Abstract
Estrogen receptor (ESR1) drives growth in the majority of human breast cancers by binding to regulatory elements and inducing transcription events that promote tumor growth. Differences in enhancer occupancy by ESR1 contribute to the diverse expression profiles and clinical outcome observed in breast cancer patients. GATA3 is an ESR1-cooperating transcription factor mutated in breast tumors; however, its genomic properties are not fully defined. In order to investigate the composition of enhancers involved in estrogen-induced transcription and the potential role of GATA3, we performed extensive ChIP-sequencing in unstimulated breast cancer cells and following estrogen treatment. We find that GATA3 is pivotal in mediating enhancer accessibility at regulatory regions involved in ESR1-mediated transcription. GATA3 silencing resulted in a global redistribution of cofactors and active histone marks prior to estrogen stimulation. These global genomic changes altered the ESR1-binding profile that subsequently occurred following estrogen, with events exhibiting both loss and gain in binding affinity, implying a GATA3-mediated redistribution of ESR1 binding. The GATA3-mediated redistributed ESR1 profile correlated with changes in gene expression, suggestive of its functionality. Chromatin loops at the TFF locus involving ESR1-bound enhancers occurred independently of ESR1 when GATA3 was silenced, indicating that GATA3, when present on the chromatin, may serve as a licensing factor for estrogen–ESR1-mediated interactions between cis-regulatory elements. Together, these experiments suggest that GATA3 directly impacts ESR1 enhancer accessibility, and may potentially explain the contribution of mutant-GATA3 in the heterogeneity of ESR1+ breast cancer.
With the emergence of transcription factor mapping using ChIP-seq, it has become evident that the composition of distal cis-regulatory regions has a major impact on transcriptional potential (Bulger and Groudine 2011). The histone landscape and transcription factor complexes at enhancer elements govern the genomic circuitry that defines cell fate (Heintzman et al. 2009; Visel et al. 2009; Kim et al. 2010), and these can be greatly influenced by chromatin accessibility (John et al. 2011). Estrogen receptor alpha (ESR1) is intensively studied for its causal role in human breast cancer and is successfully targeted by antihormonal therapy (Jordan 2007). The heterogeneity of the disease in terms of expression profiles, clinical grade, and overall survival implies that distinct ESR1 binding and transcriptional targets may dictate clinical outcome (Ross-Innes et al. 2012). ESR1 occupies distant enhancers (Carroll et al. 2005; Lin et al. 2007; Welboren et al. 2009) and via DNA-looping brings enhancers in spatial proximity of promoters of regulated genes (Pan et al. 2008). However, many predicted estrogen receptor elements (EREs) in the human genome do not correspond to observable ESR1-binding events, and there are many more binding events than regulated genes (Joseph et al. 2010), although the ESR1-mediated transcriptome may be larger than initially thought (Hah et al. 2011). Recent findings suggest that noncoding transcription may be occurring at ESR1 enhancer elements (Hah et al. 2011), although these regions can still simultaneously be involved in chromosomal loops (Fullwood et al. 2009). ESR1-induced long-range intrachromosomal interactions reveal that genes strongly up-regulated by estrogen are mostly positioned in proximity to the ESR1 anchoring events, whereas down-regulated targets tend to be in the intervening chromatin loops (Fullwood et al. 2009). Current research indicates that additional factors regulate enhancer accessibility and transcriptional potential in ESR1-positive breast cancer cells. Whether these enhancer regions are involved in localized transcription of noncoding RNAs or in long-distance chromatin interactions, or both, it is necessary to understand what allows these regulatory regions to be bound by ESR1 to facilitate transcriptional activity.
Forkhead and GATA3 DNA motifs are enriched within ESR1-binding regions (Carroll et al. 2005; Lin et al. 2007). In combination with expression profiling of breast cancers (Perou et al. 2000; Sorlie et al. 2003), this has led to the discovery and characterization of GATA3 and FOXA1, two transcription factors functionally linked with ESR1. Depletion of FOXA1 abolishes ESR1-binding capacity and transcriptional activity (Carroll et al. 2005), while cell-specific FOXA1 distribution can explain part of the differential ESR1-binding profiles in various breast cancer cell lines (Hurtado et al. 2011). Overexpression of FOXA1 and GATA3 in ESR1-negative cells can, to a certain extent, render them estrogen responsive, demonstrating their ability to impact ESR1 binding (Kong et al. 2011). GATA3, a key developmental factor for mammary gland cell fate in mouse models (Kouros-Mehr et al. 2006; Asselin-Labat et al. 2007), is often mutated in human breast cancer, unlike ESR1 (Usary et al. 2004; Chanock et al. 2007; Arnold et al. 2010). However, a global understanding of the mechanistic role of GATA3 in mediating ESR1 function in breast cancer cells has been largely unexplored.
We have utilized genome-wide mapping and expression microarray analysis, in the presence and absence of GATA3, to gain global insight into the events that dictate enhancer accessibility and ESR1-binding potential. Here, we report that the majority of ESR1-binding events following exposure to estrogen are determined by the composition of enhancer elements prior to hormonal stimulation. The histone landscape and the cofactor composition in unstimulated cells predefine ERE accessibility, subsequent ESR1-binding affinity, and the downstream transcriptional program. These events are directly modulated by GATA3, which appears to function as a critical regulator of transcription factor binding, chromatin structure, and long-range genomic communication.
Results
GATA3 associates with the chromatin prior to ESR1 recruitment
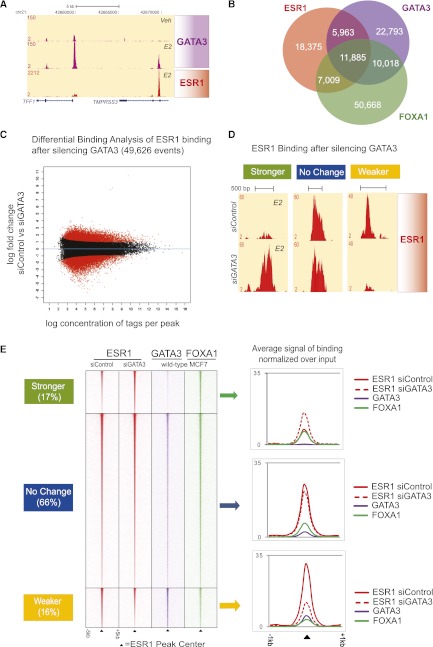
GATA3 has been previously shown to bind in MCF7 breast cancer cells in a ligand-independent manner (Kong et al. 2011). When overexpressed in combination with FOXA1 and ESR1 in an ESR1-negative, estrogen-unresponsive breast cancer cell line, GATA3 can induce ESR1 binding and transcriptional activity (Kong et al. 2011). We confirmed by ChIP-seq (five independent replicates) in MCF7 breast cancer cells that GATA3 binding is almost exclusively estrogen independent, with only 3.5% of GATA3-binding events acquired after estrogen treatment (Supplemental Fig. 1A, averaged log concentration of reads in Supplemental Fig. 1B). An example of GATA3 binding is shown at the TFF1 gene, an ESR1-E2 up-regulated target (Fig. 1A). At the promoter of TFF1 and the upstream enhancer in the TMPRSS3 gene GATA3 binding is induced by estrogen, whereas GATA3 binding inside the TFF1 gene (far left) is not altered by hormonal treatment. GATA3, FOXA1, and ESR1 are involved in a biologically and functionally important network and appear to cooperate in mediating the estrogen response at the transcriptional level, a hypothesis that is further supported by the fact that these three factors are requisite features of ESR1+ breast cancers (Perou et al. 2000). At a genome-wide level, approximately a quarter of all ESR1-binding events are co-occupied by GATA3 and FOXA1 (Fig. 1B). We mapped 5963 GATA3–ESR1 shared regions that did not recruit FOXA1 and 7009 FOXA1–ESR1 cobound regions not shared with GATA3, possibly due to the lack of a forkhead or GATA DNA-binding motif, respectively (Supplemental Fig. 1C). Integration with ChIA-PET data that described global ESR1-mediated chromatin loops (Fullwood et al. 2009) implied that the GATA3–ESR1 regions (7%) are more often involved in transcriptionally active chromatin interaction networks compared with the FOXA1–ESR1 bound regions (4%) (Supplemental Fig. 1D), suggesting that GATA3 may be mediating the estrogenic response.
Figure 1.
Silencing of GATA3 results in redistribution of ESR1 binding globally. (A) GATA3 binding at the TFF1 locus in unstimulated and E2-treated MCF7 cells. (B) Venn diagram showing overlap in ESR1, FOXA1, and GATA3 binding in MCF7 cells. (C) MA plot of Differential Binding Affinity (DBA). Analysis using edgeR of ESR1 binding in siControl versus siGATA3 conditions of three biological replicates. The universe of ESR1-binding events consists of 49,626 peaks, which were used in the DBA analysis. The x-axis shows the log concentration of sequenced tags per peak. The y-axis represents the log fold change of siControl over siGATA3. A third of all ESR1-binding events showed significant changes in affinity at FDR < 0.1. The binding events plotted red above 0 on the y-axis represent Weaker ESR1 binding in siGATA3 (16%), whereas the red at the bottom represents events with Stronger ESR1 affinity in siGATA3 (17%). (D) A representative example of the redistributed ESR1 binding in siGATA3 MCF7 cells. (E, left) Heatmaps of the ChIP-seq data sets centered on the ESR1 peaks and their averaged signal intensity (right) show that the Weaker ESR1 events are cobound by GATA3, suggesting that at these regions GATA3 mediates ESR1–chromatin associations. However, GATA3 binding did not overlap at regions where ESR1 binding was gained following silencing of GATA3.
Silencing of GATA3 globally redistributes ESR1-binding events
Since GATA3 associates with chromatin prior to estrogen treatment, we investigated its potential role in modulating ESR1 recruitment. For the first time, we explored what effect modulation of GATA3 would have on ESR1 and FOXA1 binding, chromatin properties, and gene-expression profiles within an ESR1+ breast cancer context. We specifically silenced GATA3 in MCF7 cells using multiple independent siRNAs, and confirmed that GATA3 was effectively inhibited (Supplemental Fig. 2A). Silencing of GATA3 did not influence ESR1 protein levels (Supplemental Fig. 2A). We subsequently mapped ESR1 binding globally in three biological replicates to identify changes with statistical confidence (Fig. 1C), resulting in a total of 49,626 ESR1-binding events.
We found that GATA3 silencing induced a redistributed ESR1-binding profile at approximately one-third of all ESR1-binding events. At a FDR < 0.1, 17% of all ESR1-binding events increased in binding intensity (8243 binding events were stronger: ESR1 Stronger-siGATA3, presented in green), 67% did not change (33,637 events ESR1 No change, presented in blue), and 16% of ESR1-binding events had decreased binding affinity (7751 binding events of ESR1 Weaker-siGATA3, presented in yellow) (Fig. 1C; see example in Fig. 1D). This partial redistribution of ESR1 binding does not appear to be cell line-specific, since GATA3 silencing in another ESR1+ breast cancer cell line, ZR75-1, recapitulates these findings, resulting in both stronger and weaker ESR1-binding events (Supplemental Fig. 2B). Given the wealth of genomic information available for MCF7 cells, all subsequent experiments were conducted using this cell line. Motif analysis of known transcription factor binding domains in the redistributed ESR1-binding events revealed that all three classes are enriched for EREs, indicative of genuine and direct ESR1 binding (Supplemental Fig. 2C).
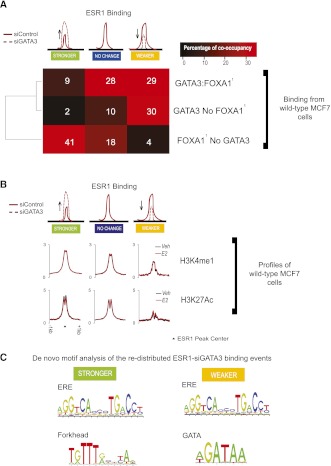
There is, however, an important difference between the two categories of differentially bound ESR1 events, as shown in Figure 1E. Sites with decreased ESR1-binding affinity (Weaker) when GATA3 is silenced tend to be cobound with GATA3 in wild-type cells. In contrast, enhancers that gain ESR1-binding affinity (Stronger) in siGATA3 conditions have limited GATA3 binding in wild-type MCF7 cells. This is shown by the averaged binding signal of GATA3 in the distinct ESR1 categories (Fig. 1E), as well as by the percentage of co-occupancy in Figure 2A (GATA3 binds 59% of the Weaker ESR1–siGATA3 vs. 11% of the Stronger ESR1–siGATA3). This suggests that while at the “Weaker” sites ESR1 binding is dependent on the presence of GATA3, other factors contribute to the increased ESR1-binding affinity at the “Stronger” sites in the absence of GATA3. We next investigated the mechanisms driving these categories of differentially bound enhancers as well as those that do not change their ESR1-binding characteristics in the absence of GATA3.
Figure 2.
GATA3-dependent ESR1-binding events show distinct transcription factor co-occupancy and histone modifications. (A) A binary co-occupancy analysis showing the percentage of the ESR1 categories affected by depletion of GATA3 normally co-occupied by GATA3 and/or FOXA1. GATA3 and FOXA1 binding is from wild-type MCF7 cells. (Data from Hurtado et al. 2011.) (B) Averaged signal intensity of H3K4me1 and H3K27Ac active histone marks in unstimulated (Veh) and estrogen (E2)-treated cells centered on the different categories of ESR1 binding influenced by silencing of GATA3. (C) De novo motif analysis (Weeder).
Silencing GATA3 inhibits ESR1 binding at enhancers normally co-occupied by GATA3 in regions depleted of active histone marks
We first considered the properties of the enhancers in wild-type MCF7 cells that harbor ESR1-binding events that become weaker following depletion of GATA3. A binary occupancy comparison showed that these sites have an elevated level of co-occupancy with GATA3, either alone (30%) or together with FOXA1 (29%), but are only rarely (4%) co-occupied with FOXA1 without GATA3 (Fig. 2A). To gain further insight into the structure of the chromatin at these sites, we performed ChIP-seq in unstimulated, wild-type MCF7 cells (siControl) against two histone marks, H3K4me1 and H3K27Ac, that discriminate poised from transcriptionally active cis-regulatory elements (Heintzman et al. 2009; Creyghton et al. 2010). We discovered that sites which lose ESR1-binding affinity in the absence of GATA3 are depleted of active histone. De novo motif analysis revealed an ERE and a GATA DNA-binding motif in this category (Fig. 3C), suggesting that GATA3 modulates these enhancers directly. Together, these data show that for these “Weaker” ESR1-binding regions GATA3 (1) possesses a role similar to the pioneer factor FOXA1, since its depletion results in reduced ESR1 binding, and (2) that GATA3 is required for ESR1 to interact with chromatin lacking active histone marks. GATA factors have previously been shown to possess pioneer factor functions capable of associating with inaccessible chromatin without assistance from additional proteins (Cirillo et al. 2002), and this appears to be the case for ESR1+ breast cancer cells as well.
Figure 3.
Silencing GATA3 modulates enhancer accessibility by redirecting FOXA1, EP300, H3K4me1, and H3K27Ac histone marks prior to ESR1-E2 recruitment. (A) FOXA1, EP300, H3K4me1, and H3K27Ac were mapped by ChIP-seq, following silencing of GATA3 or siControl. Silencing of GATA3 results in an altered histone profile at enhancer elements prior to ESR1 recruitment (Veh) at the same regions that will harbor the redistributed ESR1 binding upon E2 stimulation. (B) An example of the changes in the histone landscape and EP300 recruitment at TFF3, an E2 up-regulated gene. Silencing of GATA3 results in a gain of H3K4me1, EP300, and subsequent H3K27Ac in unstimulated cells, which parallels the E2-dependent gain in ESR1-binding affinity. (C) Average ChIP-seq signal intensity of FOXA1 and EP300 binding in unstimulated (Veh) MCF7-siGATA3 and siControl cells, centered on the redistributed ESR1-siGATA3 events. (D) Average ChIP-seq signal intensity of H3K4me1 and K3K27Ac binding in unstimulated (Veh) MCF7-siGATA3 and siControl cells, centered on the redistributed ESR1–siGATA3 events.
Silencing GATA3 increases ESR1 binding at FOXA1-bound enhancers and promoters highly enriched with active histone marks
A substantial number (8243) of sites exhibit increased ESR1-binding affinity when GATA3 is silenced. These binding events are rarely cobound by GATA3 in wild-type MCF7 cells (11%), but exhibit elevated levels of FOXA1 co-occupancy (50%) (Fig. 2A). The high occurrence of Forkhead motifs (Supplemental Fig. 2C), as well as the de novo motif discovery (Fig. 2C) in the Stronger ESR1–siGATA3 sites, suggests that FOXA1 is the factor that drives the enhanced ESR1 binding at these sites in the absence of GATA3. In addition, these events are centered in regions of accessible chromatin environment as indicated by the high levels of both histone marks (H3K4me1 and H3K27Ac) (Fig. 2B). Moreover, these sites are more likely than those from the other categories to occur at promoter proximal regions, representing ∼7% versus ∼4% for all ESR1-binding events (Supplemental Fig. 2D). As assessed by the active histone marks, these data show that the enhanced ESR1 binding, when GATA3 is depleted, occurred at potentially transcriptionally active regions co-occupied by FOXA1; and the lack of GATA3 co-occupancy indicates that its role in suppressing ESR1 binding at these sites is indirect.
Silencing GATA3 has no effect on ESR1 binding at enhancers normally co-occupied by GATA3 and/or FOXA1
The binding at most (∼66%) ESR1-binding regions remained unaffected when GATA3 was specifically silenced (Fig. 1C). FOXA1 and GATA3 co-occupancy analysis indicates a pattern similar to the Weaker ESR1–siGATA3 binding events, but with greater levels of FOXA1 cobinding (46%) (Fig. 2A). A striking difference is that the unaffected enhancers are in regions of more accessible chromatin, similar to the Stronger ESR1–siGATA3 binding events (Fig. 2B). This implies that ESR1 binding at enhancers with active histone marks is less dependent on GATA3, but is more dependent on FOXA1.
Taken together, these findings decouple the functions of GATA3 and FOXA1 in their abilities to tether ESR1 to the chromatin. Specifically, GATA3 is more likely to enable ESR1 binding in chromatin of limited active histone composition, while FOXA1 binding parallels ESR1 binding in both accessible and inaccessible regions, consistent with previous work showing that the majority of ESR1-binding events decrease when FOXA1 is specifically silenced (Hurtado et al. 2011).
GATA3 functions upstream of FOXA1 and EP300 in breast cancer cells
A recent study (Joseph et al. 2010) has shown that at FOXA1-bound enhancers there is no further gain in active histone marks upon ESR1 recruitment, suggesting that at FOXA1-bound cis-elements containing an ERE motif, ESR1 accessibility is predefined prior to estrogen stimulation. In addition, we have recently shown that EP300 is a global cofactor for ESR1 in breast cancer cells (Zwart et al. 2011) and can be chromatin bound prior to E2 stimulation. In order to investigate the role of GATA3 in shaping enhancer accessibility we mapped FOXA1, EP300 binding, and the active marks H3K4me1 and H3K27Ac in siControl and siGATA3 conditions in both unstimulated and E2-treated cells. For FOXA1, we performed the ChIP-seq experiments only in unstimulated cells, since FOXA1 has been shown to be unchanged by estrogen treatment (Lupien et al. 2008) and ESR1 depletion (Hurtado et al. 2011). We find that in cells with intact GATA3 (siControl), EP300 is redistributed upon estrogen stimulation from regions not bound by ESR1 to regions co-occupied by ESR1–EP300, showing that estrogen induces recruitment of EP300 only to a subset of cis-regulatory elements (Supplemental Fig. 3A,B), which correlates with the transcriptional regulation of estrogen gene targets (Supplemental Fig. 3C). At these enhancers there is limited histone remodeling upon estrogen treatment (Supplemental Fig. 3E), and similarly, there is limited histone mark changes at the regions that become depleted of EP300 following estrogen treatment (Supplemental Figs. 3F, 4A).
However, in MCF7 unstimulated siGATA3 cells, enhancer composition is dramatically affected, and this coincides with the redistributed ESR1-binding profile upon estrogen stimulation. Figure 3, A and C show the patterns of FOXA1 and EP300 binding in the presence and absence of GATA3, as well as changes observed in the histone marks H3K4me1 and H3K27Ac. For example, at the TFF3 locus GATA3 silencing results in a dramatic increase of the levels of the active histone marks and EP300 recruitment in serum-starved conditions and coincides with the stronger ESR1 binding upon E2 stimulation (Fig. 3B). Globally, at the 8243 “Stronger” ESR1 events that increased in binding affinity after GATA3 silencing, we found a coincident gain in FOXA1-binding signal and recruitment of the coactivator EP300 (Fig. 3C) together with an enrichment of the active histone marks (Fig. 3D). At “Weaker” regions with decreased ESR1-binding affinity following GATA3 depletion, we found a notable decrease in both FOXA1 and EP300 cofactor binding and a depletion of the histone marks, suggesting that at these loci GATA3 mediates FOXA1 and ESR1 binding. Mapping of FOXA1 binding in siGATA3 unstimulated MCF7 cells revealed 48,686 novel binding events, which suggests that GATA3 is, to a certain extent, capable of influencing FOXA1 recruitment (Supplemental Fig. 6). In addition, these results show that the differences between the categories of ESR1-binding sites in terms of transcription factor co-occupancy and active chromatin are present prior to stimulation with estrogen (and hence, prior to ESR1 binding) (Supplemental Fig. 5).
Together, these experiments suggest that the contribution of GATA3 in preparing cis-elements for the estrogen-induced ESR1 binding appears to be distinct from that of FOXA1. In wild-type MCF7 cells, ESR1-bound enhancers co-occupied by ESR1 and GATA3, lacking FOXA1, have lower levels of the active histone marks H3K4me1 and H3K27Ac than those with FOXA1 co-occupancy (Supplemental Fig. 4B), indicating that GATA3 facilitates ESR1 binding in chromatin depleted of these two active histone modifications, independent of FOXA1. A model depicting these events is shown in Supplemental Figure 4C. Given that silencing of GATA3 redirected both FOXA1 and EP300 in unstimulated cells (Fig. 3A,C), it is suggested that GATA3 can act upstream of FOXA1 in mediating ESR1-binding preferences.
Silencing of GATA3 results in changes of the gene-expression program that correlate with the redistributed ESR1-binding events
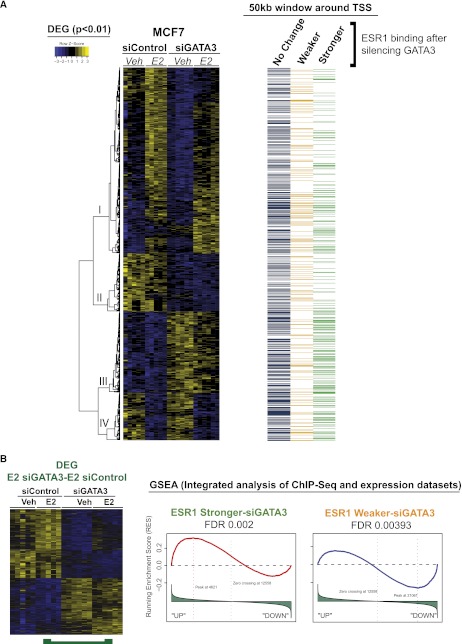
Microarray expression analysis in MCF7 cells transfected with siControl or siGATA3 in either untreated or estrogen-stimulated conditions enabled us to examine the functional impact of GATA3-driven redistribution of ESR1 binding (Fig. 4; Supplemental Fig. 7). We performed four different pairwise comparisons (Supplemental Fig. 7B) and combined all differentially expressed genes in a visual map presented in Figure 4A. We correlated the “Stronger” and “Weaker” events representing gain or loss of ESR1-binding affinity in the absence of GATA3, with the differential expressed genes using a 50-Kb window around the transcription start sites by Gene Set Analysis Enrichment (GSEA) (Fig. 4B; Supplemental Fig. 7C–F). A 50-kb window was chosen since ESR1 binding was shown to be enriched within 50 kb of target genes (Carroll et al. 2006). Independent hypergeometric testing (data not shown) agreed with the significant association of the binding events with the gene-expression changes identified by the GSEA.
Figure 4.
MCF7 cells depleted of GATA3 have an altered E2 transcriptional program that correlates with the redistributed ESR1 binding. (A) Integration of the Differentially Expressed Genes (DEG) with the redistributed ESR1 ChIP-seq data sets. A 50-kb window around the transcription start sites (TSS) of all DEG targets was overlapped with the altered ESR1 binding in siGATA3 conditions. ESR1 stronger binding is enriched near genes that are overexpressed in siGATA3 compared with siControl (bottom of gene cluster I, cluster III, and cluster IV). ESR1 Weaker events are in the vicinity of genes down-regulated by siGATA3 (Gene clusters 1 and II). Also included (right) is information showing whether each gene has an ESR1-binding event within 50 kb of the TSS. Included is ESR1 binding that is gained (green), lost (yellow), or not changed (blue) following GATA3 silencing. (B) Gene Set Analysis Enrichment (GSEA) reveals that ESR1 Stronger events after GATA3 silencing are enriched near up-regulated genes, whereas ESR1–Weaker siGATA3-binding events correlate with transcriptional down-regulation in siGATA3-treated cells.
We found a set of genes that are normally estrogen regulated, but in the absence of GATA3 are differentially transcribed without the requirement of ligand (E2) (Fig. 4B; Supplemental Fig. 7E). This is reminiscent of the binding of FOXA1, EP300, and the enrichment of active histone marks, which also occur in an estrogen-independent manner. The genes differentially regulated when GATA3 was silenced can be divided into two clusters, based on whether they increase or decrease in expression in the absence of GATA3. We find that the up-regulated genes are significantly correlated with the “Stronger” ESR1-binding events that occur in the absence of GATA3, whereas the down-regulated genes are significantly correlated with the “Weaker” ESR1-binding events (Fig. 4B; Supplemental Fig. 7E).
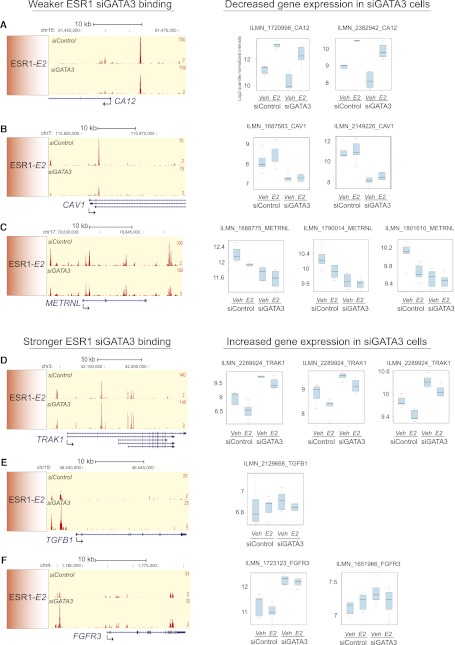
We also identified a set of genes normally up-regulated by estrogen stimulation that exhibit limited induction in the absence of GATA3 (Fig. 4A, cluster I; Supplemental Fig. 7C). Within 50 kb of the transcription start sites of these genes, we see a significant enrichment of the “Weaker” ESR1 binding when GATA3 is silenced (Supplemental Fig. 7C). This suggests that regions with loss in ESR1-binding affinity can no longer induce gene activation of the adjacent gene targets to the same extent. Individual examples of how the fluctuations in ESR1-binding affinity following GATA3 silencing correlates with changes in gene expression are shown in Figure 5 (average of six biological replicates per sample group). Weaker ESR1 binding in the vicinity of CA12 (Fig. 5A), CAV1 (Fig. 5B), and METRNL (Fig. 5C), correlates with decreased gene expression in siGATA3 MCF7 cells, whereas Stronger ESR1 binding near TRAK1 (Fig. 5D), TGFB1 (Fig. 5E), and FGFR3 (Fig. 5F) correlates with increased transcription.
Figure 5.
Redistribution of ESR1 binding after GATA3 silencing correlates with altered gene transcription. Weaker ESR1–siGATA3 binding correlates with decreased gene expression at the (A) CA12, (B) CAV1, and (C) METRNL genes. Stronger ESR1–siGATA3 binding correlates with increased gene transcription of the (D) TRAK1, (E) TGFB1, and (F) FGFR3 genes. (Left) Snapshots of ESR1 binding in siControl and siGATA3 estrogen conditions; (right) log2 quantile normalized averaged intensities of six biological replicates with the corresponding Illumina probes. For the METRNL gene we omitted the probes ILMN_2342066 and ILMN_2258004 due to space restrictions; however, they agreed with the other probes.
In addition, we found that the GATA3-dependent gene expression signature (Fig. 4A, clusters I–IV) correlated with changes of the active histones marks through the promoter and gene body (1 kb upstream of and 4 kb downstream from TSS) (Supplemental Fig. 8). For instance, genes down-regulated in siGATA3 cells (Fig. 4A, cluster II) have overall reduced levels of the active histone marks H3K4me1 and H3K27Ac, whereas up-regulated genes from cluster III gained active histone marks. This indicates that the histone composition within the open reading frame of genes can successfully represent their transcriptional state.
Together, the significant correlations of the redistributed ESR1-binding profile with the altered gene expression indicates that the observed changes in transcription factor binding affinity are functional and transcriptionally relevant.
GATA3 participates in E2-induced long-range chromatin interactions
Loss of functional GATA3 results in changes in ESR1, FOXA1, and EP300 binding, plus H3K4me1 and H3K27Ac histone marks, which correlated with altered gene expression. Except for ESR1 binding, all of these changes are seen in the absence of estrogen treatment, when ESR1 is not on the chromatin. We speculated that these global genomic changes may be attributed to transcriptional chromatin loops that form in ESR1+ breast cancer cells (Fullwood et al. 2009). We experimentally assessed this using chromosomal conformation capture (3C) assays within the TFF gene cluster (Supplemental Fig. 9). We focused on the TFF locus because it provides a unique snapshot of an ESR1-regulated microenvironment. TFF1 and TFF3 are classic ESR1 target genes, strongly estrogen induced, which in siGATA3 conditions become two of the most differentially up-regulated genes (Supplemental Fig. 10A). We assessed chromatin interactions between regions that contained GATA3-dependent ESR1-binding events or were bound by other transcription factors such as FOXA1 or CTCF. Importantly, at this locus we identified two ESR1-binding events that become stronger in siGATA3 conditions (Supplemental Fig. 9A, anchors 2 and 10). While many of these genomic regions can form chromatin loops in wild-type (siControl) MCF7 cells (Supplemental Fig. 10B,C), when GATA3 was silenced, we find an ESR1 and hormone-independent gain in the chromatin loops that are formed between these specific loci (Supplemental Fig. 9B–D). These results suggest that estrogen–ESR1 may be dispensable for the formation of intrachromosomal loops when GATA3 is not present at this locus. Since GATA3 appears to modulate long-range chromatin interactions, loss of GATA3 may result in different chromatin loops forming, explaining the indirect gain of ESR1 and FOXA1 binding in siGATA3 conditions.
Discussion
FOXA and GATA family members are crucial for hormone-driven cancers. In breast cancer, GATA3 and FOXA1 are genes that define an ESR1+ breast tumor (Perou et al. 2000; Sorlie et al. 2003), possibly due to their requirement in eliciting ESR1-mediated transcription and cell growth. Supportive evidence suggests that GATA3 and FOXA1 are sufficient to partially restore a functional ESR1 pathway in cells that are normally ESR1 negative (Kong et al. 2011). The global analysis presented here suggests that GATA3 mediates ESR1 and FOXA1 binding at ERE elements embedded in chromatin depleted of active histone modifications, highlighting a distinct role for GATA3 relative to FOXA1. In addition, we show that GATA3 can act upstream of FOXA1 in mediating ESR1 binding by modulating enhancer composition, as indicated by the altered cofactor binding and distribution of active histone marks (Fig. 3). One possible explanation for this is that GATA3 binding is required for maintaining optimal chromatin conditions at specific loci for FOXA1-binding capacity (direct GATA3- and FOXA1-binding events). Another is that GATA3 mediates chromatin loops between FOXA1-bound enhancers and other cis-regulatory elements (indirect FOXA1-binding regions), which may explain why silencing of GATA3 influences FOXA1 and ESR1 at specific regions. However, it is clearly evident that there are functional consequences of GATA3 silencing that link fluctuations of transcription factor binding and deposition of active histone marks with changes in gene expression programs (Fig. 4).
Furthermore, since ESR1 and its cooperating transcription factors bind predominantly at distal enhancers, it is likely to elicit its transcriptional effect through DNA looping to proximal promoters or “transcription factories” as indicated by studies of higher genomic organization (Li et al. 2012). Here we show that GATA3 participates in the formation of three-dimensional chromatin interactions in breast cancer cells (Supplemental Figs. 9, 10), supporting similar conclusions about GATA factors in blood cells (Vakoc et al. 2005; Jing et al. 2008). In our system and in the presence of GATA3, interactions are formed between ESR1, GATA3, and FOXA1-bound enhancer elements at the TFF locus, suggesting that GATA3 participates in chromatin loops, but simultaneously blocks potential interactions until ESR1 is recruited. However, in the absence of GATA3, these interactions become ESR1 independent and are likely to mediate the elevated expression of TFF1 and TFF3.
Based on the observation that ESR1 redistribution correlated with cofactor binding, chromatin state, and gene transcription, as well as our experimental evidence at the TFF locus, we can speculate that enhancers brought in the proximity of anchoring events may become more accessible due to an “opening” of the surrounding chromatin. In contrast, enhancers moved away from anchoring neighborhoods may become less accessible and transcriptionally active, as suggested by the ESR1 Weaker–siGATA3-binding events (Figs. 3, 4).
Silencing of FOXA1 in prostate cancer cells reprograms AR binding, with a loss of many AR-binding events and a simultaneous acquisition of numerous new AR-binding events (Sahu et al. 2011; Wang et al. 2011). This resembles our observations for GATA3 and its ability to both positively and negatively impact ESR1 recruitment in breast cancer cells. A distinct difference is that in breast cancer cells lacking GATA3, we observed substantial epigenetic changes at distal enhancers, whereas in the prostate cells, enhancer accessibility was not affected when FOXA1 was silenced (Sahu et al. 2011; Wang et al. 2011). In general, forkhead and GATA members appear to facilitate nuclear receptor access to their DNA response elements. In breast cancer cells accessibility at cis-regulatory elements is predetermined by the presence of specific histone modifications such as H3K4me1-2 (Lupien et al. 2008; Wang et al. 2011) and H3K27Ac (this study). FOXA1 mediates nuclear receptor binding in both breast and prostate cancer cells, whereas GATA3 is mostly limited to the breast. However, another member of the GATA family, GATA2, is highly expressed in prostate (Bohm et al. 2009) and involved in AR function (Wang et al. 2007). Although FOXA and GATA factors are crucial for nuclear receptor function in breast and prostate they may have evolved tissue-specific functions, such as the ability for FOXA1 to positively and negatively influence AR binding in prostate cancer cells, a scenario observed for GATA3 and ESR1 in breast cancer cells.
In the clinical setting, GATA3 is one of the most differentially expressed genes in ESR1-positive versus ESR1-negative breast cancers and is a prognostic marker for aggressive disease and poor survival (Mehra et al. 2005). Interestingly, GATA3 is mutated in breast cancers with a frequency of ∼10% (The Cancer Genome Atlas Network 2012). Based on our findings that GATA3 depletion redistributes ESR1 binding, we speculate that breast cancers with GATA3 mutations may have an altered ESR1-binding profile and transcriptional program. As presented here, silencing of GATA3 resulted in substantial changes in the presence of histone marks, transcription factor binding, and gene expression, even in hormone-depleted conditions. If GATA3 mutations in breast tumors render GATA3 nonfunctional, it is possible that similar to our findings, there may be an altered ESR1-mediated transcriptional program. The TFF1 and TFF3 genes, shown to be up-regulated by GATA3 silencing (Supplemental Fig. 10A), are two of the most significant prognostic genes for bone metastases (Smid et al. 2006). The altered higher genomic organization at this locus following silencing of GATA3 recapitulates an E2-induced program, but importantly, in an ESR1-independent manner. Since the majority of luminal breast cancers remain ESR1 positive even in distant metastases (Harrell et al. 2006; Hoefnagel et al. 2010), we can speculate that changes in the sequence fidelity and function of factors, such as GATA3, may contribute to the heterogeneity observed in ESR1+ breast disease.
Our findings help define the hierarchy of different transcription factors in the ESR1-mediated transcriptional program (Supplemental Fig. 10D). In the presence of GATA3, ESR1 binding is mediated by GATA3 and FOXA1 to the cis-regulatory elements that drive transcription of the ESR1 target genes. However, in the absence of GATA3, chromatin accessibility at potential ESR1-binding elements may be altered, resulting in a rewired ESR1-binding profile and expression program. Our data indicate that GATA3 is one of the central components of the ESR1 complex that determines the binding potential and transcriptional targets in breast cancer cells.
Methods
Cell culture
MCF7 cells were cultured in DMEM and ZR-75-1 cells in RPMI media (Invitrogen) at 10% FCS. For hormonal deprivation, cells were cultured for 3 d in phenol-red free DMEM or RPMI, respectively, supplemented with 5% charcoal-treated serum. E2 (Sigma) was added at a final concentration of 100 nM, for 3 h for ChIP-seq and 3C experiments, 6 h for the microarray expression experiment.
siRNA transfections
We used three different siRNAs against GATA3: ON-TARGET plus siRNA J-0033781-09 and two additional custom-made siRNAs targeting the 3′ UTR of the RefSeq NM_002051. The sense sequences of the custom made ones are siGATA3-Custom-1: AAACUAGGUCUGAUAUUCAUU and siGATA3-Custom-2: CUUUAUUGCAUCUGGGUAGUU. AllStars Negative Control siRNA (Cat nr: 1027281, Qiagen) was used as a negative control. All siRNA experiments were at a final concentration of 50 nM. Transfections were conducted using Lipofectamine 2000 (Invitrogen). For the microarray expression experiment we used the three siRNAs against GATA3 separately; for the ChIP-seq and 3C we used a pool of them to achieve better knockdown and eliminate potential nonspecific effects by using a single siRNA.
ChIP-seq library preparation, Illumina Sequencing, and enrichment analysis
MCF7 cells were transfected with siRNA and cultured in hormone-deprived conditions for a further 3 d. Chromatin Immunoprecipitations and library preparations were performed as previously described (Schmidt et al. 2009). Sequences generated by the Illumina GAIIx genome analyzer were aligned against NCBI Build 36.3. For more details regarding antibodies and data analysis, please refer to the Supplemental Information for materials and methods.
Microarray experiment
MCF7 cells were transfected with siRNA and cultured in hormone-deprived conditions for a further 3 d. Cells were subsequently treated with 100 nM estrogen (E2) or control (Veh) for 6 h. We performed two independent biological experiments, each using three different siRNAs against GATA3 individually (six siGATA3 replicates in total). For siControl we used RNA from five biological replicates.
Chromosome-conformation-capture (3C) assays
3C assays were performed as previously described (Nativio et al. 2009) with minor modifications (see Supplemental Information). The siRNA transfections procedure was the same as for the ChIP-seq experiments shown in Supplemental Figure 4A. E2 stimulation was performed for 3 h. Primer sequences and a detailed protocol are provided in the Supplemental section.
Data access
The microarray data and ChIP-seq data from this study have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession nos. GSE39623 and GSE40129, respectively.
Acknowledgments
We thank all members of the Carroll lab for their constructive input, especially Rosalind Launchbury for technical assistance. We thank Aisling Redmond, Jessica Robinson, Adele Murrell, and Wayne Tilley for critically reading the manuscript. We thank Silvia Halim for help with depositing the data. We thank Yoko Ito for help with the 3C experiments and Dominic Schmidt for scripting, James Hadfield and members of the sequencing facility for their genomic support, and Matthew Eldridge and Stewart MacArthur for bioinformatics support. J.S.C. is supported by an ERC Starting Grant and an EMBO Young Investigator Award.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.139469.112.
References
- Arnold JM, Choong DY, Thompson ER, Waddell N, Lindeman GJ, Visvader JE, Campbell IG, Chenevix-Trench G 2010. Frequent somatic mutations of GATA3 in non-BRCA1/BRCA2 familial breast tumors, but not in BRCA1-, BRCA2- or sporadic breast tumors. Breast Cancer Res Treat 119: 491–496 [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. 2007. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9: 201–209 [DOI] [PubMed] [Google Scholar]
- Bohm M, Locke WJ, Sutherland RL, Kench JG, Henshall SM 2009. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene 28: 3847–3856 [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Burdett L, Yeager M, Llaca V, Langerod A, Presswalla S, Kaaresen R, Strausberg RL, Gerhard DS, Kristensen V, et al. 2007. Somatic sequence alterations in twenty-one genes selected by expression profile analysis of breast carcinomas. Breast Cancer Res 9: R5 doi: 10/1186/bcr1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9: 279–289 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL 2011. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145: 622–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell JC, Dye WW, Allred DC, Jedlicka P, Spoelstra NS, Sartorius CA, Horwitz KB 2006. Estrogen receptor positive breast cancer metastasis: Altered hormonal sensitivity and tumor aggressiveness in lymphatic vessels and lymph nodes. Cancer Res 66: 9308–9315 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefnagel LD, van de Vijver MJ, van Slooten HJ, Wesseling P, Wesseling J, Westenend PJ, Bart J, Seldenrijk CA, Nagtegaal ID, Oudejans J, et al. 2010. Receptor conversion in distant breast cancer metastases. Breast Cancer Res 12: R75 doi: 10.1186/bcr2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA 2008. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol Cell 29: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC 2007. Chemoprevention of breast cancer with selective oestrogen-receptor modulators. Nat Rev Cancer 7: 46–53 [DOI] [PubMed] [Google Scholar]
- Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, Pan YF, Li G, Lim M, Thomsen JS, et al. 2010. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol Syst Biol 6: 456 doi: 10.1038/msb.2010.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SL, Li G, Loh SL, Sung WK, Liu ET 2011. Cellular reprogramming by the conjoint action of ERα, FOXA1, and GATA3 to a ligand-inducible growth state. Mol Syst Biol 7: 526 doi: 10.1038/msb-2011.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z 2006. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127: 1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. 2012. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148: 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, et al. 2007. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet 3: e87 doi: 10.1371/journal.pgen.0030087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132: 958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R, Varambally S, Ding L, Shen R, Sabel MS, Ghosh D, Chinnaiyan AM, Kleer CG 2005. Identification of GATA3 as a breast cancer prognostic marker by global gene expression meta-analysis. Cancer Res 65: 11259–11264 [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet 5: e1000739 doi: 10.1371/journal.pgen.1000739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YF, Wansa KD, Liu MH, Zhao B, Hong SZ, Tan PY, Lim KS, Bourque G, Liu ET, Cheung E 2008. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem 283: 32977–32988 [DOI] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. 2000. Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu B, Laakso M, Ovaska K, Mirtti T, Lundin J, Rannikko A, Sankila A, Turunen JP, Lundin M, Konsti J, et al. 2011. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J 30: 3962–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT 2009. ChIP-seq: Using high-throughput sequencing to discover protein-DNA interactions. Methods doi: 10.1016/j.meth.2009.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smid M, Wang Y, Klijn JG, Sieuwerts AM, Zhang Y, Atkins D, Martens JW, Foekens JA 2006. Genes associated with breast cancer metastatic to bone. J Clin Oncol 24: 2261–2267 [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usary J, Llaca V, Karaca G, Presswala S, Karaca M, He X, Langerod A, Karesen R, Oh DS, Dressler LG, et al. 2004. Mutation of GATA3 in human breast tumors. Oncogene 23: 7669–7678 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA 2005. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M 2007. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell 27: 380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. 2011. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474: 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG 2009. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J 28: 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart W, Theodorou V, Kok M, Canisius S, Linn S, Carroll JS 2011. Oestrogen receptor-co-factor-chromatin specificity in the transcriptional regulation of breast cancer. EMBO J 30: 4764–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]