Abstract
Over the past 80 years, physiological research has moved progressively in a reductionist direction, providing mechanistic information on a smaller and smaller scale. This trend has culminated in the present focus on “molecular physiology,” which deals with the function of single molecules responsible for cellular function. There is a need to assemble the information from the molecular level into models that explain physiological function at cellular, tissue, organ, and whole organism levels. Such integration is the major focus of an approach called “systems biology.” The genome sequencing projects provide a basis for a new kind of systems biology called “data-rich” systems biology that is based on large-scale data acquisition methods including protein mass spectrometry, DNA microarrays, and deep sequencing of nucleic acids. These techniques allow investigators to measure thousands of variables simultaneously in response to an external stimulus. My laboratory is applying such an approach to the question: “How does the peptide hormone vasopressin regulate water permeability in the renal collecting duct?” We are using protein mass spectrometry to identify and quantify the phosphoproteome of collecting duct cells. The response to vasopressin, presented in the form of a network model, includes a general downregulation of proline-directed kinases (MAP kinases and cyclin-dependent kinases) and upregulation of basophilic kinases (ACG kinases and calmodulin-dependent kinases). Further progress depends on characterization and localization of candidate protein kinases in these families. The ultimate goal is to use multivariate statistical techniques and differential equations to obtain predictive models describing vasopressin signaling in the renal collecting duct.
Keywords: aquaporin-2, signaling networks, collecting duct, phosphoproteomics, mass spectrometry, Nedd4-2
Vasopressin Action in the Kidney
The most distal segments of the renal tubule, the connecting tubules and collecting ducts, have variable water permeability that is controlled in response to the peptide hormone vasopressin, allowing water excretion to vary physiologically (46). These renal tubule segments express three molecular water channels called “aquaporins” (57). These are aquaporin-2 (AQP2), aquaporin-3 (AQP3) and aquaporin-4 (AQP4). AQP2 is present in the apical plasma membrane and in endosomes (6, 20, 56). The latter two aquaporins are seen chiefly in the basolateral plasma membrane (16, 71). Although there is evidence for regulation of the basolateral water channels by vasopressin (15, 16), it seems clear that the regulation of water permeability of the collecting duct epithelium is mediated largely through vasopressin's effects on AQP2 (57).
There are at least two modes of water permeability regulation in the kidney corresponding to two processes that control the amount of active AQP2 in the apical plasma membrane. The first mode is “short-term regulation” occurring over a period of 5–30 min as a result of the regulation of trafficking of AQP2-containing membrane vesicles to and from the apical plasma membrane in response to vasopressin (55). The second mode is “long-term regulation” occurring over a period of hours to days as a result of regulation of whole cell AQP2 abundance by vasopressin (13, 28, 56). Both modes depend on the binding of vasopressin to V2-type vasopressin receptors (gene symbol: Avpr2)1 present in the basolateral membrane. The downstream signaling events that are pertinent to the two modes of regulation are, however, largely undiscovered. In this review, I discuss application of systems biology techniques to the discovery of the signaling processes that are triggered by V2 receptor occupation in the renal collecting duct.
Reductionist Versus Integrative Approaches to Biology
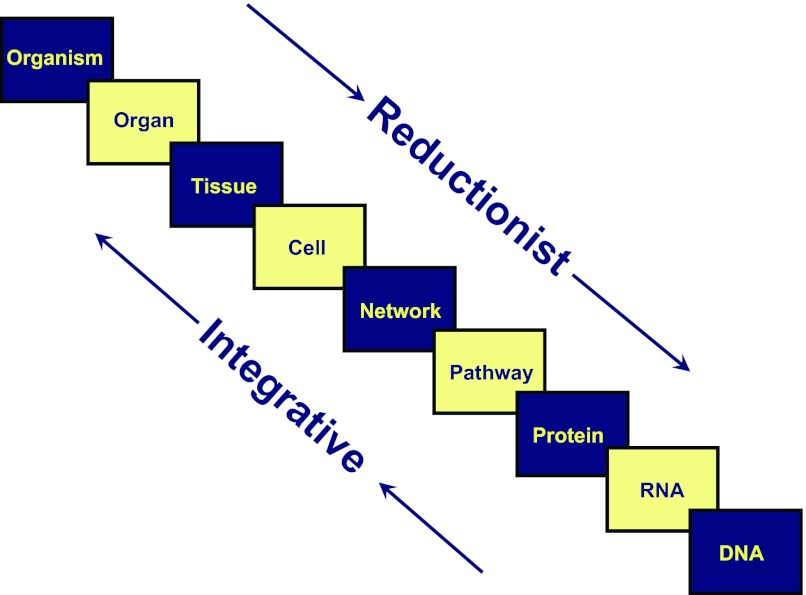
Over the past 80 years, research in the field of physiology (and biological study in general) has proceeded in a reductionist direction, dealing with smaller and smaller structural levels (Fig. 1). Taking renal physiology as an example, in the 1930s, whole animal or whole person studies were the major sources of information employing clearance techniques. This allowed information to be gleaned about the three major processes determining excretion, namely glomerular filtration, secretion, and reabsorption (69). The rise of micropuncture (21) and the isolated perfused tubule technique (7) during the 1960s changed the focus of renal physiology to a tissue level, providing information about each of the nephron segments or about the glomerulus. Application of these techniques together with biochemically based techniques such as transport measurements in isolated brush border vesicles (54) provided a wealth of detail about transport mechanisms. This knowledge paved the way for the next major era in renal physiology in the 1980s and 90s, during which cDNAs for many of the important renal transporters were cloned and sequenced. This development switched the emphasis from a tissue and cellular level down to a molecular level. Success in cloning many of these cDNAs was greatly abetted by the development of the Xenopus oocyte expression system (12). This expression cloning approach allowed cDNA pools and ultimately single cDNAs to be selected on the basis of functional assays that were designed utilizing the findings of isolated perfused tubule experiments, micropuncture experiments, and isolated brush-border vesicle experiments done in the preceding decades. Thus by the mid- to late 1990s renal physiologists had obtained the primary structure of the proteins that mediate the most important transport processes along the nephron. The cDNAs could then be expressed in cultured cell lines or in Xenopus oocytes and studied, either in native form or mutated form, to gain a greater understanding of the relationship between protein structure and function. Thus, by the end of the 20th century there had been a spectacular expansion of information about transport in the kidney based on a progression of knowledge from reductionist approaches.2
Fig. 1.
Concept figure illustrating the different levels of integration commonly investigated in physiological studies. Physiological research over the past 80 years has progressed toward the analysis of smaller and smaller structural elements including single molecules (bottom right). The object of systems biology methods is to integrate information from molecular levels toward the whole organism level, using computational tools such as modeling using differential equations and multivariate statistical analyses to identify network structures.
Because of the drive toward reductionism in physiology, we have achieved a high level of understanding of structure and function of many of the individual proteins that mediate important physiological processes including transport along the renal tubule. However, knowledge of molecular structure and function does not necessarily imply a mastery over the physiological knowledge needed to understand disease processes in humans. Most human disease is not easily explained as an effect of a single dysfunctional gene and protein, but rather often represents complex phenomena that depend on nonlinear integration of the functional properties of many proteins. Even so-called “monogenic” diseases have polygenic consequences. Likewise, normal physiological phenomena at the organism or organ level can rarely be understood from knowledge about a single protein. Therefore, there is a need to develop methods to integrate information about the molecular functions of individual proteins and their molecular interactions to create predictive models of cellular, tissue, organ, and whole organism function (Fig. 1). The ultimate goal is to understand disease processes and to discover how to treat them. Thus, there is a need for integration, i.e., learning how many small pieces can assemble into larger ones to create complex emergent behavior. This integration process has been referred to as “systems biology.”
Systems Biology and Physiology
Data-poor systems biology.
Systems biology is not a new approach. Starting in the 1960s and 70s with the advent of mainframe computers, various investigators used differential equation-based mathematical modeling to integrate data from the literature. The differential equations were constructed to represent physical principles such as mass balance, energy balance, and various laws that represent the relationships between driving forces and movements of various substances, e.g., nonequilibrium thermodynamics (42). Typically, data from a myriad of sources were used in parameter estimation in these models, a labor intensive process that could only be done manually, assigning one parameter at a time. This type of systems biology has been referred to by Westerhoff and Palsson (74) as “data-poor” systems biology. Despite its laborious nature, this approach has been very productive. For example, models of the kidney concentrating mechanism (75) have been useful in generating hypotheses that could be tested experimentally (26). My own mathematical modeling of the kidney medulla [done as part of my PhD dissertation (44)] led to the conclusion that a specialized urea transport mechanism was required in the inner medullary portion of the collecting duct to explain the observed cortico-medullary urea gradient. This observation from mathematical modeling stimulated a series of studies using isolated perfused inner medullary collecting ducts that ultimately demonstrated the predicted transporter (10, 11, 45, 65), eventually leading to the cloning of corresponding cDNAs (41, 68, 76) and gene knockouts of collecting duct urea transporters in mice (17, 18).
Data-rich systems biology.
With the advent of genome sequencing projects for various animals, plants, and microorganisms at the beginning of the current century, systems biology has moved to a so-called “data-rich” format (74). As a result of these sequencing projects, we can now limit our view to a finite set of protein-coding genes (approximately 21,000 in humans, mice, and rats) when looking for protein species that may explain a particular physiological phenotype. This translates to a finite set of molecular hypotheses. More importantly, the genome sequence information has made possible a set of technologies that are capable of providing genome- or proteome-wide read-outs of physiological experiments. These large-scale data acquisition techniques include protein mass spectrometry (58), hybridization-based expression microarrays (3), and deep sequencing of nucleic acids (27). Each of these modalities allows the experimenter to identify the cellular responses to physiological stimuli comprehensively across the entire proteome, transcriptome, or epi-genome. Such experimental approaches are becoming relatively straightforward and can be readily applied to a variety of physiological problems at the cellular level.
To some extent such experiments can be interpreted through direct perusal of the data. For example, a protein mass spectrometry study of renal inner medullary collecting duct cells isolated from rat kidneys showed that vasopressin changes the phosphorylation of the aquaporin-2 water channel at four sites near the carboxyl terminus of the protein (30, 34), a finding that has spawned numerous follow-up studies both within our own laboratory and in others. Additional studies revealed the presence of three vasopressin-regulated phosphorylation sites in the vasopressin-regulated urea transporter UT-A (5).
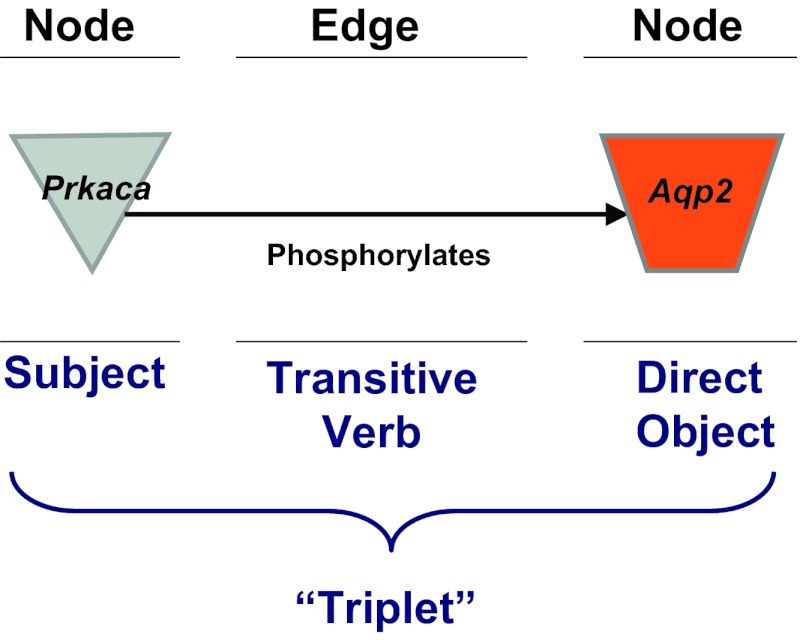
However, most of the information present in large-scale data sets from these technologies cannot be so easily interpreted, and requires computational approaches aimed at achieving large-scale data integration to create models describing the interactions among the relevant proteins. In a particular physiological experiment involving a given physiological perturbation, the large-scale integration task often consists of two major elements: 1) analysis of the responding proteins, transcripts, or genes within a data set to determine how the responding entities are related to one another functionally; and 2) analysis of the responding proteins, transcripts, or genes to determine their relationships to information that has been obtained in previous studies. The latter analysis typically results in the generation of a so-called “network” consisting of “nodes” and “edges” (Fig. 2). For a protein network, the nodes represent individual proteins and the edges are the relationships between proteins. A pair of nodes and edges that connect them can be viewed as a grammatical sentence known as a “triplet,” where one node is the subject, the edge is the verb, and the other node is the direct object. In the example shown in Fig. 2, the sentence represented is “Protein kinase A catalytic subunit phosphorylates aquaporin-2.” Large numbers of triplets can be merged into directed graphs like that shown in Fig. 3 depicting the vasopressin-signaling network for the renal inner medullary collecting duct based on phosphoproteomic studies (33).
Fig. 2.
Terminology used in network models of physiological processes. Individual “nodes,” typically corresponding to specific proteins, are connected by “edges.” The combination of two nodes and the edge connecting them can be viewed as analogous to an English sentence (subject/verb/direct object) called a “triplet.” Many such triplets can be connected to construct an interaction network like that shown in Fig. 3.
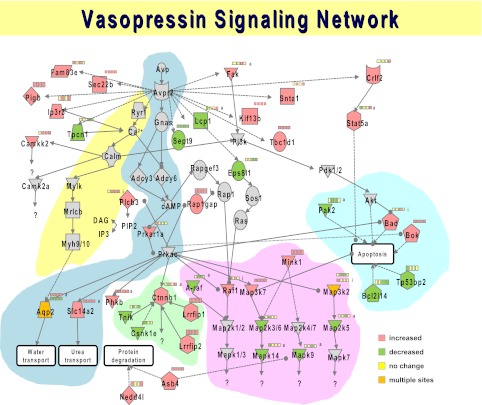
Fig. 3.
Vasopressin signaling network. Constructed from data reported by Hoffert et al. (33). Colored nodes represent proteins that contain phosphorylation sites that are changed in abundance in response to exposure of suspensions of rat inner medullary collecting ducts (IMCDs) to the vasopressin analog dDAVP: red nodes, increased phosphorylation; green nodes, decreased phosphorylation. Proteins represented by individual nodes are designated by official gene symbols. User can obtain full protein annotation for each by going to http://www.ncbi.nlm.nih.gov/protein and inserting the gene symbol. Series of four small boxes show direction of responses at 0.5, 2, 5, and 15 min following dDAVP addition (red, up; green, down; yellow, no change). Edges are drawn based on ad hoc manual curation (see text). Background highlight shows individual modules: blue, canonical vasopressin pathway; yellow, calcium-dependent vasopressin signaling; green, Wnt signaling; pink, MAP kinase signaling; turquoise, apoptosis.
To carry out large-scale data integration needed for generation of network models, databases of triplets (also known as “protein interactions”) can be gleaned from the general literature to identify possible interactions between nodes of the physiological system under study. There is a major effort within systems biology to develop computer-based text mining algorithms that can extract appropriate information (i.e., triplets) from the literature to create these databases (29). Lacking full success in this area, a number of databases have been constructed chiefly through manual curation. These include two commercially available databases within Ingenuity Pathway Analysis and Metacore software suites. These approaches and others like them are useful in organizing large data sets but continue to have significant limitations with regard to the ability to construct causal models (43, 53, 67). Consequently, the network models that can be produced with today's technologies fall short of having the predictive power of differential equation-based models. Nevertheless, network models like that shown in Fig. 3 provide a useful way to consolidate and visualize the findings of large-scale experiments.
Using Mass Spectrometry to Construct a Vasopressin-Signaling Network for the Renal Collecting Duct
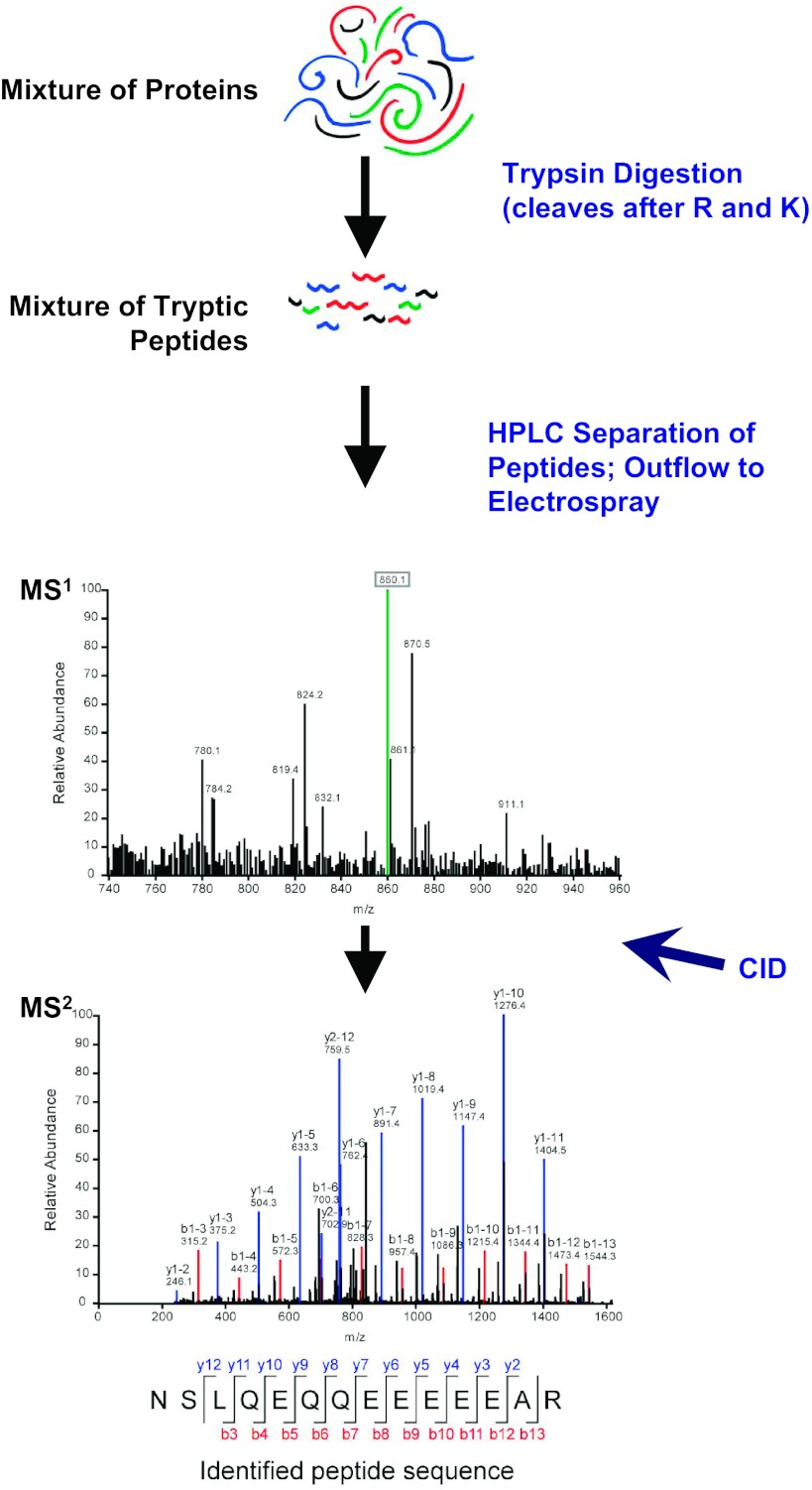
My laboratory is using systems biology approaches to identify the vasopressin-signaling network responsible for regulation of water permeability in the renal collecting duct. The studies use liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) to identify and quantify thousands of phosphorylation sites in native inner medullary collecting ducts isolated from rat kidneys (5, 33, 34) and in cultured mouse mpkCCD(clone 11) cells (62). Figure 4 shows a general scheme describing how LC-MS/MS can be used to identify thousands of peptides in a given biological sample. Application of the method to identification and quantification of phosphopeptides has been the subject of several reviews (31, 32, 59, 61).
Fig. 4.
Principal elements of protein mass spectrometry using LC-MS/MS (liquid chromatography coupled to tandem mass spectrometry) as is most commonly carried out in physiological studies. A mixture of thousands of proteins is proteolyzed to a mixture of hundreds of thousands of peptides using trypsin, which cuts after arginines and lysines. Acidified tryptic peptides can be gradually introduced to the mass spectrometer using reverse phase HPLC to stratify the peptides. The mass spectrometer measures the mass-to-charge ratios (m/z) of the peptides (MS1 level). The most abundant peptides are fragmented by collision-induced dissociation (CID), through molecular collisions with an inert gas, to produce fragmentation spectra (MS2 level) that can be mapped to theoretical tryptic peptides present in databases derived from genomic sequence data for a particular animal or plant species. This approach can identify many thousands of peptides in a single run. The method can be adapted for peptide quantification as discussed in the text.
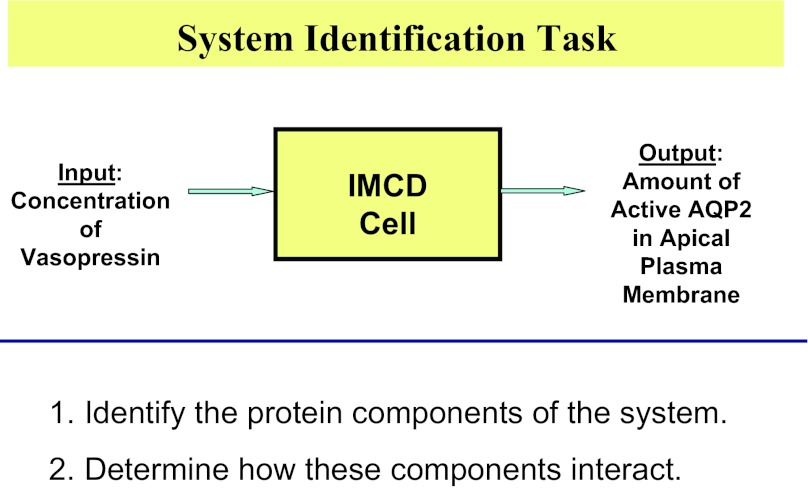
The studies aim to identify a signaling network (Fig. 5) linking the input (a change in the concentration of vasopressin in the extracellular environment) to a key output (a change in the amount of active aquaporin-2 in the apical plasma membrane). The signaling network to be identified (yellow box) is the set of proteins in the collecting duct cell that link the input to the output, carrying information via a sequence of biochemical changes in the cell. We can identify two subtasks (Fig. 5, bottom): 1) identification of the protein components of the system, i.e., the proteins expressed in collecting duct cells including their posttranslational modifications; and 2) determination of how these components interact.
Fig. 5.
The system identification task discussed in this paper. The studies seek to identify a network of proteins (yellow box) that connect the changes in the input (altered vasopressin concentration in extracellular fluid) to changes in the output [change in the amount of active aquaporin-2 (AQP2) in the apical plasma membrane of collecting duct cells]. The system identification task can be divided into two subtasks listed at the bottom.
Identification of system components.
The former subtask is the easiest of the two. Application of standard transcriptomics (from microarray studies or RNA-seq) and proteomics analysis has identified most of the gene products expressed in native inner medullary collecting duct cells and in mpkCCD(clone 11) cells. This information has been placed on publicly accessible databases described by Huling et al. (37) and available at http://helixweb.nih.gov/ESBL/Database/ or https://intramural.nhlbi.nih.gov/labs/LKEM_G/LKEM/Pages/--TranscriptomicandProteomicDatabases.aspx. Collecting duct cells appear to express approximately 8,000 out of the 21,000 protein coding genes in the genome, and the user can easily identify by searching these databases whether a given protein or a related protein is expressed in these cells. Indeed, these databases can be interrogated online in a variety of ways or the entire database can be downloaded by the user.
Databases enumerating phosphorylation sites that have been identified in collecting duct proteins (as well as proteins in other renal tubule segments) are available at the same URLs listed in the previous paragraph. In general, these phosphorylation sites are modifications of the side-chains of serines, threonines, or tyrosines through covalent attachment of a phosphate group. The phosphate addition produces characteristic changes in the mass-to-charge ratio of peptides that can be recognized by the mass spectrometer. The databases have been derived from a number of LC-MS/MS-based studies profiling phosphorylation sites observed in collecting duct cells (5, 33, 34, 36, 62, 79) and other kidney cells (19, 23).
Protein phosphorylation databases provide useful tools for the study of cell signaling. However, other types of posttranslational modifications are important in cell signaling (including glycosylation, acetylation, methylation, S-nitrosylation, SUMO-ylation, and ubiquitylation) and their identification could provide key information regarding the nature of vasopressin action in the collecting duct. However, protein phosphorylation plays a preeminent role in vasopressin signaling and so has drawn most of our attention.
Determination of how system components interact.
The second subtask described below (Fig. 5, bottom) requires that LC-MS/MS be adapted to allow quantification of proteins and phosphorylation sites. The quantification techniques available include label-free methods and labeling approaches such as Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) and isobaric Tags for Relative and Absolute Quantitation (iTRAQ). The technical details of these approaches have been reviewed previously (8, 58, 59) and are beyond the scope of this paper. We used quantitative phosphoproteomics methodologies to determine what phosphorylation sites undergo changes in abundance in response to vasopressin (using the V2 vasopressin-receptor selective analog dDAVP) both in suspensions of native inner medullary collecting ducts (4, 33, 34, 36) and in cultured collecting duct mpkCCD cells (62). The proteins that bear the changes can then be considered elements (nodes) in the vasopressin signaling network (Fig. 3), although they are not necessarily involved in regulation of aquaporin-2 or water permeability. Indeed, elements of Fig. 3 point to potential roles of vasopressin in processes that may be unrelated to regulation of transport. For example, a set of proteins that contain vasopressin-regulated phosphorylation sites are involved in regulation of programmed cell death or apoptosis (Fig. 3, turquoise background: Bad, Bok, Pak2, Bcl2l14, and Tp53bp). The direction of these phosphorylation changes suggested the hypothesis that vasopressin inhibits apoptosis, which was confirmed by direct experiments (33). Other regions of Fig. 3 are enriched for proteins in the Wnt signaling pathway (green background: Ctnnb1, Lrrfip1, Lrrfip2, Csnk1e, Tnik, and Phkb), and pathways involved in the regulation of various MAP kinases (pink background) including p38 (Mapk14) and Jnk2 (Mapk9). The phosphorylation changes seen for p38 and Jnk2 are consistent with inactivation of these kinases by vasopressin as seen previously for additional MAP kinases, viz. Erk1 and Erk2 (60). Finally, the results showed changes in phosphorylation of two proteins involved in regulation of protein degradation, namely Nedd4-2 (Nedd4l) and Ankyrin repeat and SOCS box protein 4 (Asb4). Nedd4-2 is a well-characterized ubiquitin E3 ligase in the renal collecting duct that is involved in the regulation of the epithelial sodium channel ENaC (70). Since, the inner medullary collecting duct exhibits little or no sodium transport activity (64) and only very low levels of ENaC expression (25), we speculate that Nedd4-2 may be involved in regulation of water or urea transport in this segment. In general, it can be assumed that a subset of the vasopressin-induced phosphorylation events depicted in Fig. 3 play roles in the regulation of aquaporin-2. The identification of these phosphorylation changes will require additional studies using systems techniques (described in the remainder of this review) to dissect different pathways within the overall network.
Identification of Protein Kinases Responsible for Regulated Phosphorylation in the Collecting Duct
The phosphoproteomics studies discussed in the last section identified a large number of phosphorylation sites in the collecting duct that exhibit altered abundance in response to vasopressin. These sites and their associated proteins represent nodes in the vasopressin signaling network (Fig. 3). The edges drawn in Fig. 3 are tentatively identified based on our reading of the literature and did not employ any of the available databases for automatic generation of a network architecture. Hence further studies are needed to assemble these nodes into a causal network that can explain how vasopressin regulates aquaporin-2 location and expression in the renal collecting duct. The general question under consideration is therefore how to draw the correct edges representing causal interactions between nodes. One way to address this is to identify the protein kinases and protein phosphatases involved in regulating the level of phosphorylation at each of the regulated sites. Our current focus is on the protein kinases.
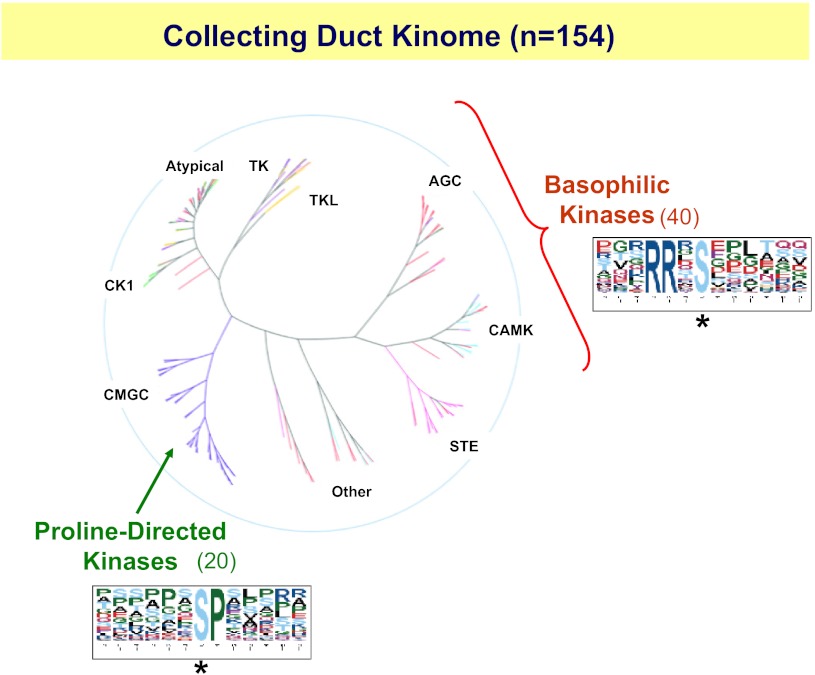
Approximately 518 protein kinases are expressed in mammalian genomes (49). Only a fraction of these protein kinases are expressed in renal collecting duct cells. Collecting duct kinases have been identified in transcriptomic profiling experiments using Affymetrix expression arrays. This approach has provided a comprehensive list of genes expressed in native inner medullary collecting duct cells (73) and in mpkCCD cells (77). Among the genes expressed in both cell types are those that code for 154 protein kinases (Fig. 6). Thus, through transcriptomic profiling we have reduced the list of protein kinases that are candidates for roles in vasopressin signaling by 70% (from 518 to 154). These kinases can be classified into distinct families based on sequence similarities (Fig. 6). In the following, we discuss a strategy for narrowing down the list of protein kinases to the few that are likely involved in vasopressin signaling.
Fig. 6.
The collecting duct kinome consists of 154 distinct protein kinases out of the 518 that exist in the whole genome. These kinases can be classified according to sequence similarity into distinct groups indicated in black. Vasopressin-induced changes in phosphorylation consist largely of increases in phosphorylation at sites recognized by basophilic kinases (AGC and CAMK families, sequence logo at right) and decreases in phosphorylation at target sites for proline-directed kinases (CMGC family including MAP kinases and cyclin-dependent kinases, sequence logo at bottom). This limits candidates for kinases involved in vasopressin signaling to 40 basophilic kinases and 20 proline-directed kinases. Asterisks below sequence logos indicate phosphorylated amino acids.
It is widely accepted that prediction of what protein kinases phosphorylate particular protein targets depends on two types of constraints (48): 1) linear target motif specificity based on the amino acid sequence just upstream and downstream from the phosphorylation site in the target protein; and 2) requirement for physical propinquity of the kinases and their prospective targets in the same subcellular compartment.
Linear target motif specificity.
A phosphoproteomic study by Rinschen et al. (62) provided key information that narrows the possible kinases involved in vasopressin signaling to fewer than the 154 protein kinases specified in Fig. 6. This study identified 273 phosphorylation sites that were increased and 254 phosphorylation sites that were decreased in response to dDAVP exposure across the mpkCCD cell proteome. When the sequences surrounding the sites that were increased were analyzed together it was found that there was a predominance of basic amino acids (lysines and arginines) upstream from the phosphorylated amino acid. Such sites are compatible with the conclusion that vasopressin activates so-called “basophilic” protein kinases. Basophilic kinases include those from the AGC protein kinase family (AGC in Fig. 6) and those from the calmodulin-activated kinase family (CAMK in Fig. 6). This narrows the potential kinases that are activated by vasopressin to the 40 kinases from the two families that are expressed in collecting ducts. In contrast, when the sequences surrounding the 254 phosphorylation sites that were decreased by vasopressin were analyzed, a predominance of sites with a proline in the amino acid following the phosphorylated amino acid was found. This consensus is compatible with the conclusion that vasopressin reduces the activity of so-called “proline-directed” protein kinases, which include those from the MAP kinase family and cyclin-dependent kinase families (CMGC in Fig. 6). Only 20 of these are expressed in native inner medullary collecting duct cells and mpkCCD cells. From these considerations, we have narrowed the number of protein kinases that are likely to be involved in vasopressin signaling by more than 88% (from the 518 present in the genome to 60). Some of these kinases are already present in the vasopressin signaling network (Fig. 3) since they are phosphorylation targets (e.g., Camkk2, Mylk, Prkar1a, Phkb, Tnik, Pak2, Mapk9, and Mapk14).
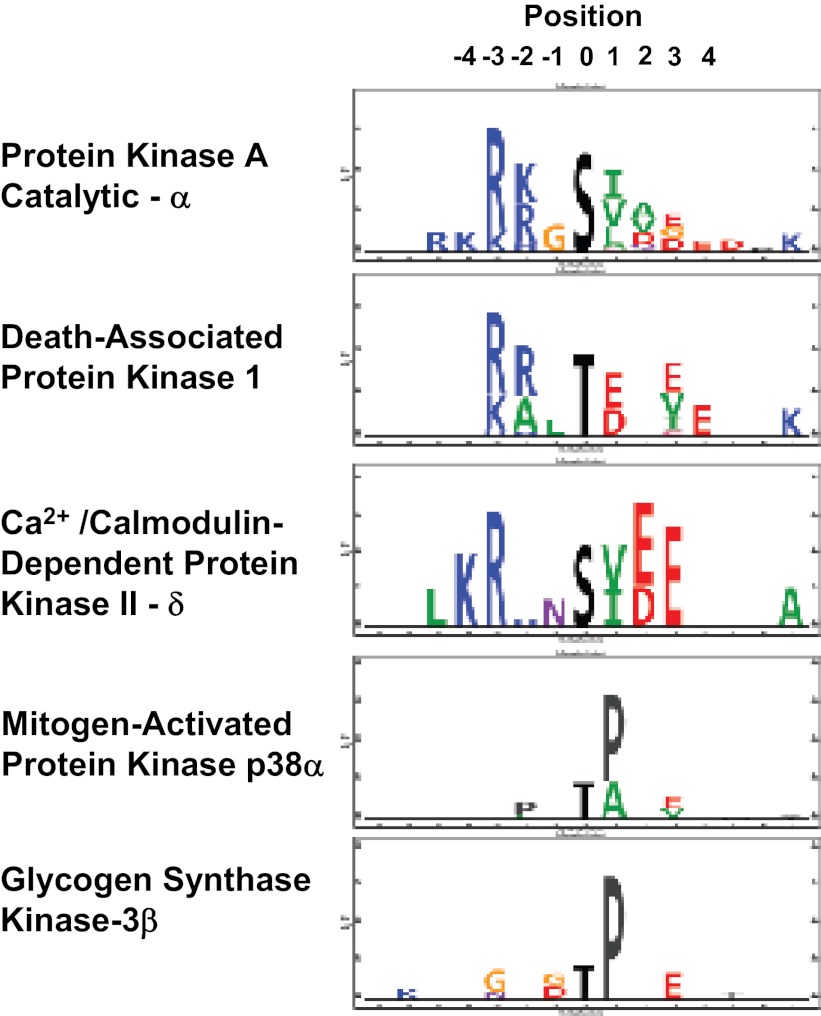
Thus, it is practical to classify protein kinases within general families based on the linear sequence surrounding their target phosphorylation sites. The question remains whether the sequence surrounding a particular phosphorylation site is ever specific enough to identify a single protein kinase that is responsible for a phosphorylation event without additional information. For example, the sequence R-R-X-pS-X (where pS indicates a phosphorylated serine) is often cited as evidence of involvement of one of the protein kinase A isoforms in the phosphorylation event. A number of experimental approaches have been used to identify target sequence preferences for specific protein kinases: 1) curation of sequences of known substrates of a given kinase from multiple literature studies (2); 2) in vitro phosphorylation of peptide arrays (38, 72); and 3) use of mass spectrometry to identify phosphopeptides from native tissues that are phosphorylated as a result of in vitro incubation with recombinant, active protein kinases (14). Many of the consensus sequences reported in the literature are summarized in a database called HPRD (Human Protein Reference Database; 22). A useful compendium of protein kinase sequence preferences derived from peptide array studies has been published as the NetPhorest Kinase Motif Atlas (52). Some data from mass spectrometry-based kinase substrate profiling (14) is shown in Fig. 7 listing target sequence preferences for three basophilic protein kinases expressed in collecting ducts {protein kinase A catalytic subunit α [Prkaca], death-associated protein kinase 1 [Dapk1], and Ca2+/calmodulin-dependent protein kinase II-δ [Camk2d] as well as two proline-directed protein kinases [mitogen-activated protein kinase p38α (Mapk14) and glycogen synthase kinase-3β (Gsk3b)]}. Comparing the first two basophilic kinases, protein kinase A (catalytic-α) and death-associated protein kinase 1, it can be seen that both have a preference for basic amino acids in positions −2 and −3 relative to the phosphorylated serine. Thus, a target sequence R-R-X-pS-X would be compatible with phosphorylation by either of these two kinases, and it may be necessary to rely on the amino acids downstream from the phosphorylation site to discriminate the two. In contrast, the preference for basic amino acids in Ca2+/calmodulin-dependent protein kinase II-δ is shifted upstream to the −3 and −4 position, overlapping the sequence preference reported for Akt and related kinases (22, 52). In general, examination of the data of Douglass et al. (10) together with the NetPhorest Kinase Motif Atlas, and the HPRD, indicates that there may be too much overlap between target sequence preferences to expect to be able to reliably assign the specific protein kinases responsible for a given phosphorylation event solely from sequence data. A comparison of the target sequence preferences for the two proline-directed kinases in Fig. 7 leads to a similar conclusion. Consequently, other approaches must be taken to reliably assign edges to the nascent vasopressin-signaling network depicted in Fig. 3.
Fig. 7.
Examples of kinase target sequence logos obtained for three basophilic kinases and two proline-directed kinases. Logos are based on data from Douglass et al. (14) obtained by use of protein mass spectrometry to identify sites phosphorylated by in vitro incubation with individual active recombinant protein kinases. The data support the view that the sequence surrounding an individual phosphorylation site cannot be expected to provide enough information to identify the kinase responsible for the phosphorylation event (see text).
Requirement for physical propinquity of the kinases and their prospective targets.
It is self-evident that a given kinase can only phosphorylate a target protein if the two come into physical contact. Thus the requirement for colocalization is a useful constraint that can be used to rule out certain kinases that would otherwise be considered candidates for phosphorylation of a given protein based on linear sequence preference (Fig. 3). The subcellular localization of the kinases and substrates can be addressed by a variety of approaches including 1) biochemical separation of subcellular fractions (63, 78) followed by either immunoblotting or mass spectrometry of the fractions to identify the protein amounts in the fractions; and 2) microscopy-based techniques including confocal immunofluorescence and immuno-electron microscopy. It is important to remember that many protein kinases are regulated through processes that involve translocation of the protein from the cytosol to a membrane-bound state (9), so that the colocalization of enzyme and substrate needs to be sought under conditions compatible with activation of the kinase. Many kinases “find” their targets by mutually binding to the same multiprotein complex. Examples include Gsk3β and β-catenin in the canonical Wnt pathway (51), protein kinase A and various substrates via AKAP family proteins (50), and the protein kinases in MAP kinase cascades through the aid of scaffold proteins (47). Hence, it may be practical to identify kinase substrates in a given cell type by immunoprecipitating the kinase under conditions in which the associated proteins are likely to remain bound and then identifying them by mass spectrometry. An important requirement for most of these approaches is the availability of high-quality antibodies or the ability to generate them. With this requirement in mind, we have produced an online antibody-design software application called NHLBI-AbDesigner (http://helixweb.nih.gov/AbDesigner/). This tool allows the user to identify regions of the protein that are predicted to be relatively immunogenic. These peptide sequences can be synthesized, linked to a carrier protein, and used to immunize rabbits or mice.
Identification of Protein Kinase Networks Using Multivariate Statistical Analysis and Protein Kinase Inhibitors
Above, I have proposed that two types of information are key to narrowing down the possible network configurations that could explain the input-output behavior of the collecting duct in its response to vasopressin: 1) determination of the target sequence specificities of every kinase expressed in collecting duct cells; and 2) determination of the subcellular locations of each kinase relative to all potential targets. Thus, once all this information has been collected, hypothetical network models can be proposed. How can these hypothetical models be tested? Two sets of tools will allow us to do the critical hypothesis testing experiments: 1) extensive collections of drugs developed as protein kinase inhibitors; and 2) a set of data analysis techniques that are classified under the general moniker of “multivariate statistical approaches.” I deal briefly with these in the following.
Protein kinase inhibitors.
Because protein kinases have long been viewed as druggable targets chiefly for therapy of malignancies, there exists a wide range of agents that can inhibit protein kinases, many with at least moderate selectivity. Since phospho-specific antibodies are available for many of the vasopressin-regulated phosphorylation sites that we have found in our mass spectrometry-based studies, it will be practical to screen protein kinase inhibitor compounds for their abilities to alter phosphorylation in collecting duct cells. For example, drugs that have been identified to alter aquaporin-2 phosphorylation at any of its four carboxyl-terminal sites can provide useful tools to probe the entire network if the drugs exhibit at least a degree of selectivity. Many of these agents have been characterized before, giving clues to which kinases they may inhibit. However, such agents can be useful even if their kinase targets are not known, or are diverse, if used in experiments involving multivariate statistical analyses.
Multivariate statistical analysis.
The simple idea is that phosphorylation events that correlate highly across a variety of perturbations are likely to be functionally related. They may for example be downstream from the same protein kinase or may be elements present in a cascade of enzymatic reactions. Conversely, phosphorylation events that correlate poorly across a variety of perturbations can be expected to be more distant in the overall signaling network. Based on this concept, experiments can be done in which proteome-wide phosphoproteomic read-outs are obtained for a battery of protein kinase inhibitor molecules at a variety of concentrations. One can then identify the phosphorylation sites that correlate most closely with sites known to be involved in regulation of water transport, e.g., the four phosphorylation sites at the COOH-terminal tail of aquaporin-2 that are believed to be involved in water channel trafficking (34, 35) or serine 552 of β-catenin which is thought to be involved in the regulation of the expression of the Aqp2 gene (66). There exist standard statistical methods for identifying correlations in large data sets, e.g., partial least squares analysis (24, 39, 40). Such approaches identify underlying relationships that allow the network to be partitioned into modules or subdomains that can be investigated experimentally. Ultimately, so-called “structural equation modeling” (1) can be employed to provide a means to map the data into directed graphs and to optimize the fit between the derived network structure and the data.
Conclusion
Here I have described the use of systems biology tools to carry out integrative study of physiological systems at a cellular level. I have illustrated the approaches with data from my own studies focusing on the question, “How does the peptide hormone vasopressin regulate water permeability in renal collecting duct cells?” These studies have exploited phosphoproteomics methodologies to reveal phosphorylation events within the cell that are triggered by vasopressin. Based on the results, we have begun to assemble vasopressin-modulated phosphoproteins into functional networks that may explain the regulation of the water channel aquaporin-2. In addition to phosphoproteins that are involved in transport regulation, the studies have revealed phosphoproteins that point to other processes in collecting duct cells that appear to be regulated by vasopressin. The studies have generated extensive lists of proteins and phosphorylation sites identified in collecting duct cells that have been placed online in the form of user-accessible databases that can be interrogated in various ways.
Both the old-style data-poor approach using differential equations to describe complex systems and the new-style data-rich approach using multivariate statistical analysis and graph theory to create relational models can be exploited to investigate physiological systems at the cellular level. Ultimately, the two types of modeling can be used together in hybrid structures to integrate large-scale experimental data with information from the literature. The ultimate goal of modeling is to expose missing information and thus to guide further experimental work. Thus, although the integrative approach described has been touted over the traditional reductionist approaches in this review, in reality the most effective overall method of attack is to use both reductionist and integrative approaches, presumably via cross-laboratory cooperation and collaboration.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author.
AUTHOR CONTRIBUTIONS
M.A.K. conception and design of the research; analyzed the data; prepared the figures; drafted the manuscript; edited and revised the manuscript; approved the final version of the manuscript.
ACKNOWLEDGMENTS
This work was funded by the Division of Intramural Research of the National Heart, Lung, and Blood Institute (Project ZO1-HL001285 to M. A. Knepper).
Footnotes
Official gene symbols are indicated by italics throughout.
One of the interesting by-products of this trend toward reductionism has been the loss of clear boundaries among traditional biological fields. Thus, by the end of the 20th century, physiological researchers were typically using the same methodologies as other biologists including biological chemists, pharmacologists, or cell biologists, often publishing in the same journals. Thus the trend toward reductionism in physiology may have had the unintended consequence of undermining the traditional field of physiology and perhaps, in some cases, the very existence of physiology departments in US medical schools.
REFERENCES
- 1. Aburatani S. Application of structure equation modeling for inferring a serial transcriptional regulation in yeast. Gene Regul Syst Bio 5: 75–88, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, Pandey A. A curated compendium of phosphorylation motifs. Nat Biotechnol 25: 285–286, 2007 [DOI] [PubMed] [Google Scholar]
- 3. Auer H, Newsom DL, Kornacker K. Expression Profiling Using Affymetrix GeneChip Microarrays. Methods Mol Biol 509: 35–46, 2009 [DOI] [PubMed] [Google Scholar]
- 4. Bansal AD, Hoffert JD, Pisitkun T, Knepper MA. Large-scale LC-MS/MS analysis of collecting duct phosphoproteome reveals three vasopressin-regulated phosphorylation sites on urea transporter (UT-A) (Abstract). J Am Soc Nephrol 20: 828A, 2009. [Google Scholar]
- 5. Bansal AD, Hoffert JD, Pisitkun T, Hwang S, Chou CL, Boja ES, Wang G, Knepper MA. Phosphoproteomic profiling reveals vasopressin-regulated phosphorylation sites in collecting duct. J Am Soc Nephrol 21: 303–315, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barile M, Pisitkun T, Yu MJ, Chou CL, Verbalis MJ, Shen RF, Knepper MA. Large scale protein identification in intracellular aquaporin-2 vesicles from renal inner medullary collecting duct. Mol Cell Proteomics 4: 1095–1106, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burg MB, Grantham JJ, Abramow M, Orloff J. Preparation and study of fragments of single rabbit nephrons. Am J Physiol 210: 1293–1298, 1966 [DOI] [PubMed] [Google Scholar]
- 8. Chaerkady R, Pandey A. Quantitative proteomics for identification of cancer biomarkers. Proteomics Clin Appl 1: 1080–1089, 2007 [DOI] [PubMed] [Google Scholar]
- 9. Chen CC. Protein kinase C alpha, delta, epsilon and zeta in C6 glioma cells. TPA induces translocation and down-regulation of conventional and new PKC isoforms but not atypical PKC zeta. FEBS Lett 332: 169–173, 1993 [DOI] [PubMed] [Google Scholar]
- 10. Chou CL, Knepper MA. Inhibition of urea transport in inner medullary collecting duct by phloretin and urea analogues. Am J Physiol Renal Fluid Electrolyte Physiol 257: F359–F365, 1989 [DOI] [PubMed] [Google Scholar]
- 11. Chou CL, Sands JM, Nonoguchi H, Knepper MA. Concentration dependence of urea and thiourea transport in rat inner medullary collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 258: F486–F494, 1990 [DOI] [PubMed] [Google Scholar]
- 12. Coady MJ, Pajor AM, Toloza EM, Wright EM. Expression of mammalian renal transporters in Xenopus laevis oocytes. Arch Biochem Biophys 283: 130–134, 1990 [DOI] [PubMed] [Google Scholar]
- 13. DiGiovanni SR, Nielsen S, Christensen EI, Knepper MA. Regulation of collecting duct water channel expression by vasopressin in Brattleboro rat. Proc Natl Acad Sci USA 91: 8984–8988, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Douglass J, Gunaratne R, Bradford D, Saeed F, Hoffert JD, Steinbach PJ, Knepper MA, Pisitkun T. Identifying protein kinase target preferences using mass spectrometry. Am J Physiol Cell Physiol 303: C715–C727, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ecelbarger CA, Nielsen S, Olson BR, Murase T, Baker EA, Knepper MA, Verbalis JG. Role of renal aquaporins in escape from vasopressin-induced antidiuresis in rat. J Clin Invest 99: 1852–1863, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ecelbarger CA, Terris J, Frindt G, Echevarria M, Marples D, Nielsen S, Knepper MA. Aquaporin-3 water channel localization and regulation in rat kidney. Am J Physiol Renal Fluid Electrolyte Physiol 269: F663–F672, 1995 [DOI] [PubMed] [Google Scholar]
- 17. Fenton RA, Chou CL, Stewart GS, Smith CP, Knepper MA. Urinary concentrating defect in mice with selective deletion of phloretin-sensitive urea transporters in the renal collecting duct. Proc Natl Acad Sci USA 101: 7469–7474, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fenton RA, Flynn A, Shodeinde A, Smith CP, Schnermann J, Knepper MA. Renal phenotype of UT-A urea transporter knockout mice. J Am Soc Nephrol 16: 1583–1592, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feric M, Zhao B, Hoffert JD, Pisitkun T, Knepper MA. Large-scale phosphoproteomic analysis of membrane proteins in renal proximal and distal tubule. Am J Physiol Cell Physiol 300: C755–C770, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fushimi K, Uchida S, Hara Y, Hirata Y, Marumo F, Sasaki S. Cloning and expression of apical membrane water channel of rat kidney collecting tubule. Nature 361: 549–552, 1993 [DOI] [PubMed] [Google Scholar]
- 21. Giebisch G, Windhager EE. Renal tubular transfer of sodium chloride and potassium. Am J Med 36: 643–669, 1964 [DOI] [PubMed] [Google Scholar]
- 22. Goel R, Harsha HC, Pandey A, Prasad TS. Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis. Mol Biosyst 8: 453–463, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gunaratne R, Braucht DW, Rinschen MM, Chou CL, Hoffert JD, Pisitkun T, Knepper MA. Quantitative phosphoproteomic analysis reveals cAMP/vasopressin-dependent signaling pathways in native renal thick ascending limb cells. Proc Natl Acad Sci USA 107: 15653–15658, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gupta S, Maurya MR, Subramaniam S. Identification of crosstalk between phosphoprotein signaling pathways in RAW 264.7 macrophage cells. PLoS Comput Biol 6: e1000654, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hager H, Kwon TH, Vinnikova AK, Masilamani S, Brooks HL, Froikiaer J, Knepper MA, Nielsen S. Immunocytochemical and immunoelectron microscopic localization of α-, β- and γ-ENaC in rat kidney. Am J Physiol Renal Physiol 280: F1093–F1106, 2001 [DOI] [PubMed] [Google Scholar]
- 26. Han JS, Thompson KA, Chou CL, Knepper MA. Experimental tests of three-dimensional model of urinary concentrating mechanism. J Am Soc Nephrol 2: 1677–1688, 1992 [DOI] [PubMed] [Google Scholar]
- 27. Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet 11: 476–486, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hayashi M, Sasaki S, Tsuganezawa H, Monkawa T, Kitajima W, Konishi K, Fushimi K, Marumo F, Saruta T. Expression and distribution of aquaporin of collecting duct are regulated by V2 receptor in rat kidney. J Clin Invest 94: 1778–1783, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirschman L, Burns GA, Krallinger M, Arighi C, Cohen KB, Valencia A, Wu CH, Chatr-Aryamontri A, Dowell KG, Huala E, Lourenco A, Nash R, Veuthey AL, Wiegers T, Winter AG. Text mining for the biocuration workflow. Database (Oxford) 2012: bas020, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffert JD, Fenton RA, Moeller HB, Simons B, Tchapyjnikov D, McDill BW, Yu MJ, Pistikun T, Chen F, Knepper MA. Vasopressin-stimulated increase in phosphorylation at ser-269 potentiates plasma membrane retention of aquaporin-2. J Biol Chem 283: 24617–24627, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoffert JD, Knepper MA. Taking aim at shotgun phosphoproteomics. Anal Biochem 375: 1–10, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoffert JD, Pisitkun T, Knepper MA. Phosphoproteomics of vasopressin signaling in the kidney. Expert Rev Proteomics 8: 157–163, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hoffert JD, Pisitkun T, Saeed F, Song JH, Chou CL, Knepper MA. Dynamics of the G protein-coupled vasopressin V2 receptor signaling network revealed by quantitative phosphoproteomics. Mol Cell Proteomics 11: M111, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA 103: 7159–7164, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoffert JD, Fenton RA, Moeller HB, Simons B, Tchapyjnikov D, McDill BW, Yu MJ, Pisitkun T, Chen F, Knepper MA. Vasopressin-stimulated increase in phosphorylation at Ser269 potentiates plasma membrane retention of aquaporin-2. J Biol Chem 283: 24617–24627, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoffert JD, Wang G, Pisitkun T, Shen RF, Knepper MA. An automated platform for analysis of phosphoproteomic datasets: application to kidney collecting duct phosphoproteins. J Proteome Res 6: 3501–3508, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huling JC, Pisitkun T, Song JH, Yu MJ, Hoffert JD, Knepper MA. Gene expression databases for kidney epithelial cells. Am J Physiol Renal Physiol 302: F401–F407, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods 1: 27–29, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol 7: 820–828, 2006 [DOI] [PubMed] [Google Scholar]
- 40. Jensen KJ, Janes KA. Modeling the latent dimensions of multivariate signaling datasets. Phys Biol 9: 045004, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karakashian A, Timmer RT, Klein JD, Gunn RB, Sands JM, Bagnasco SM. Cloning and characterization of two new isoforms of the rat kidney urea transporter: UT-A3 and UT-A4. J Am Soc Nephrol 10: 230–237, 1999 [DOI] [PubMed] [Google Scholar]
- 42. Katchalsky A, Curran PF. Nonequilibrium Thermodynamics in Biophysics. Cambridge, MA: Harvard Univ. Press, 1965 [Google Scholar]
- 43. Kirouac DC, Saez-Rodriguez J, Swantek J, Burke JM, Lauffenburger DA, Sorger PK. Creating and analyzing pathway and protein interaction compendia for modelling signal transduction networks. BMC Syst Biol 6: 29, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Knepper MA. Systems Modeling and Experimental Studies on Renal Urea, Sodium, Chloride, and Water Transport (Dissertation). Cleveland, OH: Case Western Reserve Univ., 1975 [Google Scholar]
- 45. Knepper MA, Sands JM, Chou CL. Independence of urea and water transport in rat inner medullary collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 256: F610–F621, 1989 [DOI] [PubMed] [Google Scholar]
- 46. Knepper MA, Valtin H, Sands JM. Renal actions of vasopressin. In: Handbook of Physiology. The Endocrine System. Bethesda, MD: Am Physiol Soc, 2000, sect. 7, vol. III, chapt. 13, p. 496–529 [Google Scholar]
- 47. Kolch W, Calder M, Gilbert D. When kinases meet mathematics: the systems biology of MAPK signalling. FEBS Lett 579: 1891–1895, 2005 [DOI] [PubMed] [Google Scholar]
- 48. Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Systematic discovery of in vivo phosphorylation networks. Cell 129: 1415–1426, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science 298: 1912–1934, 2002 [DOI] [PubMed] [Google Scholar]
- 50. McSorley T, Stefan E, Henn V, Wiesner B, Baillie GS, Houslay MD, Rosenthal W, Klussmann E. Spatial organisation of AKAP18 and PDE4 isoforms in renal collecting duct principal cells. Eur J Cell Biol 85: 673–678, 2006 [DOI] [PubMed] [Google Scholar]
- 51. Merkel CE, Karner CM, Carroll TJ. Molecular regulation of kidney development: is the answer blowing in the Wnt? Pediatr Nephrol 22: 1825–1838, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R. Linear motif atlas for phosphorylation-dependent signaling. Sci Signal 1: ra2, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Muller T, Schrotter A, Loosse C, Helling S, Stephan C, Ahrens M, Uszkoreit J, Eisenacher M, Meyer HE, Marcus K. Sense and nonsense of pathway analysis software in proteomics. J Proteome Res 10: 5398–5408, 2011 [DOI] [PubMed] [Google Scholar]
- 54. Murer H, Hopfer U, Kinne R. Sodium/proton antiport in brush border membrane vesicles isolated from rat small intestine and kidney. Biochem J 154: 597–604, 1976 [PMC free article] [PubMed] [Google Scholar]
- 55. Nielsen S, Chou CL, Marples D, Christensen EI, Kishore BK, Knepper MA. Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc Natl Acad Sci USA 92: 1013–1017, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nielsen S, DiGiovanni SR, Christensen EI, Knepper MA, Harris HW. Cellular and subcellular immunolocalization of vasopressin-regulated water channel in rat kidney. Proc Natl Acad Sci USA 90: 11663–11667, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nielsen S, Frokiaer J, Marples D, Kwon TH, Agre P, Knepper MA. Aquaporins in the kidney: from molecules to medicine. Physiol Rev 82: 205–244, 2002 [DOI] [PubMed] [Google Scholar]
- 58. Pandey A, Mann M. Proteomics to study genes and genomes. Nature 405: 837–846, 2000 [DOI] [PubMed] [Google Scholar]
- 59. Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. Tandem mass spectrometry in physiology. Physiology (Bethesda) 22: 390–400, 2007 [DOI] [PubMed] [Google Scholar]
- 60. Pisitkun T, Jacob V, Schleicher SM, Chou CL, Yu MJ, Knepper MA. Akt and ERK1/2 pathways are components of the vasopressin signaling network in rat native IMCD. Am J Physiol Renal Physiol 295: F1030–F1043, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rigbolt KT, Blagoev B. Quantitative phosphoproteomics to characterize signaling networks. Semin Cell Dev Biol. http://dx.doi.org/10.1016/j.semcdb.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 62. Rinschen MM, Yu MJ, Wang G, Boja ES, Hoffert JD, Pisitkun T, Knepper MA. Quantitative phosphoproteomic analysis reveals vasopressin V2-receptor-dependent signaling pathways in renal collecting duct cells. Proc Natl Acad Sci USA 107: 3882–3887, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sachs AN, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. LC-MS/MS analysis of differential centrifugation fractions from native inner medullary collecting duct of rat. Am J Physiol Renal Physiol 295: F1799–F1806, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sands JM, Nonoguchi H, Knepper MA. Hormone effects on NaCl permeability of rat inner medullary collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 255: F421–F428, 1988 [DOI] [PubMed] [Google Scholar]
- 65. Sands JM, Nonoguchi H, Knepper MA. Vasopressin effects on urea and H2O transport in inner medullary collecting duct subsegments. Am J Physiol Renal Fluid Electrolyte Physiol 253: F823–F832, 1987 [DOI] [PubMed] [Google Scholar]
- 66. Schenk LK, Bolger SJ, Luginbuhl K, Gonzales PA, Rinschen MM, Yu MJ, Hoffert JD, Pisitkun T, Knepper MA. Quantitative proteomics identifies vasopressin-responsive nuclear proteins in collecting duct cells. J Am Soc Nephrol 23: 1008–1018, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shmelkov E, Tang Z, Aifantis I, Statnikov A. Assessing quality and completeness of human transcriptional regulatory pathways on a genome-wide scale. Biol Direct 6: 15, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smith CP, Lee WS, Martial S, Knepper MA, You G, Sands JM, Hediger MA. Cloning and regulation of expression of the rat kidney urea transporter (rUT2). J Clin Invest 96: 1556–1563, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smith HW. The Kidney: Structure and Function in Health and Disease. New York: Oxford Univ. Press, 1951 [Google Scholar]
- 70. Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J 16: 6325–6336, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Terris J, Ecelbarger CA, Marples D, Knepper MA, Nielsen S. Distribution of aquaporin-4 water channel expression within rat kidney. Am J Physiol Renal Fluid Electrolyte Physiol 269: F775–F785, 1995 [DOI] [PubMed] [Google Scholar]
- 72. Turk BE, Hutti JE, Cantley LC. Determining protein kinase substrate specificity by parallel solution-phase assay of large numbers of peptide substrates. Nat Protoc 1: 375–379, 2006 [DOI] [PubMed] [Google Scholar]
- 73. Uawithya P, Pisitkun T, Ruttenberg BE, Knepper MA. Transcriptional profiling of native inner medullary collecting duct cells from rat kidney. Physiol Genomics 32: 229–253, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Westerhoff HV, Palsson BO. The evolution of molecular biology into systems biology. Nat Biotechnol 22: 1249–1252, 2004 [DOI] [PubMed] [Google Scholar]
- 75. Wexler AS, Kalaba RE, Marsh DJ. Three-dimensional anatomy and renal concentrating mechanism. I. Modeling results. Am J Physiol Renal Fluid Electrolyte Physiol 260: F368–F383, 1991 [DOI] [PubMed] [Google Scholar]
- 76. You G, Smith CP, Kanai Y, Lee WS, Stelzner M, Hediger MA. Cloning and characterization of the vasopressin-regulated urea transporter. Nature 365: 844–847, 1993 [DOI] [PubMed] [Google Scholar]
- 77. Yu MJ, Miller RL, Uawithya P, Rinschen MM, Khositseth S, Braucht DW, Chou CL, Pisitkun T, Nelson RD, Knepper MA. Systems-level analysis of cell-specific AQP2 gene expression in renal collecting duct. Proc Natl Acad Sci USA 106: 2441–2446, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang Y, Mircheff AK, Hensley CB, Magyar CE, Warnock DG, Chambrey R, Yip KP, Marsh DJ, Holstein-Rathlou NH, McDonough AA. Rapid redistribution and inhibition of renal sodium transporters during acute pressure natriuresis. Am J Physiol Renal Fluid Electrolyte Physiol 270: F1004–F1014, 1996 [DOI] [PubMed] [Google Scholar]
- 79. Zhao B, Knepper MA, Chou CL, Pisitkun T. Large-scale phosphotyrosine proteomic profiling of rat renal collecting duct epithelium reveals predominance of proteins involved in cell polarity determination. Am J Physiol Cell Physiol 302: C27–C45, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]