Abstract
Using antibodies prepared against a unique region (exon 22–24) of rat K+-Cl− cotransporter-2 (KCC2), we confirmed that the ∼140-kDa KCC2 protein is exclusively expressed in rat brain, but in chicken, we observed strong reactivity not only with the ∼140-kDa KCC2 protein in brain but also a slightly larger ∼145-kDa protein in heart. In silico analysis showed that while exon 22 of KCC2 is unique to this isoform in therian mammals, it is retained in KCC2's closest paralog, KCC4, of lower vertebrates, including chicken. To eliminate potential cross-reactivity with chicken KCC4, the antibodies were preadsorbed with blocking peptides prepared over the only two regions showing significant sequence identity to chicken KCC4. This completely eliminated antibody recognition of exogenously expressed chicken KCC4 but not of the ∼145-kDa protein in chicken heart, indicating that chicken heart expresses KCC2. Real-time PCR confirmed robust KCC2 transcript expression in both chicken brain and heart. Chicken heart expressed predominantly the longer KCC2a splice variant consistent with the larger ∼145-kDa protein in chicken heart. Immunofluorescence microscopy revealed prominent plasma membrane KCC2 labeling in chicken ventricular cardiomyocytes. We hypothesize that KCC2 is an important Cl− extrusion pathway in avian cardiomyocytes that counters channel-mediated Cl− loading during high heart rates with β-adrenergic stimulation. While KCC2 is absent from mammalian cardiomyocytes, understanding the role that the other KCC isoforms play in Cl− homeostasis of these cells represents a nascent area of research.
Keywords: chloride, homeostasis, KCC2, KCC4, avian, Cl− channels
the K+-Cl− cotransporter (KCC) is a member of the cation chloride cotransporter (CCC) gene family and mediates the obligatorily coupled, electroneutral movement of K+ and Cl− across the plasma membrane. Under normal physiological conditions, KCC functions as a net K+Cl efflux pathway that uses the favorable K+ chemical gradient to drive Cl− out of the cell against its chemical gradient. Before its characterization at the molecular level, KCC was most often studied in single cells such as red cells where it was implicated in the regulation of cell volume following swelling by promoting the efflux of K+, Cl−, and osmotically obliged water. Few researchers could have predicted the genetic and functional diversity that is now evident among the KCC isoforms. Within the vertebrate CCC family, the KCCs exhibit the greatest genetic diversity with four separate isoforms, KCC1–4. Further genetic diversity of the KCCs is manifest as a result of alternative splicing, including alternate promoters and alternate first exons (30, 41). Studies using KCC1−/− and KCC3−/− mice have provided strong evidence that KCC3 is the primary isoform involved in cell volume regulation following hyposmotic swelling (8, 10, 39). In addition to cell volume regulation, key functional roles have been described for the KCCs in epithelia and neurons. For example, in acid-secreting epithelial cells (e.g., renal distal nephron α-intercalated cells), KCC4 appears to operate as a basolateral Cl− efflux pathway that helps recycle Cl− entering via anion exchange (7). A more novel function is evident in the central nervous system (CNS), where the developmental upregulation of KCC2 is associated with maturation of postsynaptic GABAergic inhibition in central neurons (38). That is, KCC2 is largely responsible for lowering intracelluar [Cl−] during neuronal maturation so that the Cl− reversal potential (ECl) is more negative than the resting membrane potential (22, 32, 38). Hence, KCC2 is critical for the development of fast hyperpolarizing synaptic inhibition mediated by GABAA receptors. The KCC2 protein also appears to play a morphogenic role, independent of its ion transport activity, in dendritic spine formation (16, 20, 28). Since KCC2 has been implicated in the development of both inhibitory and excitatory neurotransmission, it is emerging as a key “synchronizing factor” in the developing nervous system (28).
KCC2 stands out among the KCC isoforms not only in terms of its unique operation in neurons but also in terms of its structure. KCC2 contains an exon (exon 22 encoding 41 amino acids) that has long been believed to be unique to this isoform of the KCCs. This exon represents a highly charged intracellular region of the protein with a number of serine and threonine residues, leading us to propose the likelihood of it serving as a unique regulatory region (33). Indeed, we have identified a protein kinase-C phosphorylation site encoded by this exon (Ser-940 of rat KCC2) that appears to be involved in modulating the membrane trafficking of KCC2 (27). To better understand the functional role of the unique carboxy-terminal region of KCC2 encoded by exon 22, we undertook an in silico analysis of this region. During our analysis, we discovered that while the sequence of exon 22 is unique to KCC2 in therian mammals (i.e., human, rat, and mouse), it is nonetheless present in KCC4 of lower vertebrates, including fish, birds, and a prototherian mammal (i.e., platypus). This led us to the discovery that a set of KCC2 antibodies that we had previously prepared against a fusion protein encoding most of exon 22 of rat KCC2 (46) strongly recognized a ∼145-kDa protein in chicken heart. The biochemical and molecular studies presented in this report demonstrate that a single splice variant of KCC2 (KCC2a) is robustly expressed in the plasma membranes of cardiomyocytes throughout both chicken ventricles. We speculate that K+-Cl− cotransport is critical for effective intracellular Cl− homeostasis especially under conditions of elevated heart rate following β-adrenergic stimulation, when channel-mediated Cl− loading [via cystic fibrosis transmembrane conductance regulator (CFTR) and Ca+2-activated Cl− channels] is predicted to be most prominent.
MATERIALS AND METHODS
Selective tissue harvest.
Tissues from both rats and chickens were obtained and used according to approved protocols by the Institutional Animal Care and Use Committee at the University of California, Davis. Fertilized chicken eggs, adult chickens, and adult rats were obtained from a local source. Chicken eggs were maintained in a humidified incubator at 37°C, and tissues were isolated before embryonic day 18. Chicken embryos were euthanized by decapitation, and adult chickens and rats were euthanized by CO2 inhalation.
RNA isolation and cDNA synthesis.
Tissues were isolated by dissection and immediately homogenized in a guanidinium thiocyanate solution or quickly frozen in liquid nitrogen. Total RNA was isolated from chicken and rat tissues using the guanidinium thiocyanate method (11) and RNeasy Fibrous Tissue Midi kit (Qiagen). Poly(A)+ RNA was isolated using the PolyATtract system (Promega), following treatment of total RNA with DNase I. Complementary DNA was generated from 100 ng of mRNA or 1 μg total RNA using RETROscript (Ambion) with oligo(dT) primers, random decamers, or gene-specific primers. Oligonucleotide primers used for real-time PCR are listed in Table 1.
Table 1.
Combinations of gene-specific primers for semiquantitative real-time PCR
| Chicken | Forward and Reverse Primers (5′-3′) |
|---|---|
| KCC1 | TCCCATGATGCCAGACTTGT |
| ACTCTCTCCAGACCCTCAGT | |
| KCC2 | GAGTCGGCCCCTGAGAAG |
| AAGTCCTTGATGCCCTCGG | |
| KCC2a | GAGAGCCGCCCGCAC |
| TCCAGGTCTGTGCTGTTGA | |
| KCC2b | AACCTGACGGACTGCGAG |
| GGGCTGGTGTCCATCTCC | |
| KCC4 | GTTCTTGTACCATCTCCGCC |
| GGGATCGCTGTTCCATCATT | |
| GAPDH | CTTTCCGTGTGCCAACCCCC |
| GCCCATCAGCAGCAGCCTTC |
Cloning chicken KCC1 (Slc12a4).
The full-length sequence of chicken (Gallus gallus) KCC1 mRNA (XM001234004) was identified in the NCBI database. With the use of sequence information from chicken KCC1 (XM001234004), three sets of forward and reverse primers were synthesized and used to amplify fragments from double-stranded cDNA isolated from chicken brain or heart (see RNA isolation and cDNA synthesis). Each reaction used a final volume of 50 μl, containing 2.5 μl of the double-stranded chicken brain or heart cDNA, 5 μM of each primer, 50 mM KCl, 10 mM tris(hydroxmethyl)aminomethane, Tris·HCl (pH 8.3, 25°C), 1.5 mM MgCl2, 0.125 mM dNTP, and 3 U of Taq DNA polymerase (Invitrogen). Twenty-five cycles of PCR were performed for each reaction, consisting of incubation of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C From the three reactions, three separate PCR products of appropriate size were obtained and subcloned into pCR 2.1 TOPO (Invitrogen). The full-length version of chicken KCC1 was cloned into the expression vector pJB20.
Cloning chicken KCC2 (Slc12a5).
The predicted sequence of chicken (Gallus gallus) KCC2 mRNA (XM001236721) was identified in the NCBI database. We noted that this sequence correctly encoded exons 2–26. The first 136-bp of this sequence, however, shared little identity to KCC2 of other vertebrates, and we reasoned it contained incorrect sequence. To identify the correct sequences for the first two exons (exon 1a and exon 1b) of chicken KCC2, we searched the chicken KCC2 genomic sequence upstream of the designated KCC2 gene (Slc12a5; Chromosome 20; GeneID: 777252) and were able to correctly identify chicken KCC2 exon 1a. We were unable to identify chicken KCC2 exon 1b, which must be within the as yet unsequenced region between exon 1a and exon 2. To clone the 3′-end of KCC2b, we performed a multiple sequence alignment of KCC2 exon 1b from zebra finch (Taeniopygia guttata, EST: FE716540), human, mouse, and rat. The first 20-bp of this exon are remarkably well conserved among these vertebrates. Hence, we used this sequence to prepare a forward primer to amplify the 3′-end of chicken KCC2b. The remaining portions of chicken KCC2 were cloned from four separate PCR products. Each PCR reaction was performed using identical conditions to those described above for cloning of chicken KCC1. Full-length versions of chicken KCC2a and KCC2b were cloned into the expression vector pJB20.
Cloning chicken KCC4 (Slc12a7).
The full-length predicted sequence of chicken (Gallus gallus) KCC4 protein (XP001233067) was identified from NCBI using the BLAST search program (1) and rat KCC2 as the queried sequence. With the use of sequence information from the chicken KCC4 cDNA (NM001006371), four sets of forward and reverse oligonucleotide primers were synthesized and used to amplify fragments from double-stranded cDNA from whole chicken brain or heart (see RNA isolation and cDNA synthesis). Each PCR reaction was performed using similar conditions to those described above for cloning of chicken KCC1. From three of the reactions, three separate PCR products of appropriate size were obtained. In a fourth reaction, two distinct PCR products were obtained, corresponding to cDNA with (S1) and without (S2) exon 22 (117 bp) from the chicken Slc12A7 (KCC4) gene. All five reaction products were subcloned into pCR 2.1 TOPO (Invitrogen). To follow protein production in expression experiments, we used PCR mutagenesis to add the c-myc epitope (EQKLISEEDL) to the amino terminus of chicken KCC4-S1 and KCC4-S2. The full-length versions of c-myc tagged chicken KCC4-S1 and KCC4-S2 were cloned into the expression vector pJB20.
RT-PCR and semiquantitative real-time PCR.
RT-PCR used to determine KCC2 expression in various chicken tissues were conducted in a volume of 50 μl containing 2 μl of cDNA (see RNA isolation and cDNA synthesis), 5 μl 10 × AmpliTaq Gold PCR buffer (Applied Biosystems), 4 μl 10 mM dNTP, 4 μl 50 mM MgCl2, 1.5 U AmpliTaq Gold DNA polymerase (Applied Biosystems), 4 μl of 10 μM gene-specific forward and reverse primers, and 26.7 μl diethylpyrocarbonate-treated water. PCR amplification was done using the following program: 95°C for 10 min; 30 cycles of 95°C for 30 s, 60°C for 15 s, 72°C for 15 s; and final extension at 72°C for 7 min. Ten microliters of the PCR reactions were then run on a 2% agarose gel containing ethidium bromide.
Semiquantitative real-time-PCR was performed with an Applied Biosystems 7900HT Fast Real-Time PCR system with SYBR green fluorescent label. Samples with a final volume of 12 μl contained the following: 0.4 μl of cDNA generated using random decamers, 6 μl 2× Power SYBR Green PCR master mix (Applied Biosystems), 2 μl of each 4.8 μM gene-specific forward and reverse primers (see Table 1), and 1.6 μl diethylpyrocarbonate-treated water. Samples were run in triplicate in 384-well plates. Cycling parameters were as follows: 50°C for 2 min; 95°C for 10 min; 40 cycles of 95°C for 15 s, and 60°C for 1 min. After the PCR amplification, a melting curve was created for each sample to verify the specificity of the reactions. All data were compared with those from a housekeeping gene (GAPDH). The crossing threshold (Ct) for each sample was calculated using the Sequence Detection System (SDS) Software v2.3. Outlier values with a difference between Ct and Ct-mean >0.5 were excluded from further data analysis. The kinetic PCR efficiency correction value (E) for each tissue and primer set was determined by completing real-time PCR runs using serial dilutions of cDNA over a range of 2 μl to 2 × 10−7 μl. By plotting the corresponding Ct values for each dilution, calculating the slope of the best fit line, we determined E from the equation, E = dilution factor(−1/slope). The relative mean normalized expression levels of transcript were determined using the following equation: (Eref)Ct−ref/(Etarget)Ct−target with GAPDH used as the reference.
Antibody production and purification.
We have previously described the production and purification of a fusion protein (termed B22) along with the generation and characterization of rabbit polyclonal antibodies (rb-B22-KCC2) against this purified fusion protein (46). Antisera to B22 were generated in guinea pigs (gp-B22-KCC2) by the Polyclonal Antibody Production Program (Comparative Pathology Lab Services of the School of Veterinary Medicine, University of California, Davis). Both rb-B22-KCC2 and gp-B22-KCC2 were purified by affinity chromatography using the B22 fusion protein coupled to agarose beads using previously published methods (46).
Protein analysis.
Methods used for preparing membranes, for deglycosylating membrane protein, and for protein analysis by SDS-PAGE and Western blotting have been previously published (46).
Immunocytochemistry.
The polyclonal antibody to human cardiac troponin I (raised to 2 peptides residues 39–52 and 195–209) was kindly provided by Dr. Aldrin Gomes (University of California, Davis) and was made by 21st Century Biochemicals. Mouse monoclonal antibodies to chicken Na,K-ATPase-α-subunit were obtained from the Developmental Studies Hybridoma Bank (α6). Cryosections (10 μm) of formalin-fixed adult chicken heart (left ventricle) were treated with 1% SDS in PBS for 5 min to unmask antigenic sites and then incubated sequentially in blocking solution (5% goat serum, 2% BSA, 25 mM glycine, 3 mM EDTA, and 0.1% Tween-20, in PBS, pH 7.4, 1 h), primary antibodies (in blocking solution, overnight), and fluorophore-conjugated secondary antibodies (in PBS plus 0.1% Tween-20), with three rinses between each step. Coverslips were mounted over Gelvatol containing the nuclear stain ToPro-3 (Molecular Probes). Fluorescence images were acquired with a Zeiss LSM-510 confocal microscope through a ×25 water immersion (0.8 NA Plan Neofluar) objective.
Stable expression in HEK-293 cells.
The human embryonic kidney cell line HEK-293 was maintained in DMEM (Cellgro) supplemented with 10% FBS, penicillin (50 U/ml), and streptomycin (50 μg/ml) in a humidified incubator (5% CO2 at 37°C). For stable cell line expression, constructs were transfected into HEK-293 cells by calcium phosphate precipitation using previously described methods (34). After 3 wk of growth in 900 μg/ml geneticin (GIBCO-BRL), single resistant colonies were amplified and screened by Western blot with a c-myc epitope monoclonal antibody (15) or KCC2 rabbit polyclonal antibodies and by 86Rb influx assay.
RESULTS
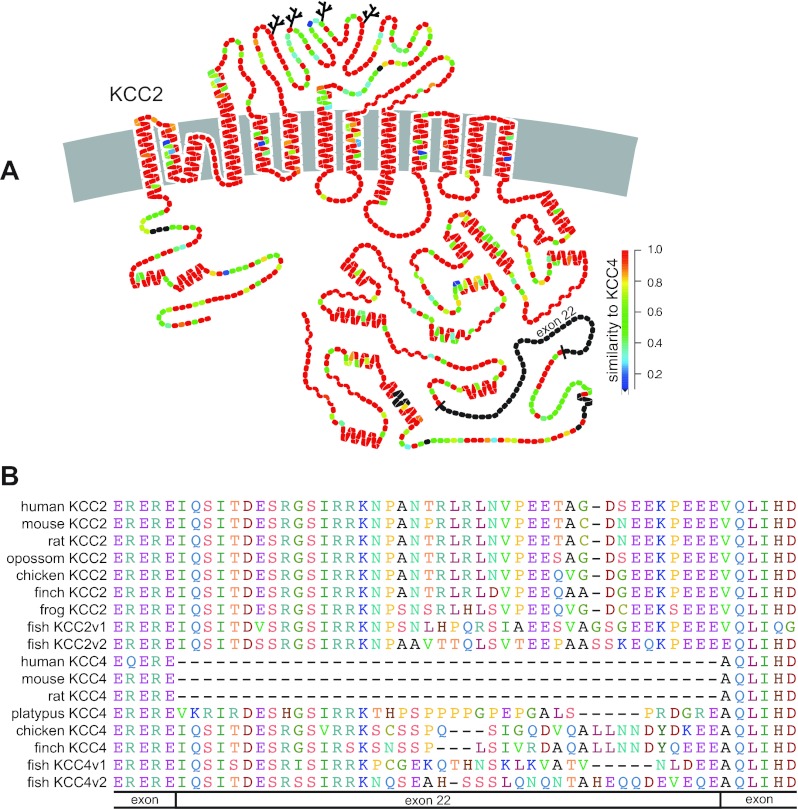
KCC2 stands apart from the other three K+-Cl− cotransporter isoforms both functionally and structurally. Functionally, KCC2 plays a key role in the regulation of intracellular [Cl−] of mature neurons and is instrumental in neuronal development and fast synaptic inhibition. Structurally, the KCC2 gene contains an exon (exon 22) encoding 41 amino acids that has long been believed to be unique to this isoform of the K+-Cl− cotransporters (Fig. 1A). To better understand the potential functional role of exon 22, we undertook an in silico analysis of this region of KCC2. During our analysis, we identified a similar exon in KCC4 of lower vertebrates, including chicken (Gallus gallus), finch (Taeniopygia guttata), and teleost fish (Danio rerio; Fig. 1B). Remarkably, KCC4 of platypus (Ornithorhynchus anatinus), a prototherian mammal, also contained an exon similar to that of exon 22 of KCC2.
Fig. 1.
Structure of K+-Cl− cotransporters. A: hypothetical model of human K+-Cl− cotransporter-2 (KCC2) where branched lines are potential N-linked glycosylation sites and secondary structural elements are displayed as helices (α-helical) and wavy lines (β-sheet). Color of individual amino acid residues indicate degree of similarity on a per residue basis between human KCC2 and human KCC4: red residues are identical and black residues are absent from human KCC4 (figure was kindly provided by Bliss Forbush using DNAPLOT). B: amino acid alignment of vertebrate KCC2 and KCC4 isoforms over exon 22 of KCC2.
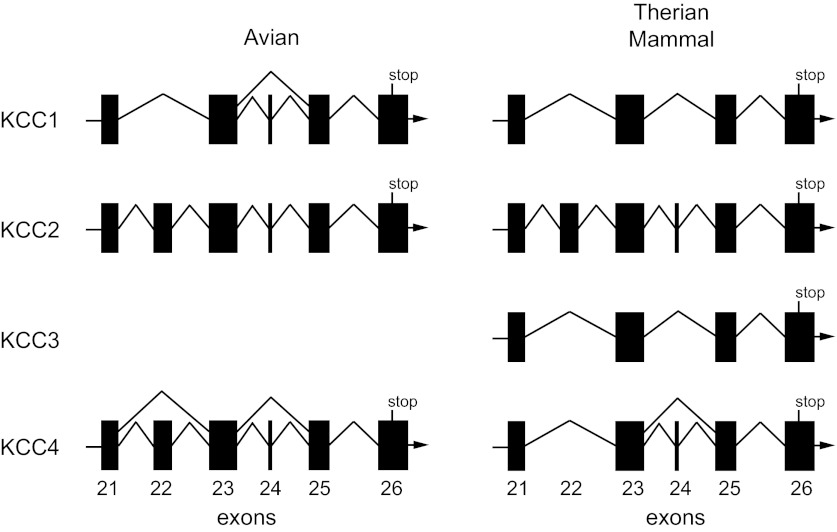
Analysis of the chicken KCC4 gene (Slc12a7; Chromosome 2; GeneID: 420797) revealed the presence of 26 exons. Exons 2–26 of chicken KCC4 share significant identity and alignment with the corresponding exons of KCC2 (Slc12a5) of therian mammals (e.g., human, rat, and mouse). This latter finding is reasonable as KCC4 is the closest paralog to KCC2. The lack of identity and alignment of exon 1 between KCC4 of chicken and KCC2 of therian mammals is not surprising given that this is a highly variable region among the KCCs and subject to alternative splicing (i.e., alternate first exons) in KCC2 (41). Analysis of the KCC4 gene of therian mammals showed that while the sequence similar to exon 22 of KCC2 is still identifiable within the intron between the bordering exons, it no longer represents coding sequence. Hence, KCC4 from therian mammals is encoded by only 25 exons (Fig. 2). Sequence analysis of the genomes of numerous vertebrates (amphibians, birds, and mammals) revealed the exon corresponding to exon 22 of KCC2 is absent from vertebrate KCC1 (Slc12a4) and KCC3 (Slc12a6) genes. While KCC3 was evident in an amphibian (Xenopus tropicalis) genome, neither its gene nor transcript was apparent in the Ensembl or NCBI databases for chicken or finch. Furthermore, numerous attempts by us to clone KCC3 transcripts using RT-PCR in chicken brain, heart, and colon proved unsuccessful. These findings lead us to conclude that the KCC3 gene has undergone genetic deletion in the avian vertebrate class. Furthermore, we conclude that exon 22 is not unique to KCC2. While it is absent from KCC1 and KCC3 genes of vertebrates, it is present in KCC4 of a prototherian mammal (platypus) and lower vertebrates, including birds and teleost fish. As we will discuss further below, we also noted that a small 15-bp exon (exon 24 in chicken KCC4) is subject to alternative splicing in avian KCC1, avian KCC4, and mammalian KCC4 (Fig. 2). Taken together, our findings indicate that within the vertebrates the KCC isoforms have undergone significant evolution, exhibiting class-specific exon deletion, alternative splicing of exons, and apparent gene deletion.
Fig. 2.
Avian and mammalian gene structure of exons (KCC2 equivalent of exons 21–26) encoding the intracellular carboxyl-terminus of the K+-Cl− cotransporters (Slc12a4–7).
Biochemical characterization of K+-Cl− cotransport protein in chicken heart.
We have previously prepared rabbit polyclonal antibodies (rb-B22-KCC2) against a fusion protein containing most of the sequence encoded by exons 22–24 of rat KCC2 (46). This region of KCC2 was chosen for antibody production because of its highly antigenic nature (i.e., high surface probability) and poor sequence conservation with rat KCC1. The rb-B22-KCC2 antibodies broadly recognize the KCC2 protein in brains of vertebrates from mammals to elasmobranchs (46). In therian mammals, these antibodies have neither shown reactivity with proteins outside the CNS nor shown cross-reactivity with any other KCC isoform. Although these antibodies have been determined to be KCC2-specific in therian mammals, they have not been extensively tested in lower vertebrates. Because exon 22 of KCC2 is absent from the other KCC isoforms and the sequence within exon 23 of KCC2 is poorly conserved among the KCC isoforms, the lack of cross-reactivity of rb-B22-KCC2 with the other KCC isoforms is not surprising (Figs. 1 and 2). Sequence corresponding to the small 15-bp exon 24 of KCC2 is absent from both KCC1 and KCC3 of therian mammals (Fig. 2). While this small exon of KCC4 is often not included in database sequences for KCC4 of therian mammals, we have been able to identify it in the KCC4 gene (Slc12a7) of human, mouse, and rat. We suspect that the sporadic appearance of this 15-bp exon in database sequences is due in part to its small size, but more importantly, its inclusion in transcripts probably represents a minor alternatively splice variant of KCC4. This hypothesis is supported by the low percentage of KCC4 ESTs that include this exon. Moreover, we rarely found this exon in clones from RT-PCR performed over this region using gene-specific primers for mouse KCC4 or chicken KCC4 (data not shown).
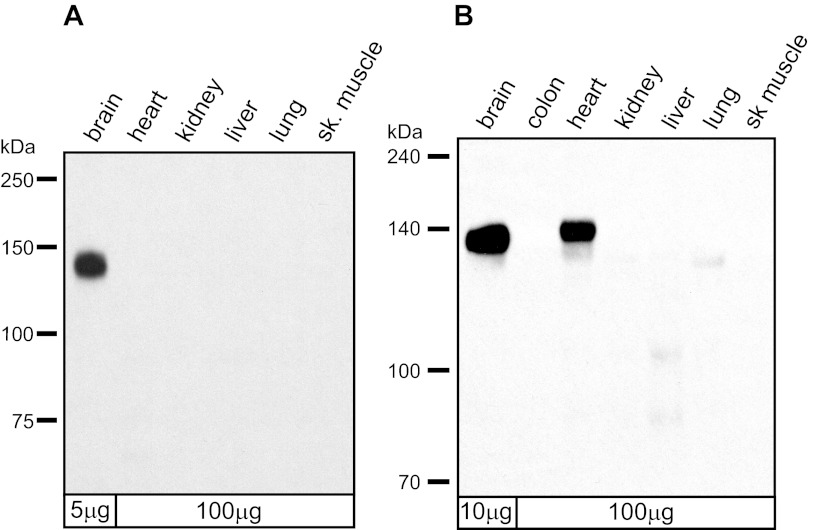
It is noteworthy that the first 15 amino acids encoded by exon 22 of rat KCC2 and KCC4 of lower vertebrates share remarkably high identity, whereas the remaining residues of the exon are poorly conserved among the vertebrate classes (Fig. 1B). This prompted us to test whether rb-B22-KCC2 might recognize chicken KCC4. Since KCC2 is believed to be a neuron-specific isoform in therian mammals and KCC4 is found in multiple mammalian tissues, we performed a Western blot with rb-B22-KCC2 against membranes prepared from multiple tissues of rat and chicken (Fig. 3). As we have shown previously, rb-B22-KCC2 recognized a single ∼140-kDa protein in rat brain (46). No protein bands were recognized by rb-B22-KCC2 in any other rat tissue even at 20-fold higher protein loads. In membranes isolated from adult chicken tissues, however, rb-B22-KCC2 strongly recognized a 140- to 145-kDa protein present in brain and heart but not other tissues. We have little doubt that the protein recognized in chicken brain is KCC2. Given that KCC2 is believed to be neuron-specific and that rat KCC2 and chicken KCC4 are identical in sequence over an extended region of exon 22 over which the rb-B22-KCC2 antibodies were prepared (Fig. 1B), we postulated that the protein recognized in chicken heart was KCC4.
Fig. 3.
Western blot analysis of membranes prepared from rat (A) and chicken (B) tissues. Rat and chicken brain membranes were loaded at 5 and 10 μg, respectively, whereas membranes from all other tissues were loaded at 100 μg. Western blot was probed with rb-B22-KCC2 antibodies (1:2,000).
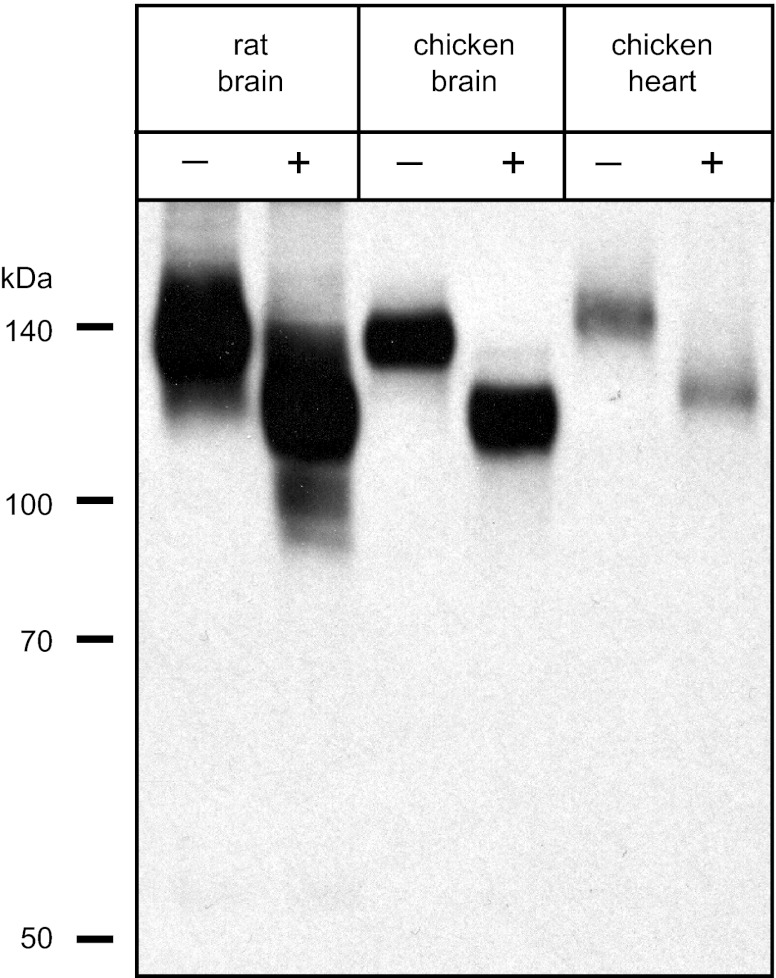
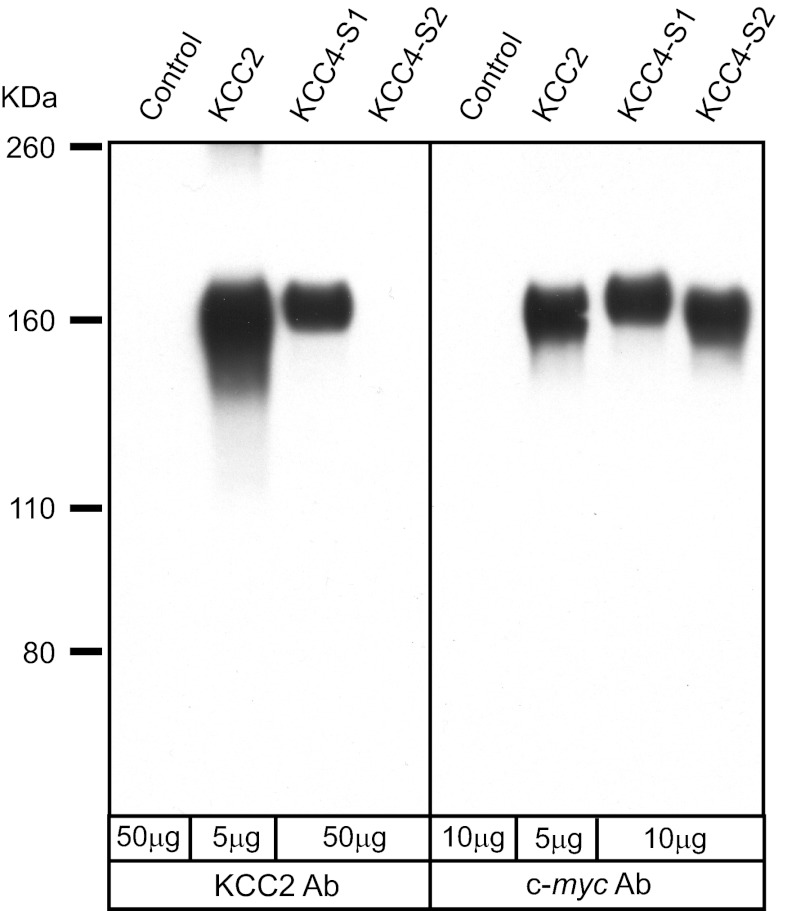
Importantly, the protein recognized in chicken heart exhibited a slightly larger mass (2–5 kDa) than that in chicken brain. To evaluate whether this size difference reflected differences in core protein mass as opposed to differential glycosylation, we treated membranes from rat brain, chicken brain, and chicken heart with N-glycosidase F to remove N-linked sugar groups. We have previously demonstrated that treatment of KCC2 from rat brain with N-glycosidase F reduces its apparent mass to ∼124 kDa, which is comparable to that of the predicted core protein (46). A similar size reduction was observed for membranes from both rat brain and chicken brain (Fig. 4). When we treated chicken heart membranes with N-glycosidase F, the band detected by Western blot was reduced in size but still recognizable as slightly larger than that from chicken brain by ∼2–5 kDa. These data indicate that the size difference of protein recognized by rb-B22-KCC2 in chicken brain and chicken heart is due to differences in the core protein mass. In comparing the chicken KCC core protein masses, chicken KCC1 is ∼1 kDa smaller than KCC2 whereas chicken KCC4 is ∼2 kDa larger than KCC2. These findings support our hypothesis that the rb-B22-KCC2 antibodies were cross-reacting with KCC4 in chicken heart. To test this hypothesis, we cloned KCC4 from chicken brain and heart using a PCR strategy along with the known sequence from the NCBI database (see materials and methods). During the cloning of chicken KCC4, we noted that exon 22 was subject to alternative splicing as we obtained two different sized products in one of our PCR reactions (see materials and methods), i.e., one with and one without this exon. We termed these alternative splice variants of chicken KCC4, KCC4-S1 (with exon 22), and KCC4-S2 (without exon 22). After cloning both KCC4 variants, expression constructs of each were prepared with a c-myc epitope tag at the amino terminus and then expressed in stable HEK-293 cell lines. As noted above, the small 15-bp exon 24 was rarely identified in our PCR reactions; hence, our KCC4-S1 and KCC4-S2 constructs lacked the peptide sequence encoded by this exon. When expressed in HEK-293 cells, both KCC4-S1 and KCC4-S2 mediated significant N-ethylmaleimide-stimulated, furosemide-sensitive 86Rb influx (data not shown). Western bot analysis indicated that HEK-293 cells expressing rat KCC2 and chicken KCC4-S1 contained one major band detected by rb-B22-KCC2 (Fig. 5). In contrast, this antibody detected no protein in cells expressing chicken KCC4-S2. Our results confirm that rb-B22-KCC2 recognizes an epitope encoded by exon 22 but not exon 23 of chicken KCC4 and support the hypothesis that the protein recognized by rb-B22-KCC2 in chicken heart was KCC4.
Fig. 4.
Deglycosylation of membranes from rat brain, chicken brain, and chicken heart. Membranes from whole rat brain, chicken brain, and chicken heart were incubated with (+) or without (−) N-glycosidase F for 4 h at 37°C. Western blot was probed with rb-B22-KCC2 antibodies (1:2,000).
Fig. 5.
Cross-reactivity of the rb-B22-KCC2 antibodies with chicken KCC4. Membranes were prepared from untransfected HEK-293 cells (control) and HEK-293 cells stably expressing either the rat KCC2 protein, chicken KCC4-S1, or chicken KCC4-S2 protein. Each of the expression constructs for the rat KCC2, chicken KCC4-S1, and chicken KCC4-S2 proteins were epitope tagged with the 10-amino acid c-myc peptide. Western blots panels were probed with either the rb-B22-KCC2 antibodies (left; 1:2,000) or the c-myc peptide monoclonal antibody (right; 1:2,000).
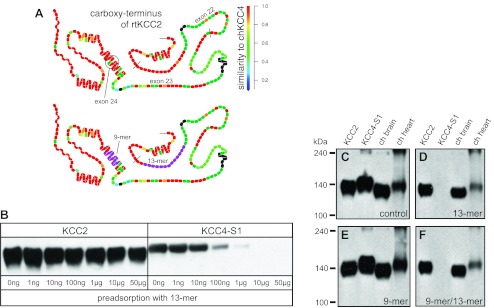
The rb-B22-KCC2 antibodies were generated against a fusion protein (rat KCC2: Ser-932 to Asn-1043) that includes only two short regions with significant identity to chicken KCC4 (Fig. 6A). The first 13 amino acids of this fusion protein are 92% identical (12 of 13 amino acids) to chicken KCC4, and the last 9 amino acids are 89% identical (8 of 9 amino acids) to chicken KCC4. To further test the hypothesis that rb-B22-KCC2 cross-reacts with KCC4 in chicken heart, two separate blocking peptides were prepared: S932ITDESRGSIRRK (13-mer; Fig. 6A) and MKPEWENLN1043 (9-mer; Fig. 6A). Membranes prepared from stable HEK-293 cells expressing rat KCC2 or chicken KCC4-S1 were then probed following preadsorption of rb-B22-KCC2 with increasing amounts of purified 13-mer peptide. Although the 13-mer peptide did not prevent KCC2 labeling, it eliminated KCC4-S1 recognition when added in amounts exceeding 1 μg (Fig. 6B). Some decrement in KCC2 signal was noted following preadsorption with the 13-mer peptide, suggesting that a subset of antibodies within the polyclonal mixture recognized the 13-mer peptide and were removed from the reactive pool. To test whether chicken heart expresses KCC4, similar blots were probed with rb-B22-KCC2 before and after preadsorption with 10 μg of the 13-mer peptide. Control blots confirmed that the rb-B22-KCC2 antibodies strongly recognizes a 140- to 145-kDa band from HEK-293 cells expressing rat KCC2 or chicken KCC4-S1 as well as chicken heart, along with a slightly smaller protein from chicken brain (Fig. 6C). The band at >240 kDa observed in some of the lanes is often seen with the KCCs and appears to represent oligomerization of these proteins (6). Preadsorption of rb-B22-KCC2 with 10 μg of the 13-mer peptide eliminated recognition of only KCC4-S1 (Fig. 6D). As noted for KCC2 in Fig. 6B, the 13-mer peptide also modestly reduced recognition of the ∼140- to 145-kDa protein for KCC2, chicken brain, and chicken heart (Fig. 6D). A similar approach was used to evaluate the effect of the 9-mer blocking peptide. Preadsorption of the rb-B22-KCC2 antibodies with 10 μg of the 9-mer peptide, either alone (Fig. 6E) or in combination with the 13-mer peptide (Fig. 6F), did not interfere with its recognition of KCC2, KCC4-S1, or KCC proteins in chicken brain and heart. These data are inconsistent with the hypothesis that the protein recognized by rb-B22-KCC2 in chicken heart is KCC4 and support the alternative hypothesis that the protein in chicken heart is, in fact, KCC2. These data establish that KCC2 protein is expressed at significant quantities in a native tissue outside the CNS of a vertebrate.
Fig. 6.
Immunoadsorption of the rb-B22-KCC2 antibodies. A: structural models of the carboxy-terminus of rat KCC2 encoded by exons 22–26; top model: degree of similarity on a per residue basis between rat KCC2 and chicken KCC4; bottom model: 9- and 13-mer peptides used in preadsorption experiments (purple residues). B: rb-B22-KCC2 antibodies (1:1,000) were first preadsorbed with increasing amounts of the 13-mer peptide (0- 50 μg) in PBS/milk overnight at 4°C. Strip blots of membranes from HEK-293 cells expressing either rat KCC2 or chicken KCC4-S1 were then probed with each of the preadsorbed fractions (final antibody concentration 1:2,000). C–F: immunoreactivity of rb-B22-KCC2 antibodies with chicken brain and chicken heart following preadsorption with 10 μg 13-mer and/or 9-mer peptides. rb-B22-KCC2 antibodies were preadsorbed with no peptide (C: control), 10 μg 13-mer peptide (D), 10 μg 9-mer peptide (E), or 10 μg both peptides (F) in PBS/milk overnight at 4°C. Strip blots of membranes from HEK-293 cells expressing either rat KCC2 or chicken KCC4-S1, chicken brain, or chicken heart were probed with each of the preadsorbed fractions (final antibody concentration 1:2,000).
Identification and quantification of KCC1, KCC2, and KCC4 transcripts in chicken tissues.
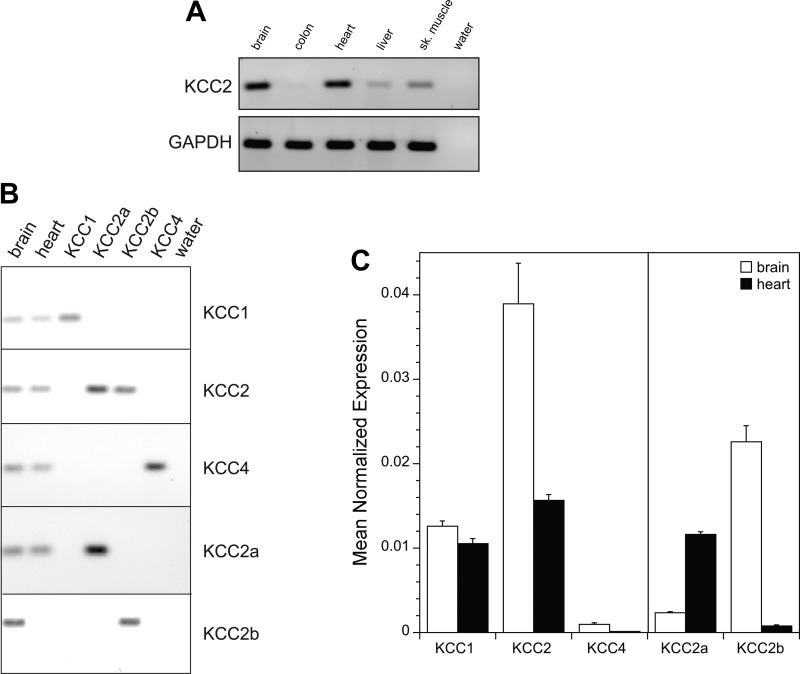
The expression of KCC2 transcript in various chicken tissues was evaluated using RT-PCR. Gene-specific primers for chicken KCC2 were designed to target sequence in adjacent exons to avoid genomic DNA artifacts. Robust KCC2 transcript expression was identified in brain and heart with much weaker expression in colon, liver, and skeletal muscle (Fig. 7A). Additional experiments were undertaken to explore the relative expression levels of the KCC isoforms in chicken heart and brain using real-time PCR. These studies required that we first identify the sequence and clone chicken KCC1 and KCC2. This was accomplished using a combination of database searching and PCR (see materials and methods). Gene-specific primer sets for chicken KCC1, KCC2, and KCC4 were designed to 1) target sequence in the region of the last 6 exons of each isoform, 2) span adjacent exons to avoid genomic DNA artifacts, and 3) avoid alternatively spliced exons (Table 1). We first tested the specificity of each primer set and confirmed the presence of a single amplicon of the predicted size on ethidium bromide-stained gels (Fig. 7B). For each tissue and primer set, the kinetic PCR efficiency was determined and used to calculate mean normalized expression levels of transcripts to a reference gene (GAPDH; see materials and methods). As shown in Fig. 7C, total KCC2 transcript was most abundant in chicken brain with lower but still considerable amounts in chicken heart. KCC1 transcript was detected at similar levels in chicken brain and heart and comparable to KCC2 in chicken heart. In contrast, negligible KCC4 transcript was detected in chicken brain and heart. These data substantiate the concept that KCC2 in chicken is expressed outside of the CNS with significant expression in heart.
Fig. 7.
KCC2 mRNA expression in chicken tissues. A: RT-PCR of KCC2 in chicken tissues. B: RT-PCR demonstrating the specificity and single amplicon production of each primer set for KCC1, KCC2, KCC4, KCC2a, and KCC2b. C: semiquantitative real-time PCR analysis of KCC1, KCC2, KCC4, KCC2a, and KCC2b in chicken brain and chicken heart. Mean normalized expression level of each transcript was determined using the equation: (Eref)Ct−ref/(Etarget)Ct−target with GAPDH used as reference and where Ct = crossing threshold (see materials and methods). Values are means ± SE of 8 runs from four separate RNA extractions (2 animals). Brain and heart means are statistically different for each isoform or splice variant tested (P < 0.05 using two sample t-test).
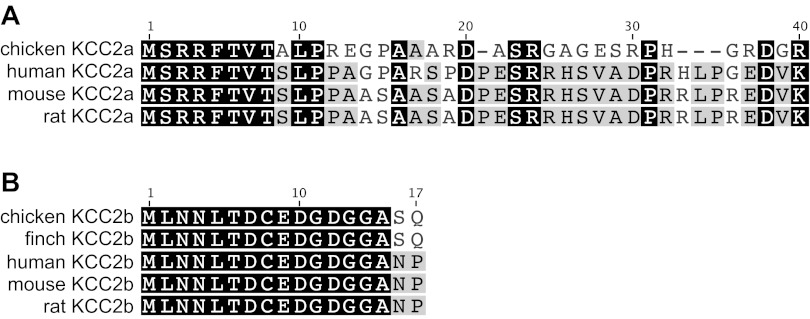
One question that still remained was the distinct size difference between the KCC2 protein observed in chicken brain and chicken heart that we consistently detected on Western blots with our KCC2 antibodies. In Fig. 4, we demonstrated that the size difference could be explained by differences in core protein mass. Uvarov et al. (41) reported that the mammalian KCC2 gene (Slc12a5) generates two proteins by using alternate promoters and alternate first exons. The first exon of the novel splice variant (termed KCC2a) in mammals encodes 40 amino acids. In contrast, the first exon of the previously known major brain KCC2 isoform (termed KCC2b) encodes only 17 amino acids. The core proteins of these two mammalian KCC2 splice variants are predicted to differ by 3 kDa (KCC2a =126.3 kDa and KCC2b = 123.6 kDa), which is remarkably similar to the size difference we detected on Western blots for KCC2 in chicken heart and brain. This suggested that the gene structure of KCC2 is similar in chicken and mammals with alternate first exons and that the longer KCC2a must predominate in chicken heart whereas KCC2b predominates in chicken brain. While the KCC2 mRNA sequence (XM001236721) identified in the NCBI database correctly coded for exons 2–26, the first 136-bp of this sequence shared little identity to that of the first exon of any vertebrate KCC2, and we reasoned that it must be incorrect sequence. To identify the alternate first exons of chicken KCC2, we searched the chicken genomic sequence upstream of the designated KCC2 gene (Slc12a5; Chromosome 20; GeneID: 777252), and we were able to identify chicken KCC2 exon 1a (Fig. 8A). However, sequence for chicken KCC2 exon 1b, which must be within the as yet unsequenced region between exon 1a and exon 2, could not be found. Since exon 1b of KCC2 is highly conserved among different vertebrates, we prepared a primer covering the first consensus 20-bp of this exon and were able to clone the remaining portion of exon 1b of chicken KCC2 using PCR. The amino acids encoded by exon 1b of KCC2 are identical between chicken and finch (Fig. 8B). As shown in the sequence alignment in Fig. 8A, the beginning of exon 1a of chicken KCC2 shares significant identity to that of mammals with residues 3–12 retaining a sequence similar to the Ste20-related proline alanine-rich kinase/oxidative stress response-1 (SPAK/OSR1) binding motif (12) that was previously noted for mammalian KCC2a (41). Expression of chicken KCC2a or KCC2b in HEK-293 cells resulted in a significant furosemide-sensitive 86Rb influx, and this activity was further stimulated after treatment with N-ethylmaleimide (data not shown). A high constitutive activity has likewise been reported for mammalian KCC2a and KCC2b (41).
Fig. 8.
Amino acid alignment of exon 1a (A) and exon 1b (B) for avian and mammalian KCC2. Amino acid similarity is shown as grayscale color with black (100% similar), gray (60–99% similar), and white (<60% similar). Scoring matrix was Blosum85 with a threshold of 2.
Chicken KCC2a encodes a core protein that is larger than chicken KCC2b by ∼2 kDa, which corresponds to the size difference we noted on Western blots for KCC2 in chicken heart and brain. To test the hypothesis that adult chicken heart and brain differentially expressed the two splice variants of KCC2 (KCC2a and KCC2b), real-time PCR was performed using primers specific for each of the two alternate first exons of chicken KCC2 (Table 1). As shown in Fig. 7C, chicken brain expressed predominantly KCC2b with KCC2a making up a minor fraction of the total KCC2 in chicken brain. In chicken heart, the only transcript that could be detected was that of KCC2a. These semiquantitative real-time PCR data are consistent with our protein data and support the hypothesis that the predominant form of KCC2 in chicken heart is KCC2a, while chicken brain expresses mainly the KCC2b splice variant.
Immunolocalization of KCC2 protein in adult chicken heart.
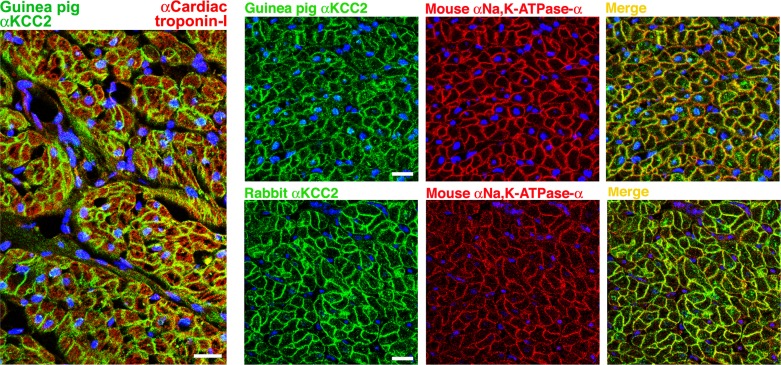
To identify the type of cells in chicken heart that express KCC2, tissues isolated from the left ventricle of adult chicken were probed with two different KCC2 antibodies prepared against the rat B22-KCC2 fusion protein, one prepared in rabbit (rb-B22-KCC2) and another in guinea pig (gp-B22-KCC2). Both rb-B22-KCC2 and gp-B22-KCC2 antibodies recognized the same bands on Western blots (data not shown). Prominent labeling was observed along the margin of cells distinguished as cardiomyocytes by colabeling with cardiac troponin-I (Fig. 9, left). In addition to this prominent plasma membrane labeling, a variable degree of punctate intracellular labeling of cardiomyocytes was noted with both KCC2 antibodies (Fig. 9). The patterns of labeling observed with the two independently generated KCC2 antibodies were similar and largely overlapping with those observed with an antibody against Na,K-ATPase-α subunit (Fig. 9, right). Similar labeling was observed in right ventricle (data not shown). To confirm the specificity of rb-B22-KCC2 and gp-B22-KCC2 to KCC2 in the chicken heart, both antibodies were preadsorbed with the 13-mer peptide shown to block recognition of KCC4-S1 (Fig. 6). The pattern of labeling with or without preadsorption was indistinguishable (data not shown). These results demonstrate that KCC2 is exceptionally abundant in the plasma membrane of cardiomyocytes throughout the chicken ventricle.
Fig. 9.
Immunolocalization of KCC2 in adult chicken heart. Left: left ventricle was labeled with a guinea pig antibody against KCC2 (green), a rabbit antibody against cardiac troponin-I (red), and the nuclear stain ToPro3 (blue); prominent KCC2 labeling is observed along the margin of red-stained cardiomyocytes. Cardiac troponin-I antibodies were kindly provided by Aldrin Gomes (University of California, Davis). Right: left ventricle was labeled with either a guinea pig antibody (top) or a rabbit antibody (bottom) against KCC2 (green), a mouse antibody against Na,K-ATPase-α (α6; red) and the nuclear stain ToPro3 (blue); note colocalization of KCC2 and Na,K-ATPase-α along the plasma membrane of cardiomyocytes. Bars = 20 μm.
DISCUSSION
In this study, biochemical and molecular analyses were employed to characterize a K+-Cl− cotransporter in avian cardiomyocytes. Using antibodies previously prepared against a rat KCC2 fusion protein (46), we demonstrated strong recognition of a 140- to 145-kDa glycoprotein in membranes prepared from chicken brain and chicken heart but not at significant levels in any other chicken tissue. Preabsorption of the antibodies with 9 and 13 amino acid peptides (prepared over the only two extended regions of the entire fusion protein displaying significant identity between rat KCC2 and the other chicken KCC isoforms) demonstrated that the antibodies specifically recognize KCC2 in chicken brain and heart on Western blots. RT-PCR and real-time PCR identified and quantified transcript expression of two alternatively spliced variants of KCC2 in adult chicken brain and heart. In earlier studies, Uvarov et al. (41) demonstrated that the mammalian KCC2 gene contains two alternate first exons each under control by separate promoters, leading to two splice variants (KCC2a and KCC2b) that differ only in the amino-terminus encoded by these two alternate first exons. Our identification of both of these KCC2 splice variants in chicken demonstrates the presence of a similar KCC2 gene structure in the avian class. In adult mouse brain, KCC2b was the predominant form whereas KCC2a made up <10% of the total KCC2 transcripts (41). Similarly, KCC2a transcript levels in adult chicken brain comprised <10% of the total KCC2 transcript levels (Fig. 9). Uvarov et al. (41) did not detect significant expression of either KCC2a or KCC2b transcripts outside the CNS and concluded that both KCC2 splice variants were restricted to CNS neurons. Significantly, we found that while KCC2b was most highly expressed in chicken brain, KCC2a was the most prevalent, if not exclusive, splice variant in chicken heart. Consistent with our finding of a slightly larger (∼2–5 kDa) KCC2 protein in heart (where KCC2a predominates) than brain (where KCC2b predominates), chicken KCC2a encodes a core protein that is ∼2 kDa larger than that encoded by chicken KCC2b. Immunofluorescent microscopy using two independently generated antibodies revealed specific expression of KCC2 protein in the plasma membrane of cardiomyocytes throughout the chicken ventricles.
Expression of KCC2 outside the CNS.
While it is generally believed that KCC2 is expressed only in the CNS, the finding of KCC2 transcript outside the CNS is not unprecedented. KCC2 transcript expression has been reported in a number of different mammalian cultured cells, including rat vascular smooth muscle cells (13), human osteoclasts (9), human lens epithelial cells (25, 26), and several human cancer cell lines (45). All of the studies to date that have detected KCC2 outside the CNS have come from studies using mammalian cultured cells. Our study, however, demonstrates that robust KCC2 transcript and protein expression is present in native tissue outside the vertebrate CNS. In the one study where data were presented examining expression of both KCC2 splice variants, KCC2a was the only variant detected in human lens epithelial cells (25). Based on our data in chicken and those of Lauf et al. (25), we suspect that KCC2b is primarily expressed in the CNS, whereas KCC2a exhibits a broader vertebrate tissue distribution.
Functional characterization of K+-Cl− cotransport in chicken cardiomyocytes.
Our finding of robust KCC2 transcript and protein in chicken cardiomyocytes is consistent with earlier evidence for KCC activity in cultured chicken cardiomyocytes (35). In this preparation, chick embryonic hearts were disaggregated with trypsin and grown as spontaneously beating sheaths of muscle around a nylon core. The chick polystrand preparation proved to be ideal for transport experiments because of the minimal extracellular diffusion distances and because of the ability to monitor accurately volume changes that follow net solute transport (21, 36). Using this preparation, Piwnica-Worms et al. (35) provided strong evidence for the existence of an electroneutral K+-Cl− cotransporter mechanism. In short, they showed that net K+ and Cl− transport and the associated volume changes were highly dependent on the combined K+ and Cl− chemical gradients. Incubation of the polystrands in high extracellular [K+] resulted in a net uptake of Cl− and K+ that fit a stoichiometry of 1:1. Furthermore, unidirectional isotopic fluxes of 42K+ and 36Cl− were furosemide sensitive and Na+ independent. Importantly, the K-Cl cotransporter activity characterized in cultured chicken cardiomyocytes by Piwnica-Worms et al. (35) was active under isotonic conditions, mediating significant net K+Cl efflux in K+-free media and net K+Cl influx in high-K+ (133 mM) media. This finding is consistent with KCC2 expression in chicken cardiomyocytes as it is the only KCC isoform to exhibit significant activity under control isotonic conditions (e.g., 29, 31, 41).
Physiological function of KCC2 in chicken cardiomyocytes.
Although the physiological functions of KCC2 in chicken cardiomyocytes remain poorly understood, K+-Cl− cotransporters have been implicated in the regulation of cell volume, intracellular [Cl−], and extracellular [K+] (19, 31, 32, 38). In addition to potential roles in regulating cell volume and extracellular [K+] in cardiomyocytes, KCC2 could contribute importantly to the control of intracellular [Cl−]. Analogous to its function in mature central neurons, we postulate that KCC2 in cardiomyocytes could serve as a Cl− extrusion pathway that helps maintain the Cl− electrochemical gradient needed for proper anion channel function in cardiomyocytes. Because KCC2a exhibits a regulatory site within its first exon (SPAK/OSR1 binding motif), the exclusive expression of KCC2a in chicken cardiomyocytes could permit differential regulation in cardiomyocytes vs. KCC2b, which predominates in central neurons. Cardiomyocytes do exhibit some characteristics with regard to Cl− homeostasis that are different from that observed in neurons. First, the level of [Cl−] in resting cardiomyocytes (CO2/HCO3− buffer: ∼20 mM, ECl = −50 mV, Refs. 24, 43, 44; HEPES buffer: ∼14 mM, ECl = −59 mV, Refs. 3, 43, 44) is much higher than that measured for most mature central neurons (∼7 mM, ECl = −70 mV, Refs. 4, 5). Second, unlike neurons, the membrane voltage of cardiomyocytes undergoes regular and sustained alterations in polarity during the course of the cardiac cycle. Hence, ECl can be either more positive (driving force for Cl− is out of the cell) or more negative (driving force for Cl− is into the cell) than the membrane voltage for extended periods of time during the cardiac cycle, thus permitting Cl− channels the ability to contribute significant inward (depolarizing Cl− efflux) as well as outward (repolarizing Cl− influx) current during the action potential (23).
Among the Cl− channels identified in cardiac cells, cAMP-activated Cl− channels (CFTR), swelling-activated Cl− channels (ClC3), and Ca+2-activated Cl− channels (CaCC) are responsible for the major anion currents that modify cardiac electrical activity (14). Currents through these Cl− channels exhibit outward rectification or are predominantly activated at depolarized voltages. Hence, under normal physiological conditions they are poised to contribute to membrane repolarization (Cl− influx) yet contribute little to diastolic depolarization. The magnitude of the effect of Cl− current (ICl) on membrane voltage (Em) will depend critically on the actual values of ECl and Em (i.e., Em − ECl = driving force for Cl− movement through the channels) and hence on [Cl−]i. In order for cardiomyocytes to regulate [Cl−]i effectively, they must employ both Cl− accumulation and Cl− extrusion transport pathways. Chloride accumulation transporters identified in the heart include Na+-K+-Cl− cotransport (NKCC1) and anion exchange (AE3, Slc26a6), both of which appear to be responsible for maintaining cardiomyocyte [Cl−]i above electrochemical equilibrium under quiescent conditions (2, 37, 40, 42–44). In contrast, Cl− extrusion transporters in cardiomyocytes have been largely ignored despite the fact that Cl− loading during the sustained depolarized plateau phase of the action potential is likely the greatest threat to Cl− homeostasis in the beating cardiomyocyte. Because of the relatively long action potential of cardiomyocytes (compared with nerve and skeletal muscle), the forces favoring net Cl− influx are sustained during the plateau phase. Chloride accumulation is of particular concern at high heart rates with β-adrenergic stimulation as the membrane voltage spends more time in the depolarized state where the outwardly rectified currents through CaCC (ICaCC activated by increased Ca+2 transient) and CFTR (ICFTR activated by increased cAMP) are enhanced (i.e., greater conductance and larger driving force). The physiological significance of enhanced ICaCC and ICFTR at high heart rates with β-adrenergic stimulation appears to be related to their ability (along with PKA activation of K+ current) to help offset the Ca2+ current-induced prolongation in action potential duration, thus permitting sympathetic stimulation to increase contractility without disturbing normal conduction patterns (17). If this Cl− load is not countered appropriately by Cl− extrusion, the resulting [Cl−]i increase will reduce the ability of ICFTR and ICaCC to help curtail action potential prolongation (18, 23). Such an elevation of [Cl−]i and the associated positive shift in the Cl− reversal potential may result in early afterdepolarizations and contribute to arrhythmogenesis (47). To maintain Cl− homeostasis, the channel-mediated Cl− influx during β-adrenergic stimulation must be countered by a Cl− extrusion mechanism that ideally is electroneutral and will therefore not disrupt normal electrical activity. Given our identification in this report of robust expression of KCC2 in chicken cardiomyocytes, we hypothesize that this cotransporter is critical for effective Cl− homeostasis especially under conditions of elevated heart rate with β-adrenergic stimulation.
In this study, we presented evidence for the presence of KCC2 in both CNS neurons and ventricular cardiomyocytes of chicken. In mammals, however, KCC2 appears to be restricted to central neurons as it exhibits no measurable expression outside the CNS. The notable appearance of KCC2 in chicken heart is yet another example of the remarkable evolution of the KCCs that has taken place within the vertebrate classes. We have identified a number of other examples of the evolution of the KCCs among the vertebrates, including class-specific exon deletion, alternative splicing of exons, and apparent gene deletion. Until additional studies are performed examining the tissue expression patterns of KCC2 in more mammals, birds, and lower vertebrates, it is difficult to speculate whether there has been an evolutionary gain of KCC2 in avian cardiomyocytes or evolutionary loss of KCC2 in mammalian cardiomyocytes. It is possible that the presence of KCC2 may correlate with the physiological demands of heart tissue (e.g., heart rate and force generation). While the role of K+-Cl− cotransport is well established in neuronal Cl− homeostasis, the importance of the KCCs in Cl− homeostasis of cardiomyocytes represents a nascent area of research.
GRANTS
This work was supported by National Institute of Neurological Disorders and Stroke Grant NS-36296 and by institutional funds.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: S.P.A., C.L., and J.A.P. performed experiments; S.P.A., C.L., and J.A.P. analyzed data; C.L. and J.A.P. interpreted results of experiments; C.L. and J.A.P. prepared figures; C.L. and J.A.P. edited and revised manuscript; C.L. and J.A.P. approved final version of manuscript; J.A.P. conception and design of research; J.A.P. drafted manuscript.
ACKNOWLEDGMENTS
We are indebted to Drs. Peter Cala, Jonathan Widdicombe, and Don Bers for many helpful discussions and critical readings of the manuscript. We thank Aldrin Gomes for providing the cardiac troponin-I antibodies.
REFERENCES
- 1. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acid Res 25: 3389–3402, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez BV, Kieller DM, Quon AL, Markovich D, Casey JR. Slc26a6: a cardiac chloride-hydroxyl exchanger and predominant chloride-bicarbonate exchanger of the mouse heart. J Physiol 561: 721–734, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baumgarten CM, Fozzard HA. Intracellular chloride activity in mammalian ventricular muscle. Am J Physiol Cell Physiol 241: C121–C129, 1981 [DOI] [PubMed] [Google Scholar]
- 4. Berglund K, Schleich W, Krieger P, Loo LS, Wang D, Cant NB, Feng G, Augustine GJ, Kuner T. Imaging synaptic inhibition in transgenic mice expressing the chloride indicator, Clomeleon. Brain Cell Biol 35: 207–228, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berglund K, Schleich W, Wang H, Feng G, Hall WC, Kuner T, Augustine GJ. Imaging synaptic inhibition throughout the brain via genetically targeted Clomeleon. Brain Cell Biol 36: 101–118, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blaesse P, Guillemin I, Schindler J, Schweizer M, Delpire E, Khiroug L, Friauf E, Nothwang HG. Oligomerization of KCC2 correlates with development of inhibitory neurotransmission. J Neurosci 26: 10407–10419, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boettger T, Hubner CA, Maier H, Rust MB, Beck FX, Jentsch TJ. Deafness and renal tubular acidosis is mice lacking the K-Cl cotransporter, KCC4. Nature 416: 874–878, 2002 [DOI] [PubMed] [Google Scholar]
- 8. Boettger T, Rust MB, Maier H, Seidenbecher T, Schweizer M, Keating DJ, Faulhaber J, Ehmke H, Pfeffer C, Scheel O, Lemcke B, Horst J, Leuwer R, Pape HC, Völkl H, Hübner CA, Jentsch TJ. Loss of K-Cl co-transporter KCC3 causes deafness, neurodegeneration and reduced seizure threshold. EMBO J 22: 5422–5434, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bräuer M, Frei E, Claes L, Grissmer S, Jäger H. Influence of K-Cl cotransporter activity on activation of volume-sensitive Cl− channels in human osteoclasts. Am J Physiol Cell Physiol 285: C22–C30, 2003 [DOI] [PubMed] [Google Scholar]
- 10. Byun N, Delpire E. Axonal and periaxonal swelling precede peripheral neurodegeneration in KCC3 knockout mice. Neurobiol Dis 28: 39–51, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chomczynski P, Sachi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159, 1987 [DOI] [PubMed] [Google Scholar]
- 12. Delpire E, Gagnon KB. Genome-wide analysis of SPAK/OSR1 binding motifs. Physiol Genomics 28: 223–231, 2007 [DOI] [PubMed] [Google Scholar]
- 13. DiFulvio M, Lincoln TM, Lauf PK, Adragna NC. Protein kinase G regulates potassium chloride cotransporter-3 expression in primary cultures of rat vascular smooth muscle cells. J Biol Chem 276: 21046–21052, 2001 [DOI] [PubMed] [Google Scholar]
- 14. Duan D. Phenomics of cardiac chloride channels: the systematic study of chloride channel function in the heart. J Physiol 587: 2163–2177, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol 5: 3610–3616, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fiumelli H, Briner A, Pusharjov M, Blaesse P, Belem BJ, Dayer AG, Kaila K, Martin JL, Vutskits L. An ion transport-independent role for the cation-chloride cotransporter KCC2 in dendritic spinogenesis in vivo. Cerebral Cortex First 2012. February 17 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17. Harvey RD. Cardiac chloride currents. News Physiol Sci 11: 175–181, 1996 [Google Scholar]
- 18. Harvey RD, Clark CD, Hume JR. Chloride current in mammalian cardiac myocytes: novel mechanism for autonomic regulation of action potential duration and resting membrane potential. J Gen Physiol 95: 1077–1102, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoffmann EK, Dunham PB. Membrane mechanisms and intracellular signaling in cell volume regulation. Int Rev Cytol 161: 173–262, 1995 [DOI] [PubMed] [Google Scholar]
- 20. Horn Z, Ringstedt T, Blaesse P, Kaila K, Herlenius E. Premature expression of KCC2 in embryonic mice perturbs neural development by an ion transport-independent mechanism. Eur J Neurosci 31: 2142–2155, 2010 [DOI] [PubMed] [Google Scholar]
- 21. Horres CR, Lieberman M, Purdy JE. Growth orientation of heart cells on nylon monofilament. J Membr Biol 34: 313–329, 1977 [DOI] [PubMed] [Google Scholar]
- 22. Hubner CA, Stein V, Hermans-Borgmeyer I, Meyer T, Ballanyi K, Jentsch TJ. Disruption of KCC2 reveals an essential role of K-Cl cotransport already in early synaptic inhibition. Neuron 30: 515–524, 2001 [DOI] [PubMed] [Google Scholar]
- 23. Hume JR, Harvey RD. Chloride conductance pathways in heart. Am J Physiol Cell Physiol 261: C399–C412, 1991 [DOI] [PubMed] [Google Scholar]
- 24. Lai ZF, Nishi K. Intracellular chloride activity increases in guinea pig ventricular muscle during simulated ischemia. Am J Physiol Heart Circ Physiol 275: H613–H619, 1998 [DOI] [PubMed] [Google Scholar]
- 25. Lauf PK, Di Fulvio M, Srivastava V, Sharma N, Adragna NC. KCC2a expression in a human fetal lens epithelial cell line. Cell Physiol Biochem 29: 13–22, 2012 [DOI] [PubMed] [Google Scholar]
- 26. Lauf PK, Misri S, Chimote AA, Adragna NC. Apparent intermediate K conductance channel hyposmotic activation in human lens epithelial cells. Am J Physiol Cell Physiol 294: C820–C832, 2008 [DOI] [PubMed] [Google Scholar]
- 27. Lee HHC, Walker JA, Williams JR, Goodier RJ, Payne JA, Moss SJ. Direct protein kinase C-dependent phosphorylation regulates the cell surface stability and activity of the potassium chloride cotransporter, KCC2. J Biol Chem 282: 29777–29784, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Li H, Khirug S, Cai C, Ludwig A, Blaesse P, Kolikova J, Afzalov R, Coleman SK, Lauri S, Airaksinen MS, Keinanen K, Khiroug L, Saarma M, Kaila K, Rivera C. KCC2 interacts with the dendritic cytoskeleton to promote spine development. Neuron 56: 1019–1033, 2007 [DOI] [PubMed] [Google Scholar]
- 29. Mercado A, Broumand V, Zandi-Nejad K, Enck A, Mount DB. A C-terminal domain in KCC2 confers constitutive K-Cl cotransport. J Biol Chem 281: 1016–1026, 2006 [DOI] [PubMed] [Google Scholar]
- 30. Mercado A, Vazquez N, Song L, Cortes R, Enck AH, Welch R, Delpire E, Gamba G, Mount DB. NH2-terminal heterogeneity in the KCC3 K+-Cl− cotransporter. Am J Physiol Renal Physiol 289: F1246–F1261, 2005 [DOI] [PubMed] [Google Scholar]
- 31. Payne JA. Functional characterization of the neuronal-specific K-Cl cotransporter: implications for [K+]o regulation. Am J Physiol Cell Physiol 273: C1516–C1525, 1997 [DOI] [PubMed] [Google Scholar]
- 32. Payne JA, Rivera C, Voipio J, Kaila K. Cation-chloride cotransporters in neuronal communication, development, and trauma. Trends Neurosci 26: 199–206, 2003 [DOI] [PubMed] [Google Scholar]
- 33. Payne JA, Stevenson TJ, Donaldson LF. Molecular characterization of a putative K-Cl cotransporter in rat brain: a neuronal-specific isoform. J Biol Chem 271: 16245–16252, 1996 [DOI] [PubMed] [Google Scholar]
- 34. Payne JA, Xu JC, Haas M, Lytle CY, Ward D, Forbush B., III Primary structure, functional expression, and chromosomal localization of the bumetanide-sensitive Na-K-Cl cotransporter in human colon. J Biol Chem 270: 17977–17985, 1995 [DOI] [PubMed] [Google Scholar]
- 35. Piwnica-Worms D, Jacob R, Horres CR, Lieberman M. Potassium-chloride cotransport in cultured chick heart cells. Am J Physiol Cell Physiol 249: C337–C344, 1985 [DOI] [PubMed] [Google Scholar]
- 36. Piwnica-Worms D, Jacob R, Horres CR, Lieberman M. Transmembrane chloride flux in tissue-cultured chick heart cells. J Gen Physiol 81: 731–748, 1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prasad V, Bodi I, Meyer JW, Wang Y, Ashraf M, Engle SJ, Doetschman T, Sisco K, Nieman ML, Miller ML, Lorenz JN, Shull GE. Impaired cardiac contractility in mice lacking both the AE3 Cl−/HCO3− exchanger and the NKCC1 Na+-K+-2Cl− cotransporter. J Biol Chem 283: 31303–31314, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rivera C, Voipio J, Payne JA, Ruusuvuori E, Lahtinen H, Lamsa K, Pirvola U, Saarma M, Kaila K. A K-Cl cotransporter, KCC2, is the switch to hyperpolarizing GABA action during neuronal maturation. Nature 397: 251–255, 1999 [DOI] [PubMed] [Google Scholar]
- 39. Rust MB, Alper SL, Rudhard Y, Shmukler BE, Vicente R, Brugnara C, Trudel M, Jentsch TJ, Hübner CA. Disruption of erythroid K-Cl cotransporters alters erythrocyte volume and partially rescues erythrocyte dehydration in SAD mice. J Clin Invest 117: 1708–1717, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shi L, Jacob R, Piwnica-Worms D, Lieberman M. Na+-K+-2Cl cotransport in cultured embryonic chick heart cells. Am J Physiol Cell Physiol 253: C721–C730, 1987 [DOI] [PubMed] [Google Scholar]
- 41. Uvarov P, Ludwig A, Markkanen M, Pruunsild P, Kaila K, Delpire E, Timmusk T, Rivera C, Airaksinen MS. A novel N-terminal isoform of the neuronal-specific K-Cl cotransporter KCC2. J Biol Chem 282: 30570–30576, 2007 [DOI] [PubMed] [Google Scholar]
- 42. Vaughan-Jones RD. Chloride activity and its control in skeletal and cardiac muscle. Phil Trans R Soc Lond B Biol Sci 299: 537–548, 1982 [DOI] [PubMed] [Google Scholar]
- 43. Vaughan-Jones RD. Nonpassive chloride distribution in mammalian heart muscle: micro-electrode measurement of the intracellular chloride activity. J Physiol 295: 83–109, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vaughan-Jones RD. Regulation of chloride in quiescent sheep-heart Purkinje fibres studied using intracellular chloride and pH-sensitive micro-electrodes. J Physiol 295: 111–137, 1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wei WC, Akerman CJ, Newey SE, Pan J, Clinch NWV, Jacob Y, Shen MR, Wilkins RJ, Ellory JC. The potassium-chloride cotransporter 2 promotes cervical cancer cell migration and invasion by an ion transport-independent mechanism. J Physiol 589: 5349–5359, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Williams JR, Sharp JW, Kumari VG, Wilson M, Payne JA. The neuronal-specific K-Cl cotransporter, KCC2: antibody development and initial characterization of the protein. J Biol Chem 274: 12656–12664, 1999 [DOI] [PubMed] [Google Scholar]
- 47. Yamawake N, Hirano Y, Sawanobori R, Hiraoka M. Arrhythmogenic effects of isoproterenol-activated Cl− current in guinea-pig ventricular myocytes. J Mol Cell Cardiol 24: 1047–1058, 1992 [DOI] [PubMed] [Google Scholar]