Dual-color total internal reflection fluorescence microscopy revealed that the N-terminal half of Srv2 (N-Srv2) directly catalyzes severing of cofilin-decorated actin filaments. N-Srv2 formed novel six-bladed structures resembling ninja throwing stars (shurikens), and N-Srv2 activities were critical for actin organization in vivo and were lethal in combination with Aip1.
Abstract
Actin filament severing is critical for the dynamic turnover of cellular actin networks. Cofilin severs filaments, but additional factors may be required to increase severing efficiency in vivo. Srv2/cyclase-associated protein (CAP) is a widely expressed protein with a role in binding and recycling actin monomers ascribed to domains in its C-terminus (C-Srv2). In this paper, we report a new biochemical and cellular function for Srv2/CAP in directly catalyzing cofilin-mediated severing of filaments. This function is mediated by its N-terminal half (N-Srv2), and is physically and genetically separable from C-Srv2 activities. Using dual-color total internal reflection fluorescence microscopy, we determined that N-Srv2 stimulates filament disassembly by increasing the frequency of cofilin-mediated severing without affecting cofilin binding to filaments. Structural analysis shows that N-Srv2 forms novel hexameric star-shaped structures, and disrupting oligomerization impairs N-Srv2 activities and in vivo function. Further, genetic analysis shows that the combined activities of N-Srv2 and Aip1 are essential in vivo. These observations define a novel mechanism by which the combined activities of cofilin and Srv2/CAP lead to enhanced filament severing and support an emerging view that actin disassembly is controlled not by cofilin alone, but by a more complex set of factors working in concert.
INTRODUCTION
Rapid turnover of filamentous actin networks is required for a number of fundamental biological processes, including cell motility, endocytosis, cytokinesis, and cell and tissue morphogenesis (Pollard et al., 2000; Pollard and Cooper, 2009; Le Clainche and Carlier, 2008). Two rate-limiting steps in the turnover of actin arrays are filament severing, which produces smaller F-actin fragments to accelerate disassembly, and nucleotide exchange on released ADP-actin monomers, which replenishes the pool of ATP-actin monomers available for new assembly. The mechanisms by which these two critical steps are controlled in vivo remain only partially understood.
Actin depolymerization factor/cofilin (herein referred to as cofilin) performs an essential role in severing and disassembling actin filaments (Bernstein and Bamburg, 2010; Bugyi and Carlier, 2010). Cofilin binds cooperatively to filaments, alters their twist and elasticity, and induces large changes in F-actin conformation (McGough et al., 1997; Galkin et al., 2011; McCullough et al., 2011). These effects cause filament fragmentation to promote disassembly (Maciver et al., 1991; Andrianantoandro and Pollard, 2006) and may catalyze subunit dissociation from filament pointed ends (Carlier et al., 1997). Recent theoretical and experimental studies indicate that severing occurs at the boundaries between cofilin-decorated and undecorated regions on the filament (Bobkov et al., 2006; De La Cruz, 2009; Suarez et al., 2011). Interestingly, the severing activity of purified cofilin alone in vitro is not particularly efficient, and even under optimized conditions, it severs only about once per micron of filament per 1000 s (Andrianantoandro and Pollard, 2006; Bugyi and Carlier, 2010). These and other observations have suggested that additional factors may be required in vivo to enhance severing and better explain the high rates of filament turnover observed in living cells (Theriot and Mitchison, 1991; Watanabe and Mitchison, 2002).
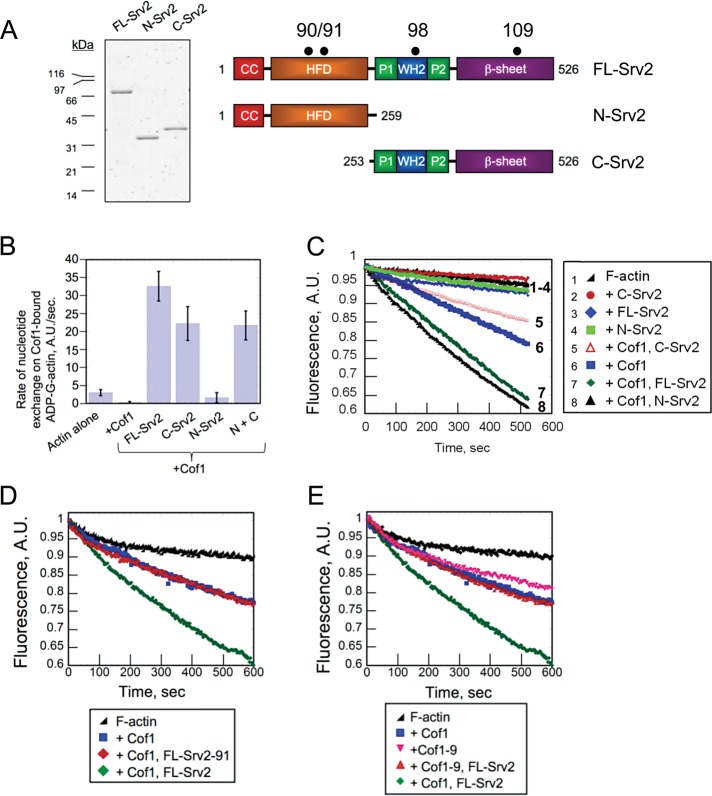
Srv2/cyclase-associated protein (CAP) is a 55-kDa actin monomer–binding protein (Figure 1A) that is well conserved in plants, animals, and fungi. The C-terminal half of Srv2/CAP (C-Srv2) has an established role in recycling cofilin-bound, ADP-actin monomers. This is accomplished by C-Srv2 both competitively displacing cofilin and catalyzing nucleotide exchange (ATP for ADP) on monomers, possibly in conjunction with its binding partner, profilin (Vojtek et al., 1991; Moriyama and Yahara, 2002; Balcer et al., 2003; Mattila et al., 2004; Bertling et al., 2007; Quintero-Monzon et al., 2009; Chaudhry et al., 2010). C-Srv2 consists of two proline-rich motifs, which bind profilin and SRC homology (SH3) domains, respectively, an intervening Wiskott-Aldrich-homology 2 (WH2) domain, and a domain rich in β-sheets (Dodatko et al., 2004). The WH2 and β-sheet domains are each sufficient to bind G-actin, and when linked together in C-Srv2, produce a high-affinity interaction with ADP-G-actin (KD = 18 nM) and a much lower affinity interaction with ATP-G-actin (KD = 2 μM; Mattila et al., 2004; Chaudhry et al., 2010). These unique binding properties are thought to be critical for the role of C-Srv2 in recycling ADP-actin monomers. Consistent with this view, specific mutations in either the WH2 (srv2-98) or the β-sheet (srv2-108) domain that impair actin binding abolish the nucleotide exchange function of C-Srv2 in vitro and partially disrupt cellular actin organization in vivo (Mattila et al., 2004; Chaudhry et al., 2010).
FIGURE 1:
Activities of N-Srv2 and C-Srv2 in nucleotide exchange on G-actin and Cof1-mediated disassembly of F-actin. (A) Coomassie Blue–stained gel and schematic of Srv2 polypeptides used for biochemical analysis. Black dots indicate locations of point mutations in the srv2 alleles used in this study (90, 91, 98, 109; Chaudhry et al., 2010; Mattila et al., 2004; Quintero-Monzon et al., 2009). (B) Effects of 100 nM FL-Srv2, N-Srv2, and C-Srv2 on rate of ε-ATP exchange on 2 μM ADP-actin monomers in the presence of 5 μM cofilin. (C–E) F-actin depolymerization assays induced by vitamin D–binding protein. Final concentrations: 2 μM F-actin, 100 nM CapZ, 50 nM Cof1 or Cof1-9, and 100 nM Srv2 polypeptides.
In contrast, the function of the N-terminal half of Srv2/CAP (N-Srv2) is not well understood. This region contains a domain composed of antiparallel helices with a 14-3-3–like fold (Ksiazek et al., 2003; Yusof et al., 2005), which we refer to as the helical folded domain (HFD). Mutations in the HFD (srv2-90 and srv2-91) have been reported to cause strong defects in cell growth (Quintero-Monzon et al., 2009). Further, this domain has been shown to bind cofilin-actin complexes but not free cofilin or free actin monomers (Moriyama and Yahara, 2002; Quintero-Monzon et al., 2009). Based on these observations, it has been proposed that N-Srv2 assists C-Srv2 in stimulating actin monomer recycling, but this model has never been tested directly.
In this study, we investigated N-Srv2 function using a combined genetic, biochemical, and structural approach, and discovered an unanticipated role for Srv2/CAP in directly catalyzing actin filament severing.
RESULTS
N-Srv2 catalyzes cofilin-mediated disassembly of actin filaments
Previous studies suggested that N-Srv2 may contribute to actin dynamics by assisting C-Srv2 in catalyzing nucleotide exchange on cofilin-bound ADP-actin monomers (Quintero-Monzon et al., 2009). We directly tested this model first by comparing the effects of purified full-length Srv2 (FL-Srv2), N-Srv2, and C-Srv2 (Figure 1A) on rate of ε-ATP exchange on cofilin-bound ADP-actin monomers. As expected, FL-Srv2 relieved the inhibitory effects of cofilin and catalyzed nucleotide exchange on ADP-actin (Figure 1B; Balcer et al., 2003). Further, C-Srv2 showed effects similar to FL-Srv2. On the other hand, N-Srv2 showed little, if any, effect and combining N-Srv2 with C-Srv2 did not alter C-Srv2 effects. These observations strongly suggest that the nucleotide exchange activity of Srv2 resides in its C-terminal half, and that N-Srv2 makes little, if any, contribution to this function. Thus our data do not support earlier models, and instead suggest that N-Srv2 makes an alternative mechanistic contribution to actin dynamics.
The two major rate-limiting steps in filamentous actin turnover are the severing/disassembly of filaments and nucleotide exchange on released monomers. Full-length yeast and human Srv2/CAP proteins have been shown to accelerate cofilin-mediated disassembly of F-actin in bulk assays in vitro (Moriyama and Yahara, 2002; Balcer et al., 2003). The mechanism underlying their effects was proposed to be indirect, via C-Srv2 recycling cofilin from ADP-actin monomers. We experimentally challenged the key prediction of this model, that is, that C-Srv2 should be sufficient to accelerate cofilin-mediated F-actin disassembly (Figure 1C). Consistent with previous reports, FL-Srv2 enhanced Cof1-mediated F-actin disassembly (Figure 1C, curve 7). However, C-Srv2 showed no effect, and instead N-Srv2 stimulated F-actin disassembly as effectively as FL-Srv2 (Figure 1C, curves 5 and 7). These data indicate that the actin disassembly–promoting activity is contained in N-Srv2 rather than C-Srv2. Importantly, no Srv2 construct had effects in the absence of Cof1 (Figure 1C, curves 2–4), and the concentration of Cof1 used (50 nM) caused minimal quenching of pyrene-F-actin (see Materials and Methods and Supplemental Figure S1).
To better understand the mechanistic basis of the N-Srv2 disassembly effects, we asked whether the activity depended on previously identified conserved surfaces on its HFD that bind cofilin-actin but not cofilin or actin alone (Quintero-Monzon et al., 2009). We directly compared the cofilin-dependent disassembly activities of FL-Srv2 and FL-Srv2-91, the latter of which carries a mutation at the conserved binding surface in the HFD (Figure 1, A and D). Unlike wild-type FL-Srv2, mutant FL-Srv2-91 failed to enhance disassembly, demonstrating that its cofilin-actin–binding surface is critical for the activity. In addition, we compared the effects of FL-Srv2 on Cof1 and Cof1-9 (Lappalainen et al., 1997), a mutant that disrupts the cognate HFD-binding site on Cof1 (Quintero-Monzon et al., 2009; Figure 1E). Cof1 and Cof1-9 showed similar F-actin severing/depolymerization activities in the absence of Srv2. However, in the presence of FL-Srv2, only the activity Cof1, and not Cof1-9, was increased. Together these results show that the ability of Srv2 to enhance Cof1-mediated F-actin disassembly requires interactions between the conserved surface on the HFD of Srv2 and its cognate binding surface on Cof1.
Total internal reflection fluorescence (TIRF) microscopy reveals that N-Srv2 enhances cofilin-mediated severing of actin filaments
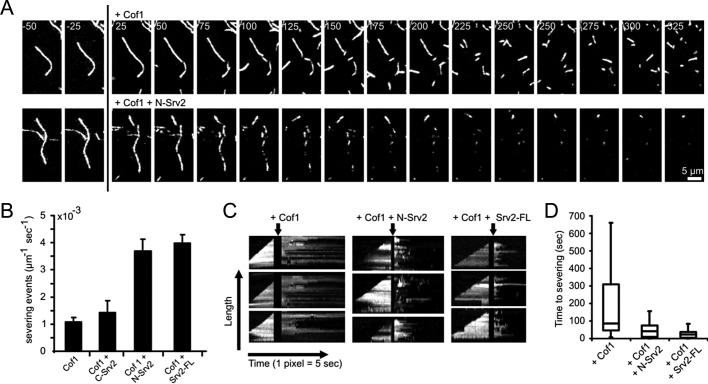
Because bulk assays do not distinguish between effects on F-actin severing versus depolymerization, we used TIRF microscopy to examine in real time the effects of Cof1 with and without Srv2 on single Oregon Green (OG)-labeled actin filaments (Figure 2A). We monitored the polymerization of immobilized filaments until their length reached ∼10 μm. Then we replaced (by flow-in) the actin monomer–containing solution with Cof1 and/or different Srv2 constructs but no actin monomers (Figure 2A, time-lapse panels). Filament severing was never observed in the absence of Cof1 (n > 100 filaments). Addition of Cof1 led to severing with a frequency of 1.1 ± 0.15 × 10−3 μm−1 s−1, similar to previous studies (Andrianantoandro and Pollard, 2006). Further addition of FL-Srv2 or N-Srv2 enhanced severing by ∼fourfold (4.0 ± 0.31 and 3.7 ± 0.44 × 10−3 μm−1 s−1, respectively), whereas C-Srv2 did not appreciably affect severing (Figure 2B and Supplemental Movie S1). FL-Srv2 and N-Srv2 effects were also analyzed in kymographs (Figure 2C), revealing that these proteins markedly reduced the time interval between Cof1 addition and severing events (Figure 2D).
FIGURE 2:
TIRF microscopy analysis of Srv2 effects on Cof1-mediated actin filament severing. (A) Filament severing by Cof1 ± Srv2 (time in seconds). Filaments were assembled from 1 μM G-actin (10% OG-labeled, 0.5% biotinylated) in flow cells and attached to biotin-PEG-(0.1% biotinylated)–coated glass slides by streptavidin. At the indicated time (vertical black line), the reaction mixture was replaced with 10 nM Cof1 ± 100 nM N-Srv2. (B) Quantification of severing efficiencies. Error bars = SD. (C) Kymographs for the time course of growth and disassembly of single filaments. (D) Average time between the addition of Cof1 ± Srv2 and filament severing. Data were obtained by analyzing kymographs as in (C). Boxes indicate 25th percentile, median, and 75th percentile of all values; error bars indicate 10th and 90th percentiles. n = 32, 34, and 36 for Cof1, Cof1+N-Srv2, and Cof1+FL-Srv2, respectively.
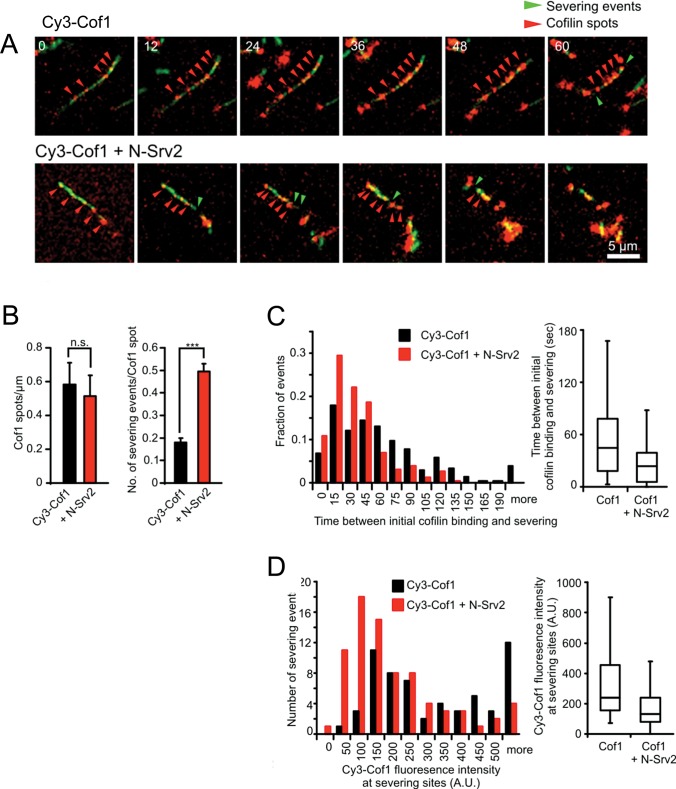
To further investigate the mechanism of N-Srv2 enhanced severing, we next used dual-color TIRF with fluorescently labeled Cof1 (Cy3-Cof1) and actin (OG-actin) to explore how N-Srv2 influences the spatiotemporal relationship between Cof1 association with filaments and severing (Figure 3). In the absence of N-Srv2, Cy3-Cof1 spots gradually accumulated on filaments (Figure 3A, red arrows, and Movie S2), and their fluorescence intensity increased over time, consistent with cooperative binding to F-actin (McCullough et al., 2011; Suarez et al., 2011). The appearance of Cy3-Cof1 spots preceded severing events (Figure 3A, green arrows). Inclusion of N-Srv2 led to a marked increase in the number of severing events per Cy3-Cof1 spot (Figure 3A, green arrows, B, and D), but without appreciably changing the number or pattern of Cy3-Cof1 spots that accumulated on the filament during the observation time (Figure 3, A and B). N-Srv2 also reduced the time interval between appearance of Cy3-Cof1 spots and severing events (Figure 3C) and reduced the levels of Cy3-Cof1 that accumulated before severing occurred (Figure 3D). Together these observations show that N-Srv2 greatly increases the efficiency of Cof1-mediated severing but without noticeably affecting Cof1 binding to actin filaments.
FIGURE 3:
Dual-color TIRF microscopy analysis of actin filament severing by Cy3-Cof1 and N-Srv2. (A) Filament severing by Cy3-Cof1 ± N-Srv2 (time in seconds). Filaments were assembled as in Figure 2A, then reaction mixtures were replaced with 10 nM Cy3-Cof1 ± 100 nM N-Srv2. Red arrowheads mark accumulations of Cy3-Cof1. Green arrowheads mark severing events. (B) Number of Cy3-Cof1-spots per micrometer of F-actin (left) and number of severing events per Cy3-Cof1-spot (right) in the presence of 10 nM Cy3-Cof1 (n = 229, black bars) and 10 nM Cy3-Cof1 + 100 nM N-Srv2 (n = 188, red bars). Error bars represent SD from three experiments. n.s., not significant; ***, p < 0.001 as determined by t test. (C) Distribution of time intervals between initial detection of Cy3-Cof1 on a filament and severing at that location, in the presence and absence of N-Srv2, determined from experiments as in (A). (C) Distribution of Cy3-Cof1 fluorescence intensity on filaments one frame prior to severing events. Determined from experiments as in (A). Boxes in box plots shown in (C) and (D) indicate 25th percentile, median, and 75th percentile of all values; error bars indicate 10th and 90th percentiles.
N-Srv2 also reduced the average length of the fragments produced by Cof1-mediated severing, consistent with an increased severing frequency (Figure S2). The remaining short, Cy3-Cof1–decorated filament fragments were stable, as recently described (McCullough et al., 2011; Suarez et al., 2011), and persisted in the presence of N-Srv2. Thus N-Srv2 does not appear to catalyze depolymerization after severing.
N-Srv2 forms shuriken structures with six symmetrical protrusions
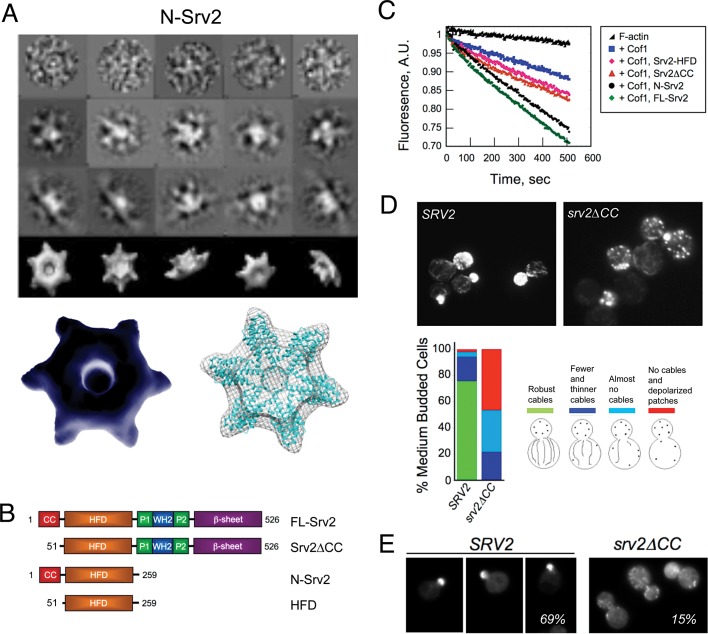
To better understand the structural basis of N-Srv2 activities, we performed electron microscopy (EM) and single-particle analysis on purified N-Srv2. Based on hydrodynamic analysis, it has been proposed that FL-Srv2 (55 kDa) hexamerizes to form 342-kDa complexes that bind actin monomers (43 kDa) with equal stoichiometry, yielding ∼600-kDa complexes (Balcer et al., 2003; Quintero-Monzon et al., 2009). Atomic structures of the HFD and β-sheet domains have been solved (Ksiazek et al., 2003; Dodatko et al., 2004; Yusof et al., 2005), but it has remained a mystery how these smaller units are spatially organized within the higher-order Srv2 complexes. To address this, we examined native N-Srv2 by negative-stain EM, and the particles were categorized into class averages in different orientations; these data were then used to generate a three-dimensional reconstruction (Figure 4A). The resolution of this structure was 25Å, as estimated by the Fourier shell correlation (FSC) between two reconstructions calculated from two half-sets of the data. The structure had a wheel-like appearance with six symmetrical protrusions or “spokes.” The resolution was sufficient to dock six monomeric HFD domains (Yusof et al., 2005, 2006) into the EM density (Figure 4A).
FIGURE 4:
Oligomerization of N-Srv2 into hexameric structures. (A) Negative-stain electron micrographs of N-Srv2 hexamers: raw images (top row), two-dimensional projections of class averages (middle two rows), and three-dimensional reconstructions (bottom row), and a three-dimensional reconstruction of N-Srv2 hexamer with six monomeric HFD domains docked into it (accession number 1S0P; Ksiazek et al., 2003). (B) Schematic of Srv2 polypeptides purified for biochemical tests. (C) Effects of 50 nM Cof1 ± 100 nM FL-Srv2, Srv2ΔCC, N-Srv2, or HFD on F-actin disassembly induced by vitamin D–binding protein, as performed in Figure 1, C–E. (D) Comparison of actin organization in SRV2 and srv2ΔCC cells grown to log phase at 25°C. Example cell images shown; phenotypes scored (n > 100 cells). (E) Comparison of GFP-Sec4 localization in SRV2 and srv2ΔCC cells. Example cell images shown; percentage of cells with GFP-Sec4 predominantly in the bud is shown in each panel (n > 100 cells).
In the crystallography and EM studies above, the HFD domain dimerized in an antiparallel manner and had distinct dimeric arrangements, leading to the proposal that monomers may be capable of “sliding” past one another and physically separating (Mavoungou et al., 2004; Yusof et al., 2005). Our EM structure supports this view, as we could readily dock HFD monomers, but not dimers, into our reconstruction (Figure 4A).
In the N-Srv2 stru cture, six HFD domains are closely tethered and spatially organized into a radial pattern, suggesting that this arrangement may be relevant to their function. Therefore we investigated the effects of deleting from FL-Srv2 and N-Srv2 a short N-terminal sequence, the CC domain (Figure 4B), which mediates oligomerization (Balcer et al., 2003; Quintero-Monzon et al., 2009). Note that although this sequence has a low predicted coiled-coil value, we have retained the “CC” nomenclature for consistency with previous studies. The resulting constructs, Srv2∆CC and Srv2-HFD (Figure 4B), exhibited markedly reduced enhanced disassembly activities compared with FL-Srv2 and N-Srv2 (Figure 4C). These data suggest that oligomerization of the HFD domain may be important for the enhanced disassembly activity. To address the physiological importance of these effects, we integrated srv2∆CC and then compared wild-type and srv2∆CC cells for actin organization (Figure 4D) and polarized distribution of GFP-Sec4-marked secretory vesicles (Schott et al., 2002; Figure 4E). In srv2∆CC, > 80% of cells showed a highly depolarized actin cytoskeleton and/or severe loss of actin cables, compared with <10% of wild-type SRV2 cells (Figure 4D). Further, the percentage of srv2∆CC cells with polarized GFP-Sec4 at the bud tip was greatly reduced (14% for srv2∆CC, 70% for SRV2) (Figure 4E).
The importance of N-Srv2 activities for in vivo function
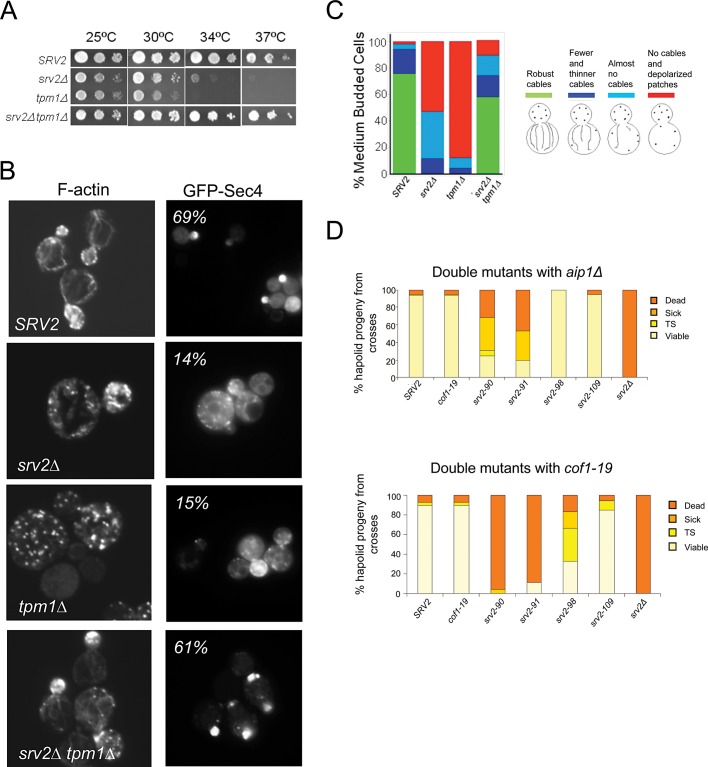
We next tested the importance of N-Srv2 enhanced severing activity in vivo. We first asked whether srv2∆ could suppress tpm1∆. Tpm1 is one of two genes in Saccharomyces cerevisiae that encode tropomyosin, a filament side-binding protein required for actin cable formation, polarized secretion, and cell growth at elevated temperatures (Liu and Bretscher, 1989, 1992; Pruyne et al., 1998). Our previous work has shown that tpm1∆ defects are partially suppressed by mutations in actin disassembly machinery (e.g., cof1-22 and aip1∆; Okada et al., 2006). Similarly, we observed here that srv2∆ strongly suppressed tpm1∆ defects in cell growth and actin organization (Figure 5, A–C) and restored GFP-Sec4 polarization to the bud tip (70% of SRV2 cells, 14% of tpm1∆ cells, 14% of srv2∆ cells, and 61% of srv2∆ tpm1∆ cells; Figure 5B). These genetic data provide strong support for Srv2/CAP functioning to promote F-actin disassembly in vivo. While some mammalian tropomyosins protect filaments from cofilin severing, purified Tpm1 does not protect yeast F-actin from Cof1 severing in vitro (Fan et al., 2008). Rather than protecting cables from Cof1 severing, Tpm1 may instead promote formin-mediated cable assembly, as has been suggested for mammalian tropomyosin and formins (Wawro et al., 2007). This model would explain why tpm1∆ cells have very short and possibly slow-growing cables, and why mutations in the actin disassembly machinery (e.g., aip1∆ and srv2∆) rescue cable length and function.
FIGURE 5:
Genetic analysis of N-Srv2 and C-Srv2 functions. (A) Suppression of tpm1Δ growth defects by srv2∆. Strains were grown to log phase, serially diluted, and grown on YPD plates for 2 d at 25°C, 30°C, 34°C, and 37°C. (B) Example images of cells grown at 25°C, fixed and stained with Alexa Fluor 488–phalloidin or showing GFP-Sec4 expressed from a low-copy plasmid in the indicated strains (Schott et al., 2002); percentage of cells with GFP-Sec4 predominantly in the bud listed in each panel (n > 100 cells). (C) Scored actin phenotypes of the same strains (n > 100 cells). (D) Genetic interactions of specific srv2 alleles with aip1∆ and cof1-19. Haploid srv2 mutants were crossed separately to aip1Δ and cof1-19. Diploids were sporulated, and tetrads were dissected (minimum: 20 tetrads, 80 spores). Resulting progeny were analyzed for cell growth at 25°C and 37°C. Shown are the percentages of haploid progeny that showed impaired growth at 37°C (temperature-sensitive, TS), grew poorly at all temperatures (sick), or did not grow at all (dead).
Finally, as a genetic test of our model that the activities in N-Srv2 rather than C-Srv2 enhance F-actin disassembly, we asked whether point mutations that separately disrupt these activities (see Figure 1A) genetically interact with AIP1, another cofilin cofactor in actin disassembly. Mutations disrupting N-Srv2 activities, srv2-90 and srv2-91, exhibited synthetic lethality and/or strong synthetic growth defects in combination with aip1∆, whereas mutations disrupting C-Srv2 activities, srv2-98 and srv2-109, did not (Figure 5D). Similar genetic interactions were observed between these srv2 alleles and cof1-19, which phenocopies aip1∆ (Figure 4D; Rodal et al., 1999). These genetic data correlate the loss of N-Srv2 activity (disrupted by srv2-90) with the appearance of striking defects in vivo actin organization and synthetic genetic interactions with aip1∆. Thus N-Srv2 biochemical activities in actin disassembly are critical for actin cytoskeleton organization in vivo, and together with Aip1 are essential for cell viability.
DISCUSSION
Rapid actin disassembly is required for cells to dynamically reorganize their cytoskeletons in response to different signals and to maintain a pool of actin monomers available for new assembly. The underlying mechanisms regulating these events are only beginning to be understood. One of the most well-defined factors in actin turnover is cofilin, which has an essential role in severing and promoting the disassembly of actin filaments (Bernstein and Bamburg, 2010). However, mounting evidence indicates that cofilin does not act alone in this capacity in vivo (see below), and instead represents only one critical piece of a larger set of machinery orchestrating actin disassembly. Our results here, in agreement with another recent study on full-length mammalian CAP1 (Normoyle and Brieher, 2012), identify Srv2/CAP as a new functional component of the disassembly apparatus. We have shown that yeast Srv2/CAP directly enhances cofilin-mediated severing of actin filaments. Further, we mapped this activity to the N-terminal half of Srv2, N-Srv2, whereas all previously described functions of Srv2/CAP in actin regulation are mediated by its C-terminal half, C-Srv2 (Figure 1; Mattila et al., 2004; Chaudhry et al., 2010). Two additional observations indicate that the enhanced filament-severing activity is novel and unrelated to the established function of the C-terminus of Srv2/CAP in recycling cofilin from actin monomers (Moriyama and Yahara, 2002; Balcer et al., 2003). First, enhanced severing activity was abolished by point mutations in N-Srv2 (e.g., srv2-90), which do not affect the cofilin-recycling activity (Quintero-Monzon et al., 2009; Chaudhry et al., 2010). Second, in the TIRF assays in which we observed enhanced filament severing (Figures 2 and 3), there was no G-actin present. The filaments were first assembled, and then the chamber was washed, while Cof1 and/or N-Srv2 were flowed in (without G-actin). These results indicate that rather than promoting severing indirectly by recycling cofilin from dissociated actin monomers, N-Srv2 acts directly with cofilin to sever actin filaments.
Based on the evolutionary conservation of the HFD domain and the conservation of its functional surfaces (targeted in srv2-90 and srv2-91) that mediate enhanced severing, this function is likely to be conserved in other systems. This view is supported by the cognate Srv2-binding surfaces on cofilin also being conserved (Quintero-Monzon et al., 2009) and by the recently reported enhanced severing activity of mammalian CAP1 (Normoyle and Brieher, 2012). Although the activity of CAP1 in the above-mentioned study was not mapped to a specific domain, it seems likely that it is mechanistically related to the activity we describe for N-Srv2. Further, enhanced severing may help explain why genetic disruptions of Srv2/CAP in diverse systems reduce rates of F-actin turnover and/or lead to an accumulation of F-actin similar to what is seen with cofilin disruptions (Baum et al., 2000; Rogers et al., 2003; Bertling et al., 2004; Noegel et al., 2004; Deeks et al., 2007).
How does N-Srv2 enhance cofilin-mediated severing of actin filaments? Cofilin binds cooperatively to filaments (McGough et al., 1997), and severing occurs predominantly at boundaries between bare and cofilin-decorated regions (De La Cruz and Sept, 2010; McCullough et al., 2011; Suarez et al., 2011). Through real-time observation using dual-color TIRF microscopy, we observed that N-Srv2 increases the efficiency of cofilin-dependent severing without altering the pattern or intensity of cofilin decoration on filaments. This argues that N-Srv2 does not simply increase cofilin recruitment to filament sides. Instead, N-Srv2 decreased the time interval between cofilin binding and severing and reduced the amount of cofilin at severing sites. F-actin is highly polymorphic, and cofilin induces major alterations in F-actin subunit contacts and conformation, which can be allosterically propagated into undecorated regions of the filament (Galkin et al., 2011). Thus it is possible that N-Srv2 induces further conformational changes in cofilin-decorated F-actin to enhance severing; however, this will require further structural analysis to resolve.
What is the structural basis for N-Srv2–enhanced severing activity? Using negative-stain, single-particle EM, we found that N-Srv2 forms hexameric structures with six symmetrical protrusions or blades, into which we could readily dock six HFD monomers (Figure 4A). Disruption of hexamerization by deletion of the CC domain strongly impaired severing activity (Figure 4C). On the basis of these results, we hypothesize that the CC domain oligomerizes and organizes the HFD domains into a symmetrical cluster of six protrusions, a structure that may optimize the exposure and spacing of the cofilin-actin binding sites on the HFD domains. Consistent with this view, the HFD alone (lacking the CC domain) showed markedly reduced activity and was previously shown to crystallize as a dimer (Ksiazek et al., 2003) in which the cofilin-actin binding sites are partially buried at the dimer interface (Quintero-Monzon et al., 2009).
Another important implication of our results is that Srv2/CAP is a bifunctional protein, catalyzing two distinct and rate-limiting steps in actin turnover: filament severing (N-Srv2) and monomer recycling (C-Srv2). These dual properties suggest that Srv2/CAP may be especially important for cellular and physiological processes that depend on high rates of F-actin turnover in which both steps in actin turnover need to be closely coordinated. Consistent with this view, RNA interference silencing of mouse CAP1 and Drosophila CAP markedly impairs leading-edge protrusion, cell motility, and endocytosis (Rogers et al., 2003; Bertling et al., 2004). Further, human CAP1 is up-regulated and required for motility in invasive pancreatic cancer cells (Yamazaki et al., 2009).
Finally, our observations provide new insights into the broader question of what cellular machinery is required to efficiently disassemble filamentous actin arrays in vivo. We found that mutations in N-Srv2 (but not C-Srv2) are synthetic lethal with aip1∆, demonstrating that while Cof1 is essential for actin disassembly in vivo (Lappalainen and Drubin, 1997), it is not sufficient, and that N-Srv2 and Aip1 share a second essential role in this process. Aip1 binds to cofilin-decorated F-actin to promote disassembly (Iida and Yahara, 1999; Okada et al., 1999, 2006; Rodal et al., 1999; Mohri and Ono, 2003; Tsuji et al., 2009) by enhancing filament severing and/or capping severed ends to block reannealing and growth (Okada et al., 2002; Balcer et al., 2003; Ono et al., 2004; Tsuji et al., 2009; Okreglak and Drubin, 2010). Thus Aip1 and N-Srv2 each bind to cofilin and depend on cofilin for their effects, supporting the view that this group of proteins works in close concert on the filament surface to promote disassembly. Cofilin is often depicted in models as functioning alone in disassembly. However, our findings, together with mounting evidence from studies on Aip1, coronin, and CAP (Brieher et al., 2006; Cai et al., 2008; Kueh et al., 2008; Gandhi et al., 2009; Normoyle and Brieher, 2012), paint a far more complex picture of the disassembly process, and suggest that cofilin represents only one piece, albeit a critical one, in a larger multicomponent apparatus that forms on the filament surface to regulate severing and disassembly.
MATERIALS AND METHODS
Yeast strains and plasmid construction
Genotypes of all yeast strains are listed in Supplemental Table S1. Plasmids for Escherichia coli expression of N-terminally 6His-tagged FL-Srv2 (residues 1–526), Srv2∆CC (residues 51–526), N-Srv2 (residues 1–259), and C-Srv2 (residues 253–526) have been described (Mattila et al., 2004; Quintero-Monzon et al., 2009). A plasmid for E. coli expression of 6His-HFD (51-259) was generated by PCR amplification of the coding sequences and subcloning into the NcoI and NotI sites of pHAT2. All plasmids were sequenced.
Protein purification
Rabbit skeletal muscle actin was purified as previously described (Spudich and Watt, 1971). All Srv2 polypeptides were expressed in E. coli BL21-RP cells grown to log phase at 37°C and induced for 16 h at 20°C with 0.4 mM isopropyl β-d-1-thiogalactopyranoside. Each protein was purified by nickel affinity and gel filtration as previously described (Quintero-Monzon et al., 2009). Cof1-9 was purified as previously described (Lappalainen et al., 1997). For fluorescence labeling of Cof1, we initially used the recently described Cof1 (D34C/C62A) mutant (Suarez et al., 2011). However, the aspartic acid residue that is converted to cysteine for labeling in this construct is a key part of the N-Srv2–binding site (Quintero-Monzon et al., 2009), and therefore N-Srv2 failed to enhance severing/disassembly of Cof1 (D34C/C62A). For this reason, we generated a new Cof1 construct for labeling, Cof1 (T46C/C62A), which did not overlap with the N-Srv2 binding site. Cof1 (T46C/C62A) was covalently labeled with Cy3-maleimide (GE Healthcare, Piscataway, NJ).
Nucleotide exchange assays
Rates of nucleotide exchange on ADP-actin monomers were determined by measuring the change in fluorescence upon incorporation of ε-ATP (Sigma-Aldrich; St. Louis, MO) over time. Briefly, 2 μM ADP-actin monomers in G-buffer lacking ATP (10 mM Tris, pH 7.5, 0.2 mM CaCl2, 0.2 mM dithiothreitol [DTT]) was mixed with proteins in Tris/NaCl buffer (20 mM Tris, pH 7.5, 50 mM NaCl) or buffer alone, and then added to 50 μM ε-ATP. The reaction was monitored at 350-nm excitation and 410-nm emission at 25°C in a fluorescence spectrophotometer (Photon Technology International, Lawrenceville, NJ). Exchange rates were determined from linear fits of the first 25 s of each reaction curve.
F-actin depolymerization assays
For monitoring rates of actin disassembly, 40 μl preformed F-actin (2 μM final, 10% pyrene-labeled) was incubated for 5 min with 20 μl of control buffer or proteins in the same buffer: capping protein/CapZ (100 nM final), Cof1 (50 nM final), and/or Srv2 polypeptides (100 nM final). At time zero, disassembly was induced by addition of 3 μM vitamin D–binding protein/human plasma Gc-globulin (Sigma-Aldrich). Fluorescence was monitored at 365-nm excitation and 407-nm emission in a fluorescence spectrophotometer (Photon Technology International). For determining effects of Cof1 on quenching of pyrene–F-actin, steady-state fluorescence was measured after addition of a range of concentrations of Cof1 (Figure S1). At the concentrations of Cof1 used in the depolymerization assays (50 nM), only ∼3% of the signal was quenched, and further addition of N-Srv2 (up to 8 μM) did not increase quenching, suggesting most of the change in fluorescence signal in the assays is due to F-actin depolymerization.
Total internal reflection fluorescence microscopy (TIRFM)
For all experiments, 24 × 60 mm coverslips (Fisher Scientific, Pittsburgh, PA) were cleaned by sonication in 1% Versaclean detergent (Fisher Scientific) for 60 min, which was followed by sonication in 1M KOH and 1M HCl for 15 min, respectively, and then >60 min sonication in ethanol. Subsequently, coverslips were extensively rinsed with ddH2O; dried in an N2 stream; layered with 200–300 μl of 80% ethanol (pH 2.0), 2 mg/ml succinimidyl (NHS) polyethylene glycol (PEG) silane, and 2 μg/ml NHS-PEG-biotin silane (Nektar, Huntsville, AL); and incubated for 16 h at 70°C. Flow cells were assembled by first rinsing the PEG-coated coverslip extensively with ddH2O, then placing four strips of double-sided tape (2.5 cm × 2 mm × 120 μm) onto the coated surface and attaching a 22 × 22 mm coverslip (Fisher Scientific). Flow cells were stored in closed polyethylene terephthalate (PET) chambers at 4°C for up to 1 wk.
OG-labeled actin was prepared as previously described (Kuhn and Pollard, 2005). Time-lapse TIRF microscopy of OG-actin filaments was performed using a Nikon-Ti200 inverted microscope equipped with a 150-mW Ar-Laser (Mellot Griot, Carlsbad, CA); for imaging Cy3-Cof1, a 5-mW He-Ne laser and a TIRF objective with a 1.49 numerical aperture were used, (Nikon, New York, NY), and an EMCCD camera (Andor Ixon, Belfast, Northern Ireland). During measurements, optimal focus was maintained using the Perfect Focus System (Nikon). Images were captured every 3–5 s. The pixel size corresponded to 0.27 μm.
Acquired image sequences were converted to 16-bit TIFF files with ImageJ (http://imagej.nih.gov/ij) using the NIS to ImageJ plug-in (Nikon). Background fluorescence for each channel was subtracted automatically using the background subtraction tool (rolling ball radius: 50 pixel) implemented in the ImageJ software.
For actin filament-severing assays, flow cells were incubated for 5 min with 20 mM Tris-HCl, 50 mM KCl, 1% bovine serum albumin (BSA at pH 8.0 [TBSA]); this was followed by a 30-s incubation with 0.1 mg/ml streptavidin in phosphate-buffered saline. Flow cells were washed with 5 chamber volumes (∼50 μl) TBSA and equilibrated with 1X TIRF buffer (10 mM imidazole, 50 mM KCl, 1 mM MgCl2, 1 mM ethylene glycol tetraacetic acid, 0.2 mM ATP, 10 mM DTT, 15 mM glucose, 20 μg/ml catalase, 100 μg/ml glucose oxidase, and 0.5% methylcellulose [4000 cP], pH 7.4). Each reaction was initiated by rapidly mixing 1 μM actin (10% OG-labeled, 0.5% biotinylated) into 1X TIRF buffer and transferring the mixture to the flow cell. After ∼4 min, the reaction mixture was replaced with TIRF buffer containing Cof1 or Srv2 polypeptides, but lacking actin monomers, by adding the mixture to one side of the flow cell and drawing it through by wicking with a piece of filter paper on the other side. Filament-severing efficiency, expressed as severing events per micrometer per second, was determined by measuring lengths of individual filaments prior to Cof1 addition using ImageJ and counting severing events over a time period of 200 s after the addition of Cof1 and/or Srv2 constructs. No severing events were observed for Srv2 polypeptides in the absence of Cof1. Kymographs were constructed with the Multiple Kymographs plug-in from ImageJ.
Dual-color TIRF experiments using Cy3-labeled Cof1 and OG-actin were essentially carried out as single-color experiments, with the exception that OG and Cy3 fluorescence was detected sequentially, with excitation times of 50 and 300 ms, respectively. Fluorescence intensities of Cy3-Cof1 spots at severing sites were obtained by integrating the fluorescence intensity in a boxed region of 4 × 4 pixels at severing sites one frame prior to when severing occurred.
EM and single-particle analysis of N-Srv2
For preparation of EM grids, a continuous carbon film was evaporated onto mica and subsequently floated onto 300-mesh copper grids (3-mm diameter), and then the grids were dried for 2 h at 60°C. Carbon-coated grids were glow-discharged in Emitech K100X (Quorum Technologies, Kent, UK) for 45 s under a 2-mA current to make them hydrophilic. Protein solution (3 μl) was applied to the grid for 10–15 s, and then the excess solution was blotted with filter paper. The grid was washed with two drops (40 μl) of buffer solution for 30 s, and then a 5-μl drop of 1% uranyl acetate was immediately applied. Grids with stained samples were then air-dried. Images were captured at a magnification of 52,000× on a 120-kV FEI G-12 Spirit instrument (FEI, Hillsboro, OR) with an electron dose of approximately 10 e/A2. With these low-dose conditions, images of N-Srv2 protein were collected under defocus 1.5 mm and saved using an FEI Eagle 4k CCD camera (FEI) with 4000 dpi resolution. Step size for this resolution was 6.35 μm, and images were binned threefold, yielding a final pixel size of 3.66 Å on a specimen.
Particles were collected manually using the program Signature (Chen and Grigorieff, 2007). A total of 1878 particles were selected, windowed into 80 × 80 pixel images, and normalized to an SD of 1. Subsequent analysis was performed in IMAGIC-5 program. Images were first band-pass–filtered using a double Gaussian filter applied in Fourier space. This filter is defined by three parameters: low-frequency, high-frequency cut-off, and remaining low-frequency transmission. Using known formulas considering pixel size (3.66 Å), particle size (∼12 nm), and expected resolution (15 Å), these three parameters were calculated for our data set: 0.05, 0.5, and 0.001, accordingly. All particles were centered using cross-correlation of each individual particle to the average and then translationally and rotationally aligned to each other. Multimeric statistical analysis (MSA) with subsequent hierarchical ascendant classification was applied, and the whole data set was divided into 35 classes. Classes with high signal-to-noise ratio were extracted and used as references for multireference analysis (MRA). After six iterations of MSA, classification, and MRA procedures, we carried out good reference images with strong sixfold symmetry (observed by eye) for three-dimensional reconstruction. Using the angular reconstitution method (Van Heel, 1987) and a back-projection algorithm, the first, rough, three-dimensional model was evolved. This model was reprojected onto a two-dimensional space for refining, using the iterative procedures described above.
Subsequent improvement of quality of three-dimensional reconstructions was performed in the Frealign program (Grigorieff, 2007). After contrast transfer function correction using defocus, angular astigmatism (calculated for each individual micrograph), and spherical aberration data, we refined Euler angles for each particle using Frealign. Resolution of final structures was defined by 0.5 FSC criterion to be 25 Å.
Three-dimensional structures were visualized and measured in Chimera (Goddard et al., 2007). We used manual docking in Chimera to check how known crystal structures of N-Srv2 monomers are arranged in a complex with sixfold symmetry.
Cell imaging
Yeast cell cultures were grown to log phase in yeast–peptone–dextrose (YPD), fixed in 2% formaldehyde for 30 min, and stained with Alexa Fluor 488–phalloidin (Invitrogen, Carlsbad, CA). Images were acquired on a Zeiss E600 microscope (Thornwood, NY) equipped with a Hammamatsu Orca ER CCD camera (Bridgewater, NJ) running Openlab software (Improvision, Waltham, MA).
Supplementary Material
Acknowledgments
We thank M. Gandhi, B. Smith, and L. Li for assistance in preparing Cy3-labeled Cof1; L. Talarico and O. Quintero for assistance with genetic analysis; T. Chohan for assistance in scoring actin phenotypes; N. Grigorieff and C. Xu for assistance with EM data collection and processing; L. Friedman for assistance with TIRF analysis; and S. Jansen for editorial help. F.C. was supported by the National Institutes of Health (NIH; T32 NS007292); D.B. was supported by the Deutsche Forschungsgemeinschaft (DFG; BR4116); O.S. was supported by the Russian Ministry of Education and Science (16.740.11.0373) and the Civilian Research and Development Foundation (CRDF; RUB1-2918-MO-07); and B.L.G. was supported by the NIH (GM063691).
Abbreviations used:
- BSA
bovine serum albumin
- CAP
cyclase-associated protein
- C-Srv2
C-terminal half of Srv2/CAP
- DTT
dithiothreitol
- EM
electron microscopy
- FSC
Fourier shell correlation
- HFD
helical folded domain
- MRA
multireference analysis
- MSA
multimeric statistical analysis
- NHS
succinimidyl
- N-Srv2
N-terminal half of Srv2/CAP
- OG
Oregon Green
- PEG
polyethylene glycol
- SH3
SRC Homology domain
- TBSA
Tris-HCl–buffered BSA, pH 8.0
- TIRF
total internal reflection fluorescence
- WH2
Wiskott-Aldrich-homology 2
- YPD
yeast–peptone–dextrose
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-08-0589) on November 7, 2012.
*These authors contributed equally to this work.
REFERENCES
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentration of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE, Goode BL. Coordinated regulation of actin filament turnover by a high-molecular weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- Baum B, Li W, Perrimon N. A cyclase-associated protein regulates actin and cell polarity during Drosophila oogenesis and in yeast. Curr Biol. 2000;10:964–973. doi: 10.1016/s0960-9822(00)00640-0. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertling E, Hotulainen P, Mattila P, Matilainen T, Salminen M, Lappalainen P. Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol Biol Cell. 2004;15:2324–2334. doi: 10.1091/mbc.E04-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertling E, Quintero-Monzon O, Mattila PK, Goode BL, Lappalainen P. Mechanism and biological role of profilin-Srv2/CAP interaction. J Cell Sci. 2007;120:1225–1234. doi: 10.1242/jcs.000158. [DOI] [PubMed] [Google Scholar]
- Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggests allosteric mechanism of cofilin function. J Mol Biol. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugyi B, Carlier MF. Control of actin filament treadmilling in cell motility. Annu Rev Biophys. 2010;39:449–470. doi: 10.1146/annurev-biophys-051309-103849. [DOI] [PubMed] [Google Scholar]
- Cai L, Makhov AM, Schafer DA, Bear JE. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134:828–842. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M-F, Laurent V, Santolini J, Melki R, Didry D, Xia G-X, Hong Y, Chua N-H, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry F, Little K, Talarico L, Quintero-Monzon O, Goode BL. A central role for the WH2 Domain of Srv2/CAP in recharging actin monomers to drive actin turnover in vitro and in vivo. Cytoskeleton. 2010;67:120–133. doi: 10.1002/cm.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JZ, Grigorieff N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J Struct Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM. How cofilin severs an actin filament. Biophys Rev. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Sept DA. The kinetics of cooperative cofilin binding reveals two states of the cofilin-actin filament. Biophys J. 2010;98:1893–1901. doi: 10.1016/j.bpj.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Rodriques C, Dimmock S, Ketelaar T, Maciver SK, Malhó R, Hussey PJ. Arabidopsis CAP1—a key regulator of actin organization and development. J Cell Sci. 2007;120:2609–2618. doi: 10.1242/jcs.007302. [DOI] [PubMed] [Google Scholar]
- Dodatko T, et al. Crystal structure of the actin binding domain of the cyclase-associated protein. Biochem. 2004;43:10628–10641. doi: 10.1021/bi049071r. [DOI] [PubMed] [Google Scholar]
- Fan X, Martin-Brown S, Florens L, Li R. Intrinsic capability of budding yeast cofilin to promote turnover of tropomyosin-bound actin filaments. PLoS One. 2008;3:e3641. doi: 10.1371/journal.pone.0003641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schröder GF, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M, Archard V, Blanchoin L, Goode BL. Coronin switches roles in actin assembly depending on the nucleotide state of actin. Mol Cell. 2009;34:364–374. doi: 10.1016/j.molcel.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Iida K, Yahara I. Cooperation of two actin-binding proteins, cofilin and Aip1, in Saccharomyces cerevisiae. Genes Cells. 1999;4:21–32. doi: 10.1046/j.1365-2443.1999.00235.x. [DOI] [PubMed] [Google Scholar]
- Ksiazek D, Bandstetter H, Israel L, Bourenkov GP, Katchalova G, Janssen KP, Bartunik HD, Noegel AA, Schleicher M, Holak T. Structure of the N-terminal domain of the adenylyl cyclase-associated protein (CAP) from Dictyostelium discoideum. Structure. 2003;11:1171–1178. doi: 10.1016/s0969-2126(03)00180-1. [DOI] [PubMed] [Google Scholar]
- Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Drubin DG. Cofilin promotes rapid actin filament turnover in vivo. Nature. 1997;388:78–82. doi: 10.1038/40418. [DOI] [PubMed] [Google Scholar]
- Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol Rev. 2008;88:489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- Liu HP, Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989;57:233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Liu HP, Bretscher A. Characterization of TPM1 disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J Cell Biol. 1992;118:285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Quintero-Monzon O, Kugler J, Moseley JB, Almo SC, Lappalainen P, Goode BL. A high affinity interaction with ADP-actin monomers underlies the mechanism and in vivo function of Srv2/cyclase-associated protein (CAP) Mol Biol Cell. 2004;15:5158–5171. doi: 10.1091/mbc.E04-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavoungou C, Israel L, Rehm T, Ksiazek D, Krajewski M, Popowicz G, Noegel A, Schleicher M, Holak T. NMR structural characterization of the N-terminal domain of the adenylyl cyclase-associated protein (CAP) from Dictyostelium discoideum. J Biomol NMR. 2004;29:73–84. doi: 10.1023/B:JNMR.0000019513.86120.98. [DOI] [PubMed] [Google Scholar]
- McCullough BR, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri K, Ono S. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J Cell Sci. 2003;116:4107–4118. doi: 10.1242/jcs.00717. [DOI] [PubMed] [Google Scholar]
- Moriyama K, Yahara I. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J Cell Sci. 2002;115:1591–1601. doi: 10.1242/jcs.115.8.1591. [DOI] [PubMed] [Google Scholar]
- Noegel AA, Blau-Wasser R, Sultana H, Müller R, Israel L, Schleicher M, Patel H, Weijer CJ. The cyclase-associated protein CAP as regulator of cell polarity and cAMP signaling in Dictyostelium. Mol Biol Cell. 2004;15:934–945. doi: 10.1091/mbc.E03-05-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normoyle KPM, Brieher WM. Cyclase associated protein (CAP) acts directly on F-actin to accelerate cofilin-mediated actin severing across the range of physiological pH. J Biol Chem. 2012;287:35722–35732. doi: 10.1074/jbc.M112.396051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Blanchoin L, Abe H, Chen H, Pollard TD, Bamburg JR. Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping ends. J Biol Chem. 2002;277:43011–43016. doi: 10.1074/jbc.M203111200. [DOI] [PubMed] [Google Scholar]
- Okada K, Obinata T, Abe H. XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF cofilin family proteins. J Cell Sci. 1999;112:1553–1565. doi: 10.1242/jcs.112.10.1553. [DOI] [PubMed] [Google Scholar]
- Okada K, Ravi H, Smith EM, Goode BL. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol Biol Cell. 2006;17:2855–2868. doi: 10.1091/mbc.E06-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak V, Drubin D. Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J Cell Biol. 2010;188:769–777. doi: 10.1083/jcb.200909176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin: a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Quintero-Monzon O, Jonasson EM, Bertling E, Talarico L, Chaudhry F, Sihvo M, Lappalainen P, Goode BL. Reconstitution and dissection of the 600 kDa SRV2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover. J Biol Chem. 2009;284:10923–10934. doi: 10.1074/jbc.M808760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott DH, Collins RN, Bretscher A. Secretory vesicles transport velocity in living cells depends on the myosin-V lever arm length. J Cell Biol. 2002;156:35–39. doi: 10.1083/jcb.200110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann AC, Guerin C, Martiel JL, De La Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–131. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- Tsuji T, Miyoshi T, Higashida C, Narumiya S, Watanabe N. An order of magnitude faster AIP1-associated actin disruption than nucleation by the Arp2/3 complex in lamellipodia. PLoS One. 2009;4:e4921. doi: 10.1371/journal.pone.0004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111–123. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- Vojtek A, Haarer B, Field J, Gerst J, Pollard TD, Brown S, Wigler M. Evidence for a functional link between profilin and CAP in the yeast S. cerevisiae. Cell. 1991;66:497–505. doi: 10.1016/0092-8674(81)90013-1. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- Wawro B, Greenfield NJ, Wear MA, Cooper JA, Higgs HN, Hitchcock-DeGregori SE. Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry. 2007;46:8146–8155. doi: 10.1021/bi700686p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K, et al. Adenylate cyclase-associated protein 1 overexpressed in pancreatic cancers is involved in cancer cell motility. Lab Invest. 2009;89:425–432. doi: 10.1038/labinvest.2009.5. [DOI] [PubMed] [Google Scholar]
- Yusof A, Hu N-J, Wlodawer A, Hofmann A. Structural evidence for variable oligomerization of the N-terminal domain of cyclase-associated protein (CAP) Proteins. 2005;58:255–262. doi: 10.1002/prot.20314. [DOI] [PubMed] [Google Scholar]
- Yusof A, Jaenicke E, Pedersen JS, Noegel AA, Schleicher M, Hofmann A. Mechanism of oligomerisation of cyclase-associated protein from Dictyostelium discoideum in solution. J Mol Biol. 2006;362:1072–1081. doi: 10.1016/j.jmb.2006.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.