Abstract
The basis of bactericidal versus bacteriostatic action of antibiotics and the mechanism of bacterial cell death are largely unknown. Related to this important issue is the essential invulnerability to killing of persisters: cells forming a small subpopulation largely responsible for the recalcitrance of biofilms to chemotherapy. To learn whether death is accompanied by changes in expression of particular genes, we compared transcription profiles of log-phase Escherichia coli treated with bactericidal concentrations of two unrelated antibiotics: ampicillin and ofloxacin. Massive changes in transcription profile were observed in response to either agent, and there was a significant overlap in genes whose transcription was affected. A small group of mostly uncharacterized genes was induced and a much larger set was transcriptionally repressed by both antibiotics. Among the repressed genes were those required for flagellar synthesis, energy metabolism, transport of small molecules, and protein synthesis.
Bacteria die of various causes. Killing by antibiotics is of particular interest to us, yet little is known about the underlying mechanism. The reason for this is the apparent obviousness of the problem. Antibiotics shut down important functions, and, not surprisingly, sooner or later a cell will die. However, there are important and unexplained variations in the timing of death. Antibiotics can be bactericidal or bacteriostatic, yet in many cases it is impossible to assign them to one of these groups on the basis of what we know about their mechanism of action. Chloramphenicol, for example, inhibits protein synthesis and is bacteriostatic, while aminoglycosides inhibit protein synthesis and are bactericidal. β-Lactams inhibit peptidoglycan synthesis, which for unknown reasons (3) elicits rapid autolysis. The picture of death is further complicated by the presence of persister cells, which constitute a small part of the population (10−6 to 10−2) and which are essentially invulnerable to killing by bactericidal antibiotics (for reviews see references 24 and 25). Persisters apparently do not express specific resistance mechanisms, since they do not grow in the presence of antibiotics, and they are not mutants. Our recent research suggests that persisters are largely responsible for the very high resistance of biofilms and stationary cultures to killing by antimicrobials (8, 37). Taken together, these facts suggest that death in bacteria might be a controlled event. Specifically, we have proposed that bactericidal antibiotics trigger a programmed cell death (PCD) response in bacteria and that persisters represent cells with disabled PCD (24). PCD in response to damage by fungicidal agents has been recently documented in unicellular Saccharomyces cerevisiae (27).
In Escherichia coli mutations in the hipAB locus have been reported to significantly increase the production of persisters without changing the MIC (5, 6, 13, 28, 29, 33, 42). Persisters generated due to these hip mutations are resistant to killing by several bactericidal factors. Among these factors are two different groups of antibiotics that affect unrelated targets: inhibitors of cell wall synthesis, such as ampicillin, and fluoroquinolones, which bind topoisomerases and cause DNA damage (13, 33, 42). In this study, we chose representatives of these two groups of drugs, ampicillin and ofloxacin, to learn whether they induce common changes in gene expression that might trigger a common process in dying bacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions
E. coli HM21 dapA6 zde-264::Tn10, used in this study, is a derivative of E. coli K-12 (28). Cells were cultured in Luria-Bertani broth supplemented with diaminopimelic acid (75 μg/ml). For growth of the log-phase culture, overnight cultures were diluted 1,000-fold in fresh media and bacteria were cultured for 3 h to a density of approximately 108 CFU/ml.
Antibiotic treatment
For killing curve experiments, ampicillin (100 μg/ml) or ofloxacin (5 μg/ml) was added to logarithmically growing cultures. Samples were taken over time, diluted, and plated for determination of cell count. All incubations were done in culture tubes, with aeration at 37°C. Ofloxacin-challenged cells were pelleted and washed with growth media before dilution and plating to avoid the carryover of ofloxacin. For DNA array experiments, log phase bacteria were incubated in the same conditions, for 30 min with ampicillin and for 60 min with ofloxacin.
RNA isolation and labeling
Bacteria (20 ml of culture for ofloxacin and untreated controls and 100 ml for ampicillin) were pelleted and lysed in 6 ml of T&C lysis solution (Epicenter Technologies). Total RNA was isolated with a MasterPure RNA purification kit (Epicenter Technologies) according to the manufacturer's instructions.
Enrichment, fragmentation, and biotinylation of mRNA were carried out as described in the GeneChip expression analysis technical manual (Affymetrix) (31). Briefly, oligonucleotide primers complementary to either 16S or 23S rRNA sequences were annealed to rRNA, and ribosomal cDNA was synthesized. In vitro-synthesized control RNAs (Bacillus subtilis dapB, lysA, trpED, and pheB) were added before enrichment. The rRNA strand of the heteroduplex was selectively degraded with RNase H, and cDNA was removed with DNase I. RNA thus enriched for mRNA was then purified and incubated at 95°C in Mg2+-containing T4 polynucleotide kinase reaction buffer for fragmentation. RNA fragments were terminally thiolated with T4 polynucleotide kinase and γ-S-ATP. Finally, biotin was cross-linked to thiol groups with (+)-biotinyl-iodoacetamidyl-3,6-dioxaoctanediamine (Pierce Chemical), and, after purification, labeled RNA was hybridized to DNA arrays. Approximately 30 μg of enriched RNA and 2 to 4 μg of labeled product were obtained from 100 μg of total RNA.
Hybridization with GeneChip arrays, staining, and scanning
The Affymetrix high-density array of the E. coli genome has been described (31, 34). Briefly, oligonucleotides are arranged in probe pairs; one is complementary to the target sequence, and the other contains a single mismatch at the central position and serves as a control for nonspecific hybridization. On the average, 15 different oligonucleotides complementary to different parts of a given gene are used to identify a gene transcript. Probe pairs complementary to a single putative transcript are organized into probe sets. An array contains probe sets for 4,403 (putative) E. coli genes, including 4,290 open reading frames, and for all intergenic regions of 40 bp or longer, as well as probes complementary to control spike transcripts and biotinylated control oligonucleotides.
Biotinylated RNA was hybridized to the array in 100 mM MES (morpholineethanesulfonic acid) buffer, pH 6.6, containing 1 M NaCl, 20 mM EDTA, 0.01% Tween 20, 100 μg of herring sperm DNA/ml, 500 μg of bovine serum albumin (BSA)/ml, and 0.5 nM control biotin-oligonucleotide 948. Hybridization was carried out at 45°C for 16 h on a rotary mixer at 60 rpm. Arrays were washed and stained according to a ProkGE-WS2 fluidics protocol using the Affymetrix fluidics station. After hybridization, arrays were washed with wash buffer A at 25°C and wash buffer B at 45°C (GeneChip expression analysis technical manual). After the posthybridization washes, arrays were incubated for 10 min at 25°C with streptavidin (10 μg/ml) in solution staining buffer containing 100 mM MES, pH 6.6, 1 M NaCl, 0.05% Tween 20, and 2 mg of BSA/ml and washed in buffer A at 30°C. Thereafter, arrays were incubated with goat immunoglobulin G (0.1 mg/ml)-biotin antistreptavidin (5 μg/ml) and fluorescently stained with streptavidin-phycoerythrin. Both incubations were for 10 min at 25°C in staining buffer. Finally, arrays were washed with wash buffer A and scanned twice at 570 nm with a resolution of 3 μm with an Affymetrix confocal laser scanner. Three independent experiments were carried out with unchallenged cells, and three were carried out with each antibiotic.
Data analysis
Arrays were analyzed with the Affymetrix Microarray Suite, version 5.0, and Data Mining Tool, version 3.0, software. All gene probe sets of a given array were scaled to a target signal of 500. Each experimental scaled probe set for a given gene was then compared to the scaled control probe set for the same gene to determine the relative change in the expression level for each transcript. Significantly increased and decreased transcripts (P ≤ 0.05) were determined with the aid of Microarray Suite, version 5.0, software. In this manner, data were obtained from pairwise comparisons of each of the three experimental arrays with each of the three control arrays. Transcripts that had been significantly changed in at least seven of nine comparisons and had average signal log2 ratios bigger than 1 were listed as having increases or decreases (http://www.atsweb.neu.edu/lewislab/microarray1.htm).
RESULTS AND DISCUSSION
Antibiotics induce massive changes in transcription profile
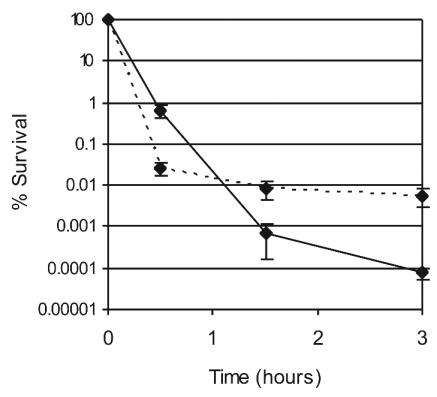
Our aim was to obtain a gene profile that would correspond to death, and therefore we used bactericidal antibiotics at levels that result in rapid killing (26, 35) (Fig. 1). Two antibiotics with unrelated mechanisms of action, ofloxacin (targeting DNA gyrase and topoisomerase), and ampicillin, inhibiting cell wall synthesis, were chosen for this study. In a recent report of genome profiling of Haemophilus influenzae in response to the fluoroquinolone ciprofloxacin (14), a 60-min incubation produced reliable results, while a shorter (30-min) incubation gave inconsistent data. A 1-h incubation was therefore chosen for ofloxacin in this study. The same incubation time resulted in a substantial lysis in the case of ampicillin, and treatment in this case was decreased to 30 min (results among independent trials gave consistent data in this case; see below).
FIG. 1.
Killing of E. coli by antibiotics. Log-phase bacteria were incubated in the presence of ampicillin (100 μg/ml; solid line) or ofloxacin (5 μg/ml; dotted line), serially diluted, and plated onto solid media for overnight incubation and estimation of survival. The values are averages obtained from three independent samples; error bars, standard deviations.
RNA was extracted after the bactericidal treatment. Since ampicillin produced substantial lysis, we increased the culture volume fivefold to obtain the same amount of RNA as in control experiments. Sharp rRNA bands and the lack of any additional bands on denaturing gel electrophoresis indicated that massive nonspecific degradation of RNA has not occurred by the time the cells were harvested (data not shown). The levels of most mRNA messages in challenged cells remained unchanged compared to the control (see below), further indicating the absence of nonspecific RNA degradation.
Isolated total RNA was enriched for mRNA by degrading rRNA (31), biotinylated, and hybridized to E. coli high-density GeneChip arrays (Affymetrix). We performed three independent experiments with both antibiotics and three control experiments with unchallenged cells. The hybridization signal from each challenged sample was compared to the baseline of each control experiment. Thus, nine comparisons were made for both antibiotics, and significant changes in transcript quantity were identified at P values ≤0.05 by the Affymetrix Microarray Suite for each experiment. Transcripts with average signal log2 ratios larger than 1 (i.e., more than twofold changes) in at least seven of nine comparisons were listed as having considerable decreases or increases and are summarized in Fig. 2. Complete lists of these genes are available at our website (http://www.atsweb.neu.edu/lewislab/microarray1.htm).
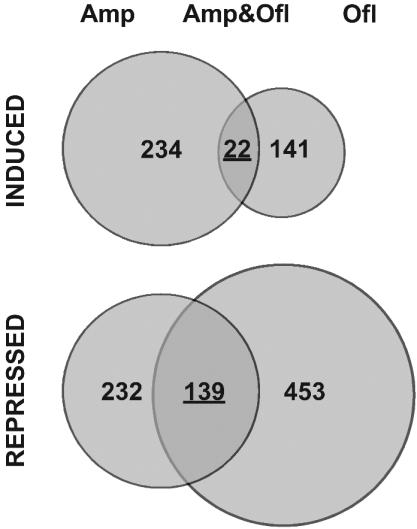
FIG. 2.
Summary of changes in transcription profile of E. coli in response to ampicillin and ofloxacin. The numbers in each circle show the number of genes either induced or repressed by these drugs twofold or more. The numbers of genes in overlap are underlined.
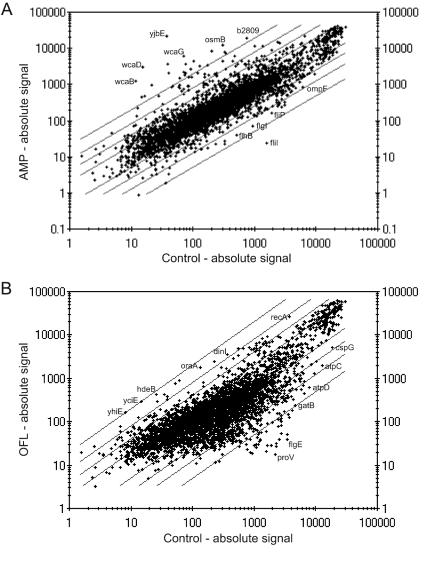
Hundreds of genes changed their level of expression in response to bactericidal treatment with antibiotics. Ampicillin induced 234 genes and repressed 232 genes (Fig. 3A); ofloxacin induced 141 genes and repressed 453 genes (Fig. 3B). Some of the observed changes were dramatic, for example, an open reading frame of unknown function, yjbE, was induced by ampicillin more than 600-fold.
FIG. 3.
Transcriptional changes in response to ampicillin (AMP; A) and ofloxacin (OFL; B). Shown is a comparison of the averaged absolute signals of control (x axis) and antibiotic-treated (y axis) cells. Diagonal lines indicate 2-, 4-, 8-, and 20-fold changes. Some of the most affected genes are shown.
Transcriptional changes caused by both antibiotics
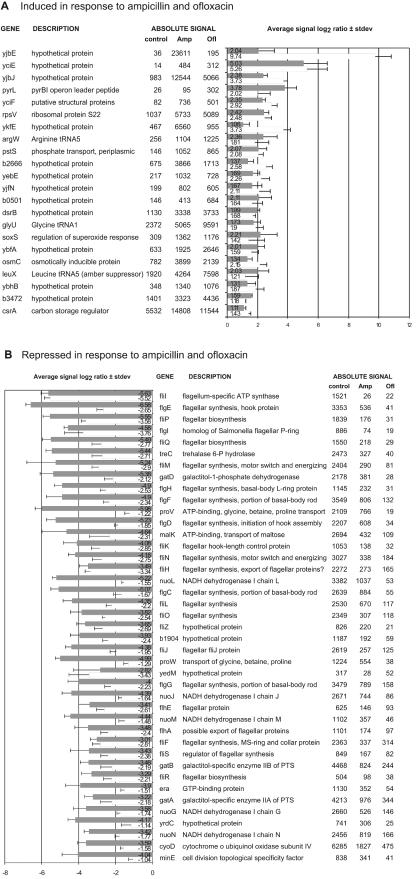
We were primarily interested in learning whether the two unrelated antibiotics would affect expression of the same genes, which could point to a possible link to cell death. Indeed, a significant number of genes (22) were induced, and an even larger number (139) were repressed by both antibiotics (Fig. 4). Most of the transcriptionally activated genes (Fig. 4A) code for (hypothetical) proteins of unknown function. Among these is yjbJ, which was previously found to be one of the most strongly induced genes in stationary phase (34), and its protein product was reported to be the most abundant stationary-phase protein (26). Five genes coding for known proteins were upregulated. One of them, rpsV, codes for ribosomal protein S22, of unknown function, and is also specifically upregulated in stationary phase (34, 41). The others encode stress response and regulatory proteins PstS, SoxS, OsmC, and CsrA. Three tRNA genes were induced, one of which was argW, the gene of arginine tRNA5, which is normally expressed at a very low level. This tRNA decodes the extremely rare AGG arginine codons, which apparently have a regulatory role (12).
FIG. 4.
Genes affected by both ampicillin (Amp) and ofloxacin (Ofl). Shown is the overlap in genes which were induced (average signal log2 ratio > 1) or repressed (signal log2 ratio < 1) in response to either drug twofold or more in at least seven of the nine comparisons of three challenged samples versus three controls. Gray bars, ofloxacin challenge; white bars, ampicillin challenge. Error bars indicate standard deviations (stdev). Absolute signals are averages of three experiments. (A) Induced genes. (B) Most strongly repressed genes.
Among the genes that were repressed by both ampicillin and ofloxacin (Fig. 4B; http://www.atsweb.neu.edu/lewislab/microarray1.htm) are numerous genes required for flagellar synthesis (39). Most of the other genes repressed by ampicillin and ofloxacin are related to energy metabolism, transport of small molecules, protein synthesis, or cell surface properties. Among the repressed genes were members of the large operons involved in oxidative phosphorylation: nuoCEFGHIJKLMN (NADH dehydrogenase), atpA through atpH (membrane-bound ATP synthase), and cyoABCD (cytochrome o ubiquinol oxidase subunits). Both drugs repressed genes encoding ATP-dependent transport systems for maltose (malE, malK, lamB, malM, and positive regulator malT) and for glycine betaine and proline (proVWX). Several other transport protein-encoding genes (mglC, tsx, oppF, nupC, msbA, exbD, and potB) were also repressed. In addition, both antibiotics repressed genes encoding outer membrane protein OmpF as well as genes gatABC, manXYZ, and treB, encoding phosphotransferase system enzymes.
Genes of several cell division regulators (gidA [glucose-inhibited division]), minE, and minD), replicative DNA helicase (dnaB), DNA methylase (hsdM), and nucleases cleaving RNA (rnb [RNase II for mRNA degradation] and rnc [RNase III for double-stranded RNA] or damaged DNA (nth encoding endonuclease III specific for apurinic and/or apyrimidinic sites) were downregulated by both antibiotics. Also, important regulatory genes cyaA, which encodes adenylate cyclase, responsible for cyclic AMP synthesis, and spoT, which is responsible for ppGpp synthesis and hydrolysis (30), were downregulated.
In general, it appears that both antibiotics lead to a decrease in the synthesis of nonessential proteins, such as flagellin, to a decrease in metabolic activity, and to suppression of cell division. It is premature to conclude from these data whether the expression profile points to a possible PCD mechanism. For example, we did not see changes in the genes coding for toxin-antitoxin proteins that have been implicated in possible PCD in E. coli, such as mazEF (32). This is perhaps not surprising, since mazEF effects on bacterial death are modest and observed only with some antibiotics that inhibit transcription or translation (32). A follow-up examination of strains with the genes affected by the two antibiotics either deleted or overexpressed will be necessary to evaluate their possible role in cell death.
Next, we consider changes that occur in response to individual antibiotics. Such data add to our understanding of bacterial responses to inhibition of two important cell functions: cell wall and DNA synthesis.
Ampicillin induces synthesis of colanic acid genes and a general stress response
Of all the genes induced by ampicillin, 146 or 62% are open reading frames of unknown function (http://www.atsweb.neu.edu/lewislab/microarray1.htm). Among the functionally characterized genes, those encoding elements of the colanic acid biosynthetic pathway showed the highest level of induction. Colanic acid is the E. coli capsular exopolysaccharide which helps bacteria resist desiccation and heat and acid shock; it is essential for biofilm formation (10) and is produced in response to hyperosmotic shock (35). All 19 genes which constitute the contiguous colanic acid gene cluster (38) were induced, as well as the transcriptional activator of colanic acid synthesis, rcsA. The regulatory gene rcsA belongs to a group of stress-inducible genes that are repressed by the H-NS protein under favorable growth conditions. In response to environmental stress (changing pH or osmolarity, cold shock, or entry into stationary phase [2]), translation efficiency and the half-life of H-NS mRNA are specifically decreased by the regulatory DsrA RNA (23). In addition, increased levels of DsrA induce another group of stress-related genes by activating translation of rpoS mRNA (23) and, as a result, a general stress response. We observed an increase in the level of DsrA following ampicillin treatment. Transcription of the hfq gene, required for regulation of both RpoS and H-NS levels by DsrA (36), was induced by ampicillin about twofold. As expected from the above analysis, a number of genes controlled by σs were induced by ampicillin. Among these were the genes osmB, osmY, osmE, and osmC, which are both stationary state and hyperosmotically inducible, as well as otsA and otsB, encoding enzymes for synthesis of the hyperosmotic protectant trehalose (40), which is required also for stationary-phase thermotolerance (16) and survival of cold shock (18). At the same time, ampicillin stimulated transcription of treF, encoding trehalase. In addition, ampicillin increased expression of galU, coding for the enzyme making UDP-glucose, which is intermediate for the synthesis of colanic acid, trehalose, lipopolysaccharide, and membrane-derived oligosaccharides and which plays a regulatory role in expression of the σs regulon (7). Ampicillin stimulated transcription of σs-dependent katE, encoding catalase, glgS, stimulating glycogen biosynthesis, bolA, controlling cell division and the shape of stationary-phase bacteria (1), the carbon storage regulator csrA, and the dps gene, encoding a stationary-phase-specific DNA protecting protein (17).
Additional stress responses induced by ampicillin
Another group of stress response genes transcriptionally activated by ampicillin were σ54 dependent. These are heat shock genes ibpA and ibpB, coding for chaperones, and psp genes, coding for phage shock proteins PspA, PspB, PspC, and PspD, which are induced in bacteria infected by filamentous phage and under several other stress conditions (21).
Ampicillin also induced heat shock genes htpX and ddg. Apart from inducing catalase, ampicillin increased transcription of sodC, encoding the periplasmic superoxide dismutase. It also induced the global regulators soxS (oxidative stress response) and marA (multiple antibiotic resistance). However, genes under the control of these regulators were not significantly affected.
Genes with a possible role in ampicillin-induced cell lysis
Since the target of ampicillin and the murein sacculus are both located in the periplasm, changes in this compartment may have a direct relation to the bactericidal effect of the drug. Ampicillin induced several genes that encode periplasmic proteins, including Spy, which is synthesized only in spheroplasts (15), and periplasmic serine protease HtrA (20). An induced gene possibly related to lysis is nlpD. It codes for an outer membrane lipoprotein which leads to abnormalities of septation and lysis when artificially overexpressed in E. coli (22). This gene is located upstream of rpoS but is not induced upon entry into stationary phase.
Ampicillin induced expression of mltC, encoding a membrane-bound lytic murein transglycosylase (Fig. 2) (11). mltC is transcribed as part of an operon together with mutY, which was also induced by ampicillin. Along with many genes induced in hyperosmotic conditions, ampicillin induced transcription of mscL, encoding a mechanosensitive channel that opens in response to hypo-osmotic shock. MscL has been shown to release certain cytoplasmic proteins (DnaK and EF-Tu) along with osmoprotectants (4) and may participate in cell lysis or release of autolysins into the periplasm.
Recently, the effect of vancomycin, another antibiotic inhibiting cell wall synthesis, in Bacillus subtilis was studied by transcriptional profiling (9). In that study, short incubation times (3 and 10 min) but relatively high doses of the antibiotic (10 times the MIC) produced changes significantly overlapping with those induced by ampicillin in E. coli. Genes encoding phage shock proteins (part of the σW regulon in B. subtilis), 33 genes of the general stress response regulon (σB regulon), and htrA, encoding “periplasmic” protease Do, were induced by vancomycin.
Genes repressed by ampicillin
A complete list of repressed genes is given in http://www.atsweb.neu.edu/lewislab/microarray1.htm. Apart from the genes that were repressed by both ampicillin and ofloxacin, 102 genes were repressed by ampicillin alone. Of these, genes involved in lipopolysaccharide synthesis are perhaps the most directly connected to ampicillin action. Both rfa genes, required for lipopolysaccharide core biosynthesis, and rfb genes, required for O-antigen synthesis, were repressed.
Ofloxacin induces SOS, the phosphate regulon, and other stress regulons
As expected, several genes of the SOS regulon were transcriptionally induced: recA and recN; sulA, encoding a cell division inhibitor; dinI, encoding an inhibitor of RecA coprotease activity and UmuD processing (43, 44); uvrB, encoding an excision repair protein; and umuD, encoding a subunit of translesion repair DNA polymerase (a complete list of genes is given at http://www.atsweb.neu.edu/lewislab/microarray1.htm). In addition to these genes, dinD, sbmC, oraA, and ssb, encoding the single-stranded DNA binding protein, and the SOS regulon genes of unknown function yebGF were induced.
Upregulation of the genes of the SOS regulon in response to ciprofloxacin, another fluoroquinolone, in H. influenzae (14) and in response to mitomycin in E. coli (19) was previously reported by genome-wide transcription profiling. While sublethal doses of ciprofloxacin induced only a small group of genes, most of them related to DNA repair, prolonged incubation with lethal doses resulted in massive changes, with a total of 140 genes induced or repressed. Except for the activation of the SOS regulon, only a few of these responses, however, overlap with those that we see in E. coli (induction of peptidyl-prolyl cis-trans isomerase B and repression of RNA polymerase subunit σE). Mitomycin C causes DNA damage by a different mechanism but also produced massive changes of transcription in E. coli after prolonged exposure (19). Apart from the induction of the SOS response, these changes did not have much in common with the response of E. coli to ofloxacin (it is not clear whether mitomycin treatment was lethal in the above-cited study).
Other genes induced by ofloxacin
Ofloxacin induced genes of several other stress regulons. Apart from soxS and csrA, which were also induced by ampicillin, ofloxacin specifically induced pspE, encoding a phage shock protein, ahpC, required for detoxification of hydroperoxides, sodA and sodB, encoding superoxide dismutases, and cspD, encoding a cold shock protein.
rfaZYJ genes involved in lipopolysaccharide biosynthesis and lpp, encoding murein lipoprotein, were induced; these genes could have a function in decreasing cell envelope permeability. Note that peptidoglycan synthesis was repressed (see below). As many as 53 genes (37%) of those induced by ofloxacin had no identified function.
Genes repressed by ofloxacin
Ofloxacin repressed a large number of genes of various function (http://www.atsweb.neu.edu/lewislab/microarray1.htm). Some of these are regulators of important cellular functions. Apart from the min genes, repressed by both ofloxacin and ampicillin, ofloxacin specifically repressed additional genes related to cell division: sdiA, encoding a regulator of ftsQAZ gene cluster; the master regulators of cell division minC and ftsZ; and ftsQ, ftsW, ftsZ, ftsJ, zipA, and ftsX. Downregulated were several genes related to peptidoglycan synthesis (murA, murE, murC, prc, and mtgA), genes encoding processive proteases (lon, hflX, and hflC) responsible for degrading denatured and/or intrinsically unstable proteins, and genes encoding protein translocase subunits (secD and secA), which are linked to cell growth and division.
Response to bactericidal antibiotics depends on drug concentration
After this study was submitted, a paper examining changes in global gene expression of E. coli in response to four bactericidal antibiotics (ampicillin, norfloxacin, rifampin, and kanamycin) was published. In that study, considerably lower drug concentrations were used, and it is not clear whether expression profiles were obtained from live or dead cells. Ten genes were repressed by all four antibiotics; among those, ompF, malK, gatB, and frdB are also repressed by bactericidal concentrations of ampicillin and ofloxacin. The authors found some similarity between changes induced by ampicillin and rifampin but practically no similarities between ampicillin- and norfloxacin-specific profiles. Similar to our results for ofloxacin incubation, the study reports induction of SOS regulon genes (recA, recN, sulA, ssb, uvrB, sbmC, oraA, and yebF) by norfloxacin. Two genes coding for ribosomal proteins, rpmG and rpsU, were also induced by both ofloxacin and norfloxacin. Norfloxacin repressed transcription of a group of genes related mostly to metabolism and transport that are repressed by both drugs in our study (atpA, atpD, atpG, cyoA, cyoC, flgH, gatC, lamB, malK, malM, manX, nuoHILMN, ompF, pykA, and treBC). Rifampin (at four times the MIC) caused repression of genes minE and minD, encoding cell division regulators, as did a bactericidal concentration of ampicillin in our study. Interestingly, the authors report that incubation with higher (four times the MIC) concentrations of kanamycin causes strong repression of the same set of Fnr-repressible genes (nuoEFGHIJKLM, cyoCD, sucA, and sucD) and flagellar gene flhA, which we see downregulated by both ampicillin and ofloxacin. yebE, induced by ampicillin and ofloxacin at highly bactericidal concentrations, was induced by kanamycin and norfloxacin. Kanamycin at four times the MIC induced several genes specifically induced by ampicillin in our study: pspABC, hfq, miaA, grpE, and htrA. The transcriptional profile induced by kanamycin at four times the MIC was more similar to what we see at higher, lytic concentrations of ampicillin than the transcription profile induced by low concentrations of ampicillin. Most of the similarities between our results and the published results listed above occur only at the highest (four times the MIC or higher) concentrations of antibiotics tested by the authors. At lower concentrations (one-half to two times the MIC) expression of these genes does not change.
The present study provides the first insight into the transcription profile of cells dying in response to two very different bactericidal agents. The fact that a cell wall inhibitor and an inhibitor of DNA gyrase and topoisomerase similarly affect transcription of a subset of genes points to their possible role in cell death. A detailed examination of these genes will help us learn whether bacterial death in response to damage is a regulated process.
Acknowledgments
This work was supported by NIH grant R01 GM61162 to K.L.
REFERENCES
- 1.Aldea, M., C. Hernandez-Chico, A. G. de la Campa, S. R. Kushner, and M. Vicente. 1988. Identification, cloning, and expression of bolA, an ftsZ-dependent morphogene of Escherichia coli.J. Bacteriol. 170:5169-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 3.Bayles, K. W. 2000. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 8:274-278. [DOI] [PubMed] [Google Scholar]
- 4.Berrier, C., A. Garrigues, G. Richarme, and A. Ghazi. 2000. Elongation factor Tu and DnaK are transferred from the cytoplasm to the periplasm of Escherichia coli during osmotic downshock presumably via the mechanosensitive channel MscL. J. Bacteriol. 182:248-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, D. S., B. Irwin, and H. S. Moyed. 1994. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 176:4081-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, D. S., A. J. Kelly, M. J. Mardis, and H. S. Moyed. 1991. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 173:5732-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohringer, J., D. Fischer, G. Mosler, and R. Hengge-Aronis. 1995. UDP-glucose is a potential intracellular signal molecule in the control of expression of σS and σS-dependent genes in Escherichia coli. J. Bacteriol. 177:413-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, M., T. Wang, R. Ye, and J. D. Helmann. 2002. Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45:1267-1276. [DOI] [PubMed] [Google Scholar]
- 10.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dijkstra, A. J., and W. Keck. 1996. Identification of new members of the lytic transglycosylase family in Haemophilus influenzae and Escherichia coli. Microb. Drug Resist. 2:141-145. [DOI] [PubMed] [Google Scholar]
- 12.Dong, H., L. Nilsson, and C. G. Kurland. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260:649-663. [DOI] [PubMed] [Google Scholar]
- 13.Falla, T. J., and I. Chopra. 1998. Joint tolerance to beta-lactam and fluoroquinolone antibiotics in Escherichia coli results from overexpression of hipA.Antimicrob. Agents Chemother. 42:3282-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gmuender, H., K. Kuratli, K. Di Padova, C. P. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagenmaier, S., Y. D. Stierhof, and U. Henning. 1997. A new periplasmic protein of Escherichia coli which is synthesized in spheroplasts but not in intact cells. J. Bacteriol. 179:2073-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengge-Aronis, R., W. Klein, R. Lange, M. Rimmele, and W. Boos. 1991. Trehalose synthesis genes are controlled by the putative σ factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli.J. Bacteriol. 173:7918-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishihama, A. 1999. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells 4:135-143. [DOI] [PubMed] [Google Scholar]
- 18.Kandror, O., A. DeLeon, and A. L. Goldberg. 2002. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. USA 99:9727-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khil, P. P., and R. D. Camerini-Otero. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44:89-105. [DOI] [PubMed] [Google Scholar]
- 20.Kim, K. I., S. C. Park, S. H. Kang, G. W. Cheong, and C. H. Chung. 1999. Selective degradation of unfolded proteins by the self-compartmentalizing HtrA protease, a periplasmic heat shock protein in Escherichia coli.J. Mol. Biol. 294:1363-1374. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, H., M. Yamamoto, and R. Aono. 1998. Appearance of a stress-response protein, phage-shock protein A, in Escherichia coli exposed to hydrophobic organic solvents. Microbiology 144(Pt. 2):353-359. [DOI] [PubMed] [Google Scholar]
- 22.Lange, R., and R. Hengge-Aronis. 1994. The nlpD gene is located in an operon with rpoS on the Escherichia coli chromosome and encodes a novel lipoprotein with a potential function in cell wall formation. Mol. Microbiol. 13:733-743. [DOI] [PubMed] [Google Scholar]
- 23.Lease, R. A., and M. Belfort. 2000. Riboregulation by DsrA RNA: trans-actions for global economy. Mol. Microbiol. 38:667-672. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, K. 2001. The riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Link, A. J., K. Robison, and G. M. Church. 1997. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis 18:1259-1313. [DOI] [PubMed] [Google Scholar]
- 27.Madeo, F., S. Engelhardt, E. Herker, N. Lehmann, C. Maldener, A. Proksch, S. Wissing, and K. U. Frohlich. 2002. Apoptosis in yeast: a new model system with applications in cell biology and medicine. Curr. Genet. 41:208-216. [DOI] [PubMed] [Google Scholar]
- 28.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moyed, H. S., and S. H. Broderick. 1986. Molecular cloning and expression of hipA, a gene of Escherichia coli K- 12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 166:399-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray, K. D., and H. Bremer. 1996. Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli.J. Mol. Biol. 259:41-57. [DOI] [PubMed] [Google Scholar]
- 31.Rosenow, C., R. M. Saxena, M. Durst, and T. R. Gingeras. 2001. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29:E112. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sat, B., R. Hazan, T. Fisher, H. Khaner, G. Glaser, and H. Engelberg-Kulka. 2001. Programmed cell death in Escherichia coli: some antibiotics can trigger mazEF lethality. J. Bacteriol. 183:2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scherrer, R., and H. S. Moyed. 1988. Conditional impairment of cell division and altered lethality in hipA mutants of Escherichia coli K-12. J. Bacteriol. 170:3321-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selinger, D. W., K. J. Cheung, R. Mei, E. M. Johansson, C. S. Richmond, F. R. Blattner, D. J. Lockhart, and G. M. Church. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18:1262-1268. [DOI] [PubMed] [Google Scholar]
- 35.Sledjeski, D. D., and S. Gottesman. 1996. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spoering, A. L., and K. Lewis. 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stirling, D. A., C. S. Hulton, L. Waddell, S. F. Park, G. S. Stewart, I. R. Booth, and C. F. Higgins. 1989. Molecular characterization of the proU loci of Salmonella typhimurium and Escherichia coli encoding osmoregulated glycine betaine transport systems. Mol. Microbiol. 3:1025-1038. [DOI] [PubMed] [Google Scholar]
- 40.Strom, A. R., and I. Kaasen. 1993. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol. Microbiol. 8:205-210. [DOI] [PubMed] [Google Scholar]
- 41.Wada, A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3:203-208. [DOI] [PubMed] [Google Scholar]
- 42.Wolfson, J. S., D. C. Hooper, G. L. McHugh, M. A. Bozza, and M. N. Swartz. 1990. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and beta-lactam antimicrobial agents. Antimicrob. Agents Chemother. 34:1938-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasuda, T., K. Morimatsu, T. Horii, T. Nagata, and H. Ohmori. 1998. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 17:3207-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yasuda, T., K. Morimatsu, R. Kato, J. Usukura, M. Takahashi, and H. Ohmori. 2001. Physical interactions between DinI and RecA nucleoprotein filament for the regulation of SOS mutagenesis. EMBO J. 20:1192-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]