrometry imaging
- NIMS
Nanostructure initiator mass spectrometry
- NIR
Near Infrared
- PC
Phosphatidylcholine
- PCA
Principal component analysis
- PESI
Probe electrospray ionization
- PI
Phosphatidylinositol
- PS
Phosphatidylserine
- SIMS
Secondary ion mass spectrometry
- SMS
Scaning mass spectrometry
- ST
Sulfatide
- TD/SI
Thermal desorption proximal probe with secondary ionization
- TLC
Thin-layer chromatography
- TM-DESI
Transmission mode desorption electrospray ionization
- TOF
Time of Flight
- WBA
Whole body autoradiograph
References
- Alberici RM, Simas RC, Sanvido GB, Romao W, Lalli PM, Benassi M, Cunha IBS, Eberlin MN. Ambient mass spectrometry: bringing MS into the “real world”. Anal Bioanal Chem. 2010;398:265–294. doi: 10.1007/s00216-010-3808-3. [DOI] [PubMed] [Google Scholar]
- Altelaar AFM, van Minnen J, Jimenez CR, Heeren RMA, Piersma SR. Direct molecular Imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal Chem. 2005;77:735–741. doi: 10.1021/ac048329g. [DOI] [PubMed] [Google Scholar]
- Andrade FJ, Shelley JT, Wetzel WC, Webb MR, Gamez G, Ray SJ, Hieftje GM. Atmospheric pressure chemical ionization source. 1 Ionization of compounds in the gas phase. Anal Chem. 2008;80:2646–2653. doi: 10.1021/ac800156y. [DOI] [PubMed] [Google Scholar]
- Augusti R, Chen H, Eberlin LS, Nefliu M, Cooks RG. Atmospheric pressure Eberlin transacetalization reactions in the heterogeneous liquid/gas phase. International Journal of Mass Spectrometry. 2006;253:281–287. [Google Scholar]
- Balog J, Szaniszlo T, Schaefer KC, Denes J, Lopata A, Godorhazy L, Szalay D, Balogh L, Sasi-Szabo L, Toth M, Takats Z. Identification of Biological Tissues by Rapid Evaporative Ionization Mass Spectrometry. Anal Chem. 2010;82:7343–7350. doi: 10.1021/ac101283x. [DOI] [PubMed] [Google Scholar]
- Brady JJ, Judge EJ, Levis RJ. Mass spectrometry of intact neutral macromolecules using intense non-resonant femtosecond laser vaporization with electrospray post-ionization. Rapid Communications in Mass Spectrometry. 2009;23:3151–3157. doi: 10.1002/rcm.4226. [DOI] [PubMed] [Google Scholar]
- Campbell DI, Ferreira CR, Eberlin LS, Cooks RG. Improved Spatial Resolution in the Imaging of Biological Tissue using Desorption Electrospray Ionization. Anal Bioanal Chem. 2012 doi: 10.1007/s00216-012-6173-6. submitted. [DOI] [PubMed] [Google Scholar]
- Carado A, Passarelli MK, Kozole J, Wingate JE, Winograd N, Loboda AV. C-60 secondary ion mass spectrometry with a hybrid-quadrupole orthogonal time-of-flight mass spectrometer. Anal Chem. 2008;80:7921–7929. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaurand P, Schwartz SA, Billheimer D, Xu BGJ, Crecelius A, Caprioli RM. Integrating histology and imaging mass spectrometry. Anal Chem. 2004;76:1145–1155. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- Chen H, Cotte-Rodriguez I, Cooks RG. Cis-diol functional group recognition by reactive desorption electrospray ionization (DESI) Chem Commun. 2006:597–599. doi: 10.1039/b516448f. [DOI] [PubMed] [Google Scholar]
- Chen HW, Gamez G, Zenobi R. What Can We Learn from Ambient Ionization Techniques? Journal of the American Society for Mass Spectrometry. 2009;20:1947–1963. doi: 10.1016/j.jasms.2009.07.025. [DOI] [PubMed] [Google Scholar]
- Chen LC, Nishidate K, Saito Y, Mori K, Asakawa D, Takeda S, Kubota T, Hori H, Hiraoka K. Characteristics of probe electrospray generated from a solid needle. J Phys Chem B. 2008;112:11164–11170. doi: 10.1021/jp803730x. [DOI] [PubMed] [Google Scholar]
- Chen LC, Yoshimura K, Yu Z, Iwata R, Ito H, Suzuki H, Mori K, Ariyada O, Takeda S, Kubota T, Hiraoka K. Ambient imaging mass spectrometry by electrospray ionization using solid needle as sampling probe. J Mass Spectrom. 2009;44:1469–1477. doi: 10.1002/jms.1632. [DOI] [PubMed] [Google Scholar]
- Cheng SC, Lin YS, Huang MZ, Shiea J. Applications of electrospray laser desorption ionization mass spectrometry for document examination. Rapid Communications in Mass Spectrometry. 2010;24:203–208. doi: 10.1002/rcm.4378. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Brodbelt JS. Transmission Mode Desorption Electrospray Ionization. Journal of the American Society for Mass Spectrometry. 2008;19:1612–1620. doi: 10.1016/j.jasms.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Gelb MH, Brodbelt JS. Surface-enhanced transmission mode desorption electrospray ionization: Increasing the specificity of ambient ionization mass spectrometric analyses. Anal Chem. 2010;82:16–18. doi: 10.1021/ac902101e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chughtai K, Heeren RMA. Mass Spectrometric Imaging for Biomedical Tissue Analysis. Chemical Reviews. 2010;110:3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello Y, Jones AD, Gunaratne TC, Dantus M. Atmospheric pressure femtosecond laser imaging mass spectrometry. Anal Chem. 2010;82:2753–2758. doi: 10.1021/ac9026466. [DOI] [PubMed] [Google Scholar]
- Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Ambient mass spectrometry. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]
- Costa AB, Cooks RG. Simulated splashes: Elucidating the mechanism of desorption electrospray ionization mass spectrometry. Chem Phys Lett. 2008;464:1–8. [Google Scholar]
- Coupland RE, Hopwood D. Mechanism of a Histochemical Reaction Differentiating Between Adrenaline- and Noradrenaline-Storing Cells in the Electron Microscope. Nature. 1966;209:590–591. doi: 10.1038/209590a0. [DOI] [PubMed] [Google Scholar]
- Dill AL, Ifa DR, Manicke NE, Costa AB, Ramos-Vara JA, Knapp DW, Cooks RG. Lipid profiles of canine invasive transitional cell carcinoma of the urinary bladder and adjacent normal tissue by desorption electrospray ionization imaging mass spectrometry. Anal Chem. 2009;81:8758–8764. doi: 10.1021/ac901028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill AL, Eberlin LS, Zheng C, Costa AB, Ifa DR, Cheng L, Masterson TA, Koch MO, Vitek O, Cooks RG. Multivariate statistical differentiation of renal cell carcinomas based on lipidomic analysis by ambient ionization imaging mass spectrometry. Anal Bioanal Chem. 2010;398:2969–2978. doi: 10.1007/s00216-010-4259-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill AL, Eberlin LS, Costa AB, Zheng C, Ifa DR, Cheng LA, Masterson TA, Koch MO, Vitek O, Cooks RG. Multivariate Statistical Identification of Human Bladder Carcinomas Using Ambient Ionization Imaging Mass Spectrometry. Chem-Eur J. 2011;17:2897–2902. doi: 10.1002/chem.201001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Dill AL, Costa AB, Ifa DR, Cheng L, Masterson T, Koch M, Ratliff TL, Cooks RG. Cholesterol Sulfate Imaging in Human Prostate Cancer Tissue by Desorption Electrospray Ionization Mass Spectrometry. Anal Chem. 2010;82:3430–3434. doi: 10.1021/ac9029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Dill AL, Golby AJ, Ligon KL, Wiseman JM, Cooks RG, Agar NYR. Discrimination of Human Astrocytoma Subtypes by Lipid Analysis Using Desorption Electrospray Ionization Imaging Mass Spectrometry. Angew Chem-Int Edit. 2010;49:5953–5956. doi: 10.1002/anie.201001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Ifa DR, Wu C, Cooks RG. Three-Dimensional Vizualization of Mouse Brain by Lipid Analysis Using Ambient Ionization Mass Spectrometry. Angew Chem-Int Edit. 2010;49:873–876. doi: 10.1002/anie.200906283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Ifa DR, Wu C, Cooks RG. Three-dimensional vizualization of mouse brain by lipid analysis using ambient ionization mass spectrometry. Angew Chem Int Ed. 2010;49:873–876. doi: 10.1002/anie.200906283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cheng L, Cooks RG. Nondestructive, Histologically Compatible Tissue Imaging by Desorption Electrospray Ionization Mass Spectrometry. Chem Biod Chem. 2011;12:2129–2132. doi: 10.1002/cbic.201100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cooks RG. Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging. Biochim Biophys Acta Mol Cell Biol Lipids. 2011;1811:946–960. doi: 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Liu X, Ferreira CR, Santagata S, Agar NYR, Cooks RG. Desorption Electrospray Ionization then MALDI Mass Spectrometry Imaging of Lipid and Protein Distributions in Single Tissue Sections. Anal Chem. 2011;83:8366–8371. doi: 10.1021/ac202016x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlin LS, Norton I, Dill AL, Golby AJ, Ligon KL, Santagata S, Cooks RG, Agar NYR. Classifying Human Brain Tumors by Lipid Imaging with Mass Spectrometry. Cancer Research. 2012;72:645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikel D, Vavrek M, Smith S, Bason C, Yeh S, Korfmacher WA, Henion JD. Liquid extraction surface analysis mass spectrometry (LESA-MS) as a novel profiling tool for drug distribution and metabolism analysis: the terfenadine example. Rapid Communications in Mass Spectrometry. 2011;25:3587–3596. doi: 10.1002/rcm.5274. [DOI] [PubMed] [Google Scholar]
- Ellis SR, Wu C, Deeley JM, Zhu X, Truscott RJW, Panhuis MIH, Cooks RG, Mitchell TW, Blanksby SJ. Imaging of Human Lens Lipids by Desorption Electrospray Ionization Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2010;21:2095–2104. doi: 10.1016/j.jasms.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Ferguson CN, Benchaar SA, Miao ZX, Loo JA, Chen H. Direct Ionization of Large Proteins and Protein Complexes by Desorption Electrospray Ionization-Mass Spectrometry. Anal Chem. 2011;83:6468–6473. doi: 10.1021/ac201390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhena AS, Harris GA, Nyadong L, Murray KK, Fernandez FM. Small molecule ambient mass spectrometry imaging by infrared laser ablation metastable-induced chemical ionization. Anal Chem. 2010;82:2178–2181. doi: 10.1021/ac902905v. [DOI] [PubMed] [Google Scholar]
- Gamaly EG, Rode AV, Luther-Davies B, Tikhonchuk VT. Ablation of solids by femtosecond lasers: Ablation mechanism and ablation thresholds for metals and dielectrics. Phys Plasmas. 2002;9:949–957. [Google Scholar]
- Girod M, Shi Y, Cheng JX, Cooks RG. Mapping Lipid Alterations in Traumatically Injured Rat Spinal Cord by Desorption Electrospray Ionization Imaging Mass Spectrometry. Anal Chem. 2010 doi: 10.1021/ac102264z. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green FM, Stokes P, Hopley C, Seah MP, Gilmore IS, O’Connor G. Developing repeatable measurements for reliable analysis of molecules at surfaces using desorption electrospray ionization. Anal Chem. 2009;81:2286–2293. doi: 10.1021/ac802440w. [DOI] [PubMed] [Google Scholar]
- Haapala M, Pol J, Saarela V, Arvola V, Kotiaho T, Ketola RA, Franssila S, Kauppila TJ, Kostiainen R. Desorption atmospheric pressure photoionization. Anal Chem. 2007;79:7867–7872. doi: 10.1021/ac071152g. [DOI] [PubMed] [Google Scholar]
- Haddad R, Milagre HMS, Catharino RR, Eberlin MN. Easy ambient sonic-spray ionization mass spectrometry combined with thin-layer chromatography. Anal Chem. 2008;80:2744–2750. doi: 10.1021/ac702216q. [DOI] [PubMed] [Google Scholar]
- Hand OW, Winger BE, Cooks RG. Enhanced silver cationization of polycyclic aromatic hydrocarbons containing bay regions in molecular secondary ion mass spectrometry. Biomedical and Environmental Mass Spectrometry. 1989;18:83–85. [Google Scholar]
- Harada T, Yuba-Kubo A, Sugiura Y, Zaima N, Hayasaka T, Goto-Inoue N, Wakui M, Suematsu M, Takeshita K, Ogawa K, Yoshida Y, Setou M. Visualization of Volatile Substances in Different Organelles with an Atmospheric-Pressure Mass Microscope. Anal Chem. 2009;81:9153–9157. doi: 10.1021/ac901872n. [DOI] [PubMed] [Google Scholar]
- Harper JD, Charipar NA, Mulligan CC, Zhang X, Cooks RG, Ouyang Z. Low-Temperature Plasma Probe for Ambient Desorption Ionization. Anal Chem. 2008;80:9097–9104. doi: 10.1021/ac801641a. [DOI] [PubMed] [Google Scholar]
- Harris GA, Galhena AS, Fernandez FM. Ambient Sampling/Ionization Mass Spectrometry: Applications and Current Trends. Anal Chem. 2011;83:4508–4538. doi: 10.1021/ac200918u. [DOI] [PubMed] [Google Scholar]
- Heeren RMA, Smith DF, Stauber J, Kukrer-Kaletas B, MacAleese L. Imaging mass spectrometry: Hype or hope? Journal of the American Society for Mass Spectrometry. 2009;20:1006–1014. doi: 10.1016/j.jasms.2009.01.011. [DOI] [PubMed] [Google Scholar]
- Hiraoka K, Nishidate K, Mori K, Asakawa D, Suzuki S. Development of probe electrospray using a solid needle. Rapid Communications in Mass Spectrometry. 2007;21:3139–3144. doi: 10.1002/rcm.3201. [DOI] [PubMed] [Google Scholar]
- Hu QZ, Noll RJ, Li HY, Makarov A, Hardman M, Cooks RG. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- Huang G, Chen H, Zhang X, Cooks RG, Ouyang Z. Rapid screening of anabolic steroids in urine by reactive desorption electrospray ionization. Anal Chem. 2007;79:8327–8332. doi: 10.1021/ac0711079. [DOI] [PubMed] [Google Scholar]
- Huang GM, Zheng OY, Cooks RG. High-throughput trace melamine analysis in complex mixtures. Chem Commun. 2009:556–558. doi: 10.1039/b818059h. [DOI] [PubMed] [Google Scholar]
- Huang MZ, Yuan CH, Cheng SC, Cho YT, Shiea J. Ambient Ionization Mass Spectrometry. In: Yeung ES, Zare RN, Yeung ES, Zare RN, Yeung ES, Zare RNs, editors. Annual Review of Analytical Chemistry. Vol. 3. Palo Alto: Annual Reviews; 2010. pp. 43–65. [DOI] [PubMed] [Google Scholar]
- Huang MZ, Cheng SC, Cho YT, Shiea J. Ambient ionization mass spectrometry: A tutorial. Anal Chim Acta. 2011;702:1–15. doi: 10.1016/j.aca.2011.06.017. [DOI] [PubMed] [Google Scholar]
- Ifa DR, Gumaelius LM, Eberlin LS, Manicke NE, Cooks RG. Forensic analysis of inks by imaging desorption electrospray ionization (DESI) mass spectrometry. Analyst. 2007;132:461–467. doi: 10.1039/b700236j. [DOI] [PubMed] [Google Scholar]
- Ifa DR, Wiseman JM, Song QY, Cooks RG. Development of capabilities for imaging mass spectrometry under ambient conditions with desorption electrospray ionization (DESI) International Journal of Mass Spectrometry. 2007;259:8–15. [Google Scholar]
- Ifa DR, Manicke NE, Dill AL, Cooks G. Latent fingerprint chemical imaging by mass spectrometry. Science. 2008;321:805–805. doi: 10.1126/science.1157199. [DOI] [PubMed] [Google Scholar]
- Ifa DR, Manicke NE, Rusine AL, Cooks RG. Quantitative analysis of small molecules by desorption electrospray ionization mass spectrometry from polytetrafluoroethylene surfaces. Rapid Communications in Mass Spectrometry. 2008;22:503–510. doi: 10.1002/rcm.3377. [DOI] [PubMed] [Google Scholar]
- Ifa DR, Jackson AU, Paglia G, Cooks RG. Forensic applications of ambient ionization mass spectrometry. Anal Bioanal Chem. 2009;394:1995–2008. doi: 10.1007/s00216-009-2659-2. [DOI] [PubMed] [Google Scholar]
- Ifa DR, Wu CP, Ouyang Z, Cooks RG. Desorption electrospray ionization and other ambient ionization methods: current progress and preview. Analyst. 2010;135:669–681. doi: 10.1039/b925257f. [DOI] [PubMed] [Google Scholar]
- Ifa DR, Srimany A, Eberlin LS, Naik HR, Bhat V, Cooks RG, Pradeep T. Tissue imprint imaging by desorption electrospray ionization mass spectrometry. Anal Methods. 2011;3:1910–1912. [Google Scholar]
- Jackson AU, Garcia-Reyes JF, Harper JD, Wiley JS, Molina-Diaz A, Ouyang Z, Cooks RG. Analysis of drugs of abuse in biofluids by low temperature plasma (LTP) ionization mass spectrometry. Analyst. 2010;135:927–933. doi: 10.1039/b920155f. [DOI] [PubMed] [Google Scholar]
- Jackson AU, Shum T, Sokol E, Dill AL, Cooks RG. Enhanced detection of olefins using ambient ionization mass spectrometry: Ag+ adducts of biologically relevant alkenes. Anal Bioanal Chem. 2010 doi: 10.1007/s00216-010-4349-5. accepted. [DOI] [PubMed] [Google Scholar]
- Judge EJ, Brady JJ, Dalton D, Levis RJ. Analysis of Pharmaceutical Compounds from Glass, Fabric, Steel, and Wood Surfaces at Atmospheric Pressure Using Spatially Resolved, Nonresonant Femtosecond Laser Vaporization Electrospray Mass Spectrometry. Anal Chem. 2010;82:3231–3238. doi: 10.1021/ac902880q. [DOI] [PubMed] [Google Scholar]
- Kauppila TJ, Talaty N, Salo PK, Kotiah T, Kostiainen R, Cooks RG. New surfaces for desorption electrospray ionization mass spectrometry: porous silicon and ultra-thin layer chromatography plates. Rapid Communications in Mass Spectrometry. 2006;20:2143–2150. doi: 10.1002/rcm.2566. [DOI] [PubMed] [Google Scholar]
- Kennedy JH, Wiseman JM. Direct analysis of Salvia divinorum leaves for salvinorin A by thin layer chromatography and desorption electrospray ionization multi-stage tandem mass spectrometry. Rapid Communications in Mass Spectrometry. 2010;24:1305–1311. doi: 10.1002/rcm.4514. [DOI] [PubMed] [Google Scholar]
- Kertesz V, Ford MJ, Van Berkel GJ. Automation of a surface sampling probe/electrospray mass spectrometry system. Anal Chem. 2005;77:7183–7189. doi: 10.1021/ac0510742. [DOI] [PubMed] [Google Scholar]
- Kertesz V, Van Berkel GJ. Scanning and surface alignment considerations in chemical imaging with desorption electrospray mass spectrometry. Anal Chem. 2008;80:1027–1032. doi: 10.1021/ac701947d. [DOI] [PubMed] [Google Scholar]
- Kertesz V, Van Berkel GJ. Improved imaging resolution in desorption electrospray ionization mass spectrometry. Rapid Communications in Mass Spectrometry. 2008;22:2639–2644. doi: 10.1002/rcm.3662. [DOI] [PubMed] [Google Scholar]
- Kertesz V, Van Berkel GJ, Vavrek M, Koeplinger KA, Schneider BB, Covey TR. Comparison of drug distribution images from whole-body thin tissue sections obtained using desorption electrospray ionization tandem mass spectrometry and autoradiography. Anal Chem. 2008;80:5168–5177. doi: 10.1021/ac800546a. [DOI] [PubMed] [Google Scholar]
- Kottke PA, Degertekin FL, Fedorov AG. Scanning Mass Spectrometry Probe: A Scanning Probe Electrospray Ion Source for Imaging Mass Spectrometry of Submerged Interfaces and Transient Events in Solution. Anal Chem. 2010;82:19–22. doi: 10.1021/ac902102x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalli PM, Sanvido GB, Garcia JS, Haddad R, Cosso RG, Maia DRJ, Zacca JJ, Maldaner AO, Eberlin MN. Fingerprinting and aging of ink by easy ambient sonic-spray ionization mass spectrometry. The Analyst. 2010;135:745– 750. doi: 10.1039/b923398a. [DOI] [PubMed] [Google Scholar]
- Lane AL, Nyadong L, Galhena AS, Shearer TL, Stout EP, Parry RM, Kwasnik M, Wang MD, Hay ME, Fernandez FM, Kubanek J. Desorption electrospray ionization mass spectrometry reveals surface-mediated antifungal chemical defense of a tropical seaweed. Proc Natl Acad Sci USA. 2009;106:7314–7319. doi: 10.1073/pnas.0812020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AL, Nyadong L, Galhena AS, Shearer TL, Stout EP, Parry RM, Kwasnik M, Wang MD, Hay ME, Fernandez FM, Kubanek J. Desorption electrospray ionization mass spectrometry reveals surface-mediated antifungal chemical defense of a tropical seaweed. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7314–7319. doi: 10.1073/pnas.0812020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue Imaging Using Nanospray Desorption Electrospray Ionization Mass Spectrometry. Anal Chem. 2012;84:141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Bjarnholt N, Hansen SH, Janfelt C. Characterization of barley leaf tissue using direct and indirect desorption electrospray ionization imaging mass spectrometry. J Mass Spectrom. 2011;46:1241–1246. doi: 10.1002/jms.2010. [DOI] [PubMed] [Google Scholar]
- Li Y, Shrestha B, Vertes A. Atmospheric pressure molecular imaging by infrared MALDI mass spectrometry. Anal Chem. 2007;79:523–532. doi: 10.1021/ac061577n. [DOI] [PubMed] [Google Scholar]
- Lin SY, Huang MZ, Chang HC, Shiea J. Using electrospray-assisted laser desorption/ionization mass spectrometry to characterize organic compounds separated on thin-layer chromatography plates. Anal Chem. 2007;79:8789–8795. doi: 10.1021/ac070590k. [DOI] [PubMed] [Google Scholar]
- Liu YY, Ma XX, Lin ZQ, He MJ, Han GJ, Yang CD, Xing Z, Zhang SC, Zhang XR. Imaging Mass Spectrometry with a Low-Temperature Plasma Probe for the Analysis of Works of Art. Angew Chem-Int Edit. 2010;49:4435–4437. doi: 10.1002/anie.200906975. [DOI] [PubMed] [Google Scholar]
- Luosujarvi L, Arvola V, Haapala M, Pol J, Saarela V, Franssila S, Kotiaho T, Kostiainen R, Kauppila TJ. Desorption and ionization mechanisms in desorption atmospheric pressure photoionization. Anal Chem. 2008;80:7460–7466. doi: 10.1021/ac801186x. [DOI] [PubMed] [Google Scholar]
- Luosujarvi L, Laakkonen UM, Kostiainen R, Kotiaho T, Kauppila TJ. Analysis of street market confiscated drugs by desorption atmospheric pressure photoionization and desorption electrospray ionization coupled with mass spectrometry. Rapid Communications in Mass Spectrometry. 2009;23:1401–1404. doi: 10.1002/rcm.4005. [DOI] [PubMed] [Google Scholar]
- Mandal MK, Chen LC, Hiraoka K. Sequential and Exhaustive Ionization of Analytes with Different Surface Activity by Probe Electrospray Ionization. Journal of the American Society for Mass Spectrometry. 2011;22:1493–1500. doi: 10.1007/s13361-011-0162-4. [DOI] [PubMed] [Google Scholar]
- Manicke NE, Wiseman JM, Ifa DR, Cooks RG. Desorption electrospray ionization (DESI) mass Spectrometry and tandem mass spectrometry (MS/MS) of phospholipids and sphingolipids: Ionization, adduct formation, and fragmentation. Journal of the American Society for Mass Spectrometry. 2008;19:531–543. doi: 10.1016/j.jasms.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Manicke NE, Dill AL, Ifa DR, Cooks RG. High resolution imaging on the LTQ orbitrap mass spectrometer by desorption electrospray ionization mass spectrometry (DESI-MS) J Mass Spectrom. 2009;45:223–226. doi: 10.1002/jms.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicke NE, Nefliu M, Wu C, Woods JW, Reiser V, Hendrickson RC, Cooks RG. Imaging of lipids in atheroma by desorption electrospray ionization mass spectrometry. Anal Chem. 2009;81:8702–8707. doi: 10.1021/ac901739s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spectrometry Reviews. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- Mugo SM, Bottaro CS. Rapid on-plate and one-pot derivatization of carbonyl compounds for enhanced detection by reactive matrix LDI-TOF MS using the tailor-made reactive matrix, 4-dimethylamino-6-(4-methoxy-1-naphthyl)-1,3,5-triazine-2-hydrazine (DMNTH) J Mass Spectrom. 2007;42:206–217. doi: 10.1002/jms.1153. [DOI] [PubMed] [Google Scholar]
- Muller T, Oradu S, Ifa DR, Cooks RG, Krautler B. Direct Plant Tissue Analysis and Imprint Imaging by Desorption Electrospray Ionization Mass Spectrometry. Anal Chem. 2011;83:5754–5761. doi: 10.1021/ac201123t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na N, Zhao M, Zhang S, Yang C, Zhang X. Development of a Dielectric Barrier Discharge Ion Source for Ambient Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2007;18:1859–1862. doi: 10.1016/j.jasms.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Nefliu M, Smith JN, Venter A, Cooks RG. Internal energy distributions in desorption electrospray ionization (DESI) Journal of the American Society for Mass Spectrometry. 2008;19:420–427. doi: 10.1016/j.jasms.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- Nemes P, Barton AA, Li Y, Vertes A. Ambient molecular imaging and depth profiling of live tissue by infrared laser ablation electrospray ionization mass spectrometry. Anal Chem. 2008;80:4575–4582. doi: 10.1021/ac8004082. [DOI] [PubMed] [Google Scholar]
- Nemes P, Barton AA, Vertes A. Three-dimensional imaging of metabolites in tissues under ambient conditions by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2009;81:6668–6675. doi: 10.1021/ac900745e. [DOI] [PubMed] [Google Scholar]
- Nemes P, Woods AS, Vertes A. Simultaneous Imaging of Small Metabolites and Lipids in Rat Brain Tissues at Atmospheric Pressure by Laser Ablation Electrospray Ionization Mass Spectrometry. Anal Chem. 2010;82:982–988. doi: 10.1021/ac902245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes P, Vertes A. Ambient mass spectrometry for in vivo local analysis and in situ molecular tissue imaging. TrAC Trends in Analytical Chemistry. 2012;34:22–34. [Google Scholar]
- Nicola AJ, Muddiman DC, Hercules DM. Enhancement of ion intensity in time-of-flight secondary-ionization mass spectrometry. Journal of the American Society for Mass Spectrometry. 1996;7:467–472. doi: 10.1016/1044-0305(96)00006-2. [DOI] [PubMed] [Google Scholar]
- Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–U1033. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- Nyadong L, Green MD, De Jesus VR, Newton PN, Fernandez FM. Reactive desorption electrospray ionization linear ion trap mass spectrometry of latest-generation counterfeit antimalarials via noncovalent complex formation. Anal Chem. 2007;79:2150–2157. doi: 10.1021/ac062205h. [DOI] [PubMed] [Google Scholar]
- Nyadong L, Harris GA, Balayssac S, Galhena AS, Malet-Martino M, Martino R, Parry RM, Wang MD, Fernandez FM, Gilard V. Combining Two-Dimensional Diffusion-Ordered Nuclear Magnetic Resonance Spectroscopy, Imaging Desorption Electrospray Ionization Mass Spectrometry, and Direct Analysis in Real-Time Mass Spectrometry for the Integral Investigation of Counterfeit Pharmaceuticals. Anal Chem. 2009;81:4803–4812. doi: 10.1021/ac900384j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikova OS, Kertesz V, Van Berkel GJ. Molecular Surface Sampling and Chemical Imaging using Proximal Probe Thermal Desorption/Secondary Ionization Mass Spectrometry. Anal Chem. 2011;83:598–603. doi: 10.1021/ac102766w. [DOI] [PubMed] [Google Scholar]
- Ovchinnikova OS, Kertesz V, Van Berkel GJ. Combining transmission geometry laser ablation and a non-contact continuous flow surface sampling probe/electrospray emitter for mass spectrometry based chemical imaging. Rapid Communications in Mass Spectrometry. 2011;25:3735–3740. doi: 10.1002/rcm.5285. [DOI] [PubMed] [Google Scholar]
- Pacholski ML, Winograd N. Imaging with mass spectrometry. Chemical Reviews. 1999;99:2977-+. doi: 10.1021/cr980137w. [DOI] [PubMed] [Google Scholar]
- Paglia G, Ifa DR, Wu C, Corso G, Cooks RG. Desorption electrospray ionization mass spectrometry analysis of lipids after two-dimensional high-performance thin-layer chromatography partial separation. Anal Chem. 2010;82:1744–1750. doi: 10.1021/ac902325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasilis SP, Kertesz V, Van Berkel GJ. Surface scanning analysis of planar arrays of analytes with desorption electrospray ionization-mass spectrometry. Anal Chem. 2007;79:5956–5962. doi: 10.1021/ac070527v. [DOI] [PubMed] [Google Scholar]
- Pasilis SP, Kertesz V, Van Berkel GJ. Unexpected analyte oxidation during desorption electrospray ionization-mass spectrometry. Anal Chem. 2008;80:1208–1214. doi: 10.1021/ac701791w. [DOI] [PubMed] [Google Scholar]
- Pasilis SP, Kertesz V, Van Berkel GJ, Schulz M, Schorcht S. HPTLC/DESI-MS imaging of tryptic protein digests separated in two dimensions. J Mass Spectrom. 2008;43:1627–1635. doi: 10.1002/jms.1431. [DOI] [PubMed] [Google Scholar]
- Pirro V, Eberlin LS, Oliveri P, Cooks RG. Interactive hyperspectral approach for exploring and interpreting DESI-MS images of cancerous and normal tissue sections. Analyst. 2012 doi: 10.1039/c2an35122f. [DOI] [PubMed] [Google Scholar]
- Pol J, Vidova V, Kruppa G, Kobliha V, Novak P, Lemr K, Kotiaho T, Kostiainen R, Havlicek V, Volny M. Automated ambient desorption-ionization platform for surface imaging integrated with a commercial fourier transform ion cyclotron resonance mass spectrometer. Anal Chem. 2009;81:8479–8487. doi: 10.1021/ac901368q. [DOI] [PubMed] [Google Scholar]
- Ricci C, Nyadong L, Fernandez FM, Newton PN, Kazarian SG. Combined fourier-transform infrared imaging and desorption electrospray-ionization linear ion-trap mass spectrometry for analysis of counterfeit antimalarial tablets. Anal Bioanal Chem. 2007;387:551–559. doi: 10.1007/s00216-006-0950-z. [DOI] [PubMed] [Google Scholar]
- Richards AL, Lietz CB, Wager-Miller JB, Mackie K, Trimpin S. Imaging mass spectrometry in transmission geometry. Rapid Communications in Mass Spectrometry. 2011;25:815–820. doi: 10.1002/rcm.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach PJ, Laskin J, Laskin A. Nanospray desorption electrospray ionization: an ambient method for liquid-extraction surface sampling in mass spectrometry. Analyst. 2010;135:2233–2236. doi: 10.1039/c0an00312c. [DOI] [PubMed] [Google Scholar]
- Sampson JS, Murray KK, Muddiman DC. Intact and top-down characterization of biomolecules and direct analysis using infrared matrix-assisted laser desorption electrospray ionization coupled to FT-ICR, mass spectrometry. Journal of the American Society for Mass Spectrometry. 2009;20:667–673. doi: 10.1016/j.jasms.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Wade AP, Enke CG, Cooks RG. Systematic Delineation of Scan Modes in Multidimensional. Mass Spectrometry Anal Chem. 1990;62:1809–1818. doi: 10.1021/ac00216a016. [DOI] [PubMed] [Google Scholar]
- Seeley EH, Caprioli RM. Molecular imaging of proteins in tissues by mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18126–18131. doi: 10.1073/pnas.0801374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley EH, Caprioli RM. 3D Imaging by Mass Spectrometry: A New Frontier. Anal Chem. 2012;84:2105–2110. doi: 10.1021/ac2032707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley JT, Ray SJ, Hieftje GM. Laser ablation coupled to a flowing atmospheric pressure afterglow for ambient mass spectral imaging. Anal Chem. 2008;80:8308–8313. doi: 10.1021/ac801594u. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Vertes A. In Situ Metabolic Profiling of Single Cells by Laser Ablation Electrospray Ionization Mass Spectrometry. Anal Chem. 2009;81:8265–8271. doi: 10.1021/ac901525g. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Nemes P, Nazarian J, Hathout Y, Hoffman EP, Vertes A. Direct analysis of lipids and small metabolites in mouse brain tissue by AP IR-MALDI and reactive LAESI mass spectrometry. Analyst. 2010;135:751–758. doi: 10.1039/b922854c. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Patt JM, Vertes A. In Situ Cell-by-Cell Imaging and Analysis of Small Cell Populations by Mass Spectrometry. Anal Chem. 2011;83:2947–2955. doi: 10.1021/ac102958x. [DOI] [PubMed] [Google Scholar]
- Sinha TK, Khatib-Shahidi S, Yankeelov TE, Mapara K, Ehtesham M, Cornett DS, Dawant BM, Caprioli RM, Gore JC. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nature Methods. 2008;5:57–59. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon EG, Schweitzer A, Stoeckli M, Prideaux B. Autoradiography, MALDI-MS, and SIMS-MS Imaging in Pharmaceutical Discovery and Development. Aaps Journal. 2010;12:11–26. doi: 10.1208/s12248-009-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Cooks RG. Reactive desorption electrospray ionization for selective detection of the hydrolysis products of phosphonate esters. J Mass Spectrom. 2007;42:1086–1092. doi: 10.1002/jms.1244. [DOI] [PubMed] [Google Scholar]
- Sripadi P, Nazarian J, Hathout Y, Hoffman EP, Vertes A. In vitro analysis of metabolites from the untreated tissue of Torpedo californica electric organ by mid-infrared laser ablation electrospray ionization mass spectrometry. Metabolomics. 2009;5:263–276. [Google Scholar]
- Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. International Journal of Mass Spectrometry. 2007;260:195–202. [Google Scholar]
- Suzuki T, Kachi T. Similarities and differences in supporting and chromaffin cells in the mammalian adrenal medullae: An immunohistochemical study. The Anatomical Record. 1996;244:358–365. doi: 10.1002/(SICI)1097-0185(199603)244:3<358::AID-AR7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kachi T. Similarities and differences in supporting and chromaffin cells in the mammalian adrenal medullae: An immunohistochemical study. Anatomical Record. 1996;244:358–365. doi: 10.1002/(SICI)1097-0185(199603)244:3<358::AID-AR7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Tawiah A, Bland C, Campbell D, Cooks RG. Solvent effects and the role of solubility in desorption electrospray Ionization. Journal of the American Society of Mass Spectrometry. 2010;21:572–579. doi: 10.1016/j.jasms.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Thunig J, Hansen SH, Janfelt C. Analysis of Secondary Plant Metabolites by Indirect Desorption Electrospray Ionization Imaging Mass Spectrometry. Anal Chem. 2011;83:3256–3259. doi: 10.1021/ac2004967. [DOI] [PubMed] [Google Scholar]
- Touboul D, Halgand F, Brunelle A, Kersting R, Tallarek E, Hagenhoff B, Laprevote O. Tissue molecular ion imaging by gold cluster ion bombardment. Anal Chem. 2004;76:1550–1559. doi: 10.1021/ac035243z. [DOI] [PubMed] [Google Scholar]
- Van Berkel GJ, Sanchez AD, Quirke JME. Thin-layer chromatography and electrospray mass spectrometry coupled using a surface sampling probe. Anal Chem. 2002;74:6216–6223. doi: 10.1021/ac020540+. [DOI] [PubMed] [Google Scholar]
- Van Berkel GJ, Ford MJ, Deibel MA. Thin-layer chromatography and mass spectrometry coupled using desorption electrospray ionization. Anal Chem. 2005;77:1207–1215. doi: 10.1021/ac048217p. [DOI] [PubMed] [Google Scholar]
- Van Berkel GJ, Kertesz V. Automated sampling and imaging of analytes separated on thin-layer chromatography plates using desorption electrospray ionization mass spectrometry. Anal Chem. 2006;78:4938–4944. doi: 10.1021/ac060690a. [DOI] [PubMed] [Google Scholar]
- Van Berkel GJ, Tomkins BA, Kertesz V. Thin-layer chromatography/desorption electrospray ionization mass spectrometry: Investigation of goldenseal alkaloids. Anal Chem. 2007;79:2778–2789. doi: 10.1021/ac0622330. [DOI] [PubMed] [Google Scholar]
- Van Berkel GJ, Kertesz V, Koeplinger KA, Vavrek M, Kong ANT. Liquid microjunction surface sampling probe electrospray mass spectrometry for detection of drugs and metabolites in thin tissue sections. J Mass Spectrom. 2008;43:500–508. doi: 10.1002/jms.1340. [DOI] [PubMed] [Google Scholar]
- Van Berkel GJ, Pasilis SP, Ovchinnikova O. Established and emerging atmospheric pressure surface sampling/ionization techniques for mass spectrometry. J Mass Spectrom. 2008;43:1161–1180. doi: 10.1002/jms.1440. [DOI] [PubMed] [Google Scholar]
- Van Berkel GJ, Kertesz V, King RC. High-Throughput Mode Liquid Microjunction Surface Sampling Probe. Anal Chem. 2009;81:7096–7101. doi: 10.1021/ac901098d. [DOI] [PubMed] [Google Scholar]
- Venter A, Sojka PE, Cooks RG. Droplet dynamics and ionization mechanisms in desorption electrospray ionization mass spectrometry. Anal Chem. 2006;78:8549–8555. doi: 10.1021/ac0615807. [DOI] [PubMed] [Google Scholar]
- Venter A, Cooks RG. Desorption electrospray ionization in a small pressure-tight enclosure. Anal Chem. 2007;79:6398–6403. doi: 10.1021/ac0708846. [DOI] [PubMed] [Google Scholar]
- Venter A, Nefliu M, Cooks RG. Ambient desorption ionization mass spectrometry. TrAC. 2008;27:284–290. [Google Scholar]
- Vickerman JC. Molecular imaging and depth profiling by mass spectrometry-SIMS, MALDI or DESI? Analyst. 2011;136:2199–2217. doi: 10.1039/c1an00008j. [DOI] [PubMed] [Google Scholar]
- Vidova V, Volny M, Lemr K, Havlicek V. Surface analysis by imaging mass spectrometry. Collection of Czechoslovak Chemical Communications. 2009;74:1101–1116. [Google Scholar]
- Vismeh R, Waldon DJ, Teffera Y, Zhao Z. Localization and quantification of drugs in animal tissues by use of desorption electrospray ionization mass spectrometry imaging. Anal Chem. 2012;84:5439–5445. doi: 10.1021/ac3011654. [DOI] [PubMed] [Google Scholar]
- Volny M, Venter A, Smith SA, Pazzi M, Cooks RG. Surface effects and electrochemical cell capacitance in desorption electrospray ionization. Analyst. 2008;133:525–531. doi: 10.1039/b717693g. [DOI] [PubMed] [Google Scholar]
- Watrous J, Hendricks N, Meehan M, Dorrestein PC. Capturing Bacterial Metabolic Exchange Using Thin Film Desorption Electrospray Ionization-Imaging Mass Spectrometry. Anal Chem. 2010;82:1598–1600. doi: 10.1021/ac9027388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous JD, Alexandrov T, Dorrestein PC. The evolving field of imaging mass spectrometry and its impact on future biological research. J Mass Spectrom. 2011;46:209–222. doi: 10.1002/jms.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston DJ. Ambient ionization mass spectrometry: current understanding of mechanistic theory; analytical performance and application areas. Analyst. 2010;135:661–668. doi: 10.1039/b925579f. [DOI] [PubMed] [Google Scholar]
- Wichmann JM, Lupulescu C, Woste L, Lindinger A. Matrix-assisted laser desorption/ionization by using femtosecond laser pulses in the near-infrared wavelength regime. Rapid Communications in Mass Spectrometry. 2009;23:1105–1108. doi: 10.1002/rcm.3974. [DOI] [PubMed] [Google Scholar]
- Wiley JS, Garcia-Reyes JF, Harper JD, Charipar NA, Ouyang Z, Cooks RG. Screening of agrochemicals in foodstuffs using low-temperature plasma (LTP) ambient ionization mass spectrometry. Analyst. 2010;135:971–979. doi: 10.1039/b919493b. [DOI] [PubMed] [Google Scholar]
- Wiseman JM, Ifa DR, Song QY, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angewandte Chemie International Edition. 2006;45:7188–7192. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- Wiseman JM, Ifa DR, Venter A, Cooks RG. Ambient molecular imaging by desorption electrospray ionization mass spectrometry. Nat Protocols. 2008;3:517–524. doi: 10.1038/nprot.2008.11. [DOI] [PubMed] [Google Scholar]
- Wiseman JM, Ifa DR, Zhu YX, Kissinger CB, Manicke NE, Kissinger PT, Cooks RG. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ifa DR, Manicke NE, Cooks RG. Rapid, direct analysis of cholesterol by charge labeling in reactive desorption electrospray ionization. Anal Chem. 2009;81:7618–7624. doi: 10.1021/ac901003u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ifa DR, Manicke NE, Cooks RG. Molecular imaging of adrenal gland by desorption electrospray ionization mass spectrometry. Analyst. 2010;135:28–32. doi: 10.1039/b919816d. [DOI] [PubMed] [Google Scholar]
- Wu C, Qian K, Nefliu M, Cooks RG. Ambient analysis of saturated hydrocarbons using discharge-induced oxidation in desorption electrospray ionization. Journal of the American Society for Mass Spectrometry. 2010;21:261–267. doi: 10.1016/j.jasms.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Wucher A, Cheng J, Winograd N. Protocols for three-dimensional molecular imaging using mass spectrometry. Anal Chem. 2007;79:5529–5539. doi: 10.1021/ac070692a. [DOI] [PubMed] [Google Scholar]