Abstract
Tumour suppressor genes encode a broad class of molecules whose mutational attenuation contributes to malignant progression. In the canonical situation, the tumour suppressor is completely inactivated through a two-hit process involving a point mutation in one allele and chromosomal deletion of the other1. Here, to identify tumour suppressor genes in lymphoma, we screen a short hairpin RNA library targeting genes deleted in human lymphomas. We functionally identify those genes whose suppression promotes tumorigenesis in a mouse lymphoma model. Of the nine tumour suppressors we identified, eight correspond to genes occurring in three physically linked ‘clusters’, suggesting that the common occurrence of large chromosomal deletions in human tumours reflects selective pressure to attenuate multiple genes. Among the new tumour suppressors are adenosylmethionine decarboxylase 1 (AMD1) and eukaryotic translation initiation factor 5A (eIF5A), two genes associated with hypusine, a unique amino acid produced as a product of polyamine metabolism through a highly conserved pathway2. Through a secondary screen surveying the impact of all polyamine enzymes on tumorigenesis, we establish the polyamine–hypusine axis as a new tumour suppressor network regulating apoptosis. Unexpectedly, heterozygous deletions encompassing AMD1 and eIF5A often occur together in human lymphomas and co-suppression of both genes promotes lymphomagenesis in mice. Thus, some tumour suppressor functions can be disabled through a two-step process targeting different genes acting in the same pathway.
Tumour suppressors may be disabled by point mutations, deletions and promoter methylation. Because mutations in one allele are often followed by deletion of the other1, somatic deletions in human cancers often pinpoint tumour suppressor genes that function as ‘drivers’ of tumour evolution. However, such deletions often encompass other genes, termed ‘passengers’, whose disruption may have no functional consequence3. They also may include haploinsufficient tumour suppressors, whose partial loss of function contributes to cancer development in the absence of mutations targeting the second wild-type allele4.
B-cell non-Hodgkin’s lymphoma is a heterogeneous group of malignancies that frequently acquire large chromosomal deletions whose biological impact is poorly understood5. We previously showed that suppression of gene function using RNA interference can mimic tumour suppressor gene inactivation, and we have developed strategies to screen for driver tumour suppressors using mouse models as a biological filter6,7. To do this for human B-cell non-Hodgkin’s lymphoma, we focused on genes present in deletions identified in several studies8–12 (Fig. 1a and Supplementary Tables 1 and 2). Although not comprehensive, this analysis identified 11 recurrent deletions (encompassing 3–103 genes) targeting a total of 323 recurrently deleted genes (Supplementary Table 3).
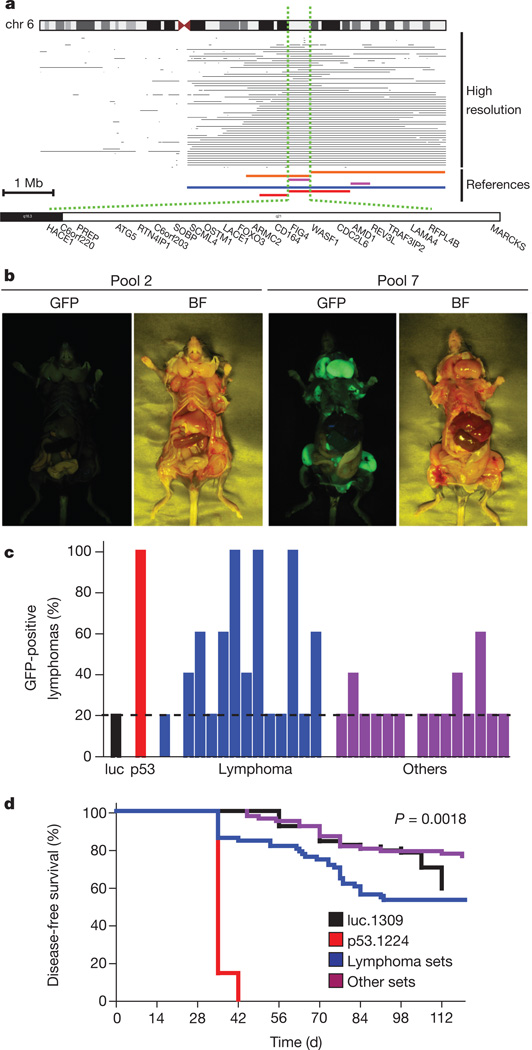
Figure 1. An in vivo shRNA screen for tumour suppressors in lymphoma.
a, Screening interval for the 6q21 deletion. Top, high-resolution data from samples from patients. Bottom, 6q21 deletions as reported in the following references: orange8; violet9; blue10; red11. The dotted green lines delimit the target interval. Representative genes are shown. b, Brightfield (BF) and GFP whole-body imaging of mice from lymphoma pools 2 and 7. c, GFP-positive lymphomas observed for mice transplanted with shRNA sets targeting genes deleted in lymphoma (blue) or in other cancers (violet). Luciferase (luc, black) and p53 (red) were used as negative and positive controls. The dotted line represents the threshold for sequencing lymphoma. Each bar represents a pool. d, Survival curves for mice transplanted with neutral control (luc.1309, black, n = 100), positive control (p53.1224, n = 100), lymphoma sets (blue, n = 70) and other sets (violet, n = 75).
We next designed a short hairpin RNA (shRNA) library targeting the mouse orthologues of deleted genes using on-chip oligonucleotide synthesis and a bar-coding strategy such that shRNAs could be amplified from the mixtures in pools of a discrete size. The pools were individually screened for their ability to increase the lymphomagenic potential of pre-malignant haematopoietic stem and progenitor cells (HSPCs) from Eμ-myc transgenic mice (Supplementary Fig. 1a), which express the Myc oncogene in the B-cell compartment and, although not producing a precise pathological match to human lymphoma, have been widely used to identify and characterize important human cancer genes13.
Using a modified syngeneic transplant model described previously14, we identified conditions such that a green fluorescent protein (GFP)-coupled p53-targeting shRNA efficiently accelerated lymphomagenesis when diluted at 1:200 with a neutral shRNA (Supplementary Fig. 1b). Although typically only about 20% of the HSPCs were transduced (as assessed by GFP fluorescence), the resulting lymphomas were GFP positive. By contrast, a control GFP-tagged shRNA targeting a neutral gene (luciferase) did not accelerate lymphomagenesis and most lymphomas that eventually arose were GFP negative (Supplementary Fig. 1c–e).Hence, GFP fluorescence provides confirmation that an shRNA confers a selective advantage during lymphomagenesis and provides a fluorescent marker to track and isolate lymphoma cells.
To determine whether lymphoma deletions are enriched for tumour suppressors specific to lymphoma, we tested in parallel equivalentsized shRNA pools targeting genes deleted in breast cancer, acute myeloid leukaemia and hepatocellular carcinoma that show only modest overlap with lymphoma (Supplementary Fig. 2). Only 20% (3/15) of the control pools accelerated lymphomagenesis whereas more than half (8/14) of the lymphoma pools scored (Fig. 1b, c). Furthermore, the lymphoma pools scored more frequently than the breast cancer, acute myeloid leukaemia and hepatocellular carcinoma sets (Fig. 1d; P = 0.0018), implying that our approach enriched for shRNAs targeting tumour suppressors and that, despite some limitations, the Eμ-myc model can accurately filter genetic interactions relevant to the human disease.
Sequencing of PCR-amplified shRNAs present in accelerated GFP-positive lymphomas was used to identify candidate ‘drivers’ of disease. Candidates were prioritized using an enrichment score based on (1) the number of shRNAs targeting the gene retrieved from the lymphomas, (2) the number of lymphomas carrying shRNAs targeting the gene and (3) the enrichment of each shRNA relative to its representation in the pool (Supplementary Table 4). Nine new tumour suppressors were subsequently validated (Supplementary Table 5), all of which (by their inclusion in the screen) were embedded within recurrent somatic deletions in human lymphoma (Supplementary Fig. 4) and for several of which there is additional evidence of mutation (AMD1, SMPD2 and CSMD1) or promoter methylation (KCTD11) in other cancers. In all cases, several shRNAs per gene were re-assayed individually and knockdown was confirmed by immunoblotting or quantitative PCR with reverse transcription (Supplementary Figs 3 and 5).
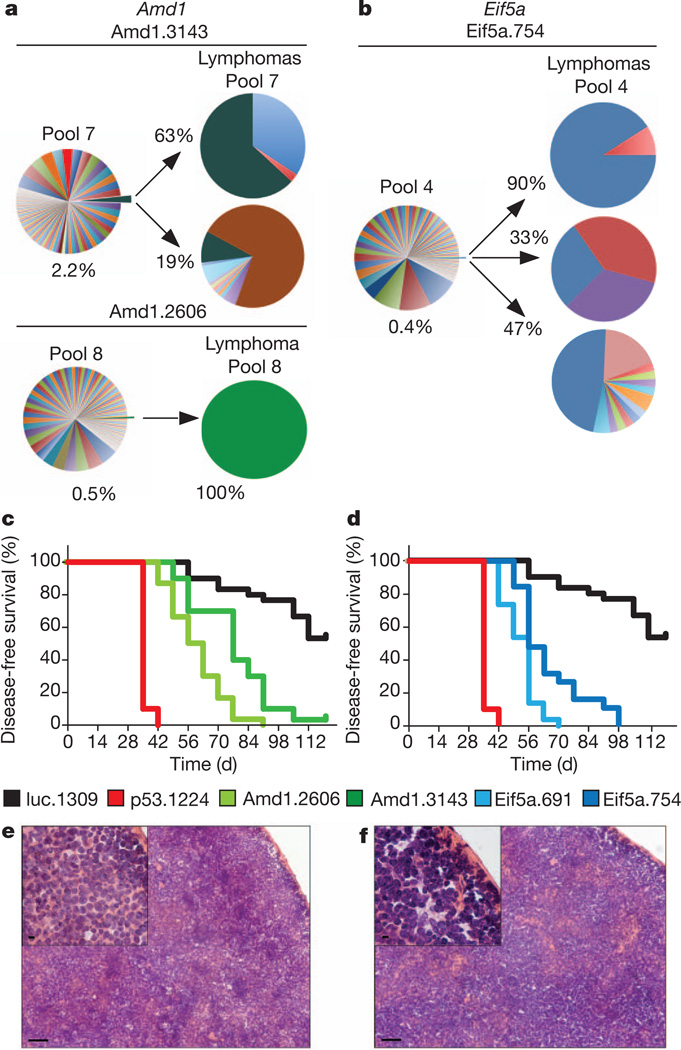
Two pairs of tumour suppressors were connected by predicted functional relationships. MED4 (13q14) and Cyclin C (6q16) are both components of the Mediator, a multi-unit complex that has been associated with transcription regulation15. Also, the candidate gene AMD1 (6q21) controls the biochemical pathway leading to a unique post-translational modification of a second candidate, eIF5A (17p) (Fig. 2a–d and Supplementary Fig. 5a, b). Owing to physical linkage, both the human AMD1 and eIF5A genes are frequently co-deleted with other genes scoring in our assay; indeed, eIF5A is adjacent to TP53 on chromosome 17 (Supplementary Fig. 6). Additionally, as is seen in lymphoma patients with TP53 mutations (ref. 16), underexpression of eIF5A protein was associated with a significantly reduced event-free survival (P < 0.01; Supplementary Fig. 7). Nonetheless, lymphomas triggered by suppression of Amd1 or Eif5a arise independently of p53 loss, because shRNAs targeting each gene alone were sufficient to trigger lymphomas with aggressive features (Fig. 2e, f) and those lymphomas expressing Eif5a shRNAs retained intact p53 function as assessed by several functional and molecular criteria (Supplementary Figs 8 and 9). These data support an emerging view that large chromosomal deletions can target several tumour suppressors whose attenuation contributes independently to tumorigenesis17.
Figure 2. Validation of eIF5A and Amd1 as tumour suppressors in lymphoma.
The distribution of shRNAs targeting Amd1 (a) or Eif5a (b) in lymphomas is compared with the corresponding pools. Percentages indicate the frequency of the shRNA in the pool and in the lymphomas. Survival curves for mice reconstituted with two shRNAs targeting Amd1 (c, n = 30 for each shRNA, P < 0.001 for both shRNAs) or Eif5a (d, n = 30 each shRNA, P < 0.001 for both shRNAs). Controls are p53.1224 (red, n = 30, P < 0.001) and luc.1309 (black, n = 30). Haematoxylin and eosin sections of spleens from mice transplanted with shRNA targeting Amd1 (e) or Eif5a (f). Disruption of tissue architecture is visible both in the spleen and lymph nodes. Scale bars, 50 µm; insets, 5 µm.
AMD1 encodes an enzyme controlling a rate-liming step for the synthesis of spermidine, which is incorporated into eIF5A in the form of the non-standard amino acid hypusine18. Interestingly, eIF5A represents one of the two known hypusine-containing proteins (the other is eIF5A2, a family member not expressed in B cells19). The fact that both Amd1 and eIF5A act as tumour suppressors suggests that the level of hypusinated eIF5A can be decreased by independent mechanisms, acting either on the total level of eIF5Aor on the enzymes controlling hypusine synthesis. The latter mechanism was unexpected because high levels of polyamines are often linked to cancer, and as ornithine decarboxylase (ODC1), which regulates another key step in spermidine synthesis (Fig. 3a), is required for Myc-induced lymphoma development20.
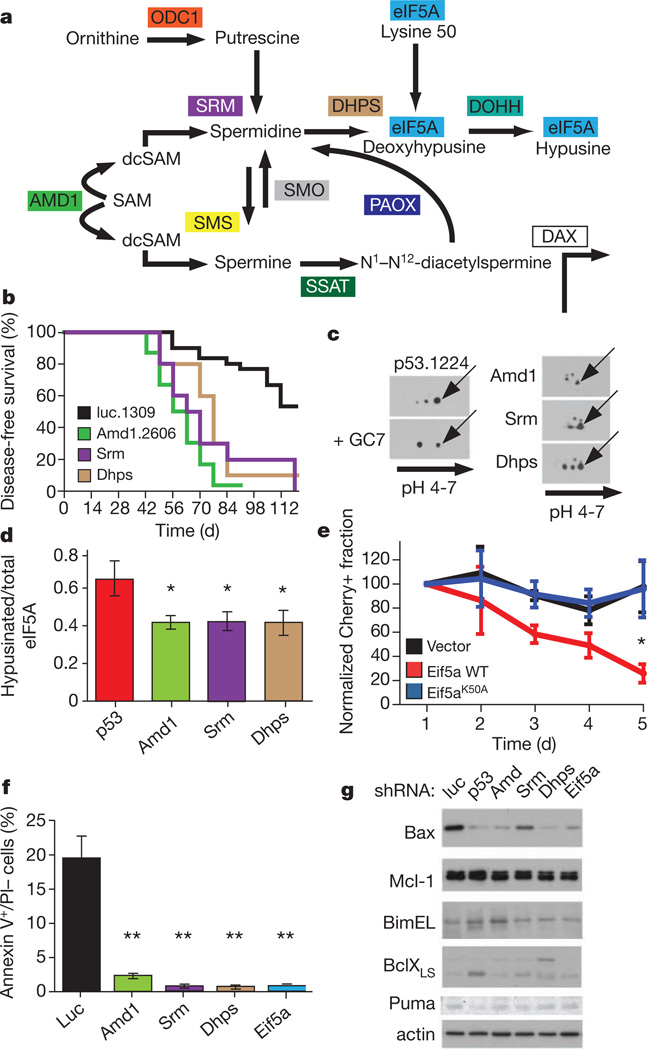
Figure 3. Biosynthesis of hypusine as a tumour suppressor pathway.
a, Schematic of the polyamine–hypusine pathway. Enzymes and compounds are indicated as follows: ODC1, ornithine decarboxylase; SRM, spermidine synthase; SMS, spermine synthase; DHPS, deoxyhypusine synthase; DOHH, deoxyhypusine hydrolase; SMO(X), spermine oxidase; PAOX, polyamine oxidase; SSAT, spermidine–spermine acetyltransferase; DAX, diamine transporter. SAM, S-adenosylmethionine; dc-SAM, decarboxylated S-adenosylmethionine. The SSAT–PAOX axis can also convert spermidine to putrescine (not shown). b, Survival curves for mice (n = 10 per pool) transduced with the following shRNAs or shRNA pools: Srm (violet, P < 0.001), Dhps (blue, P < 0.001), Amd1 (Amd1.2606, green, P < 0.001) and control (luc.1309, black). c, Two-dimensional PAGE followed by eIF5A immunoblotting of lymphomas driven by the indicated shRNAs. The p53.1224 lymphomas were treated with 10 µM N1-guanyl-1,7-diaminoheptane (GC-7) in short-term culture conditions. Arrows indicate the hypusinated form. d, Quantification of hypusinated/total eIF5A ratio for the indicated shRNAs (n = 3 per group, *P < 0.05). Error bars, s.d. e, Lymphomas generated by transduction of Eif5a shRNAs were retrovirally transduced with control vector (black), a complementary DNA encoding wild-type Eif5a (red) or a mutant complementary DNA that cannot be hypusinated because of the substitution of the modified lysine (K50A, blue). Cherry percentages were monitored for 5 days and normalized to the Cherry fraction at day 1 (100). Error bars, s.d. (n = 4 for each time point, *P < 0.05). f, Annexin V+/PI− fractions of GFP-positive B cells 3 weeks after adoptive transplant of Eμ-myc HSPCs transduced with the indicated shRNAs or shRNA pools (n = 3 per each shRNA; **P < 0.01). Error bars, s.d. g, Western blot analysis for expression of the indicated proteins in Eμ-myc pre-malignant B cells transduced with the indicated shRNAs and sorted 21 days after transplant. Cells from three mice were pooled for each shRNA.
For clarification, we performed a secondary screen by designing shRNA pools (three or four shRNAs per gene) targeting each enzyme in the polyamine pathway and testing each for their tumour-promoting activity in vivo (Fig. 3b). The results of these experiments were decisive: although shRNAs targeting peripheral activities showed no effect, those targeting three of the four enzymes responsible for hypusine synthesis showed potent tumour-promoting effects. Hence, knockdown of Srm (spermidine synthase) and Dhps (deoxyhypusine synthase), which are both essential for the hypusine modification of eIF5A, promoted lymphomagenesis in vivo (Fig. 3b and Supplementary Fig. 10).
As predicted, lymphomas harbouring each of the tumour-promoting shRNA pools showed a reduction in eIF5A hypusination relative to controls as assessed by two-dimensional polyacrylamide gel electrophoresis (PAGE) followed by eIF5A immunoblotting (Fig. 3c, d and Supplementary Fig. 11). Furthermore, reintroduction of an shRNA-resistant wild-type Eif5a cDNA inhibited the outgrowth of lymphoma cells expressing Eif5a shRNAs, and a mutant (Eif5aK50A) that could not be hypusinated21 had no effect (Fig. 3e and Supplementary Fig. 12). Finally, suppression of Eif5a attenuates spermidine-induced cell death in lymphomas driven by Amd1 knockdown while not altering sensitivity to the cytotoxic drug adriamycin (Supplementary Fig. 13). Therefore, although Amd1 and Srm may have additional cancer-relevant targets, our collective genetic and biochemical data provide strong evidence that the polyamine–hypusine axis is a potent tumour suppressor network in lymphomas.
To gain insights into the mechanism, we analysed pre-malignant B cells expressing each set of shRNAs. Although we observed no obvious effect on BrdU incorporation, S phase content and PCNA expression (data not shown), we noticed that suppression of Amd1, Srm, Dhps and Eif5a led to the reduction of apoptosis, as assessed by annexin V staining and by the absence of substantial caspase-3 cleavage (Fig. 3f and Supplementary Fig. 14). Conversely, enforced expression of a wild-type (but not mutant) eIF5A into a human line with 17p deletions also triggered apoptosis, as has been described in other contexts22 (Supplementary Fig. 15).
We next determined whether the expression of apoptotic regulators known to modulate Myc-induced lymphomagenesis (for example Bax, Bim, Puma, Bcl-2, Bcl-xL and Mcl-1) were altered in pre-malignant B cells expressing shRNAs targeting the hypusination biosynthetic pathway (Fig. 3g). Bax was the only protein consistently underexpressed (Fig. 3g) and was found reduced through a non-biased isobaric tag for relative and absolute quantitation (iTRAQ)-based proteomic analysis of pre-malignant B cells transduced with shRNAs targeting Eif5a and Amd1 (Supplementary Fig. 16). Gene ontology analysis of the 431 differentially expressed proteins identified ‘apoptosis’ as the most significantly enriched functional category (P = 1.9 × 10−4; Supplementary Table 8). Precisely how Bax is controlled by Eif5a hypusination remains to be determined; nonetheless, Bax is a known eIF5A target22 and is an established suppressor of lymphomagenesis in the Eμ-myc model23.
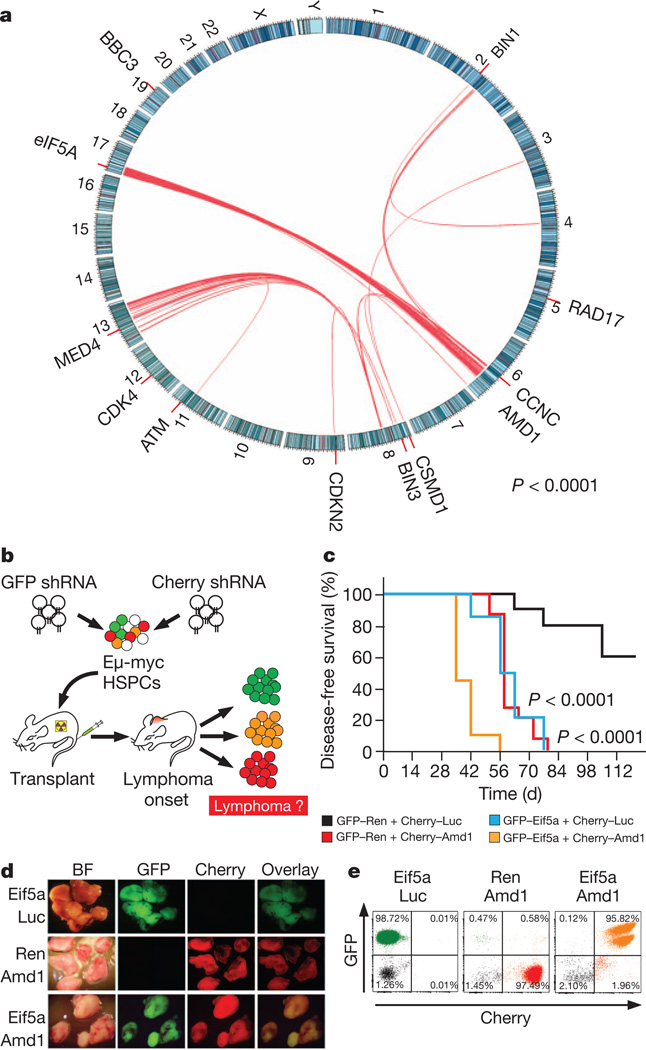
Next, we returned to our analysis of human lymphoma to search for somatic mutations or evidence of an epistatic relationship between eIF5A and AMD1. Sequencing efforts so far have only identified two non-synonymous mutations in AMD1 (ref. 24) and none in eIF5A, raising the possibility that these tumour suppressors are haploinsufficient. Accordingly, we never observed complete suppression of either eIF5A or Amd1 in lymphomas driven by their corresponding shRNAs (Supplementary Fig. 5a, b), and primary B cells with near-complete suppression had poor viability (Supplementary Fig. 17). Surprisingly, analysis of publicly available comparative genomic hybridization data of diffuse large B-cell lymphomas25 showed a significant co-association between the deletion of AMD1 (6q21) and DHPS (19p13) (Supplementary Fig. 18) and AMD1 and eIF5A (17p13) (Fig. 4a; P < 0.0001), with the 17p and 6q co-deleted samples enriched in the ABC subtype. Although other genes in the deleted regions probably also contribute to tumour phenotypes, the co-deletion of two genes in the same pathway was unexpected.
Figure 4. Loss of eIF5A andAMD1 cooperate in lymphoma progression.
a, Deletions of eIF5A (17p) and AMD1 (6q21) are significantly associated in human lymphoma. Red lines identify significant pairwise co-deletion frequency for the indicated genomic regions. Representative tumour suppressors are shown. b, Outline of the two-colour in vivo cooperation assay. c, Survival curves for the following shRNA combinations: GFP–Ren + Cherry–Luc (n = 10, black); GFP–Eif5a + Cherry–Luc (n = 15, blue); GFP–Ren + Cherry-Amd1 (n = 15, red); GFP–Eif5a + Cherry–Amd (n = 20, orange). P values refer to Mantel–Cox tests of the Eif5a–Amd1 knockdown versus Eif5a + Luc or Ren + Amd1. Bright-field (BF), GFP and Cherry imaging (d) and flow cytometry (e) for the indicated shRNA combinations. The first shRNA was tagged with GFP, the second with Cherry.
Because AMD1 and eIF5A are each essential genes26,27, we reasoned that attenuation of two haploinsufficent tumour suppressors might selectively target the anti-tumour activities of polyamine biogenesis while leaving essential functions intact. As an initial test, we co-transduced HSPCs with shRNAs targeting Eif5A and Amd1, tagged with either GFP or Cherry fluorescent markers, which allowed us to track cells transduced with one or both shRNAs (Fig. 4b). Strikingly, the co-knockdown of Eif5A and Amd1 accelerated lymphomagenesis over both individual shRNAs (P < 0.0001; Fig. 4c), and the resulting lymphomas were invariably GFP–Cherry double positive (Fig. 4e and Supplementary Fig. 19). High-resolution isoelectric point (pI) separation followed by eIF5A immunoblotting indicated that co-depletion of Amd1 and Eif5a led to lower levels of hypusinated eIF5A than single knockdown of either gene, providing a biochemical basis for the accelerated lymphomagenesis driven by co-suppression of both genes (Supplementary Fig. 20).
Using a mouse lymphoma model to distinguish driver from passenger lesions, we functionally validated nine tumour suppressors of B-cell non-Hodgkin’s lymphoma. Two of these, AMD1 and eIF5A, are involved in the biosynthesis of hypusine. This highly conserved pathway involves two enzymes (DHPS, DOHH) that specifically target the same lysine on—and apparently only on—eIF5A2. Although its action is not established28, studies in model organisms suggest that hypusinated eIF5A controls proliferation by regulating translation initiation or elongation29,30. Paradoxically, the eIF5A family member eIF5A2 can function as an oncogene in a murine hepatocellular carcinoma model6 but not in the system described here (data not shown), highlighting the contextual action of pathway components that, in pre-malignant B cells, act primarily to promote apoptosis. The fact that an offshoot pathway of polyamine biogenesis suppresses tumorigenesis may partly explain the failure of strategies to inhibit the polyamine pathway in the clinic31.
Our study also shows a previously unanticipated mechanism for disabling tumour suppressor networks, which are typically thought to be inactivated through mutation and/or loss of two alleles of the same gene. Here the ‘two hits’ arise through inactivating a single allele of two genes in the same network, whose combined effect in this case presumably reduces eIF5A function below the threshold needed to restrict cancer development while retaining sufficient polyamines for essential roles in gene regulation and membrane stability31. Although the prevalence of this mode of tumour suppressor inactivation remains to be determined, such cooperation between haploinsufficient tumour suppressors provides one strategy for minimizing the anti-proliferative output of complex networks that also control essential metabolic processes. As such interactions are impossible to identify from genomic approaches alone, our results further highlight the importance of integrating genomic analysis with functional studies to decode the complexity of cancer genomes.
METHODS
Oncogenomics data
To generate the lymphoma screening interval, deletion data were retrieved from the references and clustered if reported by several sources. Overlapping intervals (Supplementary Table 2) were used to generate the gene list (Supplementary Table 3). Murine orthologues were obtained from Biomart and were used to generate the shRNA library. Control libraries consist of shRNA pools targeting genes deleted in other cancers including the following: hepatocellular carcinoma (eight pools6); breast cancer (three pools, targeting genes frequently deleted (>5%) and included in focal deletions (<5 Mb)); acute myeloid leukaemia (four pools, targeting genes included in focal deletions at 7q32,33). SNP profiles of human lymphoma cell lines used in this study are deposited in the Gene Expression Omnibus database under accession number GSE36967.
Vectors and library construction
To generate the shRNA library, 101-base-pair oligonucleotides (including the mir30-shRNA precursor, XhoI–EcoRI sites and a pool-specific barcode) were synthesized on a 55k customized oligonucleotide array (Agilent Technologies). Pool-specific PCR products were individually cloned in the LMS vector34. The LMS–Cherry vector was constructed from the LMS vector by standard cloning techniques. Viruses were produced by transient transfection of Phoenix ecotropic packaging cell line. shRNA sequences are reported in Supplementary Table 6.
Tumour sequencing
DNA was extracted from lymphoma cell pellets using a modified Laird protocol. Briefly, lymphoma single-cell suspensions were prepared by passing lymph nodes through a 100 µM mesh. Laird buffer (0.2% SDS, 5 mM EDTA, 200 mM NaCl, 100 mM Tris, pH 8.5) supplemented with 5 µg ml−1 Proteinase K (Roche) was added to lymphoma pellets that were incubated overnight in an Eppendorf Thermomixer at 55 °C at 6.5g agitation. After centrifugation, DNA was extracted by adding a volume of isopropanol. DNA pellets were washed in 70% ethanol, air dried and resuspended in TE buffer. shRNA representation was determined by both mass Sanger sequencing (30–100 colonies per tumour) and by Solexa bar-coded deep-sequencing. For the former method, shRNAs were amplified by PCR with vector-specific primers (MSCV.Bgl2, 5′-TCCTTCTCTAGGCGCCGGAATTAG-3′ mir30.EcoRI 5′-CTAAAGTAGC CCCTTGAATTCCGAGGCAGTAGGCA-3′). The PCR products were digested and cloned in the LMS vector. shRNAs were identified by sequence alignment using the Blat algorithm35. For the latter method, a PCR-based barcoding strategy was designed. Briefly, shRNA sequences were amplified from the genomic tumour DNA with primers annealing to the shRNA loop (forward primer) and the miR30 3′-region (reverse primer). The primers also contained the P5 and P7 adaptor sequences required for sequencing on the Illumina GAII platform. Furthermore, the forward primer carried an eight-nucleotide barcode sequence between the loop- and the P5-sequence. After amplification, PCR products from individual tumour samples amplified with specific barcodes were purified, quantified and pooled. Approximately 2 × 106 50-base-pair reads were acquired for each sample. By reading 50 nucleotides into the amplicon starting from the shRNA guide strand, we were able to deconvolute the different tumour samples according to the sample-specific barcode. The specific sequences were subsequently identified by comparing the sequences with the original library sequences using the BLAT algorithm, and the relative distribution was calculated. A gene enrichment score was calculated by adding the percentages of enrichment of all shRNAs in each tumour and multiplying it by the number of lymphomas and the number of different shRNAs targeting the same gene.
Haematopoietic stem and progenitor cell isolation and in vivo adoptive transfer
Eμ-myc HSPCs were prepared from E13.5 fetal livers as described36 and retrovirally transduced with shRNA pools or individual shRNA. After short-term in vitro expansion, 6 × 105 HSPCs were transplanted by tail-vein injection into sublethally irradiated (600 rad) 6- to 8-week-old syngenic C57/BL6 females. Recipient mice were administered ciprofloxin (125 mg l−1, Sigma-Aldrich; supplemented with sucrose 20 g l−1) in the drinking water for 1 week before and 2 weeks after transplantation.
Lymphoma monitoring and analysis
Mice were examined weekly by lymph node palpation. Disease state was defined by the presence of palpable lymph nodes or paralysis. Statistical evaluation of survival curves was performed using the log-rank (Mantel–Cox) test. To assess cell immunophenotype and GFP content, single-cell suspensions were obtained by passing lymph nodes through a 100 µM mesh, treated with ACK buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 5 min and resuspended in PBS. Pe-Cy5-B220 and Pe-Cy5-IgM (Biolegend) were used for B-cell immunophenotyping. Samples were acquired using a LSR-II Flow Cytometer System (BD Biosciences). All animal experiments were performed according to federal, National Institutes of Health and local Institutional Animal Care and Use Committee guidelines. All mouse experiments were approved by the Cold Spring Harbor Laboratory Institutional Animal Care and Use Committee.
Apoptosis assays
Spleens of asymptomatic mice were dissected 3 weeks after transplant in B6/SJL mice (CD45.1+), and cells were treated with ACK buffer as described above. Half a million cells were washed in PBS, and incubated for 15 min at room temperature in 100 µl Annexin V Buffer (10 mM Hepes, 140 mM NaCl, 25 mM CaCl2, pH 7.4) with 2 µg ml−1 propidium iodide (Sigma-Aldrich) and 5 µl Pacific Blue Annexin V (Biolegend). For cleaved caspase-3 immunoblotting, spleen single-cell suspensions were first depleted of host cells using MagnaBind Streptavidin Beads (Pierce) coupled to a biotin anti-mouse CD45.2 antibody (Biolegend) according to the manufacturer’s protocol. The unbound fraction was then positively selected for B cells using a Dynabeads Mouse Pan B cell kit (Invitrogen). Correct immunophenotype was confirmed by flow cytometry for B220 expression as described above. The neutral control sample (luc) was obtained by pooling spleens from three mice.
Western blot
Lymphoma pellets were lysed in modified RIPA buffer (20 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.1% NP-40, 0.1% SDS) and protein concentration was determined using the DC Protein Assay (Biorad). Twenty micrograms per sample were subjected to SDS–PAGE and transferred to Immobilon P-membrane (Millipore). The following antibodies were used: anti-eIF5A (ab32407, Abcam; 1:2,000); anti-Med4 (ab75791, Abcam; 1:1,000); anti-Bin3 (ab67032, Abcam; 1:1,000); anti-cleaved caspase-3 (number 9664, Cell Signalling, 1:500); anti-p19Arf (Upstate, 1:500); anti-Puma (number 7467, Cell Signalling 1:1,000); HRP-anti-actin (A3854, Sigma; 1:20,000). For Csmd1, a rabbit polyclonal serum was developed by Prime Biotech. Blots were visualized using ECL (Amersham).
Two-dimensional PAGE
Lymphomas were washed in PBS and then lysed in urea lysis buffer (8 M urea, 2% Chaps, 10 mM DTT, 0.5% IPG pH 4–7 buffer (Amersham), trace of bromophenol blue) at room temperature for 20 min. Membranes were removed by centrifugation, and 75 µg of extract was loaded on Immobiline Dry Strips (pH 4–7, 13 cm) (GE Healthcare) for isoelectric focusing with an IPGphor (Amersham Pharmacia Biotech). Strips were rehydrated in sample for 11–14 h, and run at 150 V for 2 h, 500 V for 1 h, 1,000 V for 1 h and 8,000 V for 4 h. Isoelectric focusing strips were then equilibrated for 15 min in modified SDS sample buffer (50 mM Tris pH 6.8, 6 M urea, 30% glycerol, 2% SDS, 1% DTT and trace bromophenol blue), separated in the second dimension by 15% SDS–PAGE, transferred to PVDF membranes and subjected to eIF5A and β-actin immunoblotting as an internal control. GC-7 was purchased from Biosearch. Quantification of hypusinated eIF5A was performed by densitometry (Image J). Error bars, s.d.
OFFGEL fractionation
Mouse embryo fibroblasts (p53−/−) were infected with the indicated combination of shRNAs. Flow cytometry analysis indicated that GFP + Cherry + content defining the double-infected fraction was greater than 90% in all conditions. After 3 days, cells were lysed as described above and 1 mg protein was precipitated using one volume of TCA. Protein pellets were washed three times with acetic acid and resuspended in the manufacturer’s buffer (Agilent). Samples were separated on a 3100 OFFGEL Fractionator (Agilent) using high-resolution pH 4–7 OFFGEL Strips according to the manufacturer’s instructions. After fractionation was completed, fractions 4–17 were separated on a 12% SDS–PAGE, transferred to PVDF membranes and blotted for eIF5A.
eIF5A hypusination quantification
Images from films were acquired with CanoScan Lide 110 in TIFF format (600 dots per inch, black and white mode). Image processing and quantification were performed with ImageJ 1.43. After noise was removed with the Despeckle option (Process/Noise/Despeckel), images were inverted (edit/invert). For one-dimensional SDS–PAGE, the measure area was set to rectangular selection so that it would include the entire length of the band. Background was quantified by averaging 10 random rectangular selections across the image in areas where no signal was present and subtracted from the signal for both eIF5A and actin. For the hypusination profile, total eIF5A was quantified by one-dimensional SDS–PAGE as described. After two-dimensional SDS–PAGE and eIF5A immunoblotting, images were acquired and elliptical selection was used to quantify each spot. The ratio of hypusinated eIF5A to total was calculated by dividing the densitometry value of the hypusinated form by the sum of the values for all the forms.
Quantitative PCR
RNA was extracted from tumours or cells using Rneasy Kit (Qiagen). RNA (2 µg) was used for first strand synthesis (Taqman RT Reagents Kit, Applied Biosystem). Quantitative PCRs were performed in triplicate on a iCycler iQ5 (Biorad) with SYBR Green PCR MasterMix (Applied Biosystem). Beta-actin was used as normalization control. Error bars, s.d. Quantification was based on a standard curve obtained by serial dilution of the indicated control RT reaction. Primer sequences are reported in Supplementary Table 7.
Spermidine competition assay
Lymphomas (sh.Amd) were plated in B-cell media (45% Iscove’s modified Eagle medium, 45% Dulbecco’s minimal essential medium, 10% fetal bovine serum supplemented with 100 U ml−1 penicillin and streptomycin and 55 µM 2-mercaptoethanol) and partly transduced with the indicated retroviral constructs. Cells were then treated with the indicated concentration of spermidine (Sigma-Aldrich) in modified B cell media supplemented with 1 mM aminoguanidine (Sigma-Aldrich) and dialysed bovine serum (Gibco). After treatment for 24 h, cells were diluted with fresh media and allowed to recover for 72 h. Cherry-positive fractions were assessed using a LSR-II Flow Cytometer System (BD Biosciences). The adriamycin competition assay was performed in Eμ-myc Arf−/− lymphomas using GFP-tagged shRNA vectors as previously described.
Deletion association study
High-resolution lymphoma comparative genomic hybridization data were obtained from the Gene Expression Omnibus database (accession number GSE11318). Normalized comparative genomic hybridization profiles were segmented using the circular binary segmentation algorithm implemented in DNAcopy (www.bioconductor.org). Copy number gains and losses were determined by identifying individual segments with a mean log2 ratio greater than 0.2 and lower than −0.2, respectively. To identify gene pairs with significant co-deletion frequency for each gene pair in a particular gene set, we computed the P value for the observed co-deletion frequency under the assumption of independent deletion (Fisher’s exact test). When evaluating the significance of particular gene pairs, and to account for possible confounding factors such as genomic distance and deletion size, we obtained a null distribution by computing the P values for random pairs of genes residing on the same chromosomes of the tested genes; if these resided on the same chromosome, we used random pairs with a similar genomic distance as the tested genes. Significant gene pairs were determined as those having a P value less than the 95% quantile of the null distribution.
iTRAQ
Pelleted cells were lysed mechanically with 18- to 25-gauge needles in 300 µl lysis buffer (10 mM HEPES pH 8, 0.5 M EDTA, 0.5% NP-40) supplemented with phosphatase inhibitors and protease inhibitor cocktail (P2850, P5726, P8340; Sigma-Aldrich). Protein concentration was measured using BCA Protein Assay. An aliquot of 100 µg of each sample was further solubilized by adding ProteaseMax (Promega) to 0.1%. Samples were reduced with tris-(2-carboxyethyl) phosphine (5 mM) and alkylated with methyl methanethiosulphonate (10 mM). Each sample was then precipitated using a methanol/chloroform extraction. After reconstitution in 50 mM triethylammonium bicarbonate (TEAB), 0.1% ProteaseMax, each sample was digested with trypsin (1:50) overnight at 37 °C. The tryptic peptides were reduced to a final volume of 20 µl in a speed-vac, and 30 µl of 500 mM TEAB solution were added to each solution. iTRAQ labelling of peptides was according to the previous report37. After labelling, each solution was acidified by the addition of 3 µl trifluoroacetic acid and combined. Mixed samples were reduced to approximately 40 µl. Combined samples were then analysed by two-dimensional liquid chromatography MudPIT38, using a two-dimensional Vented Column Setup with a Proxeon nano-flow high-performance liquid pump39. An equivalent of about 50 µg of sample was loaded onto a tri-phasic, fused silica capillary column: 250 µm internal diameter packed with 3 cm of 5 µm Aqua C18, followed by 3 cm of 5 µm Luna SCX and 2 cm of 5 µm Aqua C18. For each salt step, peptides were eluted on an analytical column of a 100 µm internal diameter capillary with a 5 µm pulled tip and packed with 15 cm of 3 µm Aqua C18 on line with an LTQ Orbitrap Velos (Thermo Electron). Data were acquired in profile mode using the following parameters: for full-scan Fourier transform mass spectrometry, resolution = 60,000, m/z = 380–1,700 and the 10 most intense ions were fragmented with higher-collision dissociation at a normalized collision energy of 40% and an activation time of 0.1. Minimum threshold signal was at 5,000 and isolation width at 1.2. Dynamic exclusion settings were repeat count 1, repeat duration of 30, exclusion list size 500, exclusion duration 60 and exclusion mass width 10 p.p.m.
For data analysis, peaklist files were generated by Distiller (Matrix Science). Protein identification and quantification was performed on Mascot 2.3 (ref. 40) against the International Protein Index mouse database (version 3.86; 58,667 sequences; 26,399,545 residues). Methylthiolation of cysteine, amino (N)-terminal and lysine iTRAQ modifications were set as fixed modifications, methionine oxidation and deamidation as variables. Trypsin was used as cleavage enzyme and one missed cleavage allowed. Mass tolerance was set at 20 p.p.m. for intact peptide mass and 0.5 Da for fragment ions. Results were re-scored with Mascot percolator to give an overall 0.8% false discovery rate for protein identification. Protein-level iTRAQ ratios were calculated as intensity weighted, using only peptides with expectation values less than 0.05. Global ratio normalization was performed using intensity summation, with no outlier rejection. To assign fold-change significance, protein ratios in each iTRAQ channel were fitted to a three-parameter Burr distribution (a member of the log-normal distribution family). Protein ratio fold-changes greater or less than 2 s.d. were assigned as significant for up- or downregulation respectively.
Tissue microarrays
Tissue microarrays were constructed as previously published41,42, using a fully automated Beecher Instrument, ATA-27. The study cohort comprised diffuse large B-cell lymphomas (173 cases) consecutively ascertained at the Memorial Sloan-Kettering Cancer Center between 1989 and 2008. All biopsies were evaluated at Memorial Sloan-Kettering Cancer Center, and the histological diagnosis was based on haematoxylin and eosin staining. Use of tissue blocks was approved by the Institutional Review Board and Human Biospecimen Utilization Committees. Anti-eIF5A (rabbit monoclonal antibody, EP57Y, ABCAM catalogue number ab32407) was used at a 1:2,000 dilution for 30 min after heat-induced epitope retrieval with citrate buffer for 30 min, polymer from envision kit from Dako for 30 min, then 3,3′-diaminobenzidine for 5 min. TP53 was performed on an automated platform according to the manufacturer’s instructions (Ventana). For eIF5A and TP53, tumour cells were scored as follows: 0, negative; 1, less than 50% of tumour cells positive; 2, greater than 50% of tumour cells positive. Array chromosomal comparative genomic hybridization was performed in collaboration with J. Houldsworth at Cancer Genetics.
Supplementary Material
Acknowledgements
We thank J. Simon, M. Taylor, D. Grace and S. Peacock for technical assistance, S. Muller for help in tumour sequencing and members of the Lowe laboratory for discussions. We thank G. Hannon and S. Powers for advice. We thank J. Maragulia, C. Gonzalez, A. Zelenetz, L. Lopez, E. Iskidarova, M. Asher and I. Linkov for help on the tissue microarray staining and data management. This work was supported by the Don Monti Memorial Research Foundation and grants from the National Cancer Institute (CA087497) and National Institutes of Health/American Recovery and Reinvestment Act (CA148532) to S.W.L. and by a Canadian Institutes of Health Research grant (MOP-106530) to J.P. C.S. was supported by the Angel Foundation with a Curt Engelhorn fellowship and C.M. was supported by an American Association for Cancer Research-Astellas fellowship. S.W.L. is a Howard Hughes Medical Institute Investigator.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions C.S., C.M. and S.W.L. designed the study. C.S. and C.M. performed the in vivo screen and analysed data. C.S., L.L. and J.P. performed eIF5A hypusination assays and analysed data. C.R. and D.P. performed the iTRAQ analysis. C.S., J.R., S.Y. and A.K. analysed the oncogenomics data. S.W.L. supervised the project. C.S., C.M. and S.W.L wrote the paper. C.S., C.M. and I.A. performed experiments on human cell lines. J.T. analysed tissue microarrays. All authors reviewed the manuscript.
Author Information The cell line data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE36967.
The authors declare no competing financial interests
References
- 1.Knudson AG. Cancer genetics. Am. J. Med. Genet. 2002;111:96–102. doi: 10.1002/ajmg.10320. [DOI] [PubMed] [Google Scholar]
- 2.Park MH. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A) J. Biochem. 2006;139:161–169. doi: 10.1093/jb/mvj034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bignell GR, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger AH, Pandolfi PP. Haplo-insufficiency: a driving force in cancer. J. Pathol. 2011;223:137–146. doi: 10.1002/path.2800. [DOI] [PubMed] [Google Scholar]
- 5.Lenz G, Staudt LM. Aggressive lymphomas. N. Engl. J. Med. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bric A, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salaverria I, et al. Specific secondary genetic alterations in mantle cell lymphoma provide prognostic information independent of the gene expression-based proliferation signature. J. Clin. Oncol. 2007;25:1216–1222. doi: 10.1200/JCO.2006.08.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubio-Moscardo F, et al. Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood. 2005;105:4445–4454. doi: 10.1182/blood-2004-10-3907. [DOI] [PubMed] [Google Scholar]
- 10.Chen W, et al. Array comparative genomic hybridization reveals genomic copy number changes associated with outcome in diffuse large B-cell lymphomas. Blood. 2006;107:2477–2485. doi: 10.1182/blood-2005-07-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bea S, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183–3190. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mestre-Escorihuela C, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–280. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D, Wagner EF, Palmiter RD. The origins of oncomice: a history of the first transgenic mice genetically engineered to develop cancer. Genes Dev. 2007;21:2258–2270. doi: 10.1101/gad.1583307. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 15.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Young KH, et al. Mutations in the DNA-binding codons of TP53, which are associated with decreased expression of TRAIL receptor-2, predict for poor survival in diffuse large B-cell lymphoma. Blood. 2007;110:4396–4405. doi: 10.1182/blood-2007-02-072082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue W, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc. Natl Acad. Sc.i USA. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper HL, Park MH, Folk JE. Posttranslational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell. 1982;29:791–797. doi: 10.1016/0092-8674(82)90441-x. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins ZA, Haag PG, Johansson HE. Human eIF5A2 on chromosome3q25–q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics. 2001;71:101–109. doi: 10.1006/geno.2000.6418. [DOI] [PubMed] [Google Scholar]
- 20.Nilsson JA, et al. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 21.Cano VS, et al. Mutational analyses of human eIF5A-1 – identification of amino acid residues critical for eIF5A activity and hypusine modification. FEBS J. 2008;275:44–58. doi: 10.1111/j.1742-4658.2007.06172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Z, Cheng Z, Taylor CA, McConkey BJ, Thompson JE. Apoptosis induction by eIF5A1 involves activation of the intrinsic mitochondrial pathway. J. Cell. Physiol. 2010;223:798–809. doi: 10.1002/jcp.22100. [DOI] [PubMed] [Google Scholar]
- 23.Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol. Cell. Biol. 2001;21:7653–7662. doi: 10.1128/MCB.21.22.7653-7662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson TJ, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenz G, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl Acad. Sci. USA. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parreiras-e-Silva LT, et al. Evidences of a role for eukaryotic translation initiation factor 5A (eIF5A) in mouse embryogenesis and cell differentiation. J. Cell. Physiol. 2010;225:500–505. doi: 10.1002/jcp.22229. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura K, et al. Essential role of S-adenosylmethionine decarboxylase in mouse embryonic development. Genes Cells. 2002;7:41–47. doi: 10.1046/j.1356-9597.2001.00494.x. [DOI] [PubMed] [Google Scholar]
- 28.Park MH, Nishimura K, Zanelli CF, Valentini SR. Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids. 2009;38:491–500. doi: 10.1007/s00726-009-0408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson A, Hershey JW. Eukaryotic translation initiation factor (eIF) 5A stimulates protein synthesis in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2011;108:6415–6419. doi: 10.1073/pnas.1008150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saini P, Eyler DE, Green R, Dever TE. Hypusine-containing protein eIF5A promotes translation elongation. Nature. 2009;459:118–121. doi: 10.1038/nature08034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casero RA, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nature Rev. Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 32.Curtiss NP, et al. Isolation and analysis of candidate myeloid tumor suppressor genes from a commonly deleted segment of 7q22. Genomics. 2005;85:600–607. doi: 10.1016/j.ygeno.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Kratz CP, et al. Candidate gene isolation and comparative analysis of a commonly deleted segment of 7q22 implicated in myeloid malignancies. Genomics. 2001;77:171–180. doi: 10.1006/geno.2001.6636. [DOI] [PubMed] [Google Scholar]
- 34.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nature Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 35.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCurrach ME, Lowe SW. Methods for studying pro- and antiapoptotic genes in nonimmortal cells. Methods Cell Biol. 2001;66:197–227. doi: 10.1016/s0091-679x(01)66010-2. [DOI] [PubMed] [Google Scholar]
- 37.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Washburn MP. Utilisation of proteomics datasets generated via multidimensional protein identification technology (MudPIT) Brief. Funct. Genom. Proteom. 2004;3:280–286. doi: 10.1093/bfgp/3.3.280. [DOI] [PubMed] [Google Scholar]
- 39.Taylor P, et al. Automated 2D peptide separation on a 1D nano-LC-MS system. J. Proteome Res. 2009;8:1610–1616. doi: 10.1021/pr800986c. [DOI] [PubMed] [Google Scholar]
- 40.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Hedvat CV, et al. Application of tissue microarray technology to the study of non-Hodgkin’s and Hodgkin’s lymphoma. Hum. Pathol. 2002;33:968–974. doi: 10.1053/hupa.2002.127438. [DOI] [PubMed] [Google Scholar]
- 42.Hemann MT, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.