Abstract
Core duplicons in the human genome represent ancestral duplication modules shared by the majority of intrachromosomal duplication blocks within a given chromosome. These cores are associated with the emergence of novel gene families in the hominoid lineage, but their genomic organization and gene characterization among other primates are largely unknown. Here, we investigate the genomic organization and expression of the core duplicon on chromosome 17 that led to the expansion of LRRC37 during primate evolution. A comparison of the LRRC37 gene family organization in human, orangutan, macaque, marmoset, and lemur genomes shows the presence of both orthologous and species-specific gene copies in all primate lineages. Expression profiling in mouse, macaque, and human tissues reveals that the ancestral expression of LRRC37 was restricted to the testis. In the hominid lineage, the pattern of LRRC37 became increasingly ubiquitous, with significantly higher levels of expression in the cerebellum and thymus, and showed a remarkable diversity of alternative splice forms. Transfection studies in HeLa cells indicate that the human FLAG-tagged recombinant LRRC37 protein is secreted after cleavage of a transmembrane precursor and its overexpression can induce filipodia formation.
Comparative genomic studies have shown that the human and great ape genomes show a two- to threefold enrichment of interspersed segmental duplications when compared to macaque and nonprimate mammalian lineages (Bailey and Eichler 2006; She et al. 2006; Marques-Bonet et al. 2009). In these genomes, intrachromosomal segmental duplications have a mosaic structure composed of different ancestral subunits called duplicons. Among duplicons, we further identified “core duplicons” as the most abundant (top 10% of repeat-graph indices) duplicons among groups of intrachromosomal duplication blocks within a given chromosome (Jin et al. 2004; Jiang et al. 2007). Core duplicons are associated with the emergence of new genes, dramatic gene-expression differences, and structural variation (Johnson et al. 2001; Paulding et al. 2003; Ciccarelli et al. 2005; Vandepoele et al. 2005, 2009; Popesco et al. 2006; Bosch et al. 2007; Jiang et al. 2007). Due to the genomic complexity of their loci, the extensive copy number variation between and within species, and the lack of clear orthologs in model organisms of genes embedded in core duplicons, functional and genetic analyses of these genes have been particularly challenging. Emerging data suggest that some may be important in cellular proliferation and critical to the evolutionary dynamics underlying new gene formation in human and primate genomes (Wainszelbaum et al. 2008).
A core duplicon distributed throughout the q arm of human chromosome 17 led to the emergence of the LRRC37 (leucine-rich repeat containing 37) family during primate evolution (Jin et al. 2004; Jiang et al. 2007). Leucine-rich repeats (LRRs) are protein-ligand interaction motifs found in a large number of proteins with different structure, localization, and function. They are distributed across many phyla including bacteria, fungi, plants, and animals (Kobe and Kajava 2001). Many LRR-containing proteins have well-known functions in the innate immune system (Nurnberger et al. 2004), such as the Toll-like receptors (West et al. 2006), or are involved in various aspects of mammalian nervous system development (Chen et al. 2006). In both cases, the LRR motifs are important for intermolecular or intercellular interactions with exogenous factors in the immune system and/or with different cell types in the developing nervous system. The structure of LRR motifs and their arrangement in repetitive stretches of variable length generate a versatile and highly evolvable framework for the potential binding of diverse proteins and nonprotein ligands (Dolan et al. 2007).
Here, we study the evolution of the LRRC37 family in primates. We provide a comparative analysis of its organization and gene structure in primate and nonprimate mammals, particularly in human, orangutan, macaque, marmoset, and lemur genomes, as well as in dog, cow, mouse, and rat genomes. We evaluate its expression profiles in different human, macaque, and mouse tissues and investigate the subcellular location of the human protein, which suggests it is processed to the plasma membrane of cells where it is secreted, consistent with extracellular location of polypeptides derived from the rat homolog Lrrc37a2 (Hemschoote et al. 1988). Our work highlights the extraordinary dynamism in structure, copy number, organization, and expression of the LRRC37 core duplicon gene family during primate evolution.
Results
Human LRRC37 family organization
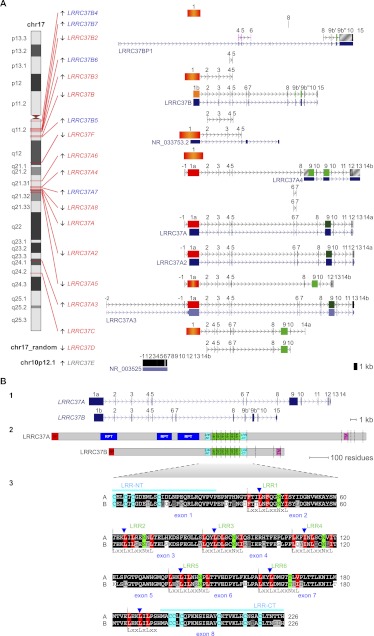
The LRRC37 family maps to 18 distinct loci in the human genome (NCBI36/hg18) on the q arm of chromosome 17. These copies derive from complete or partial duplication of the gene and contain both exons and introns. Further, a retrocopy is present on the p arm of chromosome 10 (Fig. 1A; Supplemental Table S1; Supplemental Note). We distinguished the copies in six different types (A–F) according to their location: B-type copies clustered at 17q11; F copy mapped at 17q12; A-type copies, except A3, clustered at 17q21; C copy mapped at 17q24; D copy mapped on chr17_random; and E copy is the retrocopy on chromosome 10. LRRC37A, A2, A3, and B have complete gene structure, RefSeq coding RNA annotated, and intact open reading frames (ORFs), whereas the remaining copies have partial gene structure and/or disrupted reading frames. LRRC37A, A2, and A3 differ from LRRC37B in the length of exon 1 (2612 bp in A type, exon 1a; 1690 bp in B type, exon 1b) and exon 9 (1532 bp in A type; split in two exons of 141 and 293 bp in B type, named exons 9b′ and 9b″). Terminal exons, downstream from exon 10, also differ: exons 11–14 in A type and exon 15 in B type (Fig. 1A,B).
Figure 1.
(A) LRRC37 family organization in human. The structure of complete and partial LRRC37 genes according to the reference human genome (hg18) is shown. (Left) Chromosomal band location of each locus on human chromosome 17 ideogram (only LRRC37E maps to chromosome 10 [gray] and is a retrocopy; LRRC37D does not have a chromosomal assignment). Genes are indicated as expressed (red) or not expressed (blue) based on the analysis of EST (expressed sequence tag) data. (Right) Variation in LRRC37 structures. Exons are represented as blocks connected by horizontal lines with arrowheads depicting introns. Exon 1 of A, A2, A3, and A4 copies (1a, in red) is longer than exon 1 of the B copy (1b, in orange); the B and B2 copies have exon 9 sequence split into two exons, 9b′ and 9b″. Exon 1 with deletions and/or insertions is red-orange. Exon 9 is dark green; exon 9 with deletions and/or insertions is light green. LRRC37A3 copy has one additional exon located at 5′ but lacks exon 12, and exon 13 is not transcribed. LRRC37A4 copy lacks exons 6 and 7 but carries a tandem duplication of exons 9 and 10. LRRC37A5 has a deletion of a single nucleotide at the end of the exon 1 sequence, causing a frameshift of the reading frame and the formation of a stop codon at the end of exon 1. LRRC37B2 is a fusion gene: exons 7, 8, and 9 match exons 9b′, 9b″, 10, and 15 of LRRC37B, whereas exons 4, 5, and 6 derive from a duplication of exons 4, 5, and 6 of SMAD-specific E3 ubiquitin protein ligase 2 (SMURF2) (NM_022739) (in pink), which maps at 17q24.1. Exapted introns are gray striped. RefSeq genes annotated in these loci are reported, following the same display conventions as in the UCSC Genome Browser. (B) Human LRRC37A and LRRC37B gene and protein structures. (Top panel) LRRC37A and LRRC37B cDNA predict an ORF with the methionine start codon (exon 1) and a stop codon (exon 14 and exon 15, respectively). LRRC37A encodes a predicted protein of 1700 amino acids with a molecular weight of 188 kDa. LRRC37B encodes a predicted protein of 947 amino acids with a molecular weight of 106 kDa. (Middle panel) Predicted structure of human LRRC37A and LRRC37B proteins according to SMART and LRRscan tools. Signal peptides (red), LRR motifs (green), LRR-NT and LRR-CT domains (turquoise), transmembrane helices (pink), and other repetitive motifs not associated with a known domain (blue) are shown. Exon boundaries are indicated with vertical dashed lines. (Bottom panel) An alignment of LRR regions of human LRRC37A and LRRC37B. The six LRR motifs and the final LRR are shown with the conserved leucine and asparagine residues highlighted in red and green, respectively. Boundaries between LRR motifs are marked with vertical dashed lines. LRR-NT and LRR-CT domains are shown with the conserved cysteine residues highlighted in turquoise. Exon boundaries are indicated with blue arrowheads.
Based on the ORF, the LRRC37A and LRRC37B putatively encoded proteins consist of 1700 and 947 residues and have a molecular weight of 188 and 106 kDa, respectively. They contain a predicted signal peptide and transmembrane helix encoded by exons 1 and 10, respectively. The LRRC37A protein has repetitive segments (RPT, RePeaT) encoded by exon 1 (Fig. 1B.2). The region from the end of exon 1 to exon 8 encodes six tandem extracellular LRR motifs, flanked by the LRR N- and C-terminal domains (LRR-NT and LRR-CT), according to the LRRscan program (Fig. 1B.2; Dolan et al. 2007). The motifs LRR1, LRR3, LRR4, and LRR5 are typical and match the consensus sequence LxxLxLxxNxL (where x can be any amino acid, and L positions can also be occupied by V, I, and F), whereas LRR2 and LRR6 are degenerate or atypical as they do not match perfectly the consensus sequence. Additionally, the last LRR before the LRR-CT domain contains only the first subdomain of eight residues. Notably, the LxxLxLxxNxL domain of each LRR motif spans a splicing junction (blue arrowheads in Fig. 1B.3). According to the protein structure and the extracellular nature of the LRR motifs, the LRRC37 family is thought to encode single-span transmembrane proteins with the N-terminal portion comprising the LRR region outside of the cell and a short C-terminal tail in the cytoplasm.
Evolutionary history of the LRRC37 family in primates
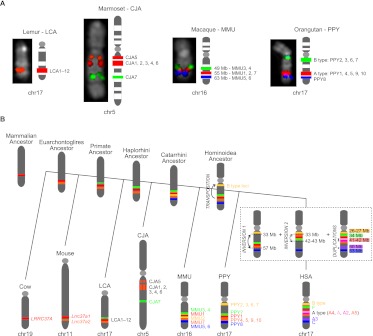
We analyzed the genomic organization of the LRRC37 family in various primate lineages, including orangutan (Pongo pygmaeus), rhesus macaque (Macaca mulatta), common marmoset (Callithrix jacchus), and ring-tailed lemur (Lemur catta) as representatives of great ape, Old World monkey, New World monkey, and prosimian lineages, respectively. Mammalian outgroup genomes included those of Mus musculus (NCBI37/mm9), Rattus norvegicus (Baylor 3.4/rn4), Canis familiaris (Broad CanFam3.1/canFam3), and Bos taurus (UMD_3.1/bosTau6). Both the mouse and the rat genomes have two tandem copies of Lrrc37 on chromosome 11 and chromosome 10, respectively. Mouse and rat Lrrc37a1 and Lrrc37a2 genomic regions show synteny to human chr17:40.9 Mbp (LRRC37A4) and chr17:42.4 Mbp (LRRC37A5) (hg18), respectively, based on the order of flanking genes. The cow genome has one copy of LRRC37 on chr19:45 Mbp and the dog genome has one complete copy on chr9:10 Mbp, both syntenic to human chr17:40.9 Mbp (Supplemental Note). This comparison of mouse, rat, dog, and cow genomes suggests that the human LRRC37A4 (chr17:40.9 Mbp) corresponds to the ancestral mammalian single-copy locus and that two tandem copies originated in the Euarchontoglires ancestor, likely corresponding to the human LRRC37A4 and LRRC37A5 (Fig. 2B).
Figure 2.
(A) Chromosomal organization of the LRRC37 family among primates. FISH results on primate metaphase chromosomes are shown for Lemur catta (LCA), Callithrix jacchus (CJA), Macaca mulatta (MMU), and Pongo pygmaeus (PPY). The following BACs were used as probes in each species: LCA chr17 (LB2-169E11 in red), CJA chr5 (CH259-152N22 in red and CH259-145C2 in green), MMU chr16 (CH250-219M3, CH250-221J22, and CH250-197J22 in red, green, and blue, respectively), and PPY chr17 (CH253-10O4, CH253-149E2, and CH253-167E20 in red, green, and blue, respectively). The corresponding LRRC37 family members are shown next to each chromosome. (B) Model of the LRRC37 family evolution. Different LRRC37 types are colored according to the human schematic. For other species, genes corresponding to syntenic locations or derived from lineage-specific duplications are colored corresponding to their human paralog. Since lemur copies are highly homogenized, we could not determine whether the ancestor of the human F copy emerged in the primate or the Haplorhini ancestor (dashed lines). Orthology relationships between human A type (A4 and A5) and marmoset 1–6 copies could not be determined (red-orange lines).
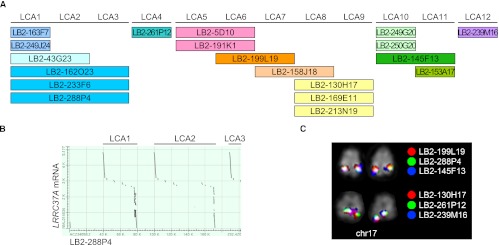
Since recently duplicated genes are frequently collapsed or missing from the draft genome assemblies (She et al. 2004; Alkan et al. 2011b), we identified large-insert clones from each primate species by genomic library hybridization. We screened BAC libraries (five- to sixfold coverage) from orangutan (CHORI-253), rhesus macaque (CHORI-250), common marmoset (CHORI-259), and ring-tailed lemur (LBNL-2) using a PCR-amplified probe (Anc409) corresponding to the exon 8/intron 8 canonical gene structure shared among most human duplicate copies. Due to the partial deletion of this locus, a second probe (Anc419, corresponding to sequence between introns 4 and 5) was used to screen the CH250 library (see below and Supplemental Note). We obtained a total of 46 (CH253), 36 (CH250), 28 (CH259), and 19 (LB-2) positive clones. Based on the genomic coverage of each library, this suggests the presence of seven, six, five, and three copies in the orangutan, macaque, marmoset, and lemur genomes, respectively. All clones were PCR amplified using conserved primers, and PCR products were sequenced. Based on the level of sequence divergence, we distinguished ten copies in the orangutan genome (PPY1–10) (Supplemental Table S2), seven copies in the macaque genome (MMU1–7) (Supplemental Table S3), seven copies in the marmoset genome (CJA1–7) (Supplemental Table S4), and twelve copies in the lemur genome (LCA1–12) (Supplemental Table S5; Supplemental Note). We identified the map location of each of these putative copies by BAC-end sequencing of clone inserts and mapping these sequences back to the human reference and respective primate genome sequence assemblies. We mapped BAC clones representing all identified copies on the respective primate metaphase chromosomes using FISH (fluorescent in situ hybridization) assays (Figs. 2A, 3C).
Figure 3.
LRRC37 family organization in Lemur catta. (A) Tandem organization of the 12 lemur copies of the LRRC37 family corresponding to the sequence from LB2 BAC clones. (B) Output of sequence alignment between LB2-288P4 (AC234058.2) and LRRC37A mRNA (NM_014834) showing the tandem organization of LRRC37 copies. (C) FISH results of representative lemur BAC clones confirm a single cluster of tandem duplicated copies on lemur chromosome 17.
We determined that the orangutan has ten copies of LRRC37 on chromosome 17 (referred to here as PPY1–10) and a retrocopy on chromosome 10 (PPY11). FISH mapping identified three locations on 17q for orangutan copies from the centromere to the telomere: a first cluster with PPY2, 3, 6, and 7; a second one with PPY1, 4, 5, 9, and 10; and a last location with PPY8 (Fig. 2A; Supplemental Fig. S2). For seven of the orangutan loci (PPY2, 4, 6, 7, 8, 9, and 10), the BAC-end sequence (BES) placements were all inconsistent with the ponAbe2 reference, suggesting potential orangutan assembly errors. Using the higher quality human genome (hg18) as a reference, we found that several of the loci map discordantly when compared to human, suggestive of human-specific duplicative transpositions. PPY1 maps to the orthologous location with respect to human A-type (LRRC37A, A3, and A4) copies; PPY2 maps to the orthologous location with respect to human chr17:26.1–26.2 and 27.5–27.6 Mbp, containing the B-type copies LRRC37B6, B3, B, and B5; and PPY6 and 7 have at least one end mapping to chr17:26–27 Mbp, hg18. PPY3, 8, and 11 map to an orthologous location with respect to human LRRC37B4, C, and E copies, respectively, whereas PPY4, 5, 9, and 10 copies map to the orthologous location of human LRRC37A5.
The rhesus macaque genome has seven copies of LRRC37 on chromosome 16 (MMU1–7) and a retrocopy on chromosome 9 (MMU8)—syntenic to human chromosomes 17 and 10, respectively. MMU1, 2, and 7 map to a cluster (chr16:55 Mbp, as annotated in rheMac2 assembly) and are the only macaque copies preserving an ORF. Intrachromosomal rearrangements shuffled human- and/or macaque-duplicated loci, and the ancestors of these copies originated both the hominid A- and B-type copies. The most likely human orthologs of MMU1, MMU2, and MMU7 are LRRC37A4, B, and A5, respectively. FISH-mapped macaque copies on orangutan and human chromosomes confirmed a transposition of the macaque locus at 55 Mbp (identified as MMU2 through sequence analysis) in the Hominoidea ancestor (Supplemental Fig. S5). This suggests an independent colonization and evolution of B-type copies at 17q12. In the human lineage, two inversions (Locke et al. 2011) and further duplications determined the current organization (Fig. 2B).
We note that MMU3 and MMU4 copies (estimated at 99.8% genomic sequence identity) are represented in the macaque assembly as a single copy (chr16:49 Mbp). A FISH cohybridization experiment using probes CH250-221J22 and CH250-269O23, corresponding to MMU3 and MMU4, shows that they colocalize in metaphase chromosomes, but both reveal close yet discrete signals in interphase nuclei (Supplemental Fig. S4). The human ortholog of MMU3 and MMU4 corresponds to LRRC37F. Similarly, MMU5 and MMU6 copies derived from a recent tandem duplication in the macaque lineage were collapsed in the macaque assembly to a single copy (chr16:63 Mbp). Their human ortholog corresponds to LRRC37C. A FISH cohybridization of CH250-219M3, CH250-221J22, and CH250-197J22, respectively representing macaque copies MMU1-2-7, 3-4, and 5-6, confirmed the order on chromosome 16 in accordance with the assembly (Fig. 2A). MMU8 is an ortholog of the human LRRC37E, both retrocopies. The macaque genome assembly indicates one additional copy of LRRC37 on chromosome 16 (chr16:56,953,955–57,000,088) containing both Anc Locus 409 and 419 regions, but none of the BAC positive clones mapped in this region.
In the marmoset genome, we identified seven copies of LRRC37 (named CJA1–7) mapping to chromosome 5 at three distinct cytogenetics bands, corresponding to calJac3 chr5:64–66 Mbp (CJA5), 76–80 Mbp (CJA1, 2, 3, 4, and 6), and 111 Mbp (CJA7) (Fig. 2A; Supplemental Fig. S3; Supplemental Table S4). The two clones containing the CJA7 copy are the only ones with concordant BES localization on both marmoset and human assemblies. Their BES localizations suggest CJA7 and human LRRC37F are orthologous genes, which was confirmed by FISH mapping as well as human-marmoset genome synteny. Most of the remaining CH259 BES mapped discordantly when comparing calJac3 and hg18 assemblies, revealing LRRC37 loci underwent intrachromosomal rearrangements, including duplications, in the marmoset lineage. According to BES locations in both genomes and FISH mapping, the two genomic regions at chr5:64–66 Mbp and 76–80 Mbp seem to be a mosaic of genomic modules corresponding to human chr17:15–20 Mbp and 39–42 Mbp and suggest that CJA1–6 are orthologous to human A-type copies. It is noteworthy that BES of the CJA4 locus map to human chr17:19 Mbp where no LRRC37 copy is present, suggestive of a lineage-specific duplication. Marmoset gene copies reveal that in the Haplorhini ancestor, besides LRRC37A4 and A5, the actual human F copy already existed (Fig. 2B).
In the Lemur catta genome we detected 12 copies of LRRC37 on chromosome 17 (syntenic to human chromosome 17) but only predicted three based on library hybridization. Remarkably, all are organized in a tandem configuration based both on large-insert clone sequencing (Fig. 3B) and FISH hybridization (Fig. 3C). No clear orthologous relationship could be traced with respect to human duplicated genes. However, the most consistent location for lemur BES corresponds to the human LRRC37C copy followed by the F and A4/A5 copies (located at human chr17 positions 63 Mbp, 34 Mbp, 40 Mbp, and 42 Mbp, respectively) (Supplemental Table S5). It is possible that in addition to the LRRC37A4 and A5 copies, the ancestors of the LRRC37C and F copies may have existed in the primate ancestor (Fig. 2B). Since no ortholog of the human LRRC37C has been found in marmoset, it may have been lost in marmoset or arose specifically in the Catarrhini ancestor. We note that the retrocopy mapping to human chromosome 10 (LRRC37E) is only identified in orangutan (PPY11) and macaque (MMU8) but is absent from marmoset and lemur, indicating that it arose in the Catarrhini ancestor.
BAC hybridization results revealed that gene structures have changed for some orthologous groups during primate evolution. For example, the human LRRC37F lacks sequence corresponding to the exon 8/intron 8 boundary (Anc409), yet MMU3, MMU4, and CJA7, which map to the orthologous location, are positive by hybridization. Thus, this deletion occurred specifically in the human lineage. Likewise, macaque MMU5 and 6 lack the region corresponding to intron 4/exon 5/intron 5 (probe Anc419) since they failed to hybridize or PCR amplify, whereas their human ortholog LRRC37C keeps this region.
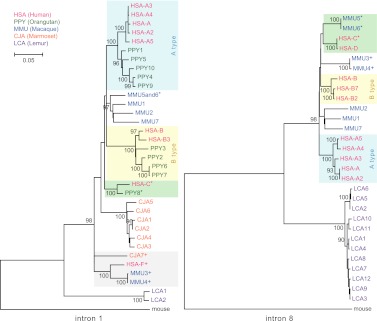
To determine the phylogenetic relationship among the various primate copies, we constructed two phylogenetic trees (maximum likelihood) using two noncoding regions corresponding to intron 1 and intron 8 (Fig. 4; Supplemental Note). We obtained sequences from reference genome assemblies (human and mouse hg18 and mm9, and CJA7 from WUGSC 3.2/calJac3), from PCR amplification and sequencing of BACs (orangutan, marmoset, and lemur), or from draft sequencing of BACs (macaque CH250, marmoset CH259, and lemur LB2) (Supplemental Note). The intron 1 phylogenetic tree included human, orangutan, macaque, marmoset, lemur, and mouse sequences, whereas the intron 8 phylogenetic tree included human, macaque, lemur, and mouse sequences. Because of the differences in gene structure, corresponding sequence was not obtained for all clones.
Figure 4.
LRRC37 family phylogeny. Maximum likelihood phylogenetic trees were constructed using noncoding regions from intron 1 (left) and intron 8 (right). Trees are drawn to scale, with branch lengths measured in the number of substitutions per site and bootstrap values (1000 replicates) shown. Human and orangutan A- and B-type copies are shaded blue and yellow, respectively; the monophyletic clade of HSA-F paralogs is gray shaded; branches with HSA-C, HSA-D, PPY8, MMU5, and MMU6 orthologous copies are shaded green. Copies clearly mapping to syntenic locations are signed by a superscript.
The topology of the phylogenetic trees confirmed the lineage-specific expansions and orthologous relationships previously inferred by BES and FISH mapping. For example, MMU3 and MMU4 are monophyletic and are part of a clade including HSA-F and CJA7 orthologs. In addition, the phylogeny confirms the orthologous relationship among HSA-C, PPY8, and MMU5-6 copies. HSA-C and HSA-D, as well as MMU5 and MMU6, are derived from two recent duplication events in the human and macaque lineages, respectively. These two clades can be inferred by the tree topology since the copies are located in two separate loci that were unlikely affected by nonallelic gene conversion and as a result did not experience strong sequence homogenization.
In contrast, the increase in copy number within the great ape lineage appears to be driven by the expansion of A and B types. Interestingly, the process appears to have occurred independently in both human and orangutan lineages, maintaining a similar balance in copy number. In humans there are five A subtypes (A, A2, A3, A4, and A5), and in orangutan there are five A subtypes (PPY1, 4, 5, 9, and 10), with at least A4 and A5 existing in their common ancestor. Comparably, there are four human (B, B2, B3, and B7) and four orangutan (PPY2, 3, 6, and 7) B-type copies that belong to the same monophyletic group. Similar lineage-specific expansions are observed in each of the primate lineages. For example, copies CJA1–6 are specific to the marmoset lineage and expanded (or homogenized) after separation from the other primate lineages (bootstrap support = 100). Similarly, we have identified multiple copies of lemur LRRC37—all of which are clustered and virtually identical at the sequence level (Figs. 3 and 4).
Evolutionary analysis of the ancestral locus
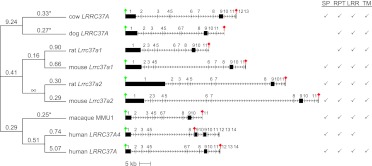
We analyzed the evolutionary conservation of the LRRC37 ancestral locus in mammals and aligned the corresponding orthologous coding sequences of cow LRRC37A, dog LRRC37A, mouse Lrrc37a1, rat Lrrc37a1, macaque MMU1, and human LRRC37A4, as well as the duplicated coding sequences of mouse Lrrc37a2, rat Lrrc37a2, and human LRRC37A. With the exception of human LRRC37A4, the exon-intron structure is conserved for exons 1–11 (Fig. 5). LRRC37 has two portions conserved differently in mammalian evolution: a variable in length, repetitive first exon and a more conserved portion corresponding to exons 2–8 containing the LRR region (Supplemental Note). We searched for evolutionary signals of natural selection on the LRRC37 ancestral protein coding sequence by using maximum likelihood models that estimate evolutionary rates for individual branches in the tree for coding sequence corresponding to exons 2–8 (Fig. 5). (Note: Due to the variable length and extensive amino acid replacement within exon 1, it was impossible to generate a high-quality sequence alignment for this portion.) Using a “one-ratio” model to test uniform dN/dS across all the branches of the phylogenetic tree, we noted that compared to a neutral model (ω = 1) the protein evolution for the LRRC37 model (exons 2 to 8) across all lineages is under negative constraint (ω = 0.4175, P-value < 0.001) (Fig. 5; Supplemental Table S6). Importantly, we find that the duplicated copies of mouse and rat (Lrrc37a2) show greater selective constraint than that of the ancestral locus (Lrrc37a1). We assessed branches for positive, neutral, and purifying selection and found that at the individual species level only the dog, cow, and macaque branches are significantly conserved (P-values 0.002, 0.04, and 0.048, respectively) (Supplemental Note).
Figure 5.
Selection of the LRRC37 ancestral locus during mammalian evolution. (Middle) The exon-intron gene structures of cow LRRC37A, dog LRRC37A, rat Lrrc37a1, mouse Lrrc37a1, rat Lrrc37a2, mouse Lrrc37a2, macaque MMU1, human LRRC37A4, and human LRRC37A are shown, with start and stop codons indicated by a green and red sign, respectively. The additional block in human LRRC37A4 exon 9 is shown in gray. (Right) Motifs identified in the translated proteins are shown: SP (signal peptide), RPT (repetitive segments), LRR (leucine-rich repeat motifs), and TM (transmembrane helix). Rat coding sequence is incomplete at 5′ and lacks the start codon. The premature stop codon in human LRRC37A4 coding sequence removes the portion encoding the transmembrane domain. (Left) Branch estimates for ω = dN/dS for exons 2–8 are shown using the free branch model in PAML. Significant branches compared to a neutral model are indicated with an asterisk.
LRRC37 family tissue-specific expression in mouse, macaque, and human
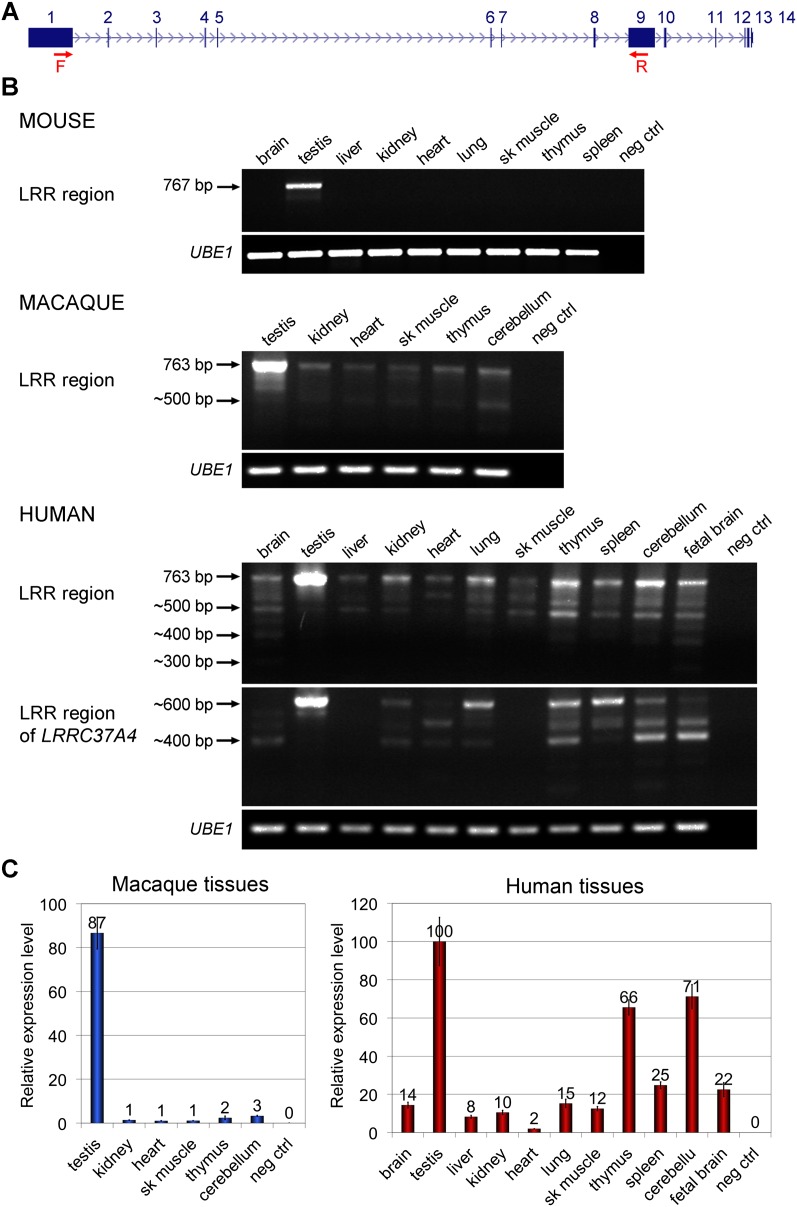
Thirteen out of 18 human loci show evidence of expression at the RNA level based on spliced and unspliced EST (expressed sequence tag) data (Fig. 1A). We investigated the mRNA expression patterns of the LRRC37 family in a panel of nine mouse, six macaque, and eleven human tissues. We designed RT-PCR assays to amplify the LRR region with degenerate forward primers designed at the end of exon 1 and degenerate reverse primers at the beginning of exon 9 (Fig. 6A) based on the canonical gene structure in each species, thus focusing on the expression of all copies having both exons 1 and 9. In mouse, both Lrrc37 genes were expressed only in the testis (Fig. 6B). In macaque, the LRRC37 family expression was predominant in the testis but with consistent, low-level expression in all other tissues tested (Fig. 6B,C). In human, the LRRC37 family showed a tissue expression profile similar to the macaque, although the intensity of expression in tissues other than the testis increased, especially for the thymus, spleen, fetal brain, and cerebellum (Fig. 6B,C). It may be noteworthy that the gene family, in general, shows higher levels of expression in pancreatic endocrine tumors (http://dcc.icgc.org), although this has not been independently confirmed (Supplemental Note). Gene expression profiling in mouse, macaque, and human suggests that a change in the regulation of mRNA expression occurred prior to the divergence of great apes and Old World monkeys likely as a result of the juxtaposition of novel regulatory machinery via segmental duplication (Bekpen et al. 2012).
Figure 6.
Tissue expression patterns of the LRRC37 family. RT-PCR and RT-qPCR results of the amplification of cDNA prepared from a panel of human tissue total RNA (Clontech) and from macaque and mouse tissue total RNA. UBE1 amplification was used as a control. (A) Human LRRC37A structure indicating the position of the primers used in RT-PCR experiments (red arrows). Forward primers were designed at the end of exon 1 and reverse primers at the beginning of exon 9 to amplify the LRR region. (B) Mouse Lrrc37a is expressed only in testis. The length of the complete LRR region of Lrrc37a2 is 767 bp. Minor products with shorter length are present. Degenerated primers have been designed for macaque and human to amplify all copies of the family. In both, the LRRC37 family is expressed in all the tissues tested, with the highest expression in testis. The expected size of the complete LRR region is 763 bp, but several products of smaller size are noted. The same forward primers and a reverse primer specific for the human LRRC37A4 copy have been tested in human tissue cDNA. There is no expression of the LRRC37A4 copy in liver and skeletal muscle. (C) Quantitative expression profiling of the LRRC37 family in macaque tissues and of LRRC37A, A2, A3, and A4 in human tissues. CT values are shown and calculated by comparative CT method (Livak and Schmittgen 2001) with UBE1 as the reference gene. In macaque, expression values are relative to heart. In human, the highest value of expression in the assay (testis) is set to 100. (Error bars) Standard error of the mean.
Alternative splicing and copy-specific expression for human LRRC37
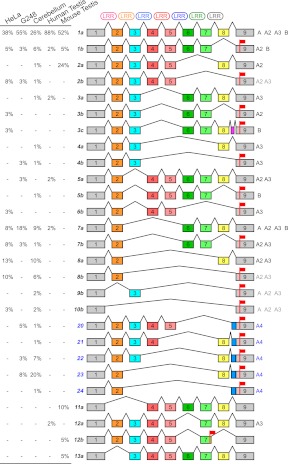
RT-PCR experiments showed a striking increase in the complexity in alternative splicing particularly among human tissues (Fig. 6). We subcloned and sequenced RT-PCR amplification products from various sources (Fig. 7; Supplemental Note) revealing extensive alternative splicing of exons encoding the LRR region. In human, we detected the expression of LRRC37A, A2, A3, A4, and B copies, with no products derived from A5 and C copies, despite having both primer annealing sites. We characterized a total of 24 different splice variants in human and six in mouse and assigned them to duplicate copies wherever possible based on diagnostic paralogous sequence differences (Fig. 7): 19 were assigned to LRRC37A, A2, A3, and B paralogs with five distinct forms corresponding to LRRC37A4. It is notable that most of this splicing diversity was observed in tissues other than the testis. We cloned and sequenced only six splice forms from both human and mouse testis.
Figure 7.
Alternative splice variants of the LRR region. The schematic depicts alternative splice variants of LRR region coding sequence based on sequencing of RT-PCR products amplified from HeLa cells, G248 lymphoblastoid cell line, human cerebellum, human testis, and mouse testis cDNA. Exons are shown (colored boxes) along with spliced introns (connecting lines), LRR motifs (top), and stop codons (red flags). The splice variants are successively numbered and, except for the ones derived from LRRC37A4, distinguished in a and b according to the presence or absence of exon 8. LRRC37A4 splice variants are indicated (blue). The observed frequency of each product is reported (percentages on left). Human paralogs presenting a specific splice variant are specified (right), with ambiguous assignments indicated in gray.
The alternative splicing creates different transcripts encoding putative LRRC37 protein isoforms with a variable number of LRR motifs. This ranges from no motifs, when exon 1 is spliced directly with exon 9, to six LRR motifs, when no exon is removed. Notably, the exclusion of exon 8 in mouse and human LRRC37A, A2, A3, and B copies removes the LRR C-terminal capping domain and shifts the ORF, introducing a premature stop codon at the beginning of exon 9. The resulting transcripts may be subjected to nonsense-mediated decay (Wilusz et al. 2001) or encode shorter putative proteins that terminate after the LRR region and lack the transmembrane domain codified by exon 10. These proteins, if expressed, may be secreted as they retain the signal peptide at the N-terminus.
Because of this potential functional difference, we categorize transcripts into two groups according to the presence or absence of exon 8: the long or a isoforms, with exon 8, and the short or b isoforms, without exon 8 and with the reading frame ending at the beginning of exon 9. The most abundant splice form in all samples tested was the complete one (1a), having all exons from 2 to 8. The mouse 12b variant has a different donor splice site in intron 7 that introduces a stop codon at the end of exon 7. The distribution of various splice products differs dramatically depending on RNA source material. In human testis, 88% of the sequenced transcripts show all exons (variant 1a), in contrast to cerebellum where this drops to 26%. In mouse testis, splice form 1a predominates with very little alternative splicing observed as suggested by the RT-PCR profile on agarose gel (Fig. 6B).
Human LRRC37A4 transcripts are unique—the gene structure lacks exons 6 and 7 and has a different splice acceptor site within intron 8 creating a longer (147 bp) exon 9 (light blue box in Fig. 7). This additional sequence generates a frameshift in the reading frame of all LRRC37A4 transcripts, which, in turn, leads to a stop codon at the beginning of exon 9 and a putative truncated version of the protein. Sequencing suggests that expression levels of different human paralogs varied depending on cell line or tissue. We designed four distinct quantitative RT-PCR assays to evaluate the expression of LRRC37A/A2, A3, A4, and B copies (Supplemental Fig. S9). Although the testis showed the highest level of expression for all copies, we note differences. For example, we find that LRRC37A4 transcripts are particularly enriched in the cerebellum (84% of the expression level of the testis), whereas LRRC37A/A2 copies are expressed highly in the thymus. LRRC37B shows the broadest pattern of expression outside of the testis including cerebellum, thymus, and fetal brain (Supplemental Fig. S9). These results are consistent with cDNA sequencing results from these tissues.
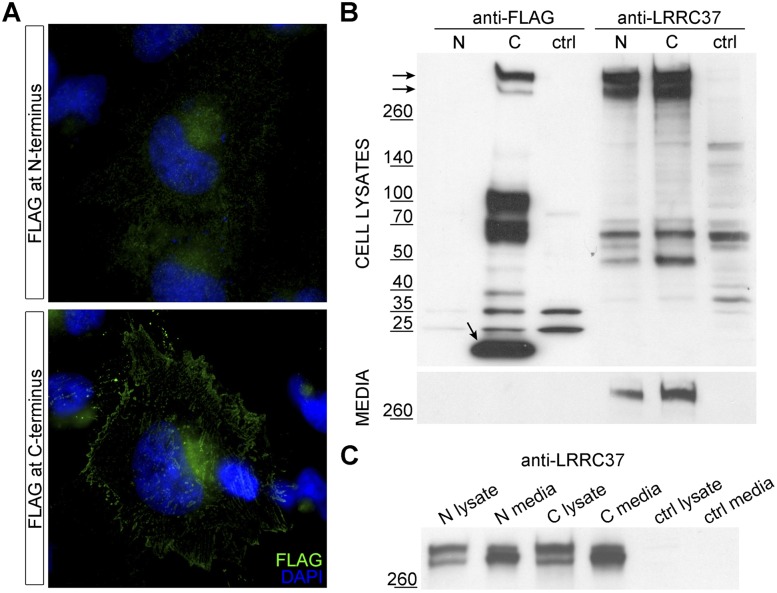
Subcellular localization of LRRC37A
To determine the subcellular localization and provide some potential functional insights into the LRRC37 family, we generated recombinant constructs carrying N-terminal or C-terminal FLAG-tagged human LRRC37A full-length coding sequence and developed polyclonal antibodies able to recognize human LRRC37 proteins. HeLa cells transiently transfected with recombinant plasmids were analyzed through immunofluorescence and Western blot using anti-FLAG and anti-LRRC37 antibodies. Immunofluorescence analysis showed that LRRC37A protein localizes to cellular vesicles, including the Golgi apparatus, and to the plasma membrane (Fig. 8A). Western blot analysis of both cell lysates and conditioned media shows two principal bands with a molecular weight >260 kDa common to both antibodies (Fig. 8B). This suggests that LRRC37A (190 kDa) has a very low electrophoretic mobility, potentially conferred by a moiety encoded by exon 1 (Supplemental Note).
Figure 8.
Subcellular localization of N- and C-terminal FLAG-tagged LRRC37A. (A) HeLa cells transiently transfected with N- and C-terminal FLAG-tagged LRRC37A constructs and probed with anti-FLAG antibody. LRRC37A tagged at the N-terminus does not show strong localization, whereas LRRC37A tagged at the C-terminus shows a plasma membrane localization (magnification, 63X). (B) Western blot of lysates and conditioned media of HeLa cells transfected with FLAG-tagged LRRC37A. Lysates and conditioned media of HeLa cells transiently transfected with N-terminal (N) or C-terminal (C) FLAG-tagged LRRC37A, probed with anti-FLAG (lanes 1–3) and anti-LRRC37 (lanes 4–6) antibodies. Nontransfected HeLa cells represent the control. (C) Conditioned media were concentrated, and a comparable amount of lysates and media was loaded. Samples were probed with anti-LRRC37 antibody.
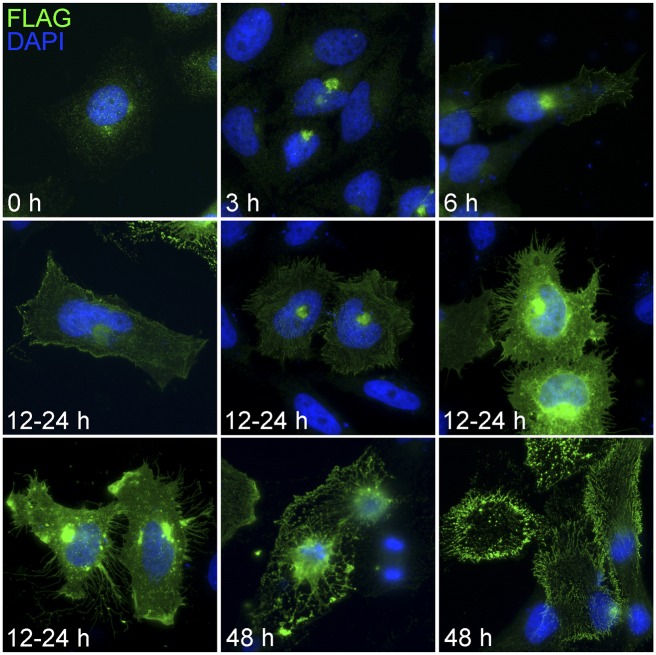
A time-course experiment (Supplemental Note) showed that C-terminal FLAG-tagged LRRC37A recombinant protein first localizes in the Golgi apparatus and then moves to the plasma membrane (Fig. 9). Our analysis indicates that the recombinant LRRC37A is routed to the endoplasmic reticulum through its signal peptide and transmembrane domain, then to the Golgi apparatus, and finally delivered to the plasma membrane by vesicles. Here, at the cell surface, it is likely cleaved within the extracellular juxta-membrane domain and released in the extracellular space (Supplemental Fig. S13; Supplemental Note). Interestingly, overexpression of LRRC37A in HeLa cells, which do not endogenously express this protein at detectable levels, causes a deformation of plasma membrane shape and the generation of filopodia-like protrusions (Fig. 9). At later stages of the expression there is an accumulation of circular anucleated elements stained by the anti-FLAG antibody, potentially as a consequence of protein overexpression (Fig. 9).
Figure 9.
Time-course experiment of FLAG-tagged LRRC37A expression in HeLa cells. HeLa cells were transiently transfected with recombinant FLAG-tagged LRRC37A and probed with anti-FLAG antibody (magnification, 40×). The pictures depict the most frequent aspects shown by transfected cells at different intervals after transfection. Note the accumulation at the plasma membrane and the formation of filopodia-like protrusions. After 48 h, cells begin to lose their integrity, and enucleated cells are observed.
Discussion
Evolutionary dynamism of LRRC37 in primates
The organization of the LRRC37 family in various genomes reveals a remarkable dynamism with a trend toward recurrent duplication in primate lineages when compared to other mammalian genomes. Both FISH and phylogenetic analyses suggest independent expansions of LRRC37 in different primate lineages. The duplication pattern becomes increasingly interspersed among great apes and associated with intrachromosomal rearrangements on hominid chromosome 17 (Zody et al. 2006). In lemur, for example, the duplication pattern is tandemly configured, reminiscent of nonprimate mammalian genomes (Tuzun et al. 2004; She et al. 2008; Liu et al. 2009). Independent duplications of both A- and B-type LRRC37 occurred specifically in the human and orangutan lineages becoming dispersed to more locations along the long arm of chromosome 17 (Figs. 1, 2) and the “cores” of more complex and larger blocks of segmental duplications composed of a greater number of duplicons (Jiang et al. 2007). We note that human LRRC37A4 and A2 loci correspond to the inversion breakpoints of the MAPT (microtubule associated protein tau) 17q21.31 region (Zody et al. 2008), one of the most structurally complex and evolutionarily dynamic regions of the human genome (Cruts et al. 2005; Stefansson et al. 2005). This duplication and restructuring has been accompanied by local changes in gene structure. The human LRRC37F copy, for example, is partial and does not retain the exon 8/intron 8 region, in contrast to its macaque orthologs (MMU3 and MMU4), which maintain the canonical gene structure. Likewise, the human LRRC37C copy retains all exons, but its macaque orthologs (MMU5 and MMU6) have lost a portion of exon 5. These data raise the possibility that different paralogs have assumed functional roles independently in different lineages as specific copies have become pseudogenized.
Expanded LRRC37 transcriptional diversity in humans
Changes in the duplication architecture of LRRC37 were accompanied by shifts in the transcriptional landscape and an overall increase in transcript diversity. In mouse, both Lrrc37 copies are expressed; in humans, we find that at least 13 of the 18 copies produce transcripts. The tissue expression profile is testis-exclusive in mouse, whereas in macaque we observe a pattern that is predominantly testis-specific but with a low level of expression in other tissues. In human, the pattern of expression becomes more ubiquitous. Although the highest level of expression still occurs in the testis, in human, we observe high levels of expression in other tissues, such as the cerebellum and thymus. Notably, the higher expression level in the cerebellum appears to have emerged in the great ape ancestor and to have been conserved over the last 15 million years of great ape evolution (Supplemental Note). We propose that a testis-specific pattern of expression was the most likely ancestral state of mRNA expression and that during primate evolution a broader landscape was acquired through the acquisition of novel regulatory elements via segmental duplication (Bekpen et al. 2012). This is consistent with the observation that rat peptides corresponding to the first exon of Lrrc37a2 are processed, secreted into the lumen of the prostate, and are androgen regulated (Heyns et al. 1982; Hemschoote et al. 1988; De Clercq et al. 1992). We point out that the macaque tissues were derived from a pig-tailed macaque (Macaca nemestrina), whereas genomic organization data were from the rhesus macaque (Macaca mulatta). Although they are closely related species, potential differences in their genomic organization may exist.
Our sequencing results also suggest remarkable diversity in LRRC37 transcriptional splice products when compared to mouse. In human, much of the alternative splicing involves exons encoding the LRRs, whose sequences have been conserved during mammalian evolution. Since LRR motifs are involved in protein-ligand interaction, the translation of these splice variants would potentially generate an expanded repertoire of proteins with the potential to interact with different ligands or with the same ligand at different strengths and affinities. From our library of sequenced products, we observed some interesting patterns. We found, for example, coordinated splicing of exons 4 and 5, as well as of exons 6 and 7, in all tissues with the exception of the testis. Removal of exon 8 disrupts the ORF and generates a stop codon at the beginning of exon 9, potentially producing two different groups of LRRC37 proteins: long isoforms, with the complete ORF, carrying the final transmembrane domain versus short isoforms, without the transmembrane domain. Similarly, a novel splice-acceptor within intron 8 of LRRC37A4 (Eichler 2001) adds 147 bp to exon 9 resulting in the formation of a premature stop codon and a protein lacking the transmembrane domain. These data suggest a complex interplay between segmental duplication and transcriptional novelty.
Functional insights into the LRRC37 protein
LRRC37 proteins, in general, are predicted to carry both a signal peptide and a transmembrane domain. Our analysis of one member of this gene family, a full-length isoform of LRRC37A2, confirms subcellular localization to both the Golgi and plasma membrane. Despite the absence of a clear cleavage or secretory signal sequence, transfection experiments in HeLa cells suggest that this isoform of LRRC37A is cleaved and released extracellularly. Interestingly, functional data about the rat ortholog Lrrc37a2 support the evidence that mammalian LRRC37 proteins are secreted. Notwithstanding, rat Lrrc37a2 experienced a peculiar expansion of a 300-bp tandem unit in the exon 1, each unit codifying for a secreted polypeptide. It is noteworthy that rat prostate cells translate a 20.6-kbp mRNA to produce at the end numerous 38-residue polypeptides. In this context, the role of the evolutionarily highly conserved LRR region remains completely unresolved.
With the exception of the first exon, the mammalian LRRC37 structure (exons 2–8) has been generally maintained for more than 100 million years of evolution and shows significant evidence of purifying selection (Fig. 5) with both rat and human proteins showing experimental evidence of secretion (Heyns et al. 1982; Hemschoote et al. 1988). The rate, however, has not been constant. We note that these estimates are based on comparisons between lineages and are unable to disentangle alternating bouts of purifying and positive selection that might appear in aggregate neutral. The human copy corresponding to the ancestral locus (LRRC37A4) has undergone some of the most radical changes in its exon-intron structure, potentially encoding the most novel protein when compared to the mammalian archetype. We propose that these changes occurred as a consequence of even greater relaxation of selection after the emergence of additional expressed copies by segmental duplication. It is interesting that the same copy acquired a higher expression in the cerebellum.
Although there is evidence that the rat ventral prostate secretes LRRC37 polypeptides (Heyns et al. 1982; Hemschoote et al. 1988; De Clercq et al. 1992), we could not detect any band in a Western blot of different human secreted fluids (seminal plasma, tears, saliva, blood plasma, breast milk, and sweat) probed with the anti-LRRC37 antibody (data not shown). Besides the lack of expression in these fluids, the possibility of the expression of a mature form devoid of the epitope has to be considered as well. Preliminary data show that the membrane remnants, forming at later stages of the overexpression, are immunofluorescently stained only by the anti-FLAG antibody and are not recognized by anti-LRRC37 antibody, confirming the proteolytic cleavage and the release in the extracellular space of the amino-terminal moiety of LRRC37A. This pattern of maturation is consistent with other secreted proteins that have a transmembrane precursor but require proteolytic cleavage to be released in the extracellular space. Many growth factors, including betacellulin (Shing et al. 1993), neuregulins (Montero et al. 2002), transforming growth factor alpha (TGF-α) (Gentry et al. 1987), stem cell factor (SCF) (Huang et al. 1992), and colony stimulating factor 1 (CSF-1) (Rettenmier and Roussel 1988), undergo this kind of maturation process. It is important that our cDNA sequence analysis also indicates the presence of splice variants encoding putative shorter proteins lacking the transmembrane domain. These isoforms, as well as LRRC37A4 proteins, may be directly secreted in the extracellular space bypassing proteolytic cleavage since they still retain the signal peptide. It is interesting that the LRRC37A family was among 87 genes recently identified as differentially expressed in dental pulp stem cell cultures from nonsyndromic cleft-lip and palate patients when compared to controls (Bueno et al. 2011). Members of a corresponding gene network were enriched in cleaved extracellular matrix proteins thought to be important during early development.
It is also intriguing that expression of LRRC37A, at least in HeLa cells, is associated with changes in cell shape in addition to the formation of filopodia-like protrusions. Our time-course analysis suggests that cells overexpressing LRRC37A generate so many protrusions that they ultimately lose their integrity and apoptose. The significance of this observation is unclear. Although this cellular phenotype might be determined by the LRRC37A protein itself, it may also be an artifact of its overexpression in transfection assays. In this context, since LRR motifs in characterized proteins are used for intermolecular or intercellular interactions, their presence suggests the LRRC37 proteins may bind some ligands. Finally, our overall observations represent only the initial step toward understanding the function of LRRC37. Separate efforts are needed to elucidate the human tissue and cell type that produce LRRC37 proteins and to obtain insights into potential ancestral and novel functions that have emerged.
Conclusions
The LRRC37 family is a member of 14 “core” duplicons that have expanded in the primate lineage since the divergence of the human lineage from other mammalian species (Jiang et al. 2007; Marques-Bonet and Eichler 2009). These cores represent seeds of genomic instability upon which the complex interspersed segmental duplication architecture of great ape and human chromosomes has evolved (Jiang et al. 2007). We have hypothesized that the selective disadvantage of this architecture predisposing the human genome to recurrent rearrangement has been offset by the emergence of novel genes with different functions and expression profiles with respect to their ancestral genes (Marques-Bonet and Eichler 2009). Our detailed analysis of the evolutionary history of the LRRC37 family provides some support for this model. The increase in copy number of the LRRC37 duplicon appears to have evolved from a tandem organization (e.g., lemur) to one that is increasingly dispersed in the hominid lineage. Although different copies have expanded in various primate lineages, the LRRC37A and LRRC37B copies have specifically expanded in humans and great apes and are preferential sites of both recurrent inversion polymorphisms (Zody et al. 2008) as well as rearrangements associated with disease (Bengesser et al. 2010). Our detailed expression and transcript analyses indicate both greater diversity and a broader expression profile in humans with marked increases in the cerebellum and thymus when compared to a testis-only expression profile in the mouse. While we can only speculate on the function of the gene, our data suggest that some members of this gene family localize to the plasma membrane and are secreted. One possibility may be that this protein family, similar to other studied proteins corresponding to core duplicons (e.g., TBC1D3), plays a role in regulating cell signaling, growth, and proliferation during development (Wainszelbaum et al. 2008; Stahl and Wainszelbaum 2009). It is also intriguing that our cell transfection studies suggest that LRRC37 may play a role in the formation of filipodia protrusions similar to what has been reported for other gene families that have expanded in the human lineage (Linardopoulou et al. 2007; Guerrier et al. 2009). A critical step forward will be accurately assessing genetic variation, including copy number changes of these and other duplicate genes with respect to human phenotypes (Sudmant et al. 2010; Alkan et al. 2011a).
Methods
Hybridization of high-density filters
Radioactive genomic hybridization of CHORI-253 (Sumatran orangutan), CHORI-250 (Indian rhesus macaque), CHORI-259 (common marmoset), and LBNL-2 (ring-tailed lemur) BAC libraries was carried out according to the protocol available at CHORI BACPAC resources (http://bacpac.chori.org/highdensity.htm). Anc409 and Anc419 probes were obtained by means of PCR amplification of human genomic DNA (Supplemental Table S7). BES (BAC end sequences) were repeat-masked (Smit et al. 1996) and mapped on human (hg18) and primate (ponAbe2, rheMac2, and calJac3) genomes using the BLAT tool at the UCSC Genome Browser (http://genome.ucsc.edu/cgibin/hgBlat).
Fluorescence in situ hybridization (FISH)
Metaphase spreads and interphase nuclei were prepared from a fibroblast cell line of Lemur catta and from lymphoblastoid cell lines of Homo sapiens, Pongo pygmaeus, Macaca mulatta, and Callithrix jacchus. FISH experiments were performed as previously described (Lichter et al. 1990). Digital images were obtained using a Leica epifluorescence microscope equipped with a cooled CCD camera. Fluorescence signals detected with Cy3, Fluorescein, and Cy5 filters and chromosomes and nuclei images detected with DAPI filter were recorded separately as grayscale images. Pseudocoloring and merging of images were performed using Adobe Photoshop software.
Phylogenetic and evolutionary analysis
A 2-kbp region from intron 1 and a 1.2-kbp region from intron 8 were PCR-amplified and sequenced from BAC clones representing different loci in each species. We reconstituted human, chimpanzee, orangutan, and macaque complete gene models based on RefSeq entries, genome assemblies, and BAC sequences. Multiple sequence alignments were performed using Clustal W (Thompson et al. 1994), and phylogenetic analyses were conducted in MEGA5 (Tamura et al. 2011). The evolutionary histories were inferred by using the maximum likelihood method based on the Kimura two-parameter model (Kimura 1980). A bootstrap test with 1000 replicates was conducted to evaluate the statistical significance of each node (Felsenstein 1985).
We aligned ancestral locus coding sequences using RevTrans (Wernersson and Pedersen 2003). Tests of selection (ω = dN/dS) were performed by maximum likelihood using PAML (Yang 1997) applying the Branch Model to calculate ω under different scenarios at different times during evolution. The likelihood ratio test (LRT) was used to assess the significance of different values of ω for different models.
Expression analyses
Mouse (wild-type mouse C57BL/6J) and macaque (Macaca nemestrina) tissue total RNA were extracted using the RNeasy Fibrous Tissue Midi Kit (Qiagen), following manufacturer recommendations. HeLa cells and G248 human lymphoblastoid cell line total RNA were extracted using the RNeasy Mini Kit (Qiagen), following manufacturer recommendations. Human tissue total RNA panels were from Clontech, Stratagene, and Capital Biosciences. 1 μg of isolated total RNA as well as of commercial human tissue total RNA (Clontech, Stratagene, and Capital Biosciences) was used for reverse transcription with Transcriptor High Fidelity cDNA Synthesis Kit (Roche) using Oligo(dT)18 primers. PCR reactions were performed with PCR Master (Roche), using standard PCR cycle conditions (initial denaturation of 3 min at 94°C; 35 cycles 30 s at 94°C, 30 s at 55°C, 1 min at 72°C; final extension 10 min at 72°C). The amplification with primers designed for UBE1 was used as a control. Quantitative gene expression profiling studies were performed using the LightCycler 480 SYBR Green System (Roche) with three replicates for each sample. The primers used are listed in Supplemental Table S7. CT values were elaborated by the comparative CT method (Livak and Schmittgen 2001). All PCR and qPCR products were sequence verified.
Sequencing
PCR products were sequenced using PCR primers or were cloned and sequenced using vector primers (Supplemental Table S7; Supplemental Note). DNA from BAC clones was extracted using the BAC Prep Protocol (Schein et al. 2004), and BAC ends were sequenced (if not available in the NCBI Trace Archive) using sp6_BAC and t7_BAC primers. Sequencing was performed on an ABI PRISM 3100 Genetic Analyzer using the BigDye Terminator v3.1 Chemistry (Applied Biosystems) according to manufacturer instructions.
Generation of FLAG-fusion constructs
Human LRRC37A coding sequence was amplified from HeLa cells cDNA using PfuTurbo DNA polymerase (Stratagene) using touchdown cycle conditions (initial denaturation 3 min at 94°C; 10 cycles 30 s at 94°C, 30 s at 65°C, 10 min at 72°C; 10 cycles 30 s at 94°C, 30 s at 60°C, 10 min at 72°C; 30 cycles 30 s at 94°C, 30 s at 55°C, 10 min at 72°C; final extension 10 min at 72°C) and subcloned in pFLAG-CMV-6c and pFLAG-CMV-5.1 mammalian expression vectors (Sigma) to generate FLAG-tagged constructs at amino and carboxyl terminus, respectively. The recombinant plasmids were transformed into chemically competent E. coli cells (Invitrogen). Recombinant plasmids were sequence verified using primers listed in Supplemental Table S7.
Generation of anti-LRRC37 antibodies
Rabbit polyclonal antibody against human LRRC37 proteins was produced by the Pocono Rabbit Farm and Laboratory Inc., using as immunogen the peptide ETLEDIQSSSLQQEA (position 228–242 of human LRRC37 protein).
Cell culture and transient transfection
HeLa cells were grown in DMEM High Glucose (Invitrogen) supplemented with 10% fetal bovine serum, 100 μg/mL streptomycin, and 100 μg/mL penicillin. FLAG-fusion constructs were extracted using the EndoFree Plasmid Maxi Kit (Qiagen), following manufacturer instructions. HeLa cells were transiently transfected with FuGENE HD Transfection Reagent (Roche), using a 6:2 ratio between reagent and DNA, according to manufacturer instructions.
Immunofluorescence
HeLa cells were seeded on coverslips and after 24 h were transfected. Transfected and nontransfected control cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min at room temperature (RT). Cells were washed with PBS and permeabilized with PBS containing 0.1% Saponin (Fluka) for 10 min at RT. Cells were then blocked with PBS, 0.1% Saponin, 3% bovine serum albumin (BSA) for 1 h at RT and probed with rabbit polyclonal anti-FLAG antibody (Abcam) overnight at 4°C. Cells were washed extensively with PBS, 0.1% Saponin, and then incubated with Alexa Fluor 488 goat anti-rabbit IgG (H+L) secondary antibody (Molecular Probes) for 30 min at RT and washed. Coverslips were mounted on slides with ProLong Gold Antifade Reagent (Molecular Probes) and observed under a Leica DMRXA2 epifluorescence microscope equipped with a cooled CCD camera (Princeton Instruments).
Western blot
Transfected and nontransfected control cells were collected 24 h after transfection and lysed in RIPA buffer (Sigma) supplemented with Complete Protease Inhibitor Cocktail Tablets (Roche). The lysates were clarified by centrifugation for 1 h at 13,000 × g. Conditioned media of the same cells were collected and ultracentrifuged at 200,000 × g for 1 h. Media were concentrated on Amicon Ultra-15 centrifugal filter devices (Millipore). Lysates and conditioned media were solubilized in SDS-PAGE sample buffer and separated by SDS-PAGE gel (4%–15%). Gels were electroblotted to Hybond ECL nitrocellulose membranes (Amersham Biosciences), which were blocked and probed with rabbit polyclonal anti-FLAG (Abcam) and anti-LRRC37 antibodies. Goat anti-rabbit HRP conjugated antibodies were from GE Healthcare. Protein expression levels were corrected for whole protein loading determined by staining membrane with Red Ponceau.
Data access
The sequence data from this study have been submitted to GenBank. This includes BAC clone sequencing of LRRC37 loci: orangutan AC206276.2, AC216100.2, AC210931.1, AC212980.2, AC212589.2, AC210533.4, and AC206550.4; macaque AC239129.2, AC239243.2, AC241896.2, AC240582.3, AC191449.5, AC241829.2, AC241249.2, AC242077.2, AC245843.1, and AC143065.2; marmoset AC239345.3, AC244374.2, and AC241431.2; lemur AC234058.2. In addition, sequenced PCR products used in the phylogenetic analysis are available from intron 1 (JQ582363-JQ582376) and intron 8 (JQ582377-JQ582394); sequenced RT-PCR products of the LRR region are available (JQ790561-JQ790788). BAC end sequences have been submitted to the Genome Survey Sequence (GSS) division (http://www.ncbi.nlm.nih.gov/nucgss) (Supplemental Table S8).
Acknowledgments
We thank NIH Intramural Sequencing Center (NISC) for BAC clone sequence data generation and T. Brown for manuscript editing. This work was supported by National Institutes of Health (NIH) grants HG002385 and GM058815 to E.E.E. and in part by the Intramural Research Program of the National Human Genome Research Institute, NIH. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.138842.112.
References
- Alkan C, Coe BP, Eichler EE 2011a. Genome structural variation discovery and genotyping. Nat Rev Genet 12: 363–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Sajjadian S, Eichler EE 2011b. Limitations of next-generation genome sequence assembly. Nat Methods 8: 61–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE 2006. Primate segmental duplications: Crucibles of evolution, diversity and disease. Nat Rev Genet 7: 552–564 [DOI] [PubMed] [Google Scholar]
- Bekpen C, Tastekin I, Siswara P, Akdis CA, Eichler EE 2012. Primate segmental duplication creates novel promoters for the LRRC37 gene family within the 17q21.31 inversion polymorphism region. Genome Res 22: 1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengesser K, Cooper DN, Steinmann K, Kluwe L, Chuzhanova NA, Wimmer K, Tatagiba M, Tinschert S, Mautner VF, Kehrer-Sawatzki H 2010. A novel third type of recurrent NF1 microdeletion mediated by nonallelic homologous recombination between LRRC37B-containing low-copy repeats in 17q11.2. Hum Mutat 31: 742–751 [DOI] [PubMed] [Google Scholar]
- Bosch N, Caceres M, Cardone MF, Carreras A, Ballana E, Rocchi M, Armengol L, Estivill X 2007. Characterization and evolution of the novel gene family FAM90A in primates originated by multiple duplication and rearrangement events. Hum Mol Genet 16: 2572–2582 [DOI] [PubMed] [Google Scholar]
- Bueno DF, Sunaga DY, Kobayashi GS, Aguena M, Raposo-Amaral CE, Masotti C, Cruz LA, Pearson PL, Passos-Bueno MR 2011. Human stem cell cultures from cleft lip/palate patients show enrichment of transcripts involved in extracellular matrix modeling by comparison to controls. Stem Cell Rev 7: 446–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Aulia S, Li L, Tang BL 2006. AMIGO and friends: An emerging family of brain-enriched, neuronal growth modulating, type I transmembrane proteins with leucine-rich repeats (LRR) and cell adhesion molecule motifs. Brain Res Rev 51: 265–274 [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, von Mering C, Suyama M, Harrington ED, Izaurralde E, Bork P 2005. Complex genomic rearrangements lead to novel primate gene function. Genome Res 15: 343–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Rademakers R, Gijselinck I, van der Zee J, Dermaut B, de Pooter T, de Rijk P, Del-Favero J, van Broeckhoven C 2005. Genomic architecture of human 17q21 linked to frontotemporal dementia uncovers a highly homologous family of low-copy repeats in the τ region. Hum Mol Genet 14: 1753–1762 [DOI] [PubMed] [Google Scholar]
- De Clercq N, Hemschoote K, Devos A, Peeters B, Heyns W, Rombauts W 1992. The 4.4-kilodalton proline-rich polypeptides of the rat ventral prostate are the proteolytic products of a 637-kilodalton protein displaying highly repetitive sequences and encoded in a single exon. J Biol Chem 267: 9884–9894 [PubMed] [Google Scholar]
- Dolan J, Walshe K, Alsbury S, Hokamp K, O'Keeffe S, Okafuji T, Miller SF, Tear G, Mitchell KJ 2007. The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genomics 8: 320 doi: 10.1186/1471-2164-8-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE 2001. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet 17: 661–669 [DOI] [PubMed] [Google Scholar]
- Felsenstein J 1985. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Gentry LE, Twardzik DR, Lim GJ, Ranchalis JE, Lee DC 1987. Expression and characterization of transforming growth factor α precursor protein in transfected mammalian cells. Mol Cell Biol 7: 1585–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F 2009. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138: 990–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemschoote K, Peeters B, Dirckx L, Claessens F, De Clercq N, Heyns W, Winderickx J, Bannwarth W, Rombauts W 1988. A single 12.5-kilobase androgen-regulated mRNA encoding multiple proline-rich polypeptides in the ventral prostate of the rat. J Biol Chem 263: 19159–19165 [PubMed] [Google Scholar]
- Heyns W, Bossyns D, Peeters B, Rombauts W 1982. Study of a proline-rich polypeptide bound to the prostatic binding protein of rat ventral prostate. J Biol Chem 257: 7407–7413 [PubMed] [Google Scholar]
- Huang EJ, Nocka KH, Buck J, Besmer P 1992. Differential expression and processing of two cell associated forms of the kit-ligand: KL-1 and KL-2. Mol Biol Cell 3: 349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Tang H, Ventura M, Cardone MF, Marques-Bonet T, She X, Pevzner PA, Eichler EE 2007. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet 39: 1361–1368 [DOI] [PubMed] [Google Scholar]
- Jin H, Selfe J, Whitehouse C, Morris JR, Solomon E, Roberts RG 2004. Structural evolution of the BRCA1 genomic region in primates. Genomics 84: 1071–1082 [DOI] [PubMed] [Google Scholar]
- Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE 2001. Positive selection of a gene family during the emergence of humans and African apes. Nature 413: 514–519 [DOI] [PubMed] [Google Scholar]
- Kimura M 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111–120 [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV 2001. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol 11: 725–732 [DOI] [PubMed] [Google Scholar]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC 1990. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science 247: 64–69 [DOI] [PubMed] [Google Scholar]
- Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ 2007. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet 3: e237 doi: 10.1371/journal.pgen.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GE, Ventura M, Cellamare A, Chen L, Cheng Z, Zhu B, Li C, Song J, Eichler EE 2009. Analysis of recent segmental duplications in the bovine genome. BMC Genomics 10: 571 doi: 10.1186/1471-2164-10-571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, et al. 2011. Comparative and demographic analysis of orang-utan genomes. Nature 469: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Eichler EE 2009. The evolution of human segmental duplications and the core duplicon hypothesis. Cold Spring Harb Symp Quant Biol 74: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. 2009. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457: 877–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero JC, Yuste L, Diaz-Rodriguez E, Esparis-Ogando A, Pandiella A 2002. Mitogen-activated protein kinase-dependent and -independent routes control shedding of transmembrane growth factors through multiple secretases. Biochem J 363: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger T, Brunner F, Kemmerling B, Piater L 2004. Innate immunity in plants and animals: Striking similarities and obvious differences. Immunol Rev 198: 249–266 [DOI] [PubMed] [Google Scholar]
- Paulding CA, Ruvolo M, Haber DA 2003. The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc Natl Acad Sci 100: 2507–2511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff GJ, Sikela JM 2006. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science 313: 1304–1307 [DOI] [PubMed] [Google Scholar]
- Rettenmier CW, Roussel MF 1988. Differential processing of colony-stimulating factor 1 precursors encoded by two human cDNAs. Mol Cell Biol 8: 5026–5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein J, Kucaba T, Sekhon M, Smailus D, Waterston R, Marra M 2004. High-throughput BAC fingerprinting. Methods Mol Biol 255: 143–156 [DOI] [PubMed] [Google Scholar]
- She X, Jiang Z, Clark RA, Liu G, Cheng Z, Tuzun E, Church DM, Sutton G, Halpern AL, Eichler EE 2004. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431: 927–930 [DOI] [PubMed] [Google Scholar]
- She X, Liu G, Ventura M, Zhao S, Misceo D, Roberto R, Cardone MF, Rocchi M, Green ED, Archidiacano N, et al. 2006. A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications. Genome Res 16: 576–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Cheng Z, Zollner S, Church DM, Eichler EE 2008. Mouse segmental duplication and copy number variation. Nat Genet 40: 909–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shing Y, Christofori G, Hanahan D, Ono Y, Sasada R, Igarashi K, Folkman J 1993. Betacellulin: A mitogen from pancreatic β cell tumors. Science 259: 1604–1607 [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P 1996–2010. RepeatMasker Open-3.2.9. http://www.repeatmasker.org
- Stahl PD, Wainszelbaum MJ 2009. Human-specific genes may offer a unique window into human cell signaling. Sci Signal 2: pe59 doi: 10.1126/scisignal.289pe59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A, Ingason A, Gudnadottir VG, et al. 2005. A common inversion under selection in Europeans. Nat Genet 37: 129–137 [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Eichler EE 2010. Diversity of human copy number variation and multicopy genes. Science 330: 641–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E, Bailey JA, Eichler EE 2004. Recent segmental duplications in the working draft assembly of the brown Norway rat. Genome Res 14: 493–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K, Van Roy N, Staes K, Speleman F, van Roy F 2005. A novel gene family NBPF: Intricate structure generated by gene duplications during primate evolution. Mol Biol Evol 22: 2265–2274 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Andries V, van Roy F 2009. The NBPF1 promoter has been recruited from the unrelated EVI5 gene before simian radiation. Mol Biol Evol 26: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Wainszelbaum MJ, Charron AJ, Kong C, Kirkpatrick DS, Srikanth P, Barbieri MA, Gygi SP, Stahl PD 2008. The hominoid-specific oncogene TBC1D3 activates Ras and modulates epidermal growth factor receptor signaling and trafficking. J Biol Chem 283: 13233–13242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernersson R, Pedersen AG 2003. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res 31: 3537–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Koblansky AA, Ghosh S 2006. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol 22: 409–437 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wang W, Peltz SW 2001. Curbing the nonsense: The activation and regulation of mRNA surveillance. Genes Dev 15: 2781–2785 [DOI] [PubMed] [Google Scholar]
- Yang Z 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK, et al. 2006. DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature 440: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zody MC, Jiang Z, Fung HC, Antonacci F, Hillier LW, Cardone MF, Graves TA, Kidd JM, Cheng Z, Abouelleil A, et al. 2008. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat Genet 40: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]