Abstract
The Xenopus embryo has provided key insights into fate specification, the cell cycle, and other fundamental developmental and cellular processes, yet a comprehensive understanding of its transcriptome is lacking. Here, we used paired end RNA sequencing (RNA-seq) to explore the transcriptome of Xenopus tropicalis in 23 distinct developmental stages. We determined expression levels of all genes annotated in RefSeq and Ensembl and showed for the first time on a genome-wide scale that, despite a general state of transcriptional silence in the earliest stages of development, approximately 150 genes are transcribed prior to the midblastula transition. In addition, our splicing analysis uncovered more than 10,000 novel splice junctions at each stage and revealed that many known genes have additional unannotated isoforms. Furthermore, we used Cufflinks to reconstruct transcripts from our RNA-seq data and found that ∼13.5% of the final contigs are derived from novel transcribed regions, both within introns and in intergenic regions. We then developed a filtering pipeline to separate protein-coding transcripts from noncoding RNAs and identified a confident set of 6686 noncoding transcripts in 3859 genomic loci. Since the current reference genome, XenTro3, consists of hundreds of scaffolds instead of full chromosomes, we also performed de novo reconstruction of the transcriptome using Trinity and uncovered hundreds of transcripts that are missing from the genome. Collectively, our data will not only aid in completing the assembly of the Xenopus tropicalis genome but will also serve as a valuable resource for gene discovery and for unraveling the fundamental mechanisms of vertebrate embryogenesis.
Xenopus is one of the major model systems for the study of vertebrate embryogenesis and basic cell biological processes. There are multiple advantages to the use of Xenopus as an experimental system, such as the availability of large abundant eggs that are easily manipulated, ready accessibility to any developmental stage, and conservation of cellular pathways between Xenopus and mammals. In the past 50 years, landmark studies on Xenopus have been critical toward our understanding of nuclear reprogramming (Gurdon et al. 1958), embryonic patterning (Harland and Gerhart 1997; De Robertis 2006), membrane channels and receptors (Kusano et al. 1977), and cell cycle control (Murray and Kirschner 1989; Murray et al. 1989; Glotzer et al. 1991).
Genomics resources for Xenopus research have emerged in the past 10–15 years. During the early days of the genomics era, several cDNA sequencing efforts, such as EST (expressed sequence tag) projects, have allowed the construction of full length cDNA clones and identification of Xenopus open reading frames (ORFs) (Gilchrist et al. 2004; Morin et al. 2006; Fierro et al. 2007). Microarrays have also been used to investigate the expression levels of annotated genes and gave some insights into transcriptome changes over development as well as expression differences between two closely related frog species, Xenopus laevis and Xenopus tropicalis (Yanai et al. 2011). In addition, forward and reverse genetic screens have uncovered mutations that affect a myriad of organogenesis and differentiation processes in Xenopus (Goda et al. 2006), while a genetic map based on simple sequence length polymorphism (SSLP) markers, which can be used to clone genes identified by mutation, has recently been generated (Wells et al. 2011).
Notably, while early developmental and molecular studies have been performed on Xenopus laevis, its closely related cousin Xenopus tropicalis has proven to be more widely used for genetic and genomic research. This is mainly because Xenopus laevis has a more complex pseudotetraploid genome, while Xenopus tropicalis has a smaller and more amenable diploid genome. Hence, the initial genome sequencing effort has been directed mostly at Xenopus tropicalis, whose genome has recently been published (Hellsten et al. 2010). Strikingly, the frog genome is highly syntenic with the human genome, with regions of synteny frequently spanning more than a hundred genes. Nevertheless, although it is largely assembled into multiple scaffolds, the Xenopus tropicalis genome is yet to be sequenced at the same depth and annotated at the same level of details and accuracy as the genomes of human and mouse. Importantly, annotations of protein-coding and noncoding genes are strikingly incomplete, including the widely used RefSeq and Ensembl annotations.
The advent of high-throughput sequencing technologies has had an enormous impact on genomics. In particular, such technologies have revolutionized studies of the transcriptome in many species from yeast to humans and have revealed tremendous amounts of complexities and gaps in our understanding of any transcriptome (Wang et al. 2009). Not only does RNA sequencing (RNA-seq) provide a more accurate measurement of expression levels, it provides single nucleotide resolution and has the ability to reveal novel splice junctions, unannotated transcripts, and allele-specific expression. Here, we present the first comprehensive study of the transcriptome of Xenopus tropicalis using RNA-seq over development from a two-cell fertilized embryo to a feeding tadpole. We report evidence for transcription of more than a hundred genes prior to the midblastula transition, when the embryonic genome is generally believed to be transcriptionally silent. We also discovered thousands of novel splicing events, including exon skipping in annotated genes, as well as thousands of unannotated, potentially noncoding transcripts. Hence, our data serve as a valuable resource for developmental biologists and the general genomics community. Furthermore, to extend the reach of our work, we have created an interactive website (http://hci.stanford.edu/∼jcchuang/frog-genes/latest/) that allows users to not only browse the heatmaps in this manuscript but to also query the expression profile of any RefSeq or Ensembl annotated gene with ease.
Results
Mapping and analysis of RNA-seq reads
To systematically examine the dynamics of the Xenopus transcriptome over development, we generated RNA-seq libraries for 23 distinct groups of stages (Fig. 1A). As biological replicates, we collected embryos from two different clutches (see Methods for detailed descriptions of the stages) and made libraries for each replicate independently. We first mapped the reads to the XenTro3 genome (JGI assembly v4.2) using SeqMap (Jiang and Wong 2008) and calculated the RPKMs (reads per kilobase per million mapped reads) for each replicate separately using RSeq (Jiang and Wong 2009). A comparison of the two replicates showed that the expression values between identical stages were highly correlated (average R2 = 0.916) (Supplemental Fig. S1). Hence, for the same group of stages, we pooled all the reads together to improve statistical power, resulting in 20–70 million reads for each group, and recalculated all the RPKMs. Of all the reads, ∼46.9% were mappable to RefSeq annotations, while ∼60.5% were mappable to Ensembl annotations (Fig. 1B). Overall, we detected 97.3% of the RefSeq genes that are annotated in the genome (or 8211 out of 8437 genes) at any time of development and 80.5% of the annotated Ensembl genes (or 16,012 out of 19,884 genes) (Fig. 1C). Furthermore, our RNA-seq results are in reasonable agreement with previously published microarray data (Yanai et al. 2011; Supplemental Figs. S2, S3; Supplemental Information). The expression levels based on the RefSeq annotation and the Ensembl annotation are given in Supplemental Files S1 and S2, respectively, while the distribution of the RPKM values over all developmental stages is displayed as a box-and-whisker plot in Supplemental Figure S4.
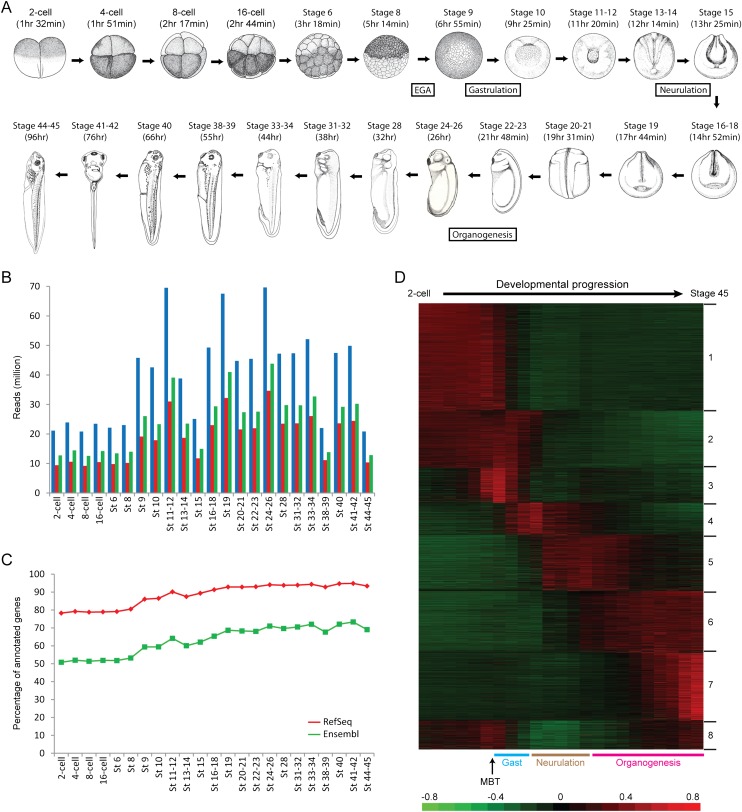
Figure 1.
Deep RNA-seq covers the majority of annotated genes and reveals dynamic temporal regulation over development. (A) The developmental stages (adapted from Nieuwkoop and Faber 1994 and reprinted with permission from Garland Science/Taylor & Francis LLC © 1994) investigated in this study. For each stage, the approximate time of occurrence after fertilization is given in brackets and is estimated from Khokha et al. (2002) or Xenbase. The beginnings of four major developmental events—embryonic genome activation (EGA), gastrulation, neurulation, and organogenesis—are also indicated in the schematic. (B) Number of sequencing reads for each of the stages. (Blue) Total number of reads; (red) number of reads that mapped to RefSeq genes; (green) number of reads that mapped to Ensembl genes. (C) Number of annotated genes that were detected at each of the stages. (Red) RefSeq genes; (green) Ensembl genes. (D) Clustered expression profiles for all detected RefSeq genes are organized by time of expression. The RPKM values were mean-centered and normalized, with each row representing a different gene. The key developmental events, namely the midblastula transition (MBT), gastrulation (Gast), neurulation, and organogenesis, are also labeled below the heatmap. The first cluster contains the set of genes whose RNAs are present at high levels in the earliest stages of development until after the embryonic genome is activated. The second cluster contains the set of transcripts that are present not only before the midblastula transition but remain expressed until the end of gastrulation. The third cluster corresponds to the early response genes that are first transcribed around the midblastula transition. The fourth cluster represents the cohort of genes that are expressed during gastrulation and neurulation, while the fifth cluster represents genes that are expressed not only during neurulation but also during early organogenesis. The sixth cluster contains the set of genes that are first transcribed at the onset of organogenesis and whose expression remains on throughout all the tadpole stages. The seventh cluster represents genes that are transcribed only at later tadpole stages. Finally, the eighth cluster contains genes that are expressed in the earliest developmental stages, repressed after the midblastula transition, and transcribed again in the late tadpole stages. Taken together, clustering analysis reveals interesting dynamics of transcript levels during Xenopus development.
We asked if the Xenopus transcriptome can be categorized into group-defining patterns. K-means clustering across all stages revealed that the majority of the annotated genes are temporally regulated over development and may be broadly classified into eight distinct clusters (Fig. 1D; Supplemental Fig. S5). To gain insights into the functional significance of the eight clusters, we performed Gene Ontology (GO) analysis using the Panther classification system (Thomas et al. 2006). The first cluster is enriched for genes associated with protein phosphorylation, meiosis, and DNA repair, including genes encoding mitogen-activated protein kinase (MAPK) signaling proteins and serine/threonine kinases. Notably, the second cluster is enriched for genes associated with RNA processing, in particular splicing, which indicates an important role for post-transcriptional regulation during the early developmental stages. The third cluster is associated with protein localization, signal transduction, and transcription from the RNA polymerase II promoter, which correlates well with activation of the embryonic genome. Strikingly, the fourth and fifth clusters are enriched for genes associated with multiple developmental processes, including anterior-posterior axis specification, segment specification, ectoderm development, and mesoderm development. In addition, the sixth cluster is strongly enriched for genes associated with translation and contains dozens of genes encoding ribosomal proteins. As evidence has recently emerged to show that ribosomes can function as regulators of key tissue patterning events during vertebrate embryogenesis (Kondrashov et al. 2011), our analysis hints at potential tissue-specific roles for the ribosome during Xenopus organogenesis. Finally, the seventh cluster is associated with steroid or lipid metabolism, response to external stimulus, and respiration, while the eighth cluster is associated with coenzyme or fatty acid metabolism and mitochondrion-related processes. The full list of GO terms for each cluster (P < 0.1) is provided in Supplemental File S3.
Identification of genes transcribed prior to the midblastula transition
Following fertilization, the beginning stages of development in many organisms are thought to be transcriptionally silent until the midblastula transition when large scale activation of zygotic genes occurs. This period of transcriptional quiescence lasts until the twelfth cell division in Xenopus embryos (approximately Stage 8.5). However, despite the general state of transcriptional repression in the earliest stages, a few genes, including the nodal-related genes xnr5 and xnr6, have been found to be activated prior to the midblastula transition (Yang et al. 2002). Here, we sought to identify genome-wide all the genes that are transcribed before the transition.
From our data sets corresponding to the pre-midblastula transition stages (two-cell to Stage 8), we detected a total of 7187 RefSeq genes. Clustering analysis reveals that these genes can be broadly divided into four clusters (Fig. 2A). The first cluster contains the set of genes whose RNAs are rapidly degraded after fertilization, and the genes may be expressed again at the approach of the transition. The second cluster corresponds to the transcripts that appear to be degraded more slowly and hence, persist for a longer period of time but are nevertheless absent by Stage 8. The third cluster represents the set of genes whose transcript levels gradually increase over development. Finally, the fourth cluster contains the genes whose expression is more sharply activated closer to the midblastula transition compared with the previous cluster. Importantly, the last two clusters appear to contain numerous genes that are transcribed prior to the transition despite the general state of transcriptional silence.
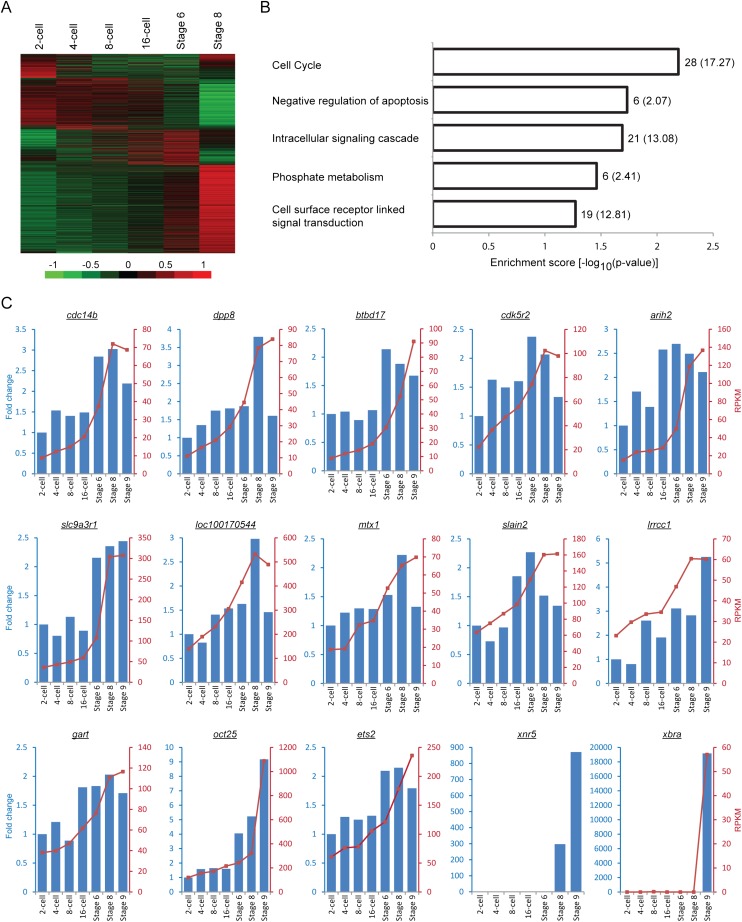
Figure 2.
Many genes are transcribed before the midblastula transition despite a general state of transcriptional repression. (A) Heatmap showing distinct expression profiles of all RefSeq genes that were detected before the midblastula transition in our RNA-seq experiments. To focus on the stages before embryonic genome activation, only the RPKM values from the two-cell stage to Stage 8 were used for mean-centering, normalization, and clustering. Each row in the heatmap represents a different gene. (B) GO analysis of the 150 early transcribed genes that passed the following three criteria: (1) the transcript level at Stage 6 is at least twofold higher than that at the two-cell stage; (2) the average RPKM at each developmental stage is at least 1; and (3) the expression profile is monotonically increasing. (C) Validations of the RNA-seq results. The RT-qPCR expression profiles (blue bars) match the RNA-seq data (red lines) closely for 13 out of the 20 genes we tested. The nodal-related gene, xnr5, serves as a positive control (its expression is activated before EGA), while xbra serves as a negative control (its expression is activated after EGA). There is no RNA-seq data for xnr5 because it is duplicated in the Xenopus genome, and the corresponding reads cannot be uniquely mapped. We also note that for the majority of the validated genes, the fold changes obtained from our RT-qPCR experiments are relatively small (two- to threefold) compared to the fold changes observed for xnr5 and xbra (greater than 100-fold).
To confidently identify a set of genes whose transcription is activated before the midblastula transition, we imposed three criteria. First, we require that the RPKM at Stage 6 is at least twofold higher than the RPKM at the two-cell stage. We chose Stage 6 instead of Stage 8 because even though embryonic genome activation occurs at Stage 8.5, the time of its onset can vary on factors like temperature changes, and we wanted to be certain that we are examining time points significantly prior to the transition. Second, we require an average RPKM of at least 1 for each gene. Third, we require that the expression level is monotonically increasing. A total of 150 genes passed all three filters. GO analysis of these genes reveals that they are associated with the cell cycle, apoptosis, signal transduction, and phosphorylation (Fig. 2B). In addition, we found that 24% of them (or 36 genes) are also activated prior to the midblastula transition in zebrafish (Aanes et al. 2011). The list of 150 genes, together with their expression profiles and associated GO terms, are provided in Supplemental File S4.
We used quantitative real-time PCR (RT-qPCR) to validate the expression patterns observed from RNA-seq (Fig. 2C). In the validation experiments, we focused on the genes with an average RPKM of at least 20 and sorted them by fold change. Of the top 10 genes (fold change ranging from 2.82 to 4.26 according to RNA-seq), we were able to validate eight of them (cdc14b, dpp8, btbd17, cdk5r2, arih2, slc9a3r1, loc100170544, and mtx1). Of the bottom 10 genes (fold change ranging from 2.00 to 2.04), we were able to validate five of them (slain2, lrrcc1, gart, oct25, and ets2). Hence, we estimate the true positive rate to be between 50% and 80%.
We recognize that our three criteria above for identifying early transcribed genes are somewhat arbitrary and may even be too stringent. In particular, our requirement for a monotonic increase in expression level may filter out bona fide genes whose RPKM values contain some measurement error. When we loosened this criterion to allow for a decrease by 1 RPKM or a 20% drop in expression level between two consecutive developmental stages while maintaining the other two filters, we obtained a total of 313 genes (Supplemental File S5). GO analysis showed that this larger gene set is still associated with phosphorylation, the cell cycle, signal transduction, and apoptosis (Supplemental Fig. S6A). We further selected 8 genes for validation by RT-qPCR and were able to confirm 6 of them (Supplemental Fig. S6B). Taken together, our analysis showed that even though the embryonic genome is generally repressed prior to the midblastula transition, the transcripts of dozens, if not hundreds, of Xenopus genes actually increase in abundance even before the onset of widespread zygotic transcription.
Detection and classification of novel splice junctions
Alternative splicing is a prevalent post-transcriptional mechanism in many higher eukaryotes to diversify the proteome. RNA-seq studies have revealed that >90% of multiexon human genes undergo alternative splicing (Wang et al. 2008; Pan et al. 2008), while 60%, 50%, and 25% of genes in Drosophila, zebrafish, and C. elegans, respectively, can be alternatively spliced (Gerstein et al. 2010; Aanes et al. 2011; Graveley et al. 2011; Ramani et al. 2011). Here, we sought to understand splicing regulation in Xenopus tropicalis on a genome-wide scale.
To detect splice junctions, we applied SpliceMap (Au et al. 2010) to our paired end RNA-seq data sets. We identified between 100,000 and 170,000 junctions at each developmental stage (Fig. 3A). The varying number of junctions detected is largely due to different sequencing coverage between the various stages (Supplemental Fig. S7A). Importantly, we were able to detect a significant number of novel splicing events at every stage (red and blue colored portions in Fig. 3A). On average, 27.4% of the total junctions identified at each developmental stage are absent from both the RefSeq and the Ensembl annotations, while 15.2% of the total detected junctions are not only novel but are also well-supported by at least two nonredundant reads (nNR > 1) (Supplemental Fig. S7B). Furthermore, the majority of the novel junctions supported by at least two reads have additional backing from existing ESTs (Supplemental Fig. S8).
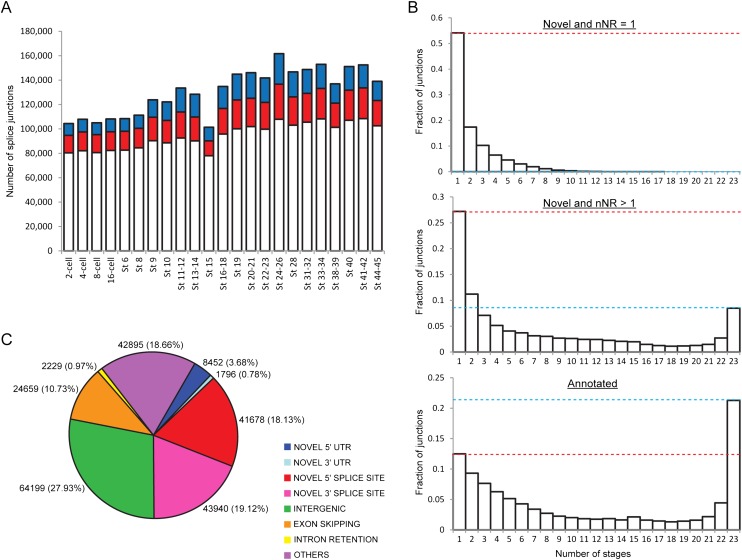
Figure 3.
Novel splice junctions of different types can be detected at all developmental stages. (A) Total number of splicing events identified at every stage. While the majority of the junctions detected at each stage are annotated in either RefSeq or Ensembl (white portions), a sizeable number of junctions are novel (colored portions). (Red) There are two or more nonredundant reads supporting the novel junction (nNR > 1). (Blue) There is only one nonredundant read supporting the novel junction (nNR = 1). (B) Plots showing the number of stages that each splice junction is detected in. Compared to the novel junctions, a greater fraction of annotated junctions is constitutively expressed. On average, each annotated junction was detected in 11.0 stages, while each strongly supported novel junction (nNR > 1) was detected in 7.7 stages and each weakly supported novel junction (nNR = 1) was detected in 2.2 stages. (Red dotted line) Height of the bar for single-occurrence junctions; (blue dotted line) height of the bar for constitutive junctions. (C) Pie chart showing the different types of novel splice junctions identified with the number of junctions (percentage in brackets) given next to each segment. Only 27.93% of the novel junctions occur in intergenic regions (in green), while the majority of them (72.07%) occur within annotated genes.
Next, we asked if annotated junctions are more likely to belong to isoforms that are either constitutively or more widely expressed, while the novel junctions tend to belong to transcripts that are expressed only at specific stages of development and hence, are more difficult to discover. To address this hypothesis, we divided our nonredundant set of splice junctions into three groups, namely, annotated junctions, novel but weakly supported junctions (nNR = 1), and novel but strongly supported junctions (nNR > 1) and counted the number of stages that each junction can be detected in (Fig. 3B). We found that the distribution of the annotated junctions is clearly different from the distribution of the novel junctions. Specifically, 21.1% of the annotated junctions were detected in all 23 stages, while 0% (nNR = 1) or 8.5% (nNR > 1) of the novel junctions were detected in all the stages. In contrast, 12.5% of the annotated junctions were detected in only one developmental stage, while 54.2% (nNR = 1) or 27.2% (nNR > 1) of the novel junctions were detected in only a single stage. Furthermore, we found that despite our deep sequencing efforts, we have not saturated the discovery of novel splicing events that occur in only one developmental stage (Supplemental Fig. S9; Supplemental Information). Hence, our data support the hypothesis that many transcripts or isoforms are currently unannotated because they are hard to detect as a result of their low or temporally restricted expression.
It is possible that some of the novel junctions that we uncovered are a result of inexact splicing. The splice site signals are short and considerably degenerate (Stamm et al. 2006); and the splicing machinery is not error-free (Hsu and Hertel 2009), thereby generating debate over the likelihood that stochastic noise contributes to much of the observed mRNA diversity arising from alternative splicing (Melamud and Moult 2009). To obtain an estimate of the percentage of novel junctions that may be products of inexact splicing, we calculated the number of novel junctions that are within three base pairs of annotated junctions. Of the 229,848 novel splicing events that we detected using SpliceMap, we found that 11,240 of them (4.89%) are shifted by three bases in either direction of annotated junctions. Although we cannot discern true functional transcripts from erroneously spliced ones, the small percentage lends support to the hypothesis that the majority of the novel junctions may have some biological impact during Xenopus development.
To gain additional insights into the novel junctions, we categorized them based on their genomic location and how they modify existing annotations (Fig. 3C; Supplemental Table S1). While about a quarter of the novel junctions lie in intergenic regions, most of them occur within annotated genes. We found that the predominant way in which existing annotations may be modified is in the creation of novel 5′ or 3′ splice sites, which together account for 37.25% of the novel junctions. This may be due to noncanonical splice sites, incorrect demarcations of splice junctions in current databases, or the presence of extra exons. Furthermore, we discovered thousands of new exon skipping events and extensions of the 5′UTR or the 3′UTR. Using reverse transcription PCR, we went on to validate the existence of novel splice junctions in nine out of 10 different transcripts (Fig. 4; Supplemental Figs. S10–S14; Supplemental Information). Taken together, our results indicate the existence of new isoforms for many known genes and also highlight the inadequacies of the present RefSeq and Ensembl annotations.
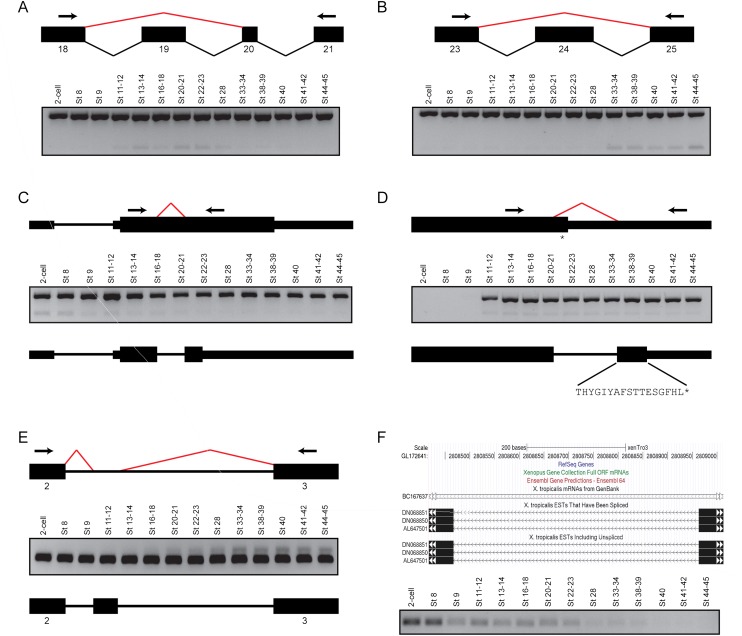
Figure 4.
Most of the novel splice junctions supported by at least two nonredundant reads can be validated by reverse transcription PCR. (A) Skipping of exon 19 in the fibronectin gene, fn1. (Upper panel) A schematic of the splicing events between exons 18 and 21 inclusive. Black boxes indicate the individual exons, while the bent lines indicate the splicing events (black, annotated; red, novel). (Arrows) Primers used for PCR. (Lower panel) Gel image of the PCR, showing two products. The upper stronger band is present in all stages and represents the annotated isoform, while the lower band represents the exon skipping event and shows a peak expression between Stages 20 and 23. (B) Skipping of exon 24 in fn1. (Upper panel) A schematic of the splicing events between exons 23 and 25 inclusive. (Arrows) PCR primers. (Lower panel) Gel image showing the results of the PCR. The upper stronger band is present in all stages and corresponds to the annotated isoform, while the lower band represents the exon skipping event. Interestingly, the product lacking exon 24 is only clearly expressed from Stage 33 onward. (C) Splicing of a facultative intron in the metabolic gene, hpdl. (Upper panel) A schematic of the gene structure of hpdl. The arrows indicate the PCR primers, while the red bent line indicates a novel splicing event detected within the coding exon of the gene. (Middle panel) Gel image showing the PCR results. The upper stronger bands represent the annotated gene product, while the lower weaker bands correspond to the novel transcript, which is present in the beginning stages of development. (Lower panel) A schematic showing that the novel splice junction creates a premature STOP codon. (D) Intron retention in the gata3 transcription factor. (Upper panel) A schematic showing the location of the novel splice junction (red bent line) and the location of the PCR primers (arrows). The newly detected splicing event removes the annotated STOP codon (asterisk). (Middle panel) Gel image of the PCR results. Both the annotated isoform (upper stronger bands) and the novel isoform (lower weaker bands) are not expressed from the two-cell stage to Stage 9 inclusive. (Lower panel) A schematic showing that the novel splice junction extends the C-terminus of the protein by 17 amino acids. (E) Extra exon in the myosin gene, myl6. (Upper panel) A schematic showing novel splicing events (red bent lines) between exons 2 and 3. (Middle panel) Gel image of the PCR results. Although the annotated isoform is strongly expressed in all the stages tested, the novel isoform containing the additional exon is only expressed after neurulation. (Lower panel) A schematic showing the location of the newly discovered exon in myl6. (F) A novel intergenic transcript between tmtc2 and slc6a15. (Upper panel) A snapshot of the UCSC Genome Browser showing the genomic locus of a splicing event detected between GL172641: 2808467 and GL172641: 2808964. Several ESTs support the detected splicing event. (Lower panel) Gel image showing the results of a PCR using primers flanking the novel splice junction. The expression of the corresponding transcript is highest at the two-cell stage and Stage 8 and it then decreases over development.
Novel transcribed regions of the genome
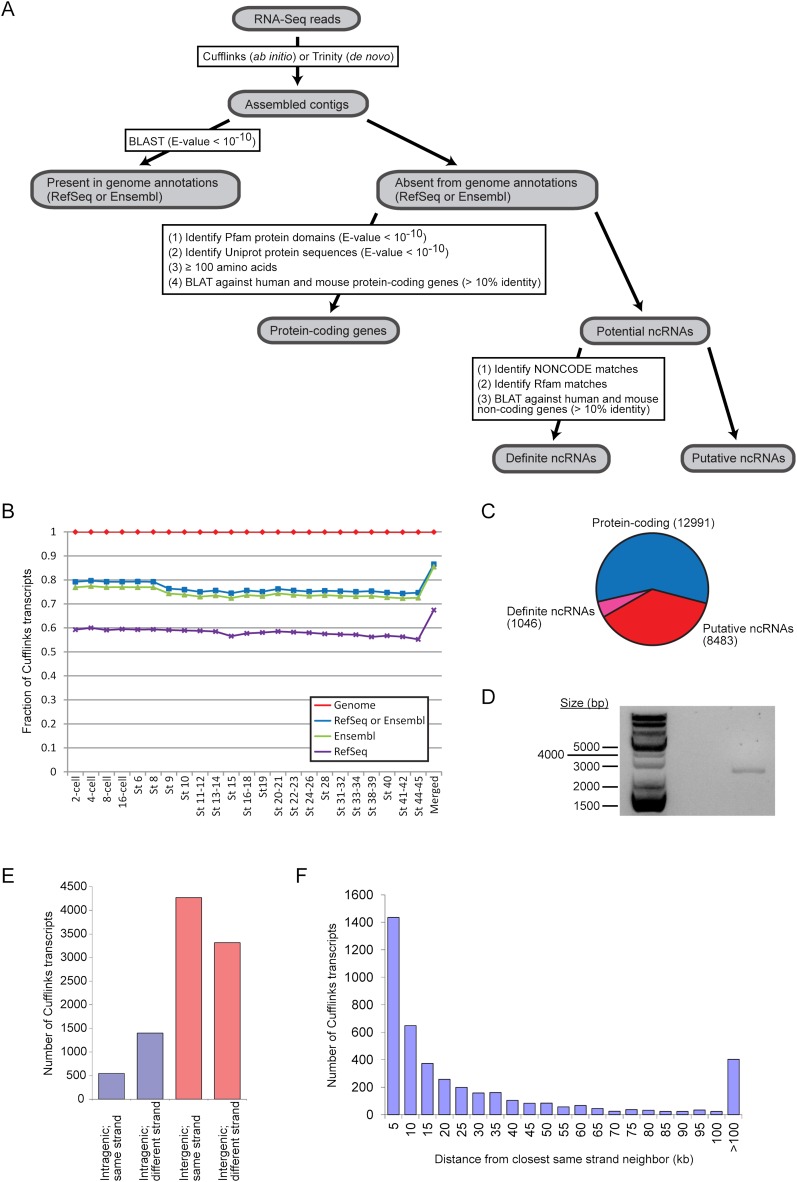
Numerous transcriptome studies have discovered that multicellular eukaryotic genomes are replete with unannotated transcripts that originate from intronic or intergenic regions (Bertone et al. 2004; Manak et al. 2006; Rinn et al. 2007; Guttman et al. 2009). In agreement with these previous studies, our splicing analysis of the frog transcriptome has uncovered thousands of novel splice junctions that do not simply modify existing gene structures but instead appear to belong to unknown gene models (Figs. 3C, 4F; Supplemental Fig. S13). Hence, we sought to assemble transcripts from our RNA-seq data in order to identify novel transcribed regions of the Xenopus genome. We also developed a filtering pipeline aimed at separating the novel transcripts into two groups, namely the protein-coding transcripts and the noncoding RNAs (Fig. 5A; Supplemental Information).
Figure 5.
Reconstruction of transcripts from RNA-seq data reveals novel transcribed regions. (A) Overview of our pipeline that defined a set of unannotated protein-coding genes and a set of novel noncoding RNAs. (B) Comparison of transcripts assembled by Cufflinks with the RefSeq (purple crosses) and Ensembl (green triangles) annotations as well as the reference genome (XenTro3, red diamonds). The blue squares correspond to Cufflinks transcripts that can be found in either the RefSeq or the Ensembl annotation. (C) A breakdown of the unannotated Cufflinks contigs into protein-coding transcripts and noncoding transcripts. (D) By reverse transcription PCR, we detected a gene product in the intergenic region between hoxc11 and hoxc12, where a long noncoding RNA, HOTAIR, is known to exist in mammals. The PCR product was further sequenced to confirm that it matched the correct genomic locus. (E) We classified the putative noncoding RNAs based on their genomic locations relative to annotated RefSeq or Ensembl genes. (F) For intergenic contigs that are on the same strand as one or both of their neighboring genes, we determined the distance of each Cufflinks transcript from its closest same-strand neighbor and binned the transcripts by this distance. In the histogram, “10 kb” means that the transcripts are between 9001 and 10,000 bp (inclusive) away from their same-strand neighbors and so on.
After transcript assembly using Cufflinks (Trapnell et al. 2010), we asked how the reconstructed contigs compare with known genes annotated in the reference genome (XenTro3) (Fig. 5B). As expected, essentially all the Cufflinks transcripts (>99.9%) can be aligned to the genome. In addition, between 74.3% and 79.7% of the transcripts assembled by Cufflinks at each developmental stage matches a gene annotated in either RefSeq or Ensembl. When we applied CuffMerge to all the individual contigs reconstructed at each stage, we obtained an overall set of 167,386 unique transcripts, of which 86.5% match a RefSeq or Ensembl gene. This higher percentage is likely a result of CuffMerge combining some shorter contigs with another transcript that already aligns to a known gene. In subsequent steps, we focused our attention on the set of merged transcripts.
Next, we examined the unannotated Cufflinks contigs (13.5% of the merged transcripts or 22,520 transcripts) for protein-coding potential. We searched existing protein databases and found 8997 matches in the Pfam database and 8252 matches in the Uniprot database (Supplemental Fig. S15A). In addition, we translated the contigs in all three forward frames and found that 8887 of them may encode peptides that are at least 100 amino acids long. We further checked the Xenopus Cufflinks transcripts against annotated human and mouse protein-coding genes and uncovered 182 transcripts that showed at least 10% identity to a human or mouse protein-coding gene. Hence, our filters identified a total of 12,991 unannotated Xenopus Cufflinks transcripts as protein-coding (Fig. 5C; Supplemental File S6), leaving 9529 transcripts as potential noncoding RNAs.
To determine how many of the putative noncoding RNAs may actually correspond to known noncoding genes, we searched the Rfam and NONCODE databases for matches to the 9529 Cufflinks transcripts that passed through the protein-coding filters (Supplemental Fig. S15B). For Rfam, we utilized the Infernal software package (Nawrocki et al. 2009) to search for matches and identified 112 contigs that have hits to one of the database's secondary structure-based covariance models (score ≥ 40). Using BLASTN, we also found 200 contigs that could align to entries in NONCODE by sequence similarity. In addition, since many long intergenic noncoding RNAs (lincRNAs) are poorly conserved by sequence but instead share conserved genomic locations (Ulitsky et al. 2011), we further searched the NONCODE database for matches by synteny and uncovered 812 intergenic contigs that shared the same neighboring genes as known noncoding RNAs. Besides the databases, we separately checked the Xenopus transcripts against annotated human and mouse noncoding genes and discovered 22 transcripts that showed at least 10% sequence identity to a known mammalian noncoding gene (Supplemental Fig. S15C). Hence, we identified a total of 1046 Xenopus Cufflinks transcripts that matched known noncoding genes in other species; and so we consider this set of 1046 transcripts to be definitely noncoding (Fig. 5C). Of these, we noted a contig that originates from the genomic locus between hoxc11 and hoxc12, where a long intergenic noncoding RNA (lincRNA), HOTAIR, is known to be present in mammals (Rinn et al. 2007). We further examined the mappings and observed that there are clusters of reads in the intergenic region between the two hox genes (Supplemental Fig. S16), which suggests that the putative Xenopus HOTAIR may be spliced. To validate the lincRNA, we performed reverse transcription PCR and obtained a product whose size is between 2000 and 3000 bp (Fig. 5D), which is similar to the length of HOTAIR in mammals. These results suggest that a HOTAIR ortholog exists beyond the mammalian lineage and can be found in the Xenopus genome.
To further analyze the noncoding RNAs, we categorized all 9529 potential noncoding Cufflinks contigs based on their genomic locations (Fig. 5E). A total of 79.6% of the contigs (or 7585 transcripts) are intergenic, while the remainder overlap with annotated RefSeq or Ensembl genes. In addition, we note that intergenic contigs that are on the same strand as one of its neighboring genes may simply be an extension of the neighboring gene and therefore are not true independent noncoding RNAs. Similarly, intronic contigs that are on the same strand as the host gene may in fact represent additional exons of that gene. Hence, we also examined the strand of the noncoding Cufflinks contigs relative to their immediate neighboring or overlapping genes. 4271 Cufflinks contigs are intergenic and on the same strand as one or both of their neighboring genes, while 3314 contigs are intergenic and on the opposite strand. Furthermore, of the 1944 contigs that are intragenic, 72.0% of them (or 1400 contigs) are antisense transcripts. In summary, a total of 4714 putative noncoding Cufflinks contigs originate from a different strand, and they certainly represent novel transcripts.
Not all intergenic contigs that derive from the same strand as one of its immediate neighboring genes are extensions of their annotated neighbors; a portion of these intergenic contigs may be genuine independent transcripts. We rationalized that the further the contig is from its nearest same-strand neighbor, the more likely the intergenic contig represents a novel stand-alone gene. Hence, we determined the distance between each intergenic Cufflinks contig and its nearest same-strand neighbor and binned the contigs by their distance from the annotated neighboring gene (Fig. 5F). The mean distance of an intergenic contig from its same-strand neighbor is 37,250 bp and the median distance is 10,605 bp. Since >95% of Xenopus introns annotated in RefSeq (Supplemental Fig. S17A) and ∼95% of introns present in the extensively annotated human genome (hg19) (Supplemental Fig. S17B) are shorter than 25 kb, we reasoned that an intergenic transcript that is >25 kb away from its nearest neighboring gene is unlikely to be part of that gene. Therefore, we estimate, with a false discovery rate (FDR) of 5%, that 1359 intergenic Xenopus contigs that have a same-strand neighbor are likely to originate from separate independent genes and represent genuine noncoding RNAs.
Collectively, our analysis has uncovered with confidence a total of 6686 unannotated noncoding transcripts in 3859 genomic loci (Supplemental File S6). Not only do they lack protein-coding potential, they also (1) match entries in NONCODE or Rfam based on sequence, synteny, or structure similarity; (2) are on a different strand from its neighboring or overlapping gene; or (3) are far away from a known protein-coding gene. We observed that the majority of these noncoding RNAs appear to be lowly expressed, with 67.6% of them (or 4519 contigs) having a maximum FPKM (fragments per kilobase per million mapped fragments) of <5 (Supplemental Fig. S18A), thereby suggesting that even deeper sequencing is required to reveal the full repertoire of noncoding RNAs in Xenopus. In addition, the noncoding transcripts that we found in this study appear to be developmentally regulated (Supplemental Fig. S18B), which indicates that they may perform stage-specific functions during embryogenesis. In summary, our results have demonstrated the diversity and the dynamic regulation of the Xenopus transcriptome.
Assembly of transcripts beyond the current Xenopus tropicalis genome
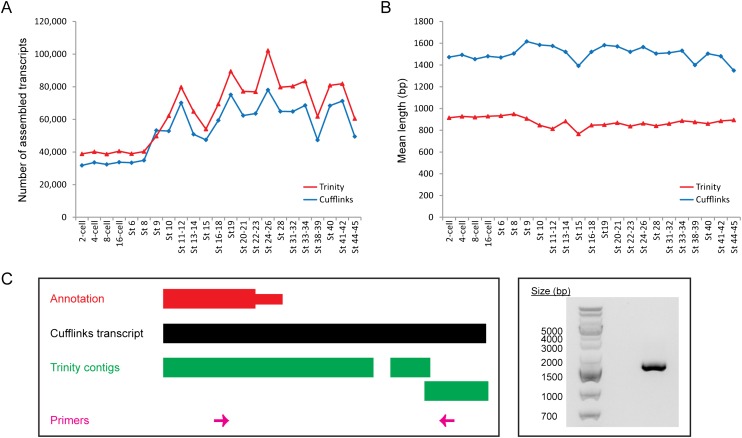
Cufflinks is a mapping-first approach and uses the reference genome as a guide to build the transcripts. Conceptually, such mapping-first methods offer superior sensitivity than de novo assemblers. However, the current version of the Xenopus tropicalis genome is still incomplete and consists of hundreds of scaffolds with scattered gaps, which may cause Cufflinks to discard unaligned reads and miss some novel transcripts. Thus, besides Cufflinks, we also assembled transcripts de novo without a reference genome by applying Trinity (Grabherr et al. 2011) to our RNA-seq data (Fig. 5A).
We compared the assembled Trinity contigs with the RefSeq and Ensembl annotations as well as the Xenopus tropicalis genome. At every developmental stage, between 47.9% and 65.8% of the contigs match either annotation (E-value < 1 × 10−10) (Supplemental Fig. S19). Interestingly, we note that a small but substantial fraction of contigs (from 1.3% to 2.4%) have sequences that could not be aligned to any part of the genome. We asked if there are any Xenopus ESTs that support these unaligned Trinity contigs. At every stage, we found between 400 and 900 contigs that contained matches in the collection of ESTs (E-value < 1 × 10−10) but are missing from the genome (Fig. 6A, gray portions). Next, we wondered if the remaining unaligned contigs with no EST support might be derived from contaminating sources. Hence, we used BLAST to remove all possible heterologous sequences from bacteria and fungi (Supplemental Fig. S20). In addition, we also discarded all contigs that matched cloning vectors or plasmids in the NCBI nucleotide collection database as another source of DNA contamination. Of all the unaligned Trinity contigs with no EST evidence, 25.2% (or 2448 out of 9733 contigs) contain matches in bacteria genomes, 0.1% (or 8 contigs) may be attributed to fungal contamination, while 3.7% (359 contigs) match cloning vectors in the NCBI database. The sequences that passed through the filters are then deemed to be noncontaminating and to represent genuine Xenopus transcripts even though they are not supported by ESTs (Fig. 6A, unshaded portions). We checked these transcripts for protein-coding potential by the same filters used for analyzing the Cufflinks sequences and found that on average 13.1% of them may encode proteins (Supplemental Fig. S21). In addition, we discovered that a small percentage of the noncontaminating unaligned Trinity contigs (from 0.3% to 7.8%) are conserved between Xenopus and humans, mice, or zebrafish (Supplemental File S7), which suggests that they might be functionally important.
Figure 6.
Hundreds of transcripts assembled de novo do not align to the reference genome. (A) Number of unaligned contigs assembled by Trinity at every developmental stage. The portions shaded in gray represent the contigs that have EST support, while the unshaded portions correspond to the contigs that do not match any existing Xenopus EST and are not derived from contaminating sources. (B) Number of unaligned Trinity contigs plotted against sequencing coverage. A linearly increasing trend is observed, indicating that deeper sequencing is required to discover even more transcribed sequences that are missing from the reference genome. (C) Validations of unaligned Trinity contigs with EST support. All the PCR products were sequenced to confirm that they were indeed the relevant contigs. The names in brackets correspond to the matching ESTs or cDNA clones, which were previously sequenced as part of either the NIH Xenopus Initiative (Klein et al. 2002; Gerhard et al. 2004) or the Sanger Xenopus tropicalis EST/cDNA project (Gilchrist et al. 2004; Carruthers and Stemple 2006). (D) Validations of unaligned Trinity contigs that have never been detected prior to this study. All the PCR products were sequenced as confirmation. The name of each contig was arbitrarily given by the program. One contig, comp29928, was most highly transcribed from Stages 13–28, while the expression of the other two contigs, comp697 and comp39806, showed a general increase over development and remained strong even in the feeding tadpole stages.
Since the discovery of novel transcripts is often a function of sequencing depth, we asked whether we have uncovered most of the transcribed sequences that are yet to be incorporated into the genome. To address this question, we plotted the number of unaligned (noncontaminating) contigs found at each stage against the corresponding number of sequencing reads (Fig. 6B). The graph showed a linearly increasing trend, suggesting that despite our efforts, many more novel transcripts may be discovered with an even higher sequencing depth. When we further divided the set of unaligned contigs into those with EST support and those without EST evidence and plotted them separately against the number of reads, we observed that, while the number of EST-supported contigs is beginning to saturate, the number of novel undocumented contigs is still increasing linearly (Supplemental Fig. S22). Taken together, our results indicate that numerous Xenopus transcripts remain to be detected and strongly suggest that even though the Xenopus tropicalis genome is close to being finished, it is still missing hundreds of transcribed sequences.
To determine if the Trinity contigs that could not be aligned to the genome are true transcripts or are simply spurious assemblies, we sought to validate some of them by reverse transcription PCR. We first picked five contigs that had matches to Xenopus ESTs and performed PCRs using cDNA templates from multiple developmental stages. PCRs for four of the contigs yielded products of the expected size (Fig. 6C). We also observed that these validated contigs are generally strongly expressed in all the stages tested, which is unsurprising since Sanger sequencing of ESTs and cDNA clones in the past covers only a small fraction of the entire transcriptome; and thus, the EST collection would be biased toward genes that are more highly or constitutively transcribed. Next, we selected another four contigs without EST matches for validations and found that PCRs for three of them yielded products of the correct size (Fig. 6D). Interestingly, all the three validated contigs that had never been observed before appeared to be developmentally regulated. In summary, we were able to confirm seven out of nine Trinity contigs that could not be aligned to the Xenopus genome; and hence, we estimate that 75%–80% of them are likely to be accurate assemblies.
Comparison of transcripts assembled with and without a reference genome
Since Cufflinks and Trinity utilize fundamentally different algorithms to reconstruct transcripts from RNA-seq reads, we wanted to compare the contigs assembled by both programs and to determine how reliable each transcript assembly may be. In almost all developmental stages, Trinity reconstructed more contigs than Cufflinks (Fig. 7A). However, while an average of 76.3% of Cufflinks transcripts matches RefSeq or Ensembl annotated genes in the Xenopus genome (E-value < 1 × 10−10) at each developmental stage (Fig. 5B), only 57.1% of Trinity contigs contain matches in either annotation (Supplemental Fig. S19). In addition, the mean length of a contig assembled by Trinity is shorter than the mean length of a transcript assembled by Cufflinks at all developmental stages (Fig. 7B; Supplemental Fig. S23). On average, a Cufflinks transcript is 1.72 times as long as a Trinity contig. Furthermore, at each stage, 71.3%–78.6% of the Trinity contigs, but only 42.5%–49.3% of the Cufflinks transcripts, are ≤ 1000 bp (Supplemental Fig. S24). Hence, even though more contigs are reconstructed by Trinity, they are generally shorter and a smaller percentage of them can be aligned to RefSeq or Ensembl annotated genes.
Figure 7.
Ab initio transcript assembly with a reference genome (Cufflinks) results in fewer but longer contigs than de novo transcript assembly without a reference genome (Trinity). (A) Number of contigs assembled by Cufflinks (blue diamonds) and Trinity (red triangles) at every developmental stage. Trinity almost always reconstructs a larger number of transcripts than Cufflinks. (B) The mean length of Cufflinks transcripts (blue diamonds) and Trinity contigs (red triangles) at each stage. On average, a transcript reconstructed by Cufflinks is 628 bp longer than a contig assembled by Trinity. (C) The Cufflinks transcript for the ccnK gene is more accurate than the corresponding Trinity contig. (Left) A schematic of the genomic locus of ccnK at the 3′ end. (Right) A gel image showing the result of a PCR performed with the primers depicted in pink at the left. The PCR product shown was further sequenced to confirm that it was indeed the intended target.
Since it utilizes the reference genome as a guide, a possible reason why Cufflinks reconstructs fewer but longer transcripts than Trinity is that Cufflinks can merge several shorter sequences that are mapped close together into a single long transcript, and it may also be able to detect splice junctions more sensitively. As examples, we examined transcripts originating from the ccnK locus (Fig. 7C) and the intergenic region between sox10 and baiap2l2 (Supplemental Fig. S25). In each case, our PCR results showed that the single long transcript reported by Cufflinks is more accurate than the multiple short contigs generated by Trinity (see Supplemental Information). Hence, Cufflinks outperforms Trinity at piecing together longer or spliced transcripts. However, as Trinity reconstructs contigs without a reference genome, it can discover novel genes that are absent from an incomplete genome. Collectively, our results indicate that both ab initio transcript assembly using Cufflinks and de novo transcript assembly using Trinity are required to obtain a more comprehensive understanding of the Xenopus transcriptome.
Discussion
We have generated the first extensive map of the Xenopus tropicalis transcriptome at single-base resolution using paired end RNA-seq. By sampling 23 distinct stages, we acquired a broad understanding of the dynamic changes in the transcriptome during development from a two-cell embryo to a feeding tadpole. We obtained at least 20 million reads for each stage and over 900 million reads in total. The deep coverage allowed us to examine the Xenopus transcriptome in unprecedented detail and to identify a large number of novel splice junctions and novel transcribed regions.
The initial stages of vertebrate development rely predominantly on maternal factors deposited in the egg, and there is minimal zygotic transcription until the embryonic genome gets activated. While this activation occurs at the late two-cell stage in mice and between the four-cell and eight-cell stage in humans, transcription is believed to be repressed in Xenopus embryos all the way until the twelfth cell division, when the midblastula transition takes place. Prior to our study, <10 genes were known to be transcribed in the beginning stages following fertilization (Yang et al. 2002; Skirkanich et al. 2011). We examined the expression of thousands of annotated genes and found that approximately 150 genes are clearly transcribed prior to the midblastula transition according to our RNA-seq data. This large collection of genes provides a rich resource for future functional studies, which will greatly enhance our understanding of the role of transcription in early development. In particular, 36 of these genes have also recently been determined to be up-regulated before the midblastula transition in zebrafish (Aanes et al. 2011). Strikingly, eight of the genes that are common between Xenopus and zebrafish, including the histone acetyltransferase hat1 and the DNA methyltransferase-associated protein dmap1, perform functions that are related to transcriptional regulation, while another three genes, namely wdr36, taf5l, and pik3r4, encode proteins that contain WD repeats. It has previously been shown that the WD repeat-containing protein, wdr5, plays an essential role in epigenetic regulation of vertebrate development by interacting with dimethylated histone3-lysine4 (H3K4me2) and mediating its transition to a trimethylated stage (H3K4me3) (Wysocka et al. 2005). Hence, wdr36, taf5l, and pik3r4, together with hat1 and dmap1, may potentially perform important epigenetic functions to prime the embryonic genome for large-scale activation at the midblastula transition in vertebrates. Future studies directed at mapping changes in histone modifications and DNA methylation marks during the earliest stages of development will serve to deepen our understanding of the interplay between chromatin architecture and transcription and will eventually allow us to decipher how a generally quiescent genome may become fully activated in order to drive further developmental events.
Surprisingly, none of the genes that have been previously reported to be transcribed before the midblastula transition (Skirkanich et al. 2011) showed up in our lists of early transcribed genes (Supplemental Files S4, S5). We examined our RNA-seq data to determine the reasons for their absence. The nodal-related gene, xnr5, is duplicated in the Xenopus genome and the corresponding short reads cannot be uniquely or reliably mapped. The homeobox-containing gene, bix4, is not demarcated in the XenTro3 genome; it is absent from both the RefSeq and the Ensembl annotations. In addition, the expression levels of xnr6, mixer, and sox17a did not meet the criterion of a twofold increase between the two-cell stage and Stage 6 (Supplemental Fig. S26). Finally, while the expression of derrière did increase by more than twofold from the two-cell to the 16-cell stage, it showed a sudden decline at Stage 6 (Supplemental Fig. S26). It is possible that the decline might be due to a measurement error as the RPKM at Stage 8 continued the increasing trend. We further note that in contrast to the report by Skirkanich et al. (2011), we did not observe a gradual increase in expression levels for any of these genes. Instead, the general trend appears to be exponential; that is, there is at most a modest degree of transcription in the beginning stages of development followed by a strong burst of transcription upon activation of the embryonic genome. Although the disparity may be due to the different species used (Xenopus tropicalis versus Xenopus laevis), further work is needed to fully understand the discrepancies.
Alternative splicing plays a widespread and important role during the development of many multicellular eukaryotes, including C. elegans (Barberan-Soler and Zahler 2008), Xenopus (Fletcher et al. 2006), and mice (Revil et al. 2010). Our global splicing analysis uncovered between 23,000 and 54,000 novel splice junctions at every developmental stage in the frog, most of which are supported by at least two nonredundant reads. Interestingly, we found that the majority of these novel splicing events are stage-specific and result in new unannotated isoforms of known genes. For example, exon skipping events in the fibronectin gene, fn1, result in two new transcript isoforms, one of which is most highly expressed from Stage 11 to Stage 28 (Fig. 4A), while the other is most strongly expressed from Stage 33 to Stage 45 (Fig. 4B). Even though some of the novel junctions might be a result of inexact splicing and may not have any biological relevance, we observed during our validation experiments that the expression of the novel isoform often does not vary together with that of the canonical isoform over development. Hence, we propose that many of the novel isoforms that we discovered here perform particular roles during certain developmental time points, and future studies are needed to unravel their specific functions.
Our extensive RNA-seq data allowed us to identify novel transcripts that are protein-coding, noncoding, or even nonexistent in the current version of the genome assembly of Xenopus tropicalis. We reconstructed transcripts from the reads using two distinct assemblers, Cufflinks and Trinity, and generated a catalog of novel transcribed regions. To discover new noncoding RNAs, we developed a series of filtering steps to remove contigs with protein-coding potential and identified a total of 9529 putative noncoding transcripts. We have higher confidence in 70.2% of them (6686 transcripts in 3859 genomic loci) as genuine noncoding genes because they contain matches in the NONCODE or Rfam databases, align to human or mouse noncoding genes, are on a different strand from its neighboring genes, encode antisense transcripts, or are located at a remote distance away from an annotated protein-coding gene. Besides uncovering new noncoding RNAs, we also found hundreds of de novo assembled contigs that could not be aligned to the current reference genome (XenTro3), which indicates that the Xenopus tropicalis genome is still unfinished. Our study will aid in the future completion and full annotation of the genome.
In summary, we used paired end RNA-seq to examine in detail the transcriptome of an important model organism, Xenopus tropicalis. We have uncovered a large number of novel transcribed regions, which will support the full assembly and annotation of the reference genome. In addition, our work has laid the foundation for the discovery and analysis of new protein-coding and noncoding genes in Xenopus and will serve as a valuable resource to be integrated into future genomic and developmental studies.
Methods
Xenopus tropicalis embryo culture and collection
The animals used in this study are out-bred Nigerian frogs from a stock maintained by the University of Virginia and were purchased directly from Nasco. Embryos were obtained by natural mating as follows: Adult female Xenopus tropicalis were injected 20–24 h before embryo collection with 10 units human chorionic gonadotropin (HCG) (Sigma). Four to five hours before embryo collection, male and female frogs were injected with 100 units and 400 units HCG, respectively, and amplexus is allowed to begin. Forty-five minutes after the onset of egg laying, embryos were collected and dejellied in 1/9 MR+ 3% cysteine. Embryos were then cultured in 1/9 MR at room temperature and were staged according to Nieuwkoop and Faber (1994). Embryos from two separate clutches were harvested and frozen at −80°C until RNA isolation. For the first clutch, embryos were collected at Stage 9, Stage 10, Stage 11, Stage 12, Stage 15, Stage 16, Stage 19, Stages 20–21, Stages 22–23, Stages 24–26, Stage 28, Stages 31–32, Stages 33–34, Stage 40, and Stages 41–42. For the second clutch, embryos were collected at two-cell stage, four-cell stage, eight-cell stage, 16-cell stage, Stage 6, Stage 8, Stage 9, Stage 10, Stages 11–12, Stages 13–14, Stages 16–18, Stage 19, Stages 20–21, Stages 22–23, Stages 24–26, Stage 28, Stages 31–32, Stages 33–34, Stages 38–39, Stage 40, Stage 41–42, and Stages 44–45. For each clutch, ∼10–15 embryos or 5–10 tadpoles were collected at every stage.
RNA isolation and RNA-seq library preparation
Total RNA was extracted from whole embryos using either the RNeasy Mini Kit (Qiagen) or the RNeasy Lipid Tissue Mini Kit (Qiagen) according to manufacturer's instructions, including the on-column DNase I treatment. The concentrations of all the RNA samples were measured using the NanoDrop (Thermo Scientific), and their integrity was determined using an Agilent 2100 Bioanalyzer. RNA-seq libraries were made from samples with a RNA Integrity Number (RIN) of at least 8.0.
The general Illumina mRNA-seq library preparation workflow was followed with some modifications. First, polyA-containing RNAs were selected using two rounds of Dynal Oligo(dT) beads (Invitrogen). The RNAs were then fragmented in 5× First-Strand Buffer (Invitrogen) at 85°C for 7–8 min. Random hexamers and SuperScript III (Invitrogen) were used to synthesize the first strand cDNA. Next, second strand cDNA synthesis was performed with dUTP in place of dTTP to mark the second strand (Parkhomchuk et al. 2009). After polishing the ends with End-It DNA End Repair Kit (Epicentre) and adding adenosine to the 3′ends using Klenow fragment (New England Biolabs), standard Illumina adapters were ligated. DNA fragments in the size range of 300–600 bp were gel extracted and treated with uracil-DNA glycosylase (New England Biolabs) before each library was amplified using Phusion DNA polymerase (Finnzymes). Libraries were quantified using the Qubit dsDNA High Sensitivity Assay Kit (Invitrogen) and sequenced on HiSeq 2000 (Illumina) to produce paired 100-bp reads.
Quantitative real-time PCR
Several genes that showed an increase in transcript levels prior to the midblastula transition according to our RNA-seq data were further tested. Reverse transcription was performed using random hexamers and the SuperScript III First-Strand Synthesis System (Invitrogen). Real-time PCR was performed using iQ SYBR Green Supermix (Bio-Rad) on the ABI 7900HT machine (see Supplemental Table S2 for the primer sequences). The cycling parameters are as follows: 95°C for 3 min followed by 40 cycles of 95°C for 15 sec, 57°C for 30 sec, and 72°C for 15 sec. With the two-cell stage as reference, fold change was calculated by normalizing Ct values in each developmental stage against the ODC gene using the 2-ΔΔCt method (Livak and Schmittgen 2001). For a gene to be considered as validated, its normalized expression level must increase by at least twofold in three out of four biological replicates. Each of the two clutches of embryos used for constructing the RNA-seq libraries constitutes a biological replicate, while embryos for the remaining two replicates were obtained from additional independent matings.
Nonquantitative reverse transcription PCR
Multiple novel splice junctions and unannotated transcripts assembled by Cufflinks or Trinity were selected for further validation. The RNA samples used to construct the RNA-seq libraries were reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen) to generate cDNA templates for the subsequent PCRs. For transcripts shorter than 1000 bp, the iQ SYBR Green Supermix (Bio-Rad) was used together with the following cycling parameters: 95°C for 3 min followed by 35 cycles of 95°C for 15 sec, 60°C for 30 sec, and 72°C for 30 sec and then followed by 72°C for 2 min. The primer sequences are given in Supplemental Table S3. For transcripts longer than 1000 bp, the Herculase II Fusion DNA Polymerase (Agilent) was used according to manufacturer's instructions for cDNA. The primer sequences and the corresponding annealing temperatures are given in Supplemental Table S4.
Expression analysis of annotated genes
The annotation-dependent expression analysis was performed using rSeq (version 0.0.7) (Jiang and Wong 2009). Two mismatches were allowed and either RefSeq or Ensembl was used as the reference annotation. To compare the transcript levels estimated by RNA-seq and microarrays, the microarray data set GSE27227 (Yanai et al. 2011) was downloaded from the Gene Expression Omnibus (GEO) database. For each gene tiled on the microarray, the transcript level was calculated as the average of the corresponding probes from all three biological replicates in the data set.
Cluster analysis
K-means clustering was performed using the software Cluster and the results were displayed using the program Java TreeView (Eisen et al. 1998). The number of clusters, K, was varied in order to find the maximum number of distinct clusters, i.e., K was chosen such that all the unique patterns were revealed, and no two clusters appeared nearly identical to one another.
RNA-seq data alignment and junction detection
SpliceMap (version 3.3.5.2) (Au et al. 2010) was used to map our RNA-seq data to the genome and to detect exon junctions. The maximum and minimum intron lengths were set as 40,000 bp and 20,000 bp, respectively. 10 hits were allowed for the 25-mer seed mapping with up to two mismatches, and four mismatches were allowed for the full read. Bowtie (version 0.12.7) (Langmead et al. 2009) was employed for the 25-mer seed mapping and the “try hard” option was used. Since the Xenopus genome (XenTro3) consists of too many individual scaffolds, which increases computing intensity of Bowtie mapping, all the scaffolds were merged into a single sequence for this mapping, and the results were then post-processed to have the correct scaffolds and coordinates assignments. We define novel junctions as the ones that are not reported in RefSeq, KnownGene, and Ensembl annotation libraries. The number of nonredundant reads (nNR, reported from SpliceMap) was used to define the reliability of the junction detections, as discussed in Au et al. (2010).
Ab initio transcript assembly and quantification
The TopHat-Cufflinks pipeline was used to predict gene isoforms from our RNA-seq data. In TopHat (version 1.2.0) (Trapnell et al. 2009), the option “min-isoform-fraction” was disabled, while “coverage-search,” “butterfly-search,” and “microexon-search” were used. The option “multi-read-correct” and “min-isoform-fraction” were enabled in Cufflinks (version 1.1.0) (Trapnell et al. 2010). The expected fragment length was set to 350 bp and the “small-anchor-fraction” was set to 0.08, which requires at least 8 bp on each side of an exon junction for our 100-bp RNA-seq data. To compare and merge the reference annotation and the isoform predictions, an in-house script was written to convert the UCSC RefSeq annotation from genephed format to GTF format in a correct sorted order. All isoform predictions from the 23 stages and the reference annotations were merged for differential analysis by Cuffdiff. In Cuffdiff, “multi-read-correct” and “time-series” analysis were enabled. The expected fragment length was set to 350 bp and the “min-alignment-count” was set to 5.
De novo transcript assembly
Trinity (trinityrnaseq_r2011-10-29) (Grabherr et al. 2011) was used to perform de novo transcriptome assembly from the RNA-seq data. The options included were “–seqType fq,” “–SS_lib_type RF,” “–CPU 10,” and “–no_run_butterfly.” Due to cluster limitations, all Butterfly commands were run in parallel after Chrysalis has finished. The Trinity pipeline was performed for reads from each of the 23 stages as well as pooled reads from the second biological replicate.
Identification of unannotated or unassembled transcripts
Each of the output Trinity and Cufflinks fasta files served as input for BLASTN (version 2.2.25) against the following databases: (1) XenTro3 reference genome; (2) XenTro3 reference genome and Xenopus ESTs; (3) RefSeq; (4) Ensembl; and (5) RefSeq and Ensembl. An E-value threshold of 1 × 10−10 was used to determine the significant hits. The fraction of BLAST matches for a stage is calculated as the number of contigs in that stage with a significant BLAST match divided by the total number of contigs in that stage.
Identification of novel noncoding RNAs
The merged Cufflinks transcripts and the Trinity contigs assembled from pool reads were used to identify novel noncoding RNAs. To eliminate potential protein-coding genes, contigs that did not have a significant BLASTN match to either RefSeq or Ensembl were processed through the following filters: (1) Hmmer (version 3.0) against PfamA and PfamB (Punta et al. 2012), with the contig sequences translated in all three forward frames (E-value ≤ 1 × 10−10): (2) BLASTX (version 2.2.25) against PfamA and PfamB (E-value ≤ 1 × 10−10); (3) BLASTX against UniProtKB/Swiss-Prot (UniProt Consortium 2012) (E-value ≤ 1 × 10−10); (4) BLAT against human and mouse protein coding genes in RefSeq (at least 10% identity); and (5) any sequence that may encode an open reading frame of at least 100 amino acids in any of the three translated forward frames (START to STOP, from the beginning of the contig to the first STOP, or from the first START after the last STOP until the end of the contig). All contigs that did not fulfill any of the above criteria were considered to be potential noncoding RNAs.
To curate a confident set of noncoding transcripts, we sought to identify the contigs that match known noncoding RNAs in other species, are on a different strand from an annotated gene, or are far away (>25 kb) from a known Xenopus gene. The following steps were taken to identify the transcripts that match noncoding RNAs in other species: (1) BLAT against human and mouse noncoding RNAs in RefSeq (at least 10% identity); (2) searched the Rfam database (Gardner et al. 2011) with Infernal (Nawrocki et al. 2009) (score ≥ 40); (3) BLASTN against the NONCODE database (Bu et al. 2012) (E-value ≤ 1 × 10−10); and (4) determined if the neighboring genes of an intergenic contig match the neighboring genes of a NONCODE-annotated noncoding RNA (i.e., contig is present in NONCODE by synteny). In addition, since contigs that are located close to one another may actually be part of the same transcript, we divided the Xenopus scaffolds into 50-kb segments and grouped the confident noncoding contigs into these segments or genomic loci.
Data access
All of the raw RNA-seq data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE37452.
Acknowledgments
We thank Amy Young and Jennefer Kohler for technical assistance and Yue Wan for critical reading of the manuscript. This work is supported by start-up funds to J.B.L. from the Stanford University Department of Genetics and an NIH Molecular Biophysics Predoctoral Research Training Grant 5T32GM008294-23 to A.L.Y. The work of K.F.A. and W.H.W. is also supported by NIH grants R01HG005717 and R01HD057970.
Author contributions: M.H.T. and J.B.L. conceived the study. M.H.T. performed experiments with guidance from A.E.W. K.F.A., M.H.T., and A.L.Y. analyzed the data with help from J.B.L. J.C. designed the interactive website. J.C.B., W.H.W., and J.B.L. provided overall supervision for the project. M.H.T. wrote the manuscript with inputs from the other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.141424.112.
References
- Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G, et al. 2011. Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res 21: 1328–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au KF, Jiang H, Lin L, Xing Y, Wong WH 2010. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res 38: 4570–4578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan-Soler S, Zahler AM 2008. Alternative splicing regulation during C. elegans development: Splicing factors as regulated targets. PLoS Genet 4: e1000001 doi: 10.1371/journal.pgen.1000001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Bu D, Yu K, Sun S, Xie C, Skogerbo G, Miao R, Xiao H, Liao Q, Luo H, Zhao G, et al. 2012. NONCODE v3.0: Integrative annotation of long noncoding RNAs. Nucleic Acids Res 40: D210–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers S, Stemple DL 2006. Genetic and genomic prospects for Xenopus tropicalis research. Semin Cell Dev Biol 17: 146–153 [DOI] [PubMed] [Google Scholar]
- De Robertis EM 2006. Spemann's organizer and self-regulation in amphibian embryos. Nat Rev Mol Cell Biol 7: 296–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro AC, Thuret R, Coen L, Perron M, Demeneix BA, Wegnez M, Gyapay G, Weissenbach J, Wincker P, Mazabraud A, et al. 2007. Exploring nervous system transcriptomes during embryogenesis and metamorphosis in Xenopus tropicalis using EST analysis. BMC Genomics 8: 118 doi: 10.1186/1471-2164-8-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RB, Baker JC, Harland RM 2006. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development 133: 1703–1714 [DOI] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. 2011. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res 39: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, et al. 2004. The status, quality, and expansion of the NIH full-length cDNA project: The Mammalian Gene Collection (MGC). Genome Res 14: 2121–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist MJ, Zorn AM, Voigt J, Smith JC, Papalopulu N, Amaya E 2004. Defining a large set of full-length clones from a Xenopus tropicalis EST project. Dev Biol 271: 498–516 [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138 [DOI] [PubMed] [Google Scholar]
- Goda T, Abu-Daya A, Carruthers S, Clark MD, Stemple DL, Zimmerman LB 2006. Genetic screens for mutations affecting development of Xenopus tropicalis. PLoS Genet 2: e91 doi: 10.1371/journal.pgen.0020091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M 1958. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182: 64–65 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R, Gerhart J 1997. Formation and function of Spemann's organizer. Annu Rev Cell Dev Biol 13: 611–667 [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SN, Hertel KJ 2009. Spliceosomes walk the line: Splicing errors and their impact on cellular function. RNA Biol 6: 526–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH 2008. SeqMap: Mapping massive amount of oligonucleotides to the genome. Bioinformatics 24: 2395–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong WH 2009. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25: 1026–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, et al. 2002. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn 225: 499–510 [DOI] [PubMed] [Google Scholar]
- Klein SL, Strausberg RL, Wagner L, Pontius J, Clifton SW, Richardson P 2002. Genetic and genomic tools for Xenopus research: The NIH Xenopus initiative. Dev Dyn 225: 384–391 [DOI] [PubMed] [Google Scholar]
- Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano K, Miledi R, Stinnakre J 1977. Acetylcholine receptors in the oocyte membrane. Nature 270: 739–741 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Manak JR, Dike S, Sementchenko V, Kapranov P, Biemar F, Long J, Cheng J, Bell I, Ghosh S, Piccolboni A, et al. 2006. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet 38: 1151–1158 [DOI] [PubMed] [Google Scholar]
- Melamud E, Moult J 2009. Stochastic noise in splicing machinery. Nucleic Acids Res 37: 4873–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Chow W, Kirkpatrick R, Butterfield YS, Young AC, Stott J, et al. 2006. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res 16: 796–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Kirschner MW 1989. Cyclin synthesis drives the early embryonic cell cycle. Nature 339: 275–280 [DOI] [PubMed] [Google Scholar]
- Murray AW, Solomon MJ, Kirschner MW 1989. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature 339: 280–286 [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR 2009. Infernal 1.0: Inference of RNA alignments. Bioinformatics 25: 1335–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J 1994. Normal table of Xenopus laevis (Daudin): A systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. Garland Science/Taylor & Francis, New York and London [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123 doi: 10.1093/nar/gkp596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. 2012. The Pfam protein families database. Nucleic Acids Res 40: D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, Nelson AC, Lee LJ, Morris Q, Blencowe BJ, Zhen M, et al. 2011. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res 21: 342–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revil T, Gaffney D, Dias C, Majewski J, Jerome-Majewska LA 2010. Alternative splicing is frequent during early embryonic development in mouse. BMC Genomics 11: 399 doi: 10.1186/1471-2164-11-399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirkanich J, Luxardi G, Yang J, Kodjabachian L, Klein PS 2011. An essential role for transcription before the MBT in Xenopus laevis. Dev Biol 357: 478–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S, Riethoven JJ, Le Texier V, Gopalakrishnan C, Kumanduri V, Tang Y, Barbosa-Morais NL, Thanaraj TA 2006. ASD: A bioinformatics resource on alternative splicing. Nucleic Acids Res 34: D46–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B 2006. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res 34: W645–W650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium 2012. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40: D71–D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DE, Gutierrez L, Xu Z, Krylov V, Macha J, Blankenburg KP, Hitchens M, Bellot LJ, Spivey M, Stemple DL, et al. 2011. A genetic map of Xenopus tropicalis. Dev Biol 354: 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872 [DOI] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW 2011. Mapping gene expression in two Xenopus species: Evolutionary constraints and developmental flexibility. Dev Cell 20: 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, Klein PS 2002. β-catenin/Tcf-regulated transcription prior to the midblastula transition. Development 129: 5743–5752 [DOI] [PubMed] [Google Scholar]