Abstract
Many eukaryotic genes possess multiple alternative promoters with distinct expression specificities. Therefore, comprehensively annotating promoters and deciphering their individual regulatory dynamics is critical for gene expression profiling applications and for our understanding of regulatory complexity. We introduce RAMPAGE, a novel promoter activity profiling approach that combines extremely specific 5′-complete cDNA sequencing with an integrated data analysis workflow, to address the limitations of current techniques. RAMPAGE features a streamlined protocol for fast and easy generation of highly multiplexed sequencing libraries, offers very high transcription start site specificity, generates accurate and reproducible promoter expression measurements, and yields extensive transcript connectivity information through paired-end cDNA sequencing. We used RAMPAGE in a genome-wide study of promoter activity throughout 36 stages of the life cycle of Drosophila melanogaster, and describe here a comprehensive data set that represents the first available developmental time-course of promoter usage. We found that >40% of developmentally expressed genes have at least two promoters and that alternative promoters generally implement distinct regulatory programs. Transposable elements, long proposed to play a central role in the evolution of their host genomes through their ability to regulate gene expression, contribute at least 1300 promoters shaping the developmental transcriptome of D. melanogaster. Hundreds of these promoters drive the expression of annotated genes, and transposons often impart their own expression specificity upon the genes they regulate. These observations provide support for the theory that transposons may drive regulatory innovation through the distribution of stereotyped cis-regulatory modules throughout their host genomes.
In recent years, a large body of work has been uncovering the complexities of transcriptional regulation in eukaryotes. The landscapes of transcription, surveyed with ever-increasing scrutiny, reveal intricate genetic architectures from which originate myriads of protein-coding and noncoding transcripts (Kapranov et al. 2007a; Djebali et al. 2012). The regulatory blueprints that orchestrate the spatiotemporal dynamics of eukaryotic transcriptomes mirror this complexity. Large-scale surveys of chromatin modifications and transcription factor occupancy in diverse organisms have started to shed light on the abundance of cis-regulatory modules (Ernst et al. 2011; Negre et al. 2011; The ENCODE Project Consortium et al. 2012; Shen et al. 2012), their relevance to development and disease (Lindblad-Toh et al. 2011; The ENCODE Project Consortium et al. 2012), and the structure of the gene regulatory networks they implement (Suzuki et al. 2009; Marbach et al. 2012). Additionally, genome-wide studies of transcription start site (TSS) usage have shown that many genes possess alternative promoters, highlighting the importance of their contribution to the diversity of gene expression patterns (Carninci et al. 2006; Suzuki et al. 2009). TSSs are of particular interest, because in addition to harboring many transcription factor binding sites (TFBSs), the promoters they are embedded in constitute the platforms where the transcriptional machinery integrates the inputs from cognate cis-regulatory elements. They are also worthy of attention from an experimental standpoint, since the quantification of transcripts coming from individual TSSs allows for precise measurements of the final output of these molecular computations.
The explosion of experimental and computational approaches in functional genomics that accompanied the advent of second-generation sequencing has been, and continues to be, the major driving force behind our progress in uncovering and understanding this regulatory complexity. For the study of TSS location and activity, however, even state-of-the-art, high-resolution techniques based on 5′-complete cDNA sequencing (Kodzius et al. 2006; Ni et al. 2010; Plessy et al. 2010) are currently lacking in multiple aspects. Here we address these issues and present RAMPAGE (RNA Annotation and Mapping of Promoters for the Analysis of Gene Expression), a very accurate 5′-complete cDNA sequencing approach that allows for the ab initio identification of TSSs at base-pair resolution, the quantification of their expression and the characterization of their transcripts. We engineered our protocol to take full advantage of the paired-end sequencing capabilities of current high-throughput platforms, thus yielding crucial transcript connectivity information. Importantly, this feature allows us to rigorously connect TSSs to the genes they drive the expression of based on direct cDNA evidence. Our method also provides much higher specificity for TSSs than current approaches, and we developed a streamlined 2-day protocol that allows the barcoding and pooling of multiple samples after the very first step, thus greatly facilitating library multiplexing and preparation. For the analysis of these data, we have developed an integrated analysis pipeline that relies on the unique features of the data to maximize TSS specificity, transcript connectivity information recovery, and quantification accuracy. At the core of this pipeline lies a novel peak-calling algorithm for TSS discovery that was specifically tailored to filter out multiple types of noise (i.e., random distortions of the underlying signal by technical factors) associated with 5′-complete cDNA sequencing.
Using this approach, we set out to profile promoter activity genome-wide throughout the life cycle of Drosophila melanogaster, so as to have a complete view of the transcriptional landscape and of the diversity of expression patterns in this model organism. This rich data set reveals that >40% of all genes are expressed from at least two promoters, underscoring the pervasiveness of this phenomenon in Drosophila. Importantly, we found that alternative promoters generally have uncorrelated expression patterns, which reveals that they most often implement independent regulatory programs. These observations suggest that the emergence of alternative promoters has been a major driving force underlying the evolutionary diversification of gene expression programs. Our analyses also uncovered a widespread role for transposons in the developmental regulation of transcription, with approximately 1300 transposon-embedded promoters driving developmentally regulated expression of diverse sets of transcripts.
Transposable elements (TEs) have been shown to influence gene expression in a variety of organisms, including plants (McClintock 1956; Lippman et al. 2004; Naito et al. 2009), Drosophila (Lipatov et al. 2005; Rouget et al. 2010), and mammals (Nigumann et al. 2002; Bejerano et al. 2006). This regulatory potential, together with the ability of transposons to disseminate stereotyped sequence modules throughout their host genomes, has led to the proposal that transposon expansion and domestication may be a powerful force underlying the assembly of complex regulatory networks (Britten and Davidson 1969; Feschotte 2008), in particular by providing promoters for host genes (Nigumann et al. 2002; van de Lagemaat et al. 2003; Faulkner et al. 2009). Their potential contribution to developmental gene expression, however, is currently only supported by modest evidence in mammals (Peaston et al. 2004; Cohen et al. 2009; Macfarlan et al. 2012) and has been reported to be extremely rare in Drosophila (Lipatov et al. 2005). Furthermore, it is unclear whether transposons actually distribute promoters with stereotyped regulatory logics through a copy-and-paste mechanism.
We found that transposons from diverse classes have been co-opted to drive the expression of hundreds of annotated genes. Many of these transposons appear to have conferred their intrinsic regulatory specificity to the genes they drive, which demonstrates that they do distribute preprogrammed regulatory modules to multiple loci. A case study of roo element long terminal repeats (LTRs) uncovered the existence of a core promoter and of a complex set of TFBSs that underlie these intrinsic regulatory properties.
Results
RAMPAGE: multiplexed paired-end sequencing of 5′-complete cDNAs
5′-Complete cDNA sequencing has proven to be a challenging task, despite significant contributions over the years from several approaches that have relied on diverse strategies. CAGE (Kodzius et al. 2006) is based on the biotinylation of the 7-methylguanosine cap of Pol II transcripts and pulldown of the 5′-complete cDNAs they are hybridized to, a technique known as “cap-trapping” (Carninci et al. 1996). CAGEscan (Plessy et al. 2010) and other approaches (Islam et al. 2011) exploit some unique features of reverse-transcriptase enzymes to add adaptors to the end of 5′-complete first-strand cDNAs during the reverse-transcription step, in a process dubbed “template-switching” (Hirzmann et al. 1993). PEAT (Ni et al. 2010) and similar techniques rely on the ligation of an RNA adaptor to the 5′ end of capped transcripts (“oligo-capping”), similarly to conventional 5′-RACE.
CAGE relies on a protocol that, although scalable, is cumbersome and requires input amounts on the order of 50 μg of total RNA. Its main limitation is the impossibility to sequence more than short 5′ tags (about 27 bases) from the cDNAs, which makes unambiguous read mapping impossible for large parts of eukaryotic genomes, precludes evidence-driven assignment of novel TSSs to gene annotations, and yields no transcript structure information. This has been a major impediment to the analysis of novel TSSs in general and of repeat-borne TSSs in particular. The specificity of CAGE for TSSs is also currently limited (please see Assessment of Assay Performance below). CAGEscan does allow paired-end sequencing of cDNA inserts, but with lower TSS specificity (Plessy et al. 2010). PEAT also allows for paired-end sequencing, although only 20 bp can be sequenced from each end due to the cloning procedure used, but this is again at the expense of specificity. Moreover, adaptor ligation is mediated by T4 RNA ligase 1, an enzyme known to have strong sequence biases (Zhenodarova et al. 1989), which is detrimental to accurate transcript representation. Finally, this complex protocol requires large quantities of starting material (∼150 μg total RNA), which is impractical for most samples.
To address these challenges, we developed RAMPAGE by modifying and combining the two orthogonal 5′-selection approaches of template-switching and cap-trapping (Fig. 1A; Supplemental Fig. S1; Methods; Supplemental Methods). In comparison to current approaches, RAMPAGE has the key advantage of yielding long paired reads as opposed to short sequence tags, while also offering greatly improved specificity for TSSs (Fig. 1B; Supplemental Fig. S2). Library preparation and multiplexing is greatly facilitated by the fact that individual samples are barcoded and pooled after the very first step of the protocol, allowing almost the entire workflow to be carried out on a single library (Supplemental Fig. S1). Additionally, all steps from the biological sample to the pooled cDNA products can be carried out in 96-well plates, and our full workflow from RNA to library can be completed in 2 d, making library preparation simple and very scalable. Input material requirements are on the order of 10- to 20-fold lower than for conventional CAGE.
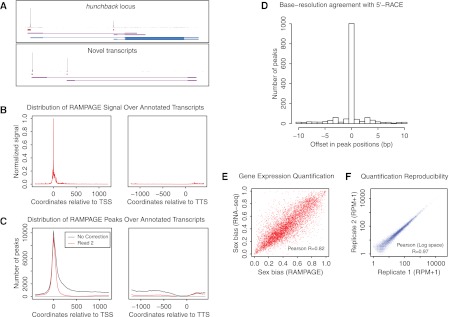
Figure 1.
RAMPAGE: specific, accurate, quantitative paired-end sequencing of 5′complete cDNAs. (A) Graphical representation of the data at the hunchback gene locus and at an unannotated locus harboring novel transcripts. For each panel, the top track shows the density of cDNA 5′ ends per position on the upper strand, which can be interpreted as a single base-resolution profile of transcription initiation activity. The second track represents the peaks (i.e., TSS clusters) called from that density profile. The third track shows the partial transcript models reconstructed ab initio from our sequencing data using Cufflinks. For the upper panel, the fourth track displays FlyBase transcript annotations. For the bottom panel, note that paired-end information allows one to infer a functional link between the two promoters, which appear to be alternative promoters for a common locus. (B) Metaprofile of signal density over all FlyBase r5.32 transcript annotations. (TSS) Transcription start site; (TTS) transcription termination site. (C) Metaprofile of peak density over annotated transcripts. (Red curve, downstream read coverage correction; black curve, no correction; all other peak-calling parameters were kept identical). (D) Histogram of the cross-correlation of TSS cluster positioning by RLM-RACE and by our method. For each cluster, we determined the positional offset (in base pairs) that maximizes the cross-correlation between the data from the two methods. (E) Comparison of RAMPAGE and standard RNA-seq performance for relative quantification of gene expression. We compared the measures of sex bias in the expression of genes obtained by the two methods. (F) Reproducibility of expression level measurements between biological replicates. ([RPM] Reads per million.)
Computational analysis of RAMPAGE data
We designed an integrated computational strategy that makes extensive use of the unique features of the data to enhance the accuracy and quality of our analysis. All analysis steps from raw sequencing data to TSS clusters (TSCs), expression level estimates, and partial transcript models can be performed in a single process for a set of samples. The complete analysis workflow is summarized in the Methods section and Supplemental Figure S3A.
The cornerstone of this pipeline is a novel peak-calling algorithm for TSS discovery that implements several noise-filtering strategies to greatly improve our ability to discriminate between true TSSs and background signal. As in other high-throughput assays, robustly detectable signal must be distinguished from a background that may have multiple possible origins. Additionally, most eukaryotic promoters do not use a single position as their TSS, but allow transcription initiation at several positions. The shapes of these TSCs vary between promoters, from sharp (a few nucleotides) to broad (≥100 nucleotides) (Carninci et al. 2006). Therefore, previous analyses of 5′-complete cDNA sequencing data have usually made use of some strategy to group individual TSSs into clusters (Carninci et al. 2006; Ni et al. 2010; Plessy et al. 2010). Building upon this work, we devised a novel approach to identify TSCs, which we define operationally as regions of statistically significant clustering of RAMPAGE 5′ end tags. Critically, our peak-calling algorithm was designed to make extensive use of paired-end information and to correct for several sources of noise inherent to 5′-complete cDNA sequencing.
First, we expect the background distribution of signal per genomic position to be overdispersed due to at least two technical factors: Failures of reverse-transcriptase to reach the 5′ end of its template are expected to be more likely at specific sites of a given transcript (e.g., strong secondary structures), and PCR duplicates in the libraries can randomly amplify the signal at individual positions. Both effects will lead to the data looking more “peaky” than the actual landscape of transcription initiation is. To attenuate these effects, we make use of an overdispersed distribution (negative binomial) to model background signal, and we remove PCR duplicates from our data sets prior to peak-calling. For our purposes, we define PCR duplicates as read pairs that share similar alignment coordinates (start, end, splice sites) and an identical reverse-transcription primer sequence (which we use as a pseudo-random single-molecule barcode).
Second, nonspecific signal coming from non-5′-complete cDNAs represents another source of background, which is complex because the amount of nonspecific signal depends on transcript abundance. In the absence of an appropriate correction, this will lead to highly expressed transcripts contributing many false-positive TSCs. To limit this effect, other investigators have made use of independent RNA-seq data to filter CAGE signal (Hoskins et al. 2011), but this approach requires the generation of additional data sets for all samples under study. Harnessing paired-end information, we make use of the fact that coverage by downstream sequencing reads (i.e., the 3′-most portion of our cDNAs) can provide us with an estimate of transcript abundance at internal (non-TSS) positions. We model background from incomplete cDNAs as linearly proportional to transcript abundance as measured by downstream read coverage and show this approach to greatly improve our ability to distinguish between true TSSs and spurious internal signal (Fig. 1C; Supplemental Fig. S3B).
These features were incorporated into a sliding window algorithm that scans the genome and assesses the significance of local signal enrichment given the null distribution. Downstream read coverage in the same window is used to correct for local transcript abundance, by subtracting from the raw signal a pseudocount proportional to this coverage. After a false discovery rate (FDR) correction (Benjamini-Hochberg), enriched windows in close proximity to each other are merged into peaks, and those are subsequently trimmed at the edges down to the first base with signal.
Our data yield rich information about transcript structure and connectivity, which allows us to connect these TSCs to annotated genes based on rigorous cDNA evidence. This is an extremely important feature, since complex transcriptional architectures (Kapranov et al. 2007b; Djebali et al. 2012) make the promoter–transcript relationships at many loci otherwise difficult to decipher. Additionally, we take advantage of the fact that the downstream portions of the inserts are distributed over broad regions of the targets to gain knowledge about medium-range transcript connectivity. In the current implementation, reads from individual TSCs are processed through Cufflinks to produce partial transcript models.
Assessment of assay performance
The combination of template-switching and cap-trapping yields libraries that are highly enriched for 5′-complete cDNAs, as can be judged from the distribution of raw signal over annotated transcripts (Fig. 1B). For individual transcript annotations, we estimate that the median proportion of 5′ tags in TSS regions is >90% (Supplemental Fig. S2). Comparisons to similar D. melanogaster data generated by CAGE or PEAT revealed a dramatic improvement in specificity over these previous methods (Supplemental Fig. S2). In turn, the peak calls are themselves extremely highly enriched over annotated TSSs (Fig. 1C). Analysis of histone modification ChIP-seq profiles confirmed that the vast majority of peaks display chromatin features characteristic of TSSs (Supplemental Fig. S4). The downstream read transcript abundance correction proves to be very effective at filtering out spurious signal in internal regions of transcripts, while having a very limited effect on sensitivity for annotated TSSs (Fig. 1C; Supplemental Fig. S3).
In terms of topological resolution, extensive comparisons on equivalent samples with a large RNA ligase-mediated 5′-RACE (RLM-RACE) data set (Hoskins et al. 2011) show very strong agreement between the two techniques (Fig. 1D). This demonstrates that RAMPAGE achieves single-base topological resolution in TSS detection, which has previously not been possible with CAGE (Hoskins et al. 2011).
Gene expression quantification accuracy was benchmarked against standard shotgun RNA-seq data from adult male and female D. melanogaster that were generated by the modENCODE consortium (Graveley et al. 2010). This comparison showed good agreement between the techniques for absolute quantification (Supplemental Fig. S5), and excellent agreement for relative quantification (Fig. 1E). Expression level estimates are very reproducible, even between full biological replicates (Fig. 1F).
TSS discovery and expression profiling throughout the D. melanogaster life cycle
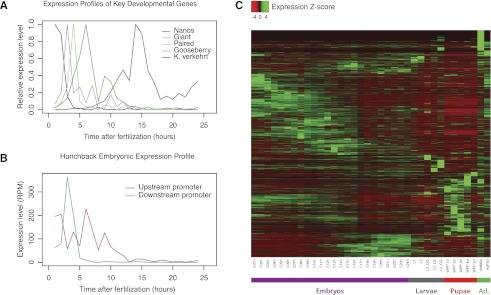
This methodological approach was used to study promoter activity dynamics throughout the life cycle of D. melanogaster (24 embryonic stages, five larval, five pupal, two adult). We sampled embryonic development, a period of fast transitions, at high temporal resolution (1 h). All sequencing data (Supplemental Table S1) were mapped to the genome with our spliced read aligner, STAR (Dobin et al. 2012). Stringent peak-calling identified 31,080 high-confidence TSCs (versus 12,454 in the most recent global study) (Hoskins et al. 2011), 76% of which could be unambiguously assigned to 12,706 annotated genes based on cDNA structure (Methods). The remaining 7421 TSCs drive novel transcripts, which we partially characterize (Fig. 1A). Of the genic TSCs, as many as 39.6% are unannotated in FlyBase r5.32. Our results are consistent with the known structure and expression dynamics of well-characterized developmental regulators (Figs. 1A, 2A), including the differential expression of alternative promoters (Fig. 2B), and represent to our knowledge the first genome-wide developmental timecourse of promoter activity (Fig. 2C).
Figure 2.
RAMPAGE recapitulates known expression profiles and establishes genome-wide promoter activity dynamics. (A) Expression profiles of well-characterized key developmental genes during embryonic development. Note the sharpness of the profiles afforded by the high temporal resolution of the timecourse. (K. verkehrt indicates krotzkopf verkehrt.) (B) Differential expression of alternative promoters (hunchback locus). Our data fully recapitulate the expression pattern for hb that has been characterized in previous work (Schroder et al. 1988). The hb mRNAs transcribed from the upstream (maternal) promoter are predominant immediately after egg laying and decay rapidly as the downstream promoter starts being expressed, displaying maximal expression 2–3 h after egg laying. The upstream promoter is active again with a second peak at 5–6 h. (C) Heatmap representing the Z-score normalized expression profiles for the 24,264 promoters we could attribute to annotated genes based on cDNA structure.
The use of alternative promoters is very common in D. melanogaster, with >40% of developmentally expressed genes having at least two promoters (Fig. 3A). In contrast, FlyBase annotations only attribute alternative promoters to 14.8% of genes (see Methods). The discovery of so many promoters with relatively shallow sequencing of complex samples and a stringent analysis indicates that alternative promoter usage is an extremely frequent phenomenon, even in a relatively simple metazoan genome. Importantly, alternative promoters tend to drive expression in uncorrelated patterns (Fig. 2B; Supplemental Fig. S6). This shows that they generally implement distinct regulatory programs, as suggested previously (Carninci et al. 2006; Rach et al. 2009). Further analysis of 1295 genes that undergo clear developmental transitions between alternative promoters revealed that these transitions occur in a great diversity of temporal patterns, throughout the entire life cycle (Fig. 3C).
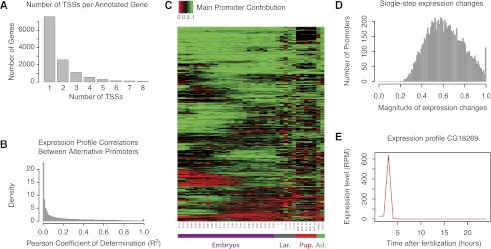
Figure 3.
Widespread differential regulation through alternative promoter usage and fast kinetics of regulatory transitions. (A) Number of TSSs detected per annotated gene. Over 40% of all expressed genes have at least two alternative TSSs. (A small number of genes are excluded from the graph [more than 10 TSSs], but these are probably affected by technical artifacts.) (B) Distribution of pairwise Pearson's coefficients of determination (R2) between the full expression profiles (36 timepoints) of alternative promoters. This gives a measure of the similarity between the expression profiles of alternative promoters. Only TSCs with a maximum expression level ≥10 RPM were included. Note the overall absence of correlation (median coefficient, 0.108). (C) Temporal dynamics of developmental transitions between alternative promoters. The heatmap represents the fraction of total expression contributed by the main promoter at each timepoint for 1295 genes that display pronounced transitions between promoters (see Methods). Note the diversity in the timing of promoter transitions. (D) Maximal fraction of the dynamic range of the profile of a given TSS spanned in a single hour during embryonic development (24 timepoints, 0–24 h). Median is 60.8%. Only genes whose expression range spans at least an order of magnitude and whose maximum expression level exceeds 10 RPM were considered in this analysis. (E) Example of a gene with fast transitions kinetics of high absolute magnitude.
The analysis of our high-resolution data shows that many genes undergo very fast transitions during embryonic development, their expression changes often spanning a large fraction of their dynamic range (median, 60.8%) within a single hour (Fig. 3D; Supplemental Fig. S7). Some of these abrupt regulatory transitions can sometimes be of a very large magnitude on an absolute scale (Fig. 3E). Functional annotation analysis of the fastest-changing genes revealed significant enrichment for categories related to transcription factor activity, tissue morphogenesis, and cell–cell contacts (Supplemental Table S4).
Role of transposons in developmental gene regulation
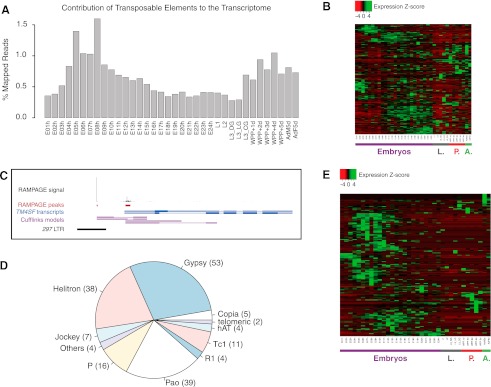
We set out to investigate the role of transposons in the developmental regulation of transcription. For certain timepoints, up to 1.6% of the transcriptome was the product of transcription initiating in TEs (Fig. 4A). Prompted by previous reports of developmental expression of transposons (Parkhurst and Corces 1987; Ding and Lipshitz 1994; Mozer and Benzer 1994), we established expression profiles for individual subfamilies (Methods). Virtually all transposon subfamilies display clear developmental regulation (Fig. 4B), in diverse patterns. This is consistent with the view that transposons have intrinsic properties governing their own expression, as shown previously for individual cases (Bronner et al. 1995; Udomkit et al. 1996; Naito et al. 2009). With regards to regulatory innovation, this makes transposons particularly interesting as a versatile toolkit of mobile regulatory modules with diverse properties.
Figure 4.
Transposable elements display developmental regulation and provide TSSs for many host genes. (A) Contribution of transcription initiating within transposable elements to the developmental transcriptome. For each time point, we report the proportion of all mapped reads (aligned uniquely or to multiple locations) for which the 5′ end lies in an annotated transposon. (B) Z-score–normalized expression profiles for all annotated classes of transposable elements. Note the developmental regulation of virtually all classes, as well as the disparity of patterns across classes. (C) A 297 LTR provides a strong alternative promoter for the TM4SF gene. (D) Subfamilies of transposable elements providing TSSs for annotated genes. The number of TSCs for each subfamily is reported in brackets (total 182). (E) Z-score–normalized expression profiles for all transposon-derived genic TSCs. The diversity of expression profiles underscores the versatility of transposons as regulatory modules.
To search for instances of transposons providing promoters for host genes, we mined our data for transposon-contained TSCs that drive the expression of annotated exons (Fig. 4C). We thus found 182 high-confidence TSCs derived from multiple classes of TEs (Fig. 4D) that drive the expression of 152 annotated genes. RNA ligase-mediated 5′-RACE on selected candidates validated our findings (Supplemental Methods; Supplemental Fig. S8). Figure 4C illustrates one such case, where a solo LTR from a 297 element provides an unannotated alternative promoter for the TM4SF gene. Their temporal patterns of expression are diverse, with subpopulations being active at any developmental stage sampled (Fig. 4E). Importantly, the expression profiles of these transposon-derived TSCs are generally uncorrelated with the profiles of alternative promoters of the same gene (Supplemental Fig. S9), which suggests that the emergence of the transposon TSCs did constitute genuine regulatory innovation. All major classes of D. melanogaster transposons are represented (LTR, LINE, DNA, Helitrons) (see Fig. 3B), although LTR retrotransposons alone—predominantly those of the gypsy and pao families—account for a little over half of all instances. Not only full-length LTR retrotransposon insertions, but also solo LTRs and other fragments, are found to provide genic TSCs.
Importantly, these 182 TSCs represent a very stringently selected set, which may lead us to underestimate the pervasiveness of the phenomenon. To obtain a more accurate estimate, we optimized our peak-calling strategy to increase sensitivity for weaker TSCs, such as those active only in rare cell types. Retaining a still stringent threshold of three or more tags in a single timepoint (see Methods), we thus discovered an additional 333 transposon-borne TSCs driving the expression of annotated genes, bringing the total number to 515. We expect that deeper sequencing and targeted examination of rare cell types will lead to dramatic revisions of this initial estimate.
Furthermore, our initial high-confidence set includes 779 transposon-borne TSCs driving the expression of novel transcripts. To provide further evidence of the biological relevance of transposon-driven developmental transcription, we sought to better characterize these nongenic transcripts. From our data, we could reconstruct Cufflinks partial transcript models for 509 of the aforementioned 779 nongenic transposon-derived promoters (total, 598 transcripts). Out of 598 transcript models, 209 are clearly spliced, showing that these transcripts often undergo post-transcriptional processing. Out of 598, at least 198 transposon-driven transcript models (from 161 promoters) contain ≥30% nontransposon sequences, which demonstrates that TE-derived promoters often drive the expression of neighboring nonrepeat regions. This is bound to be an underestimate, since our transcript models are usually partial. We hypothesize that the creation of promoters by transposons could be a very powerful evolutionary mechanism for the creation of novel noncoding RNA genes. Strikingly, 112 of the 598 nongenic transcripts are antisense to FlyBase-annotated mRNA transcripts. Another 61 overlap annotated transcripts on the same genomic strand. The abundance of such gene-overlapping transcripts points to a potentially important role of transposon-driven noncoding transcription in the regulation of gene expression.
Transposons distribute promoters with preprogrammed regulatory logics
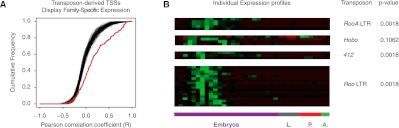
We next investigated whether the transposons that contribute TSCs to host genes have similar expression profiles to the transposon class they belong to. This would imply that transposons contribute functional modules with predetermined and stereotyped regulatory logics to host genes. We show that the 182 high-confidence transposon-derived genic TSCs overall have a clear tendency to share the expression profiles of their class of origin (Fig. 5A). This trend becomes even clearer when focusing on TSCs derived from specific classes of elements. In particular, the 18 TSCs derived from the LTRs of roo elements are expressed in temporal patterns that display compelling similarity to each other and to the overall class pattern (Fig. 5B). This observation also holds true for other classes of elements (Fig. 5B). Roo-driven expression was clearly detectable in profiles established by standard RNA-seq, indicating that these elements drive the expression of full-length genic transcripts (Supplemental Fig. S10).
Figure 5.
Transposons impart their own expression specificity upon the genes they regulate. (A) Cumulative distribution of pairwise Pearson correlation coefficients (R) between individual transposon-derived TSCs and the class of TEs they are derived from (red curve). This measures the similarity between the expression profile of a given gene-driving insertion and the overall profile of the class it belongs to. The black curves show 100 simulations in which the TSS-transposon class pairs were randomized. Permutation test (10,000 randomizations) P = 0.0001. (B) Z-score-normalized expression profiles for individual subfamilies of transposons. Bonferroni-corrected P-values from permutation tests quantify the significance of the similarity between each group of TSCs and its cognate class profile. Note that 0.0018 is the limit of the power of the statistical tests.
This is quite a striking result, since the detection of such broadly correlated patterns is only possible if a large fraction of gene-driving insertions possess the same specificity. As the analysis of certain transposon sequences has shown, however, a large number of diverse TFBS motifs can often be found throughout the length of the sequence (see, e.g., Lynch et al. 2011). Thus, different fragments derived from the same original element may confer vastly different expression specificities, or even carry out other molecular functions. For instance, different human MER20 insertions can bear, in the same cell type, chromatin profiles that are characteristic of either transcriptional enhancers, repressors, or insulators (Lynch et al. 2011). Therefore, even a strict interpretation of the copy-and-paste model does not necessarily imply simple and systematic expression profile correlations between fragments belonging to the same TE family. We conclude that our observations argue very strongly that transposons often impart their own regulatory properties upon the genes they drive the expression of.
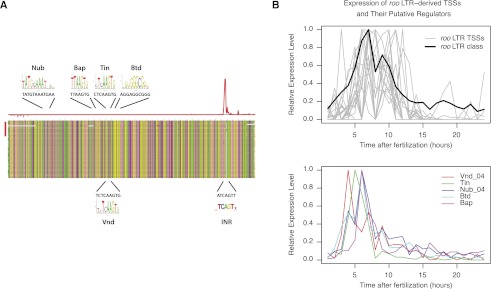
In order to identify cis-elements in the transposons that could explain these regulatory properties, we focused on roo LTR TSCs, the largest group with clear class-specific expression patterns. The analysis of multiple sequence alignments revealed little divergence among all these insertions and relative to the class consensus (Fig. 6A). The consensus LTR sequence was found to have matches to six TFBS motifs (Nub, Tin, Vnd, Btd, and Br_z4: Q-value < 0.05 for each instance; Bap: Q-value = 0.075) (Fig. 6A; Supplemental Table S5). With the exception of br, all the genes encoding transcription factors predicted to bind these motifs have expression profiles consistent with that of the roo LTRs (Fig. 6B). The analysis of endogenous truncated LTR copies is consistent with a role for these sequences in transcriptional regulation (Supplemental Fig. S11). Embryonic expression of roo transposons has previously been shown to require the mesoderm-determining genes twist and snail (Bronner et al. 1995). It is also known that the tin and vnd genes are direct targets of the TWI transcription factor (Mellerick and Nirenberg 1995; Lee et al. 1997; Yin et al. 1997) and that bap is a direct target of TIN (Zaffran et al. 2001). Additionally, we show that the TSS is at the same position in all of the LTRs of interest (Supplemental Fig. S12) and that it overlaps a canonical core promoter Initiator (INR) sequence (Fig. 6A). Overall, this analysis shows that roo LTRs possess a proper Pol II core promoter and cis-regulatory elements that can explain their expression specificity.
Figure 6.
Core promoters and cis-regulatory elements in transposable elements: roo LTRs. (A) Multiple alignment of the sequences of the 18 LTRs providing TSCs for host genes (red bar on the left) to the roo consensus (upper sequence) and to a set of full-length LTRs with high similarity to the class consensus. The histogram above shows the density of tags on the upper (red) and lower (gray) strands. The positions of various sequence motifs are depicted, along with the logo of the known motif and the actual consensus sequence of the LTR. The TFBSs for NUB and BAP and the Initiator sequence (INR) are on the upper strand; the TFBSs for TIN, VND, and BTD are on the lower strand. (B) Expression profiles of the genes encoding putative regulators of roo LTRs. nub and vnd have more than one TSS, and only the one with the expression profile most consistent with roo LTRs is shown.
Population genetics of transposon-derived genic TSCs
To explore the evolutionary implications of our observations, we used existing data (Petrov et al. 2011) on the population frequencies of many transposons, including 56 of the TSC-bearing insertions we identified (Methods; Supplemental Table S6). Of those insertions, 45 are estimated to be rare or very rare variants in the wild North-American (NA) populations studied. Notably, 42 of these rare variants were absent from the ancestral African (AF) populations the NA ones split from 10,000–16,000 yr ago—a number that again underscores the power of this mutational mechanism to continuously create standing variation for regulatory networks. Additionally, we found that 11 variants (20% of total) are either common (four) or fixed (seven) in NA populations, showing clearly that transposon-derived variants can make significant contributions to population gene pools.
Discussion
We have developed and validated a method for high-throughput, high-quality discovery of TSSs; the characterization of the transcripts that emanate from them; and the quantification of their expression. We propose this approach, which directly delineates promoter-specific expression and offers a simple workflow and optimized sample multiplexing, as an advantageous alternative to standard RNA-seq for many gene expression profiling applications. Importantly, this library preparation method will also be easily portable to other sequencing platforms with minimal alterations. This is particularly attractive as new technologies yielding greater read lengths will allow us to move toward large-scale, full-length cDNA sequencing.
We measured promoter activity throughout the life cycle of D. melanogaster, thus providing a high-quality reference data set for the community. Importantly, this data set offers particularly high temporal resolution (1 h) for the period of embryonic development. We observed a very widespread use of alternative promoters as a means to implement differential regulation in a developmental context. Our results also show that transposons contribute large numbers of developmentally expressed TSSs, and strongly support a long-hypothesized mechanism through which transposons distribute preassembled cis-regulatory modules throughout the genome. These modules appear to affect the developmental regulation of hundreds of genes and noncoding transcripts. We expect that further study of more complex genomes with higher transposon contents, such as mammalian or plant genomes, will uncover even greater numbers of such instances. Additionally, our study focused very specifically on transposons providing promoters, but these elements have been shown to have the potential to also contribute TFBSs, enhancers, silencers, insulators, or microRNA target sites (Bourque et al. 2008; Bourque 2009; Lindblad-Toh et al. 2011; Lynch et al. 2011). Overall, our observations underscore the potential of transposons as a powerful and versatile creative force in regulatory innovation.
Methods
Fly stocks and sample collections
Stocks of the y; cn bw sp strain were maintained in standard cornmeal medium bottles in a 24°C incubator. Embryo collections were performed in population cages (Flystuff, no. 59-116). Two-day-old to 7-d-old flies were left to acclimatize to the cage for at least 48 h and were regularly fed with grape juice–agar plates (Flystuff, no. 47-102) generously loaded with yeast paste. After two 2-h prelays, embryos were collected in 1-h windows and aged appropriately (24 timepoints, 0–24 h). Embryos were washed with deionized water, dechorionated for 90 sec with 50% bleach, rinsed abundantly with water, and snap-frozen in liquid nitrogen. Larvae and pupae were collected according to the method described previously (Graveley et al. 2010). For L1 and L2 stages, 2-h embryo collections were aged for 42 or 66 h; larvae were briefly rinsed with deionized water and snap-frozen. For L3 stages, embryos were transferred to bottles containing cornmeal medium supplemented with 0.05% bromophenol blue, and wandering L3 larvae were staged based on gut staining (dark, light, or clear gut) and snap-frozen. For pupae, 2-h embryo collections were transferred to standard cornmeal medium bottles; the positions of new white prepupae on the walls of the bottle were marked; and pupae were collected and snap-frozen at the desired age. For adults, 0- to 12-h-old flies were sexed and kept in vials with cornmeal medium for 5 d and then snap-frozen.
RNA extraction
Total RNA was extracted from adult flies using TRIzol (Invitrogen) according to the manufacturer's instructions and treated with DNase I (Roche). Extraction from embryos, larvae, and pupae was performed using the RNAdvance Tissue kit (Agencourt A32649) according to the manufacturer's instructions, including DNase I treatment. For the human K562 cell line, RNA was extracted using TRIzol according to the manufacturer's instructions and treated with DNase I (Roche). We systematically checked on a Bioanalyzer (Agilent) that the RNA was of very high quality. 5′Monophosphate species—mainly ribosomes—were depleted by TEX digest (Supplemental Methods).
Library preparation and sequencing
Three multiplexed libraries were prepared: one for embryos (24 barcoded samples), one for larvae and pupae (10 samples), and one for adults (two samples). The reverse-transcription was run in parallel for all samples destined to the same library, and the samples were pooled right after reverse-transcription. Our 5′-complete cDNA selection strategy relies on the combination of two orthogonal enrichment methods: reverse-transcriptase template-switching and cap-trapping. The template-switching approach is based on the ability of reverse-transcriptase to add linker sequences to the ends of 5′-complete cDNAs—preferentially if they are made from capped transcripts (Supplemental Fig. S1). Cap-trapping relies on the biotinylation of capped RNA molecules and specific pulldown of their associated 5′-complete cDNAs. The libraries were run on a DNA HS Bioanalyzer chip for quality control, quantified by quantitative PCR, and sequenced on one lane each on an Illumina GAIIx (adults, 2 × 76 bp) or HiSeq (embryos, larvae, and pupae, 2 × 101 bp). For detailed protocol and sequencing data summary, see Supplemental Methods (Supplemental Table S1).
Sequencing reads alignment
The sequences corresponding to the library identification barcode and the reverse-transcription primer were trimmed prior to mapping. Trimmed reads were mapped with STAR, with parameters described in Supplemental Tables S2 and S3. All uniquely mapping reads were kept. As a rescue strategy for multiply mapping reads, if all alignments for those reads started within an annotated transposon and overlapped the same gene annotation, the alignment starting in the closest transposon insertion was selected. All nonrescued multimappers were discarded.
Data analysis pipeline (Fig. S3A)
For details about the peak-calling algorithm, see Supplemental Methods.
PCR duplicates, defined as reads sharing the same alignment coordinates (start, end, and splice sites), were removed from the individual data sets. To avoid overcollapsing, we took advantage of the fact that the long random sequence (15-mer) of our reverse-transcription primer often primes with mismatches. We used this sequence as a pseudo-random barcode, allowing us to distinguish between true duplicates (same barcode) and independent identical inserts. All collapsed data sets were then combined prior to peakcalling. The density of cDNA 5′ ends across the genome was determined from this combined data set, as well as the density of coverage by second (i.e., downstream) sequencing reads. Peaks were called by a sliding window algorithm that assesses the significance of local signal enrichment given a null distribution. Downstream read coverage in the same window was used to correct for local transcript abundance, by subtracting from the raw signal a pseudocount proportional to this coverage. After FDR correction, significant windows in close proximity to each other were merged into peaks, and those were trimmed at the edges down to the first base with signal. (Parameters: window width 15 bases, null distribution negative binomial with k = 4, background weight 0.5, FDR 0.01, merging range 150 bases.) For a more detailed description of the algorithm, see Supplemental Methods. These peaks were connected to annotated genes based on cDNA structure information. For each peak, if we could find at least two inserts having their 5′ in the peak and overlapping an annotated exon of a gene, the peak was functionally linked to that gene. If a peak could potentially be linked to several genes, ties were broken by removing all links that were fivefold weaker than the strongest one. For quantification, the signal for each peak and each timepoint was derived from the uncollapsed data sets and normalized to data set size (defined as the total number of reads attributed to any genic TSS). We built partial transcript models by running Cufflinks separately on the set of reads coming from each peak for each given data set and collapsing all transcripts for each peak using Cuffmerge. This pipeline was implemented with scripts written in Python, including Scipy and Numpy. BEDtools (Quinlan and Hall 2010), version 2.11.2, and Cufflinks (Trapnell et al. 2010), version 1.0.3, were used for some analyses, and plotting was done in R. All scripts are available upon request.
Comparison with 5′-RACE
Our adult flies RAMPAGE data (replicate 2, sexes pooled) was compared to the modENCODE adult flies 5′-RACE data set (see Supplemental Information). For each RAMPAGE peak that was ≤500 bases wide and for which there were five or more tags in each data set (exactly in the peak for RAMPAGE, in the peak ± 10 bases for RACE), we determined the positional offset that maximizes the cross-correlation between the two signals.
Comparison with RNA-seq
The modENCODE 5-d-old adult flies RNA-seq data (Graveley et al. 2010) were mapped with STAR, and the expression of annotated genes was quantified using Cufflinks. Sex bias = Male expression (reads per million, RPM)/(Male expression + Female expression). All genes for which the sum of (Male expression + Female expression) in RAMPAGE was ≥20 RPM were considered for this analysis.
Reproducibility between biological replicates
RAMPAGE libraries were generated for two independent batches of adult D. melanogaster females, and sequenced on separate flowcells on Illumina GAIIx sequencers (8.3 M and 16.7 M million reads). The second data set was randomly subsampled to match the size of the first one. Both data sets were mapped in parallel with the same parameters, duplicates were collapsed, and the data sets were pooled prior to peak-calling (window width 15 bases, null distribution negative binomial with k = 1.0, read 2 background weight 0.8, FDR = 0.001%, merging distance 150 bases). Expression values for this common set of intervals were derived from each uncollapsed data set.
Alternative promoters in FlyBase
The number of distinct TSSs was counted for all FlyBase r5.32 mRNA and ncRNA transcript annotations for which we could detect expression in our data set. Since our peak-calling algorithm merges windows closer than 150 bp, we also merged together annotated TSSs within 150 bp of each other, for the fairness of the comparison.
Identification of weaker peaks
Weaker peaks were identified by calling peaks from the individual (noncombined) collapsed data sets, to increase sensitivity for briefly expressed peaks. We also used slightly less stringent parameters (window width 10 bases, null distribution negative binomial with k = 5, no downstream read background correction, FDR 0.05, merging range 150 bases) and retained all peaks supported by at least three independent tags. To filter out contributions from background signal in the body of transcripts, we discarded any peaks that overlapped annotated exons. We then combined the peaks from all data sets and merged any peaks closer than 50 bp using BEDtools (mergeBed). Peaks were attributed to genes based on evidence from at least one cDNA.
Genome annotations
Transcript annotations were obtained from FlyBase (release 5.32). Analyses performed involved all transcripts annotated as “mRNA” or “ncRNA.” Transposable element RepeatMasker annotations were downloaded from the UCSC Genome Browser. We corrected the annotation of the DNAREP1_DM element to “Helitron,” based on analysis by Kapitonov and Jurka (2007).
Correlation of expression profiles between alternative TSSs
All genic TSSs having a maximum expression level of at least 5 RPM were considered. We computed Pearson's coefficient of determination (R2) for all possible pairs of alternative promoters.
Developmental transitions between alternative promoters
For all genes with maximum expression ≥10 RPM for five or more consecutive timepoints that had at least two alternative promoters, we computed the fraction of the total gene expression at each individual timepoint that was contributed by the main TSS (defined as the one that contributes the largest proportion of the total expression over the whole time series). This metric is represented as a heatmap for 1295 genes that underwent clear transitions between alternative promoters (difference ≥0.5 between the maximum and minimum of the main promoter fraction). (Note that a default value of 0.5 (black) was attributed to all timepoints where total gene expression <10 RPM.)
Analysis of fast-regulation genes
All genes with maximum expression levels ≥10 RPM during embryonic development were considered for this analysis (full set). The fastest-changing genes were defined as those that overall undergo ≥10-fold expression level variations and display single-step variations of ≥85% of their full dynamic range. Functional category enrichment in the fast gene set relative to the full set was assessed using the DAVID database tools (Huang et al. 2009).
TSSs in transposons
BEDtools (intersectBed) was used to search for TSSs ovelapping transposons, and we retained all TSSs that overlapped a transposon over at least 50% of their length.
Transposon subfamily profiles
Transposon subfamily profiles were established by considering all alignments (from uniquely or multiply mapping reads) starting within any insertion of the class, weighed by the inverse of the number of alignments for the read. These profiles were normalized to the total number of transposon-derived reads in each data set.
Expression profile comparisons between TE-derived TSCs and transposon classes
The expression profiles of transposon-overlapping TSCs were paired to their cognate transposon class profile, and Pearson's correlation coefficient was computed for every such pair. The statistical significance of the overall similarity between profiles was assessed by a permutation test (following the recommendations of Phipson and Smyth 2010) in which the TSC profiles were paired to random transposon class profiles (or, alternatively, to random genic TSC profiles). The same strategy was applied to transposons coming from individual classes. In that case, we conducted the permutation tests on all classes for which there were at least three TSCs by pairing the individual TSCs to random transposons, and the P-values were adjusted for multiple testing by Bonferroni correction.
Roo LTR sequence analysis
We retrieved the sequences of the 18 roo LTR insertions bearing genic TSCs, of all other annotated insertions with length ≥420 bp and RepeatMasker alignment score ≥4000 (chrUextra excluded, 50 insertions). Multiple sequence alignments were generated using MUSCLE (default parameters) on the EMBL website and visualized using Jalview. Consensus transposon sequences were downloaded from FlyBase. The LTR sequence we used corresponds to the first 429 bp of the roo consensus (see FlyBase). We used FIMO (Grant et al. 2011) to search for matches to TFBS motifs from the Jaspar Core Insecta database (Bryne et al. 2008), using default parameters and a fourth-order Markov background model derived from the whole genome. A custom script was used to search for matches to previously characterized core promoter motifs (TATA, INR, INR1, DPE, DPE1) (FitzGerald et al. 2006).
Population genetics data analysis
We used the genotyping data for FlyBase-annotated transposon insertions from Petrov et al. (2011). Each transposon-contained TSC was attributed to a FlyBase transposon annotation if it fully overlapped one of them (108 insertions). Allele frequency data were available for 56 insertions.
Data access
Data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE36213.
Acknowledgments
We thank Richard McCombie and the CSHL sequencing team for sequencing, Chris Zaleski and Sonali Jha for data handling, Felix Schlesinger and Carrie Davis for discussions, Gregory Hannon and Emily Hodges for discussions and reagents, and Alexander Gann, Joshua Dubnau, Adrian Krainer, and Robert Martienssen for discussions. Financial support was provided by the Watson School of Biological Sciences and the Florence Gould Foundation (P.B.).
Author contributions: P.B. and T.R.G. designed experiments; P.B. performed experiments and analysis; A.D. provided advice for analysis; C.P. and P.C. contributed intellectually to protocol development; and P.B. and T.R.G wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.139618.112.
References
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D 2006. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441: 87–90 [DOI] [PubMed] [Google Scholar]
- Bourque G 2009. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev 19: 607–612 [DOI] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH 1969. Gene regulation for higher cells: A theory. Science 165: 349–357 [DOI] [PubMed] [Google Scholar]
- Bronner G, Taubert H, Jackle H 1995. Mesoderm-specific B104 expression in the Drosophila embryo is mediated by internal cis-acting elements of the transposon. Chromosoma 103: 669–675 [DOI] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MHE, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A 2008. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res 36: D102–D106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, et al. 1996. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics 37: 327–336 [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engstrom PG, Frith MC, et al. 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635 [DOI] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL 2009. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. Gene 448: 105–114 [DOI] [PubMed] [Google Scholar]
- Ding D, Lipshitz HD 1994. Spatially regulated expression of retrovirus-like transposons during Drosophila melanogaster embryogenesis. Genet Res 64: 167–181 [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi AM, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 41: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR 2012. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 489: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571 [DOI] [PubMed] [Google Scholar]
- Feschotte C 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C 2006. Comparative genomics of Drosophila and human core promoters. Genome Biol 7: R53 doi: 10.1186/gb-2006-7-7-r53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS 2011. FIMO: Scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2010. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirzmann J, Luo D, Hahnen J, Hobom G 1993. Determination of messenger-Rna 5′-ends by reverse transcription of the cap structure. Nucleic Acids Res 21: 3597–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Landolin JM, Brown JB, Sandler JE, Takahashi H, Lassmann T, Yu C, Booth BW, Zhang D, Wan KH, et al. 2011. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res 21: 182–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S 2011. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 21: 1160–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V, Jurka J 2007. Non-autonomous family of Helitrons: A consensus sequence. Repbase 7: 313 [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007a. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR 2007b. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 8: 413–423 [DOI] [PubMed] [Google Scholar]
- Kodzius R, Kojima M, Nishiyori H, Nakamura M, Fukuda S, Tagami M, Sasaki D, Imamura K, Kai C, Harbers M, et al. 2006. CAGE: Cap analysis of gene expression. Nat Methods 3: 211–222 [DOI] [PubMed] [Google Scholar]
- Lee YM, Park T, Schulz RA, Kim Y 1997. Twist-mediated activation of the NK-4 homeobox gene in the visceral mesoderm of Drosophila requires two distinct clusters of E-box regulatory elements. J Biol Chem 272: 17531–17541 [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatov M, Lenkov K, Petrov DA, Bergman CM 2005. Paucity of chimeric gene-transposable element transcripts in the Drosophila melanogaster genome. BMC Biol 3: 24 doi: 10.1186/1741-7007-3-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, et al. 2004. Role of transposable elements in heterochromatin and epigenetic control. Nature 430: 471–476 [DOI] [PubMed] [Google Scholar]
- Lynch VJ, Leclerc RD, May G, Wagner GP 2011. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet 43: 1154–1158 [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Roy S, Ay F, Meyer PE, Candeias R, Kahveci T, Bristow CA, Kellis M 2012. Predictive regulatory models in Drosophila melanogaster by integrative inference of transcriptional networks. Genome Res 22: 1334–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B 1956. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol 21: 197–216 [DOI] [PubMed] [Google Scholar]
- Mellerick DM, Nirenberg M 1995. Dorsal-ventral patterning genes restrict NK-2 homeobox gene expression to the ventral half of the central nervous system of Drosophila embryos. Dev Biol 171: 306–316 [DOI] [PubMed] [Google Scholar]
- Mozer BA, Benzer S 1994. Ingrowth by photoreceptor axons induces transcription of a retrotransposon in the developing Drosophila brain. Development 120: 1049–1058 [DOI] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR 2009. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1232 [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma LJ, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T, Corcoran DL, Rach EA, Song S, Spana EP, Gao Y, Ohler U, Zhu J 2010. A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat Methods 7: 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Matlik K, Speek M 2002. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79: 628–634 [DOI] [PubMed] [Google Scholar]
- Parkhurst SM, Corces VG 1987. Developmental expression of Drosophila melanogaster retrovirus-like transposable elements. EMBO J 6: 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606 [DOI] [PubMed] [Google Scholar]
- Petrov DA, Fiston-Lavier AS, Lipatov M, Lenkov K, Gonzalez J 2011. Population genomics of transposable elements in Drosophila melanogaster. Mol Biol Evol 28: 1633–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Smyth GK 2010. Permutation P-values should never be zero: Calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol 9: Article39 doi: 10.2202/1544-6115.1585 [DOI] [PubMed] [Google Scholar]
- Plessy C, Bertin N, Takahashi H, Simone R, Salimullah M, Lassmann T, Vitezic M, Severin J, Olivarius S, Lazarevic D, et al. 2010. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat Methods 7: 528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rach EA, Yuan HY, Majoros WH, Tomancak P, Ohler U 2009. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol 10: R73 doi: 10.1186/gb-2009-10-7-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M 2010. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 467: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder C, Tautz D, Seifert E, Jackle H 1988. Differential regulation of the two transcripts from the Drosophila gap segmentation gene hunchback. EMBO J 7: 2881–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Forrest ARR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJL, et al. 2009. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet 41: 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udomkit A, Forbes S, McLean C, Arkhipova I, Finnegan DJ 1996. Control of expression of the I factor, a LINE-like transposable element in Drosophila melanogaster. EMBO J 15: 3174–3181 [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat LN, Landry JR, Mager DL, Medstrand P 2003. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19: 530–536 [DOI] [PubMed] [Google Scholar]
- Yin Z, Xu XL, Frasch M 1997. Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development 124: 4971–4982 [DOI] [PubMed] [Google Scholar]
- Zaffran S, Kuchler A, Lee HH, Frasch M 2001. biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev 15: 2900–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhenodarova SM, Kliagina VP, Maistrenko FG, Pustoshilova NM, Smolianinova OA 1989. Substrate specificity of T4 RNA-ligase. The effect of the nucleotide composition of substrates and the size of phosphate donor on the effectiveness of intermolecular ligation. Bioorg Khim 15: 478–483 [PubMed] [Google Scholar]