Abstract
Background
Patients with chronic Chlamydia-induced reactive arthritis (ReA) often show a remitting-relapsing disease phenotype. We have some information regarding bacterial and host responses to one another during active disease but no information for quiescence. We present the first molecular genetic insight into the behavior of bacterium and host during remitting ReA.
Methods
Synovial biopsies were procured from the knees of 4 patients with quiescent ReA by the Parker-Pearson technique. Nucleic acids prepared from them were analyzed by real time PCR and RT-PCR and results compared with data averaged from the knee synovial tissue samples of 10 patients with active ReA.
Results
Real time PCR indicated that bacterial load in remitting samples was ~20% of that in active disease samples. Transcripts from the hsp60 gene were equal to or higher than those seen in active disease. mRNA from the paralog hsp60 genes were equal to/lower than those of active disease. Host mRNA encoding IL-10, TNFα, IFNγ were 4-fold lower than those in active disease samples, while MCP-1, RANTES mRNA levels were equal to or higher.
Conclusions
Bacterial load in synovial tissue of patients with remitting disease is lower than that of active disease, but messengers encoding pro-inflammatory proteins are equal to/higher than those of active disease. Transcription in the host is attenuated for cytokines and chemokines. These initial results demonstrate that organism is present and metabolically active in synovium during the remitting phase of chronic Chlamydia-induced ReA, and that the genetic events characterizing quiescence are complex.
Keywords: Chlamydia trachomatis, reactive arthritis, synovitis, inflammation
Introduction
The obligate intracellular pathogen Chlamydia trachomatis is the most prevalent sexually transmitted bacterial infection in the US and elsewhere1. This bacterium also is the etiologic agent for trachoma in some underdeveloped nations, a disease which remains an important cause of treatable blindnesse.g., 2. Significantly, genital infection with this organism can engender a number of severe disease sequelae, including a chronic inflammatory arthritis, blockage of the fallopian tubules leading to ectopic pregnancy, and others 2–4. As is the case with any organism, details underlying the pathogenic processes engendered by C. trachomatis are a function of the biology of the organism.
All chlamydiae undergo a transcriptionally controlled biphasic developmental cycle which is initiated when elementary bodies, the extracellular form of the organism, attach to a target host cell and are brought into a membrane-bound inclusion in the host cell cytoplasm. In the inclusion, elementary bodies undergo a development process resulting in production of reticulate bodies, the metabolically active form of the organism. After several rounds of cell division, and near the termination of the cycle, most reticulate bodies dedifferentiate back to the elementary body form; these are released to the extracellular milieu by host cell lysis or exocytosis. The cycle requires about 50 hours for completion5 for review.
This classic description of chlamydial biology was derived from study of culture systems that utilized permissive cell lines, that is, cell lines supporting passage of the organism through the entire developmental cycle. Under some circumstances, including those relevant to synovial infection, that cycle is arrested at a late point, obviating production/release of new elementary bodies. This state is referred to as “persistence”, and the arrest in the cycle which engenders this state is transcriptionally governed4,6,7. Importantly, persistently infecting chlamydiae display an unusual profile of gene expression, with transcript levels from some genes attenuated and others up-regulated3,4.
Patients with acute or chronic Chlamydia-induced arthritis harbor persistently infecting chlamydiae in synovial tissue, and these individuals often show a remitting-relapsing disease phenotype3,4,7,8. While we have some understanding of the metabolic behavior of persistently infecting chlamydiae in the synovium during active disease, we know essentially nothing of the genetic, metabolic, or physiologic characteristics of the infecting organism, or of the host response to it, during quiescent disease. Here we present the first molecular genetic insights regarding the behavior of persistent chlamydiae and the host immune system during the remitting phase of Chlamydia-induced reactive arthritis.
Patients and Methods
Patient samples
Not surprisingly, it is difficult to obtain samples from patients in the remitting/quiescent phase of chronic Chlamydia-induced reactive arthritis. However JDC was able to obtain synovial biopsies from 4 consenting patients, under an approved protocol; these were patients 1, 2, 3, and 7 in a continuing study. With one exception, all were single samples from individuals with quiescent disease; the exception was 2 samples from one patient, one of which (sample 7A) was obtained during quiescence and the second during slightly increased disease activity (sample 7B, procured from same joint 2.5 yr after sample 7A). All samples were obtained by the Parker-Pearson technique as in our earlier studiese.g., 8,9, snap-frozen immediately, and sent to APH’s laboratory for processing and analysis. No patient had any sign of synovitis in knees or any other joints at the time of sample procurement, with the exception of the 7B sample. Available demographic and other data for these patients is summarized in Table 1. In addition to the data provided in Table 1, patients 2 and 7 were HLA B27 positive, patient 1 was negative, and patient 3 was not assessed for that genotype. Interestingly, the two patients with the longest disease duration also had the longest remission time.
Table 1.
Demographic and related information for patients studied
| Patient | Gender/Age (yr) | Diagnosis* | Disease Duration (yr) | Remission& (mo) | Reactivation? | Joint Biopsied | Current Therapy | Previous Therapies |
|---|---|---|---|---|---|---|---|---|
| 1 | F/43 | ReA | 6.0 | 6 | yes | left knee | NSAID, prednisone methotrexate | same |
| 2 | F/50 | ReA | 3.5 | 3 | yes | right knee | hydrocodone/acetominophen | NSAID, etanercept, sulfasalazine, methotrexate |
| 3 | M/58 | ReA | 9.0 | 96 | no | left knee | none | NSAID, etanercept, methotrexate, sulfasalazine |
| 7 | M/57 | ReA | 34 | 96 | yes | right knee | naproxen, rifampin erythromycin | NSAID, sulfasalazine, multiple antibiotics |
Abbreviation is: ReA, reactive arthritis
Duration of remission in the joint biopsied
Molecular analyses
The frozen synovial biopsy samples were processed for DNA and/or RNA by the hot phenol method, as described by use.g, 10,11. We assessed relative bacterial load in total DNA samples from the synovial biopsies using our standard highly quantitative real time PCR systemse.g.,11. cDNA was produced from pure RNA from the samples by standard reverse transcription methods, following DNase treatment of an aliquot from each sample. Analyses of relative transcript levels from chlamydiae and host were done as described by us, using our primer systems given in Table 211. Results of the DNA and various RNA analyses were compared with bacterial load and transcript values averaged from the knee synovial tissue samples of 10 patients in the active disease phase of chronic Chlamydia-induced arthritis. Available demographic and related information for these 10 patients is provided in Table 3. These patients were chosen for inclusion for comparison to those in remitting disease (i.e., as “controls”) because all had disease durations consistent with chronic disease, all were known to be PCR-positive for C. trachomatis DNA, and all had been studied for the characteristics examined here in previous work from this group.
Table 2.
Primer systems used in this study
| Ct 16S rRNA us 5′-ctgcagcctcccgtaggagtctgggc-3′; ds 5′-aagcttttcttaacaatgcaaatgag-3′ |
| CT110 us 5′-tcatgttgtcggaagctc-3′; ds 5′-tcatgtttgtcggcaagctc-3′ |
| CT604 us 5′-tttatctagcgcagtgggtt-3′; ds 5′-tattctaccgttcctgtttcc-3′ |
| CT755 us 5′-aactcgatcacgctatacaca-3′; ds 5′-gtgttgagtttagcgttttgaga-3′ |
| 18S us 5′-gctggaattaccgcggct-3′; ds 5′-cggctaccacatccaaggaa-3′ |
| IL-10 us 5′-ctgtgaaaacaagagcaagg-3′; ds 5′-aactcactcatggctttgttag-3′ |
| TNFα us 5′-cctgccccaatccctttatt-3′; ds 5′-ccctaagcccccaattctct-3′ |
| IFNγ us 5′-acagttgtcaacaatatttggaa-3′; ds 5′-aacttgaatgtgtcaggtgac-3′ |
| MCP-1 us 5′-gtcacctgctgctataacttc-3′; ds 5′-tgctgctggtgattcttcta-3′ |
| RANTES us 5′-cctcgctgtcatcctcat-3′; ds 5′-acttgccactggtgtagaaa-3 |
Table 3.
Demographic and related information for control patients with chronic reactive arthritis
| Patient | Gender/Age (yr) | Diagnosis* | Disease Duration (mo) |
|---|---|---|---|
| A | M/28 | ReA | 60 |
| B | F/36 | RS | 20 |
| C | F/60 | RS | 48 |
| D | F/55 | ReA | 30 |
| E | F/57 | RS | unknown |
| F | F/48 | ReA | 44 |
| G | F/62 | ReA | 16 |
| H | M/57 | RS | 13 |
| I | F/53 | ReA | 11 |
| J | F/33 | ReA | 15 |
Abbreviations are: ReA, reactive arthritis; RS, Reiter’s syndrome
Results
Bacterial load and levels of selected chlamydial transcripts during quiescent reactive arthritis
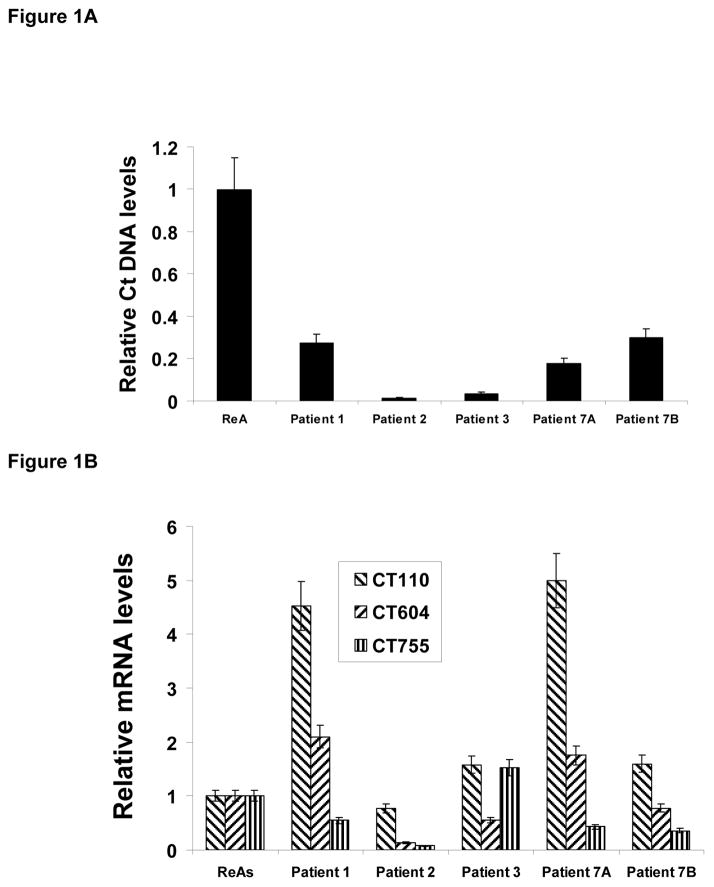
We first assessed whether chlamydiae were present in synovial tissue in patients with remitting disease, and if so what the bacterial load was in the samples compared to values for load averaged from congruent freezer library samples from 10 patients with well characterized active Chlamydia-induced arthritis (Table 3). Standard PCR analyses indeed showed that the organism was present in each remitting sample (data not shown), and as shown in Figure 1A the relative level of C. trachomatis DNA/5 mg tissue was lower by at least 5-fold in the quiescence samples compared to that averaged from active arthritis samples. Reactive arthritis elicited by chlamydiae is an inflammatory disease, and the C. trachomatis genome includes 3 genes encoding non-identical copies of the proinflammatory hsp60 protein, designated Ct110, Ct604, and Ct755 in the genome sequence12. A previous report indicated that each of the three is highly expressed during active infection, with the Ct604 gene showing the highest transcript levels11. Interestingly, that report also showed that in persistently infecting chlamydiae, Ct604 mRNA levels were high while expression from Ct755 was strongly down-regulated. The data provided in Figure 1B show that Ct755 is transcribed at an even lower level in quiescent disease compared to that in congruent samples from patients with active disease. Ct604 is transcribed at about the same relative level in quiescent and active samples, but Ct110, the authentic groEL gene, is expressed at extremely high relative levels in two samples and at levels at or above those seen in active disease in two other quiescent samples. Thus, absence of the organism or attenuation of expression from Ct110 and/or Ct604 does not explain the low/absent level of active disease characteristic of remission.
Figure 1.
Relative level of chlamydial DNA and transcript levels from selected chlamydial genes in synovial tissues of patients with quiescent ReA. Comparison bacterial load (Panel A) and relative transcript levels from hsp60-encoding genes (Panel B) are given relative to that/those averaged from samples from 10 patients in active disease. Determinations done by real time PCR targeting the chlamydial 16S rDNA (Panel A) or real time RT-PCR targeting 16S rRNA (Panel B). Normalization to host 18S rDNA or chlamydial 16S rDNA, respectively. Samples run independently twice in triplicate each. Standard error shown.
Relative transcript levels from cytokine and chemokine genes during quiescent reactive arthritis
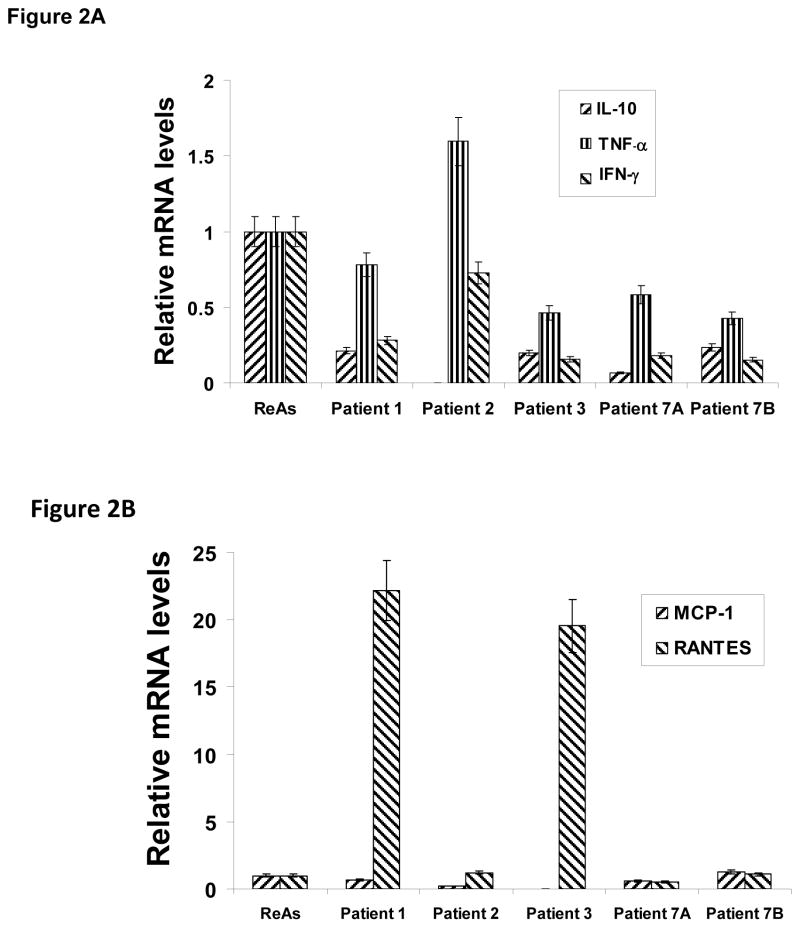
In an earlier report, we identified the proinflammatory mediators IL-10, TNFα, IFNγ as being in rough balance in synovial tissue samples from patients with active disease13. The data given in Figure 2A indicate that relative transcript levels from the host genes encoding these cytokines are somewhat lower in quiescent than in active synovial tissue samples, but the balance among them is similar to that of active disease. Thus, a significantly altered host response for these three gene products cannot provide the sole explanation for the attenuated inflammatory response of quiescent disease. Interestingly, in 3/5 samples mRNA encoding MCP-1 and RANTES were at the same relative levels as those in patients with active disease. In contrast, in 2 samples from patients with quiescent disease relative RANTES mRNA levels showed strong increases over those of active samples; MCP-1 remained low in those 2 samples. Thus, as with the cytokine transcript data, chemokine transcript levels alone do not provide a full explanation for differences in inflammation in the synovial tissues of patients with quiescent Chlamydia-induced arthritis.
Figure 2.
Relative transcript levels from selected host cell genes in synovial tissues of patients with quiescent ReA. Relative transcript levels from three proinflammatory cytokine genes (Panel A; IL-10 blue, TNFα red, IFNγ yellow) and two chemokine genes (Panel B; MCP-1 blue, RANTES red) compared to those averaged from samples from 10 patients with active disease. Determinations done by real time RT-PCR targeting the specific genes. Normalization to chlamydial 16S rDNA. Samples run independently twice in triplicate each. Standard error shown.
Discussion
C. trachomatis, the causative agent in Chlamydia-induced arthritis, exists in synovial tissues in the persistent infection form, rather than the form characteristic of active infection2–4,7 for review. In this state, chlamydiae display an unusual transcript profile compared to that seen during normal active infection4,11,14,15, and we hypothesized that if the organism is indeed present in synovial tissue during the remitting disease phase, then the significantly lower level of inflammation that characterizes the joint during quiescent phase arthritis must result from an attenuated host immune response to some as yet uncharacterized different pattern of bacterial gene products. As we expected, the organism was present in all patients during quiescent disease, and the overall level of bacterial load observed during quiescent disease was lower than that seen in active disease. Interestingly, the organism was present in synovial tissue samples from patients with extremely long periods of remission (Table 1).
Importantly, the initial data presented here regarding mRNA produced from selected bacterial and host genes during quiescence collectively indicate a far more complex pattern of events than predicted. Transcript levels from the highly immunogenic hsp60-encoding genes, and presumably translation products from those mRNA, are not strongly attenuated during remitting disease, as we expected them to be; rather, they are for the most part roughly comparable to transcript levels from these genes seen during active disease. Similarly, messenger levels from host genes encoding important proinflammatory cytokines exist in relative balance in persistent chlamydiae during quiescent disease as in active disease, although the relative level of those transcripts (and presumably translation products from them) are attenuated somewhat over those of active disease. Transcript levels from the host MCP-1-and RANTES-encoding genes reflect those observed during active disease as well, although in some samples studied the level of the latter are higher than in active disease. We do not have MRI or other imaging data from the patients studied here to be able to assess in a quantitative manner the level of inflammation present in these samples. However, given the level of chlamydial messengers specifying hsp-60 proteins, and of host transcripts encoding proinflammatory mediators, it is likely that some level of inflammation was present. In turn this suggests that remitting disease in Chlamydia-induced arthritis may not mean complete lack of joint inflammation, but rather devolution of disease to subclinical levels. More study, and additional relevant patient samples, will be required to expand these initial observations before meaningful generalizations can be made.
We do not know the full panel of bacterial gene products responsible for eliciting the powerful inflammatory response to persistent chlamydial infection in the synovial tissue of patients with Chlamydia-induced arthritis. However, the initial data presented here clearly indicate that the attenuated inflammation seen during remitting disease vs. that of active disease cannot be a function of simple differences in the production of strongly immunogenic bacterial molecules and the host response to them. These initial observations do suggest that the sequence of genetic events underlying the transitions from active to quiescent and back to active disease is complex and multifactorial. Disease in 3 of the 4 patients studied here did eventually reactivate to end the remission period, but disease in the remaining individual (patient 3) has not done so. It would be a significant advance if we could predict if, and if so when, disease will remit in patients with Chlamydia-induced arthritis, and when relapse to active disease will take place as well, since an understanding of the molecular genetic details of the latter transition might suggest therapeutic interventions to prevent that relapse. Indeed, along this same line it would be a significant advance if we could predict which patients with acute Chlamydia-induced arthritis will progress to chronic disease. We previously demonstrated that the persistently infecting synovial chlamydiae in both acute and chronic disease belong to ocular rather than genital strain groups10. Regardless, whether the continued, low level presence of the inciting pathogen during quiescent reactive arthritis indicates that clinicians should not terminate antibiotic or other therapies remains to be determined. We are continuing to define the genetic and biochemical underpinnings of the cycling between the active and quiescent disease phases of Chlamydia-induced arthritis, and future studies must include not only focus on other relevant gene products but also on detailed histopathologic examination of synovial samples from the quiescent phase.
Acknowledgments
This work was supported by grants AR-42541 (APH) and AR-53646 (JDC) from the US National Institutes of Health.
References Cited
- 1.www.cdc.gov/std/Chlamydia/STDFact-Chlamydia.htm
- 2.Whittum-Hudson JA, Hudson AP. Human chlamydial infections: persistence, prevalence, and prospects for the future. Nat, Sci, et Soc. 2005;13:371–382. [Google Scholar]
- 3.Carter JD, Inman RD, Whittum-Hudson J, Hudson AP. Chlamydia and chronic arthritis. Ann Med. 2011 doi: 10.3109/07853890.2011.606830. in press. [DOI] [PubMed] [Google Scholar]
- 4.Gérard HC, Whittum-Hudson JA, Carter JD, Hudson AP. Molecular biology of infectious agents in chronic arthritis. In: Espinoza L, editor. Rheumatic Disease Clinics of North America. Vol. 35. Elsevier/Saunders; Philadelphia PA: 2009. pp. 1–19. [DOI] [PubMed] [Google Scholar]
- 5.Abdelrahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol Rev. 2005;29:949–59. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Schoborg RV. Chlamydia persistence a – tool to dissect Chlamydia-host interactions. Microbes Infect. 2011;13:649–62. doi: 10.1016/j.micinf.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inman RD, Whittum-Hudson JA, Schumacher HR, Hudson AP. Chlamydia-associated arthritis. Curr Opin in Rheumatol. 2000;12:254–62. doi: 10.1097/00002281-200007000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Inman RD. Reactive and enteropathic arthritis. In: Klippel JH, Stone JH, Crofford LJ, White PH, editors. Primer on the Rheumatic Diseases. Springer Science and Business Media; New York NY: 2007. pp. 217–223. [Google Scholar]
- 9.Schumacher HR, Kulka JP. Needle biopsy of the synovial membrane – experience with the Parker-Pearson technique. New Eng J Med. 1972;286:416–19. doi: 10.1056/NEJM197202242860807. [DOI] [PubMed] [Google Scholar]
- 10.Gerard HC, Stanich JA, Whittum-Hudson JA, et al. Patients with Chlamydia-induced arthritis have ocular (trachoma), not genital, serovars of C. trachomatis in synovial tissue. Microb Pathogen. 2010;48:62–68. doi: 10.1016/j.micpath.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gérard HC, Whittum-Hudson JA, Schumacher HR, Hudson AP. Differential expression of the three Chlamydia trachomatis hsp60-encoding genes in active vs persistent infection. Microb Pathogen. 2004;36:35–39. doi: 10.1016/j.micpath.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Stephens RS, Kalman S, Lammel C, et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–59. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 13.Gérard HC, Wang Z, Whittum-Hudson JA, et al. Cytokine and chemokine mRNA produced in synovial tissue chronically infected with C trachomatis and Chlamydia pneumoniae. J Rheumatol. 2002;29:1827–35. [PubMed] [Google Scholar]
- 14.Gérard HC, Krauß-Opatz B, Rudy D, et al. Expression of Chlamydia trachomatis genes required for DNA synthesis and cell division in active vs. persistent infection. Mol Microbiol. 2001;41:731–41. doi: 10.1046/j.1365-2958.2001.02550.x. [DOI] [PubMed] [Google Scholar]
- 15.Gérard HC, Freise J, Rudy D, et al. Chlamydia trachomatis genes whose products are related to energy metabolism are expressed differentially in active vs. persistent infection. Microb Infect. 2002;4:13–22. doi: 10.1016/s1286-4579(01)01504-0. [DOI] [PubMed] [Google Scholar]