Abstract
Antibiotic efflux is an important mechanism of resistance in pathogenic bacteria. Here we describe the identification and characterization of a novel chromosomally encoded multidrug resistance efflux protein in Staphylococcus aureus, MdeA (multidrug efflux A). MdeA was identified from screening an S. aureus open reading frame expression library for resistance to antibiotic compounds. When overexpressed, MdeA confers resistance on S. aureus to a range of quaternary ammonium compounds and antibiotics, but not fluoroquinolones. MdeA is a 52-kDa protein with 14 predicted transmembrane segments. It belongs to the major facilitator superfamily and is most closely related, among known efflux proteins, to LmrB of Bacillus subtilis and EmrB of Escherichia coli. Overexpression of mdeA in S. aureus reduced ethidium bromide uptake and enhanced its efflux, which could be inhibited by reserpine and abolished by an uncoupler. The mdeA promoter was identified by primer extension. Spontaneous mutants selected for increased resistance to an MdeA substrate had undergone mutations in the promoter for mdeA, and their mdeA transcription levels were increased by as much as 15-fold. The mdeA gene was present in the genomes of all six strains of S. aureus examined. Uncharacterized homologs of MdeA were present elsewhere in the S. aureus genome, but their overexpression did not mediate resistance to the antibacterials tested. However, MdeA homologs were identified in other bacteria, including Bacillus anthracis, some of which were shown to be functional orthologs of MdeA.
Eukaryotic and bacterial cells may produce diverse efflux proteins which confer resistance to toxic compounds or drugs by lowering their effective cytoplasmic concentration (for reviews, see references 1 and 27). In pathogenic bacteria, multiple drug resistance (MDR) proteins are important resistance mechanisms which protect these species from antibiotics and antiseptics. In gram-positive bacteria, which lack the added permeability barrier conferred by the outer membrane of gram-negative bacteria, MDR proteins are especially important (22). MDR proteins found in gram-positive bacteria may be subdivided into the major facilitator superfamily (MFS), the small multidrug resistance family (SMR), ABC transporters, and the multidrug and toxic compound extrusion (MATE) family (27).
Staphylococcus aureus is an important cause of community-acquired disease and nosocomial infection (21), and antibiotic resistance is a major problem for antimicrobial therapy (13, 24, 31). S. aureus strains may be resistant to antimicrobials by production of MDR proteins, such as NorA. A member of the MFS group, NorA (10, 34, 37) is a chromosomally encoded protein with 12 transmembrane segments (TMS). NorA acts as an efflux protein for multiple compounds, with certain fluoroquinolones being among the best substrates for transport (26), and its efflux activity is inhibited by reserpine, a plant alkaloid (11). Recombinant NorA protein has recently been functionally reconstituted into proteoliposomes, where it functioned as a pump in response to an artificial proton gradient (38). The norA gene has been inactivated by three groups, all of whom confirmed the importance of the NorA product for quinolone resistance (7, 12, 36). In addition to NorA, other efflux proteins such as QacA and QacB (members of the MFS family) and QacC (a member of the SMR group) have been reported in S. aureus, and their respective genes are located on plasmids (1). Finally, a potential ATP-dependent transporter has been identified in the S. aureus genome by sequence analysis (35), but to our knowledge this protein has not been functionally characterized.
Recently, Kaatz and colleagues (12) suggested the existence of a non-NorA multidrug efflux transporter in S. aureus. Additionally, they observed that the genome of S. aureus contains at least 10 open reading frames (ORFs) whose products show significant residue identity to NorA. If some or all of these were functional efflux proteins, then a gene library of S. aureus in an expression vector would likely contain clones resistant to the substrates for these efflux proteins. We have recently constructed an overexpression library of the entire set of ORFs encoded by the S. aureus genome (unpublished data). This library allows us to determine the target of new antibiotics, since the clone expressing the target will show an increased MIC of the respective compound. Furthermore, the library allows us to screen for novel endogenous resistance mechanisms, which become apparent on overexpression. We describe here the identification and characterization of a novel efflux protein, MdeA (multidrug efflux A), and its promoter mutants. Homologs of MdeA in other bacteria were also characterized and shown to be functional efflux pumps. A phylogeny is presented for this new subfamily of MDR proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions
S. aureus RN4220 is a plasmid-free and prophage-free laboratory strain (14). The following strains were from the GlaxoSmithKline (GSK) culture collection: S. aureus strain Buttle, Salmonella enterica serovar Typhimurium CN648, Staphylococcus haemolyticus J225, and Bacillus cereus BRL1244. The strains Bacillus subtilis 168 and Enterobacter cloacae ATCC 13047 were from the American Type Culture Collection, Manassas, Va. The shuttle plasmid pYH4 consists of a pE194-based replicon from S. aureus (30), a ColE1 origin for Escherichia coli, and a regulated promoter system (Pxyl/tet) (39) to control gene transcription (unpublished data). The plasmid pCR-Blunt (Invitrogen, Carlsbad, Calif.) was used for cloning PCR products. Bacteria were grown in Luria broth or cation-adjusted Mueller-Hinton broth (CAMHB; BBL Microbiology Systems, Cockeysville, Md.) as required, supplemented with ampicillin (100 μg/ml) for pCR-Blunt and pLZ113 and their derivatives in E. coli, erythromycin (5 μg/ml) for pYH4 and derivatives in S. aureus, or kanamycin (5 μg/ml) for pLZ113 and its derivatives in B. subtilis.
Antimicrobial compound testing and determination of MICs
Compounds were from Sigma Chemical Co., St. Louis, Mo., unless indicated otherwise. MICs were determined in 96-well microtiter plates by serial twofold dilution of antibacterials in CAMHB following the NCCLS recommended guidelines (25). The MIC was the lowest concentration of an antibacterial that showed no visible growth after incubation at 37°C for 18 to 24 h, with a starting inoculum of 105 CFU/ml. Uptake and efflux assays for ethidium bromide were conducted essentially as previously described (12). Briefly, cells were grown overnight in CAMHB, diluted 1/50 the next day in fresh CAMHB, and cultured at 37°C with vigorous shaking to exponential phase (optical density at 600 nm of 0.6 to 0.7). The cells were washed and resuspended in 20 mM HEPES buffer, pH 7.0, and used immediately for uptake or efflux assays as described elsewhere (12) with ethidium bromide at 10 μg/ml. The effect of carbonyl cyanide .-chlorophenylhydrazone (CCCP) was assessed by treating cell suspensions, previously normalized for cell number, with 100 μM CCCP for 5 min at room temperature prior to use directly in the assay. The effect of reserpine was determined by treating normalized cell suspensions with the compound at a 10-μg/ml final concentration for 45 min at 37°C. The cells were then harvested by brief centrifugation and washed in HEPES buffer, and the cell densities were finally standardized in HEPES buffer.
Isolation and characterization of spontaneous resistant mutants
S. aureus RN4220 cells were plated onto tryptic soy agar (TSA) plates containing 2 times the MIC of SB-698197-EJ, a proprietary quinoline-methanol (QM) compound which is a substrate of the MdeA efflux protein. After a 24-h incubation at 37°C, resistant colonies were purified once on SB-698197-EJ-containing plates and once on TSA plates. The resistant mutants occurred at a frequency on the order of 10−8. The mdeA gene and its flanking regions were amplified by PCR from the mutants and their parent RN4220 and sequenced directly.
Molecular cloning techniques
Standard procedures were used for molecular cloning in E. coli (28) and the PCR. Genes encoding proteins with significant residue identity to MdeA were cloned from S. haemolyticus (SHA1), B. subtilis (YccA/LmrB/NP388149), B. cereus (RZC03923), E. cloacae (ENCL54535), and S. enterica serovar Typhimurium (AAL21700/EmrB). The respective genes were amplified from chromosomal DNA of the organism in question, using primers designed to include the native ribosome binding site and the entire ORF. The products were cloned into a regulated xyl/tet expression vector for gram-positive bacteria, pLZ113 (39). Primers to amplify genes for MdeA homologs were from Integrated DNA Technologies, Inc., Coralville, Iowa. Primers were as follows, with clamps and restriction enzyme recognition sites added to facilitate recloning shown in lowercase: S. enterica serovar Typhimurium forward primer (FP), cccgggaagcttCTGAGGTGCGTGTGATGC, and reverse primer (RP), cccggggaattcTTAGTGCGCGCCGCCACCG; S. haemolyticus FP, aaatttcccgggGTAGATAAGAGGGGAATAATATG, and RP, aaatttaagcttAGGAGGTCCGAAGGTGAGCGTTATGC; B. cereus FP, cccgggaagcttGTAAGAAACAGGAGGAGAATC, and RP, gggaaagaattcCACCCAAAACAAGTGTTACG; B. subtilis FP, aaatttgatatcCATGCAGTTGATAGAGAGGGG, and RP, gggaaagaattcTTAATGATCTACTTTAACGCGTTTCATAAAG; E. cloacae FP, aaatttaagcttAGGAGGTCCGAAGGTGAGCGTTATGC, and RP, gggaaagaattcCTAGTGCGCCCCACCGCCGCC. Site-directed mutagenesis was carried out using the QuikChange XL kit from Stratagene (La Jolla, Calif.) following the manufacturer's instructions. Competent cells of E. coli strain DH5α were from Invitrogen. B. subtilis strain 168 was transformed by natural competence as previously described (2).
RNA isolation and primer extension
Total S. aureus RNA was isolated by the modified method of Chuang and colleagues (3). Briefly, a 10-ml overnight culture of S. aureus was homogenized by a FastPrep FP120 machine (Savant; Bio101, Inc., Vista, Calif.) in FastRNA tubes containing 400 μl of beads (Bio101, Inc.). The released RNA-containing supernatant was extracted with phenol, phenol-chloroform, and finally chloroform-isoamyl alcohol and then precipitated with isopropanol. The crude RNA pellet was dissolved in water and treated with DNase (Promega, Madison, Wis.) twice. The integrity of isolated RNA was checked by electrophoresis on a 1% formaldehyde gel and quantified by spectrophotometer reading.
Primer extension analysis was performed as described by Huang and Schell (8) with the following modifications. The primer (TCAAAATCTTTCATAATACTTGG) was 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase, based on the manufacturer's protocol (primer extension system; Promega). Fifty micrograms of RNA was annealed with 2.8 × 107 cpm (1 pmol) of 32P-labeled primer in 2× avian myeloblastosis virus primer extension buffer (Promega) by heating at 58°C for 30 min and then placing at room temperature for 1 h. Extension was performed at 42°C for 30 min by addition of 1 μl (5 units) of avian myeloblastosis virus reverse transcriptase (Promega). The transcription product was analyzed on a 6% acrylamide gel containing 7 M urea. A DNA sequencing reaction (Thermo Sequenase cycle sequencing kit; U.S. Biochemicals) was electrophoresed in parallel with the primer extension reaction product. Briefly, 120 ng of PCR product containing the promoter region of the S. aureus strain Buttle mdeA gene was used as the template. A PCR was run in the presence of [α-33P]dideoxynucleoside triphosphates and 8 U of Thermo Sequenase polymerase for 30 cycles. After electrophoresis, the gel was soaked for 15 min in a solution consisting of 5% acetic acid and 15% methanol to remove urea, dried, and exposed to X-ray film (Kodak BioMax MR) with an intensifying screen at −80°C overnight.
Quantitative transcription analysis
One-milliliter aliquots of overnight cultures of S. aureus strains grown in tryptic soy broth was harvested by centrifugation, snap-frozen in a dry ice-ethanol bath, and stored in liquid nitrogen until processed. Total RNA was isolated and reverse transcribed in three separate reactions using commercial kits following the manufacturers' instructions (FastRNA-Blue kit from Bio101; DNA-Free from Ambion, Austin, Tex.; and SuperScript first-strand cDNA synthesis kit from Invitrogen). The resulting cDNA was pooled and subjected to SybrGreen quantitative reverse transcription-PCR using two sets of primer pairs amplifying regions of the mdeA ORF from 996 to 1,168 bp (FP and RP, 5′-TATGGCGATTGTTGTTTTTACTAC-3′ and 5′-AACCGTGTGCATTCATTTCTGG-3′, respectively) and 1,072 to 1,227 bp (FP and RP, 5′-GTTTATGCGATTCGAATGGTTGGT-3′ and 5′-AATTAATGCAGCTGTTCCGATAGA-3′, respectively). SybrGreen PCRs were set up in triplicate using the TaqMan PCR core reagent kit (Applied Biosystems, Foster City, Calif.) according to the instructions provided. Integrity of the PCRs was confirmed by the presence of single, correctly sized bands by agarose gel electrophoresis. Quantitative data obtained for mdeA were normalized to 16S rRNA as an endogenous control and calibrated as described previously (39).
Sequence analysis and phylogeny
Reciprocal BLASTP searches were performed, and the top 500 hits were aligned in CLUSTAL W (33). The resulting trees showed S. aureus MdeA in a monophyletic group of gram-positive bacteria (with the exception of one gram-negative representative, Erwinia chrysanthemi) that was sister taxa to a clade of gram-negative bacteria including the known efflux protein EmrB from E. coli. Sequences from the members of the gram-positive clade as well as an outgroup sequence (EmrB of E. coli) were extracted from public databases, proprietary GSK databases (S. haemolyticus and Staphylococcus epidermidis), Baylor College of Medicine (www.hgsc.bcm.tmc.edu) (Enterococcus faecium), and Integrated Genomics (www.integratedgenomics.com) (B. cereus). Transmembrane helices in proteins were predicted with the TMHMM 2.0 algorithm (15). Sequences were aligned using the MACAW multiple protein alignment software package (29), followed by manual refinements using the Se-Al alignment editor (A. Rambaut; http://evolve.zoo.ox.ac.uk/Se-Al/Se-Al.html). Parsimony and distance analyses were performed using PAUP* 4.0b (D. L. Swofford, Sinauer Associates, Sunderland, Mass.), and maximum likelihood analyses were performed using the ProML program in the PHYLIP package (version 3.6a3; J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle). Bootstrap support values were obtained with 1,000 replicates for maximum parsimony and neighbor-joining analyses and with 100 replicates for the maximum likelihood analysis.
Bayesian analyses were performed using the MrBayes (9) with 500,000 generations, sampling frequency of 100 generations, four Markov chains, random starting trees, and a burn-in of 50,000 generations.
RESULTS
Overexpression of MdeA confers resistance to a range of antibacterial compounds
We have recently constructed an ORF expression library of the genome of S. aureus strain Buttle in a shuttle expression vector pYH4 in S. aureus strain RN4220 (unpublished data). Over 2,300 of the annotated ORFs (∼2,480) in the genome were individually cloned into pYH4, placing their expression under the control of the regulated xyl/tet promoter (39) and a strong S. aureus ribosome binding site (sequence, AGGAGG). Part (∼1,700 ORFs) of this library was screened by plating on inhibitory concentrations of a number of antibacterial compounds to identify genes involved in their susceptibility. An ORF clone encoding MdeA was identified during this screening due to its ability to confer increased resistance to multiple compounds, including mupirocin, fusidic acid, and several GSK proprietary compounds. MIC determinations showed that the presence of pYH4-mdeA in the cell conferred a 4- to 16-fold increase in the MIC of fusidic acid and mupirocin (Table 1) as well as some GSK compounds (data not shown), relative to those for the pYH4 vector control strain. Sequence analysis of mdeA indicated it encodes a multidrug efflux protein (see below).
TABLE 1.
MdeA confers resistance to a range of antimicrobial agents.
| Compound | MIC (μg/ml)
|
||
|---|---|---|---|
| RN4220 (pYH4) | RN4220 (pYH4-MdeA) | RN4220 (pYH4-NorA) | |
| Ethidium bromide | 4 | 64 | ≥128 |
| Benzalkonium chloride | 1 | 4 | 8 |
| Dequalinium chloride | 1 | 2 | 8 |
| Pentamidine isethionate | 16 | 16 | ≥256 |
| Virginiamycin | 0.5 | 8 | 0.5 |
| Novobiocin | 0.13 | 1 | 0.5 |
| Mupirocin | 0.03 | 0.5 | 0.06 |
| Fusidic acid | 0.06 | 0.25 | 0.13 |
| Norfloxacin | 0.5 | 0.5 | 32 |
Values tabulated are MICs in micrograms per milliliter and are the averages of three determinations. In addition, MdeA did not confer significant resistance to the following antibacterials: bacitracin, ceftazidime, chloramphenicol, erythromycin, gentamicin, imipenem, kanamycin, linezolid, oxacillin, phosphomycin, piperacillin, streptomycin, tetracycline, tiamulin, trimethoprim, vancomycin, ciprofloxacin, levofloxacin, gatifloxacin, tosufloxacin, grepafloxacin, sparfloxacin, and moxifloxacin.
To determine the substrate specificity of MdeA, we measured the MIC for strain RN4220 containing pYH4-mdeA against more than 30 antibacterials representing various classes of antibiotics. We found that the pYH4-mdeA clone also led to a significant increase (more than twofold) in the MICs of four compounds: ethidium bromide, benzalkonium chloride, virginiamycin, and novobiocin (Table 1), compared to those for the strain harboring the vector pYH4 alone. Notably, the mdeA clone (pYH4-mdeA) did not confer significant resistance to any fluoroquinolones tested. In contrast, the norA clone on pYH4, which encodes the known S. aureus efflux protein NorA, conferred resistance to fluoroquinolones (Table 1). In addition, pYH4-norA also conferred resistance to the divalent cationic compound pentamidine, but pYH4-mdeA did not. However, both pYH4-mdeA and pYH4-norA conferred resistance to ethidium bromide, benzalkonium chloride, and novobiocin.
The resistance phenotype conferred by pYH4-mdeA is probably due to overexpression of MdeA from the pYH4 expression vector. No increase in MdeA expression was detectable in the strain with pYH4-mdeA compared with the strain with pYH4 alone by Coomassie blue staining of total cell lysates separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). However, quantitative transcription analysis of mdeA using real-time PCR detection showed that the level of transcription of mdeA was elevated 320-fold in cells harboring pYH4-mdeA (without adding the inducer, anhydrotetracycline [ATc]) compared to those with pYH4 alone. Attempts to further increase MdeA production by adding ATc inducer to the growth medium resulted in severe inhibition in cell growth and colony formation. Performing MIC tests in the presence of inducer (as low as 10 ng/ml) also resulted in no visible cell growth. However, the ATc inducer did not have an obvious inhibitory effect on the strain with the pYH4 vector until its concentration was greater than 0.5 μg/ml. Thus, our results indicated that the increased copies of mdeA and the leaky level of xyl/tet promoter activity in the absence of inducer (39) were sufficient to express phenotypically significant levels of MdeA, and further induction with inducer resulted in cell toxicity to the host cell.
MdeA is a member of the MFS
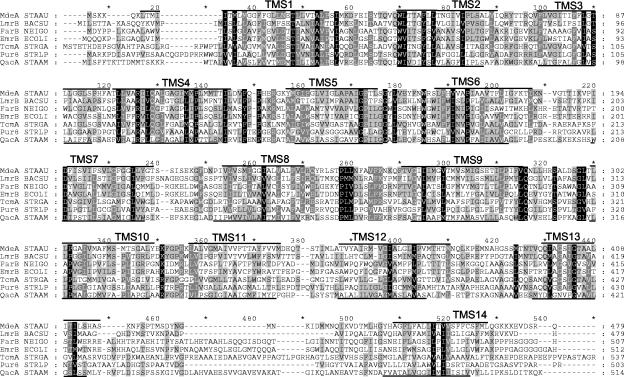
The mdeA gene of S. aureus strain Buttle is 1,437 bp long and encodes a predicted protein of 52,236 Da. The corresponding gene in the S. aureus strain MU50 genome (18) is NP372939, which encodes a protein that is 99% identical to MdeA of strain Buttle. Comparison of MdeA with protein sequence databases revealed significant residue identity to hypothetical and known MDR proteins. Among the top 100 hits in a standard BLASTP search (June 2002), there were only 5 experimentally proven efflux or MDR proteins: LmrB of B. subtilis (O35018 [reference 16]; 37% identity); FarB of Neisseria gonorrhoeae (AAD54074 [19]; 24% identity); EmrB of S. coli (P27304 [20]; 24% identity); TcmA of Streptomyces glaucescens (P39886 [6]; 24% identity); and Pur8 of Streptomyces anulatus (P42670 [32]; 25% identity). These proteins confer resistance to hydrophobic compounds or antibiotics (LmrB, TcmA, Pur8, and EmrB) or antibacterial fatty acids (FarB). The QacA protein (23) of S. aureus, at 23% identity to MdeA, was the closest related, functionally characterized staphylococcal efflux protein. Multiple alignment of MdeA and these six known MDRs (Fig. 1) demonstrated conservation of many of the motifs identified in bacterial MDR proteins (reviewed by Konings and colleagues [27]). MdeA was predicted to have 14 TMS, and the alignment of the MdeA and QacA sequences illustrated the coincidence of the predicted and experimentally demonstrated TMS regions, respectively (Fig. 1).
FIG. 1.
Multiple alignment of MdeA and related MDR proteins. Identical and conserved residues in some or all sequences are indicated by dark and light shading, respectively. Lines above and below the aligned blocks indicate the predicted (MdeA) and experimentally determined (QacA) locations of TMS.
The vast majority of the other homologs identified by BLAST searching were hypothetical or uncharacterized proteins of either S. aureus or other bacteria, most of which had been annotated as MDR proteins but had not been experimentally proven as such. The closest homolog was one of 77% identity in S. haemolyticus (locus SHA1). Closely related proteins (25 to 40% identity) were also present in other gram-positive bacteria, including S. epidermidis, B. subtilis, B. anthracis, Bacillus halodurans, Listeria sp., Streptococcus pyogenes, Streptococcus mutans, Enterococcus faecium, Lactococcus lactis, Corynebacterium glutamicum, Clostridium acetobutylicum, and Oenococcus oeni.
MdeA subfamily phylogenetic relationships
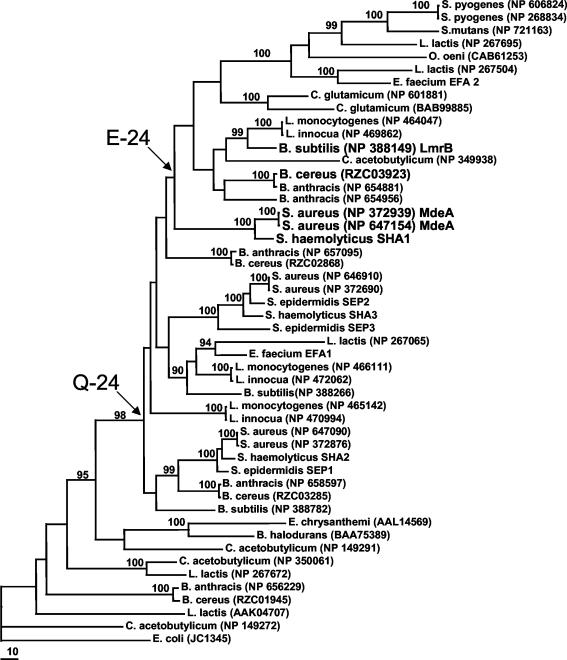
Figure 2 depicts a maximum likelihood phylogram of the 48 closest relatives of S. aureus MdeA as well as the outgroup EmrB from E. coli. In addition, Fig. 2 shows support values for nodes where a variety of phylogenetic methods were in agreement. On the majority of trees, S. aureus MdeA appeared at the base of a group containing the known efflux protein LmrB from S. subtilis as well as a highly supported subclade (100% with all phylogenetic methods) containing S. pyogenes, S. mutans, S. oeni, S. lactis, and S. faecium.
FIG. 2.
Phylogeny of MdeA and related proteins. The maximum likelihood phylogram shows the relatedness of MdeA and 48 other homologous proteins with EmrB of E. coli included as an outgroup. Branch lengths are proportional to the amount of sequence change, and bootstrap support values are indicated by percentages at nodes where the majority of phylogenetic methods yielded support above 90%. The proteins shown to have efflux activity are shown in larger font (MdeA from S. aureus, LmrB from B. subtilis, SHA1 from S. haemolyticus, and RZC03923 from B. cereus). Arrows point to clades that contain either a Q or an E at position 24 (relative to S. aureus MdeA). Where available, accession codes for publicly available sequences are indicated. For sequences without accession numbers, a code of species abbreviation followed by a number (i.e., S. haemolyticus locus 1 is coded as SHA1) distinguishes the different hypothetical proteins.
MdeA is a proton-driven efflux pump
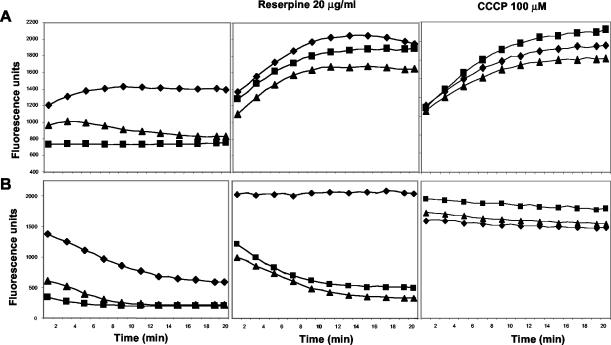
Introduction of either pYH4-mdeA or pYH4-norA plasmids into RN4220 cells rendered them similarly less susceptible to ethidium bromide (Table 1). Thus, the efflux activity of the MdeA protein was studied and compared with that of the NorA efflux pump by measuring uptake and efflux of ethidium bromide in S. aureus strain RN4220 carrying the respective plasmids. The presence of pYH4-mdeA or pYH4-norA in the strains led to decreased uptake or enhanced efflux of ethidium bromide relative to that of the pYH4 vector-alone control strain (Fig. 3). The rates of efflux of the strains harboring either cloned gene were similar to that of the vector-containing control, but the initial and final levels of fluorescence were significantly lower.
FIG. 3.
MdeA is a proton-driven efflux pump. Uptake (A) and efflux (B) of ethidium bromide by study strains are shown. Symbols: ⧫, RN4220/pYH4; ▪, RN4220/pNorA; ▴, RN4220/pMdeA. The data are the means of duplicate experiments.
Pretreatment of the cells with reserpine, a plant alkaloid that inhibits NorA-mediated efflux, resulted in uptake of ethidium bromide in the MdeA-overexpressing cells and NorA-expressing cells at levels similar to that of the pYH4 control (Fig. 3) and in a manner significantly different for the corresponding untreated cells. Efflux of cells overexpressing either MdeA or NorA was significantly inhibited by reserpine compared to the respective untreated controls, but efflux in pYH4-containing cells was totally abolished, suggesting that the increased protein levels in MDR-overexpressing cells partially mitigated the reserpine inhibition.
Proton-driven efflux pumps are effectively inhibited by uncouplers, compounds which collapse the transmembrane potential. Pretreatment with CCCP of cells overexpressing either MdeA or NorA resulted in ethidium bromide uptake and efflux rates similar to those in pYH4 cells (Fig. 3). This suggests that the efflux activity of the MdeA protein is dependent on proton-motive force and an energized membrane.
Spontaneous mutants with mutations in the mdeA promoter increased mdeA transcription and resistance to MdeA substrates
Since overexpression of MdeA from the pYH4 vector conferred an MDR phenotype, this prompted us to investigate if S. aureus is able to spontaneously mutate in mdeA to result in an MDR phenotype. We used a proprietary QM compound to select spontaneous resistant mutants, because this compound is a substrate of MdeA but appears not to have a specific target. Two different mutants (based on resistant levels RM3 and RM8) were thus isolated. These mutants had two- to fourfold increases in the MIC of the compound used in the mutant selection and also had increased MICs of seven other compounds (Table 2). The spectrum of compounds overlapped directly with that of pYH4-MdeA (Table 1) except that the mutants also conferred resistance to lincomycin, suggesting possible mutations in the mdeA gene. Thus, the mdeA gene and its flanking regions were PCR amplified from the mutants and sequenced to compare them with the sequences from the parental mdeA gene of strain RN4220. Sequence comparison revealed no mutations in the mdeA coding region. However, single point mutations (C→A) and (G→T), 53 and 84 bp upstream of the mdeA translation start site, were found in QM-RM3 and QM-RM8, respectively (Fig. 4A).
TABLE 2.
MIC values for mutants with similar resistance pattern as the MdeA-expressing clone.
| Compound | MIC (μg/ml)
|
||
|---|---|---|---|
| RN4220 | QM-RM 3 | QM-RM 8 | |
| Virginiamycin | 0.5 | 1 | 2 |
| Lincomycin | 0.25 | 0.5 | 1 |
| Novobiocin | 0.125 | 0.25 | 0.25 |
| Mupirocin | 0.03 | 0.06 | 0.125 |
| Fusidic acid | 0.06 | 0.06 | 0.125 |
| Ethidium bromide | 4 | 8 | 16 |
| Benzalkonium chloride | 1 | 2 | 2 |
Values tabulated are MICs in micrograms per milliliter and are the averages of three determinations.
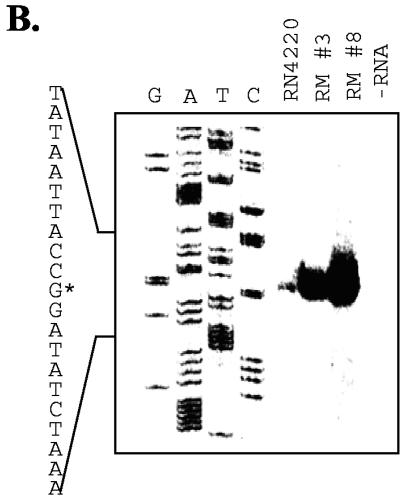
FIG. 4.
Sequence and primer extension analysis of the mdeA promoter region. (A) Promoter region of mdeA from S. aureus RN4220 showing the stop codon (TAA, in bold) of the gene upstream of mdeA, the intergenic region, and the beginning of the mdeA structural gene. Indicated are base substitutions in resistant mutants isolated (see text), inferred −35 and −10 promoter elements, and the mapped transcription start site (+1 and right-angled arrow). (B) Primer extension reactions on RNA extracted from the wild-type and resistant mutant strains were performed with the same primer used for the DNA sequencing reaction on the mdeA gene. The DNA sequence shown on the right is the coding strand from +10 to −10 relative to the transcriptional start site (+1), marked by an asterisk.
To map the mutation position relative to the mdeA transcript, we determined the transcription start site of mdeA in strain RN4220 by primer extension. The results indicated that mdeA transcription starts around a G residue (Fig. 4B), 51 bp upstream of the mdeA translation start site. A GTGCTA-17 bp-TATAAT promoter motif homologous to the E. coli σ70 −35 and −10 promoter consensus sequences was positioned 4 bp upstream of the experimentally determined mdeA transcription start site and 54 bp upstream of the translation initiation codon (Fig. 4A). Thus, in the case of mutant 8, the mutation in the −35 sequence from GTGCTA to TTGCTA increased the match to the consensus −35 sequence TTGACA for E. coli. In mutant 3, the mutation was at −2 between the −10 sequence and the transcription start site. The rationale for affecting promoter activity is not readily apparent. However, mutations at similar positions (−1, −2, or −3) in the lmr promoter have been reported to increase lmr transcription and confer lincomycin resistance in S. subtilis (17). In summary, our data showed that the mutations are located upstream of the mdeA transcript start site. This is in contrast to mRNA stabilization mutations observed in norA (4).
Primer extension analysis was also performed on the two mutants with elevated resistance to MdeA substrates. The results indicated that mdeA gene transcription in these two mutants started from the same position as in the wild-type parent strain (Fig. 4B). However, the mdeA transcription levels in the mutants were drastically increased. Quantification of primer extension products showed a 10- and 15-fold increase in mdeA transcripts from mutants 3 and 8, respectively. Similar results were also obtained with TaqMan analysis of mdeA transcripts (six- to sevenfold increase [data not shown]). Thus, our results indicated that the spontaneous mutants with increased resistance to MdeA substrates had undergone mutations in the mdeA promoter region which resulted in increased transcription in mdeA.
Functional characterization of MdeA homologs in S. aureus
BLAST analysis indicated that the S. aureus genome potentially encodes 14 proteins which are homologous to MdeA (sequence identity range, 20 to 37%), including NorA. None of the corresponding clones had been detected when the ORF expression library was screened for drug resistance with a variety of compounds. Individual clones from the library expressing five of the MdeA homologs that are most related to MdeA (NP372690, NP372968, NP371960, NP372876, and NP372693) were tested for ethidium bromide efflux and were not significantly different from vector plasmid controls (data not shown). Determination of the MICs of eight antimicrobial compounds, including a range of quaternary ammonium compounds and hydrophobic moieties, also showed no significant increase compared to that for strain RN4220/pYH4 (data not shown). It was noted that none of these MdeA homologs had a charged amino acid in TMS1; the position of Glu24 in TMS1 of MdeA was occupied by a Gln residue. The acidic residue in TMS1 is essential for the function of many MDR proteins (reviewed by Zheleznova et al. [40]). Site-directed mutagenesis was used to change the Gln at position 29 in NP372968 and at position 26 in NP372690 to a Glu residue. This mutation, however, did not change the ethidium bromide uptake or efflux properties of the cells expressing the mutated proteins (data not shown).
Functional characterization of MdeA homologs from other bacteria
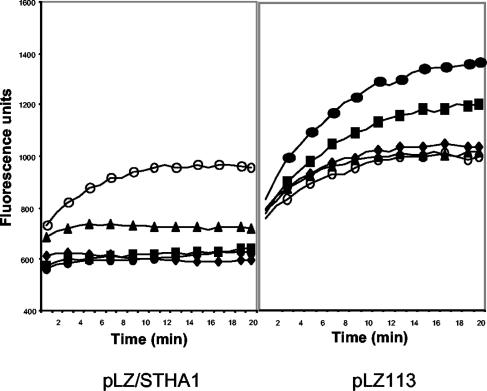
B. subtilis strains harboring cloned MdeA homologs were tested for ethidium bromide uptake relative to that of a pLZ113 vector control strain. None of the recombinants had reduced uptake (data not shown) except for the strain with the cloned MdeA homolog from S. haemolyticus. This strain showed an inducer concentration-dependent reduction in ethidium bromide uptake (Fig. 5), whereby maximal induction was achieved by treatment with 50 ng of ATc/ml for 1 h prior to the uptake assay. Higher concentrations of inducer did not decrease uptake. Complete absence of inducer resulted in uptake of ethidium bromide at the same rate as the vector control strain. Exposure of B. subtilis/pLZ113 to higher concentrations of ATc increased ethidium bromide uptake, suggesting that the inducer alone stresses cells or otherwise permeabilizes them. None of the other strains with cloned MdeA homologs displayed uptake rates significantly different from that of the vector control strain, over a range of inducer concentrations (data not shown).
FIG. 5.
The MdeA homolog of S. haemolyticus reduces ethidium bromide uptake. Uptake of ethidium bromide by B. subtilis cells harboring pLZ113 or pLZ113/SHA1 is indicated. The cultures were induced with ATc at the following concentrations: 0 (○), 0.01 (▴), 0.05 (⧫), 0.1 (▪), and 0.2 (•) μg/ml.
To investigate if the cloned MdeA homologs had efflux activity against a wider panel of compounds, MIC determinations were carried out for the five recombinant strains and vector control strain with nine compounds at two inducer concentrations. The presence of the cloned genes from E. cloacae and S. enterica serovar Typhimurium did not affect the MIC of any of the compounds tested (data not shown). In fact, these strains grew poorly overnight at higher inducer concentrations, in the absence of any antimicrobial agents, while the vector control strain was unaffected. This suggests that the cloned genes in these two recombinant plasmids were expressed.
Among the other three recombinant strains, only the S. haemolyticus protein conferred increased resistance to ethidium bromide, consistent with the uptake studies. The presence of the pLZ113/SHA1 plasmid increased the ethidium bromide MIC fourfold (Table 3), and the MIC was affected only when the xyl/tet promoter was induced. Again, this is consistent with the observed dependence for induction in the uptake assay (Fig. 5). The B. subtilis strain with the cloned S. haemolyticus MdeA homolog also had a fourfold increase in the MICs of virginiamycin, lincomycin, and erythromycin relative to those for the pLZ113 vector control strain. Similarly, the presence of the cloned MdeA homologs from B. cereus and B. subtilis gave a four- to eightfold increase in the MICs of lincomycin and virginiamycin but had a relatively minor effect on the other drugs (Table 3). In the presence of 10 and 50 ng of ATc inducer/ml, the growth of both clones was severely inhibited and, thus, the MIC could not be determined in the presence of inducer.
TABLE 3.
MIC values for B. subtilis cells expressing cloned MdeA homologs.
| Compound |
B. subtilis strain 168 isolate and ATc concn (ng/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| pLZ113
|
pLZ113/SHA1
|
PLZ113/RZC03923
|
PLZ113/LmrB
|
|||||
| 0 | 10 | 50 | 0 | 10 | 50 | 0 | 0 | |
| Ethidium bromide | 4 | 4 | 2 | 4 | 4 | 16 | 2 | 4 |
| Virginiamycin | 16 | 16 | 8 | 16 | 32 | 64 | 128 | 64 |
| Mupirocin | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.125 | 0.5 |
| Ciprofloxacin | 0.063 | 0.031 | 0.016 | 0.063 | 0.03 | 0.03 | 0.125 | 0.063 |
| Erythromycin | 0.03 | 0.03 | 0.03 | 0.03 | 0.063 | 0.125 | 0.125 | 0.063 |
| Lincomycin | 16 | 16 | 8 | 16 | 32 | 64 | >64 | 64 |
Values tabulated are MICs in micrograms per milliliter and are the averages of three determinations.
DISCUSSION
This study has demonstrated the presence of a novel MDR protein, MdeA, in the major human pathogen S. aureus. MdeA contains 479 amino acid residues with 14 predicted TMS. Sequence alignments show it belongs to the MFS and is most closely related, among known efflux proteins, to LmrB of S. subtilis and EmrB of S. coli. Analysis of ethidium bromide efflux and uptake provided direct evidence that MdeA is an efflux pump. The uncoupler CCCP completely abolished the uptake and efflux activity of the MdeA-expressing strain, suggesting that MdeA is a proton-driven efflux pump. Further experiments to test valinomycin or nigericin inhibition of MdeA in inverted vesicles will be required to definitively prove this.
However, MdeA overexpression as indicated by a >300-fold increase in mdeA transcription conferred only a modest MIC increase (2- to 16-fold) for a limited number of antimicrobial compounds, indicating that it is not a very strong conveyor of resistance to the antibiotics tested. In fact, phylogenetic analysis suggested that the MdeA protein is very ancient and widespread and could possess a different function other than drug efflux. Similarly, the contribution of the MdeA efflux pump in mupirocin resistance is likely to be insignificant, because the highest MIC conferred by the maximum expression of MdeA was 0.5 μg/ml, which is still considered as susceptible. High-level resistance to mupirocin in S. aureus is mainly due to the acquisition of a second copy of the isoleucyl-tRNA synthetase gene (5). Naturally occurring resistance due to MdeA has never been reported; therefore, it is also very unlikely that the use of mupirocin will select out MdeA to confer multiple drug resistance.
To examine relationships among the 48 MdeA homologs in detail, we constructed phylogenetic trees and included the E. coli known efflux protein EmrB as an outgroup. Immediately apparent were several gene duplication events in the evolutionary history of these bacteria. In addition, most of the duplication events appeared to have preceded speciation events and occurred in the common ancestor of these bacteria. For example, the three S. aureus proteins did not group together and, instead, there were three S. aureus clades each including a wide variety of gram-positive species. However, some species such as S. mutans only have one copy of the gene, suggesting that S. mutans lost two or more of its copies.
Since the MdeA protein in S. aureus was shown to have efflux activity, the question arose as to whether or not the other two S. aureus homologs display efflux activity. The corresponding ORF clones for these two MdeA homologs were individually tested in S. aureus RN4220 for drug resistance and efflux activities. We have been unable to detect any drug pump activity with these ORF-expressing strains. Since many of the homologs of these two proteins were annotated as possible sugar transporters, we suspect that these two S. aureus MdeA homologs could be sugar transporters instead of drug efflux pumps.
As a first attempt to functionally explore related genes from other bacteria, the closest relative of S. aureus MdeA from S. haemolyticus was shown to be an active efflux pump for ethidium bromide and to confer increased resistance on B. subtilis cells to ethidium bromide and selected MLS antibiotics (erythromycin, lincomycin, and virginiamycin). A homolog from B. cereus which is 96% identical to a protein in B. anthracis and from S. subtilis did not affect ethidium bromide efflux when expressed in B. subtilis but significantly increased the MICs of virginiamycin, erythromycin, and lincomycin. Interestingly, all these tested pump proteins contained a glutamic acid (E) in the relative position 24 and grouped with S. aureus MdeA in the phylogenetic analyses. Thus, these results seem to suggest that proteins from the clade with an E at relative position 24 are active drug pumps, whereas proteins from sequences with a Q in relative position 24 are not active drug pumps. This leads to speculation that species such as S. mutans have retained only the copy of the MdeA subfamily gene that has efflux activity. Furthermore, with the exception of B. anthracis and C. glutamicum (where relatively recent gene duplication events have occurred), there is only one copy of the protein with the glutamic acid at relative position 24 in any given organism. It also appears that the sequences with a Q at relative position 24 are more basal (older) than the lineages with an E, suggesting that the drug pumps in the MdeA subfamily evolved from proteins with functions other than drug efflux. Therefore, further functional analysis of mdeA is required to define its exact physiological functions.
Acknowledgments
This work was partly supported by a research grant from the Defense Advanced Research Projects Agency (N65236-97-1-5810).
The content of this publication does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
The availability of bacterial genome sequences from The Institute for Genomic Research, Baylor University, and Integrated Genomics Inc. is gratefully acknowledged.
REFERENCES
- 1.Borges-Walmsley, M. I., and A. R. Walmsley. 2001. The structure and function of drug pumps. Trends Microbiol. 9:71-79. [DOI] [PubMed] [Google Scholar]
- 2.Bron, S. 1990. Plasmids, p. 139-174. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, United Kingdom.
- 3.Chuang, S. E., D. L. Daniels, and F. R. Blattner. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fournier, B., Q. C. Truong-Bolduc, X. Zhang, and D. C. Hooper. 2001. A mutation in the 5′ untranslated region increases stability of norA mRNA, encoding a multidrug resistance transporter of Staphylococcus aureus. J. Bacteriol. 183:2367-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbart, J., C. R. Perry, and B. Slocombe. 1993. High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob. Agents Chemother. 37:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guilfoile, P. G., and C. R. Hutchinson. 1992. The Streptomyces glaucescens TcmR protein represses transcription of the divergently oriented tcmR and tcmA genes by binding to an intergenic operator region. J. Bacteriol. 174:3659-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh, P. C., S. A. Siegel, B. Rogers, D. Davis, and K. Lewis. 1998. Bacteria lacking a multidrug pump: a sensitive tool for drug discovery. Proc. Natl. Acad. Sci. USA 95:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, J., and M. A. Schell. 1995. Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol. Microbiol. 16:977-989. [DOI] [PubMed] [Google Scholar]
- 9.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 10.Kaatz, G. W., S. M. Seo, and C. A. Ruble. 1993. Mechanisms of fluoroquinolone resistance in Staphylococcus aureus. J. Infect. Dis. 163:1080-1086. [DOI] [PubMed] [Google Scholar]
- 11.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaatz, G. W., S. M. Seo, L. O'Brien, M. Wahiduzzaman, and T. J. Foster. 2000. Evidence for the existence of a multidrug efflux transporter distinct from NorA in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1404-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kluytmans, J., A. van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature (London) 305:709-712. [DOI] [PubMed] [Google Scholar]
- 15.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 16.Kumano, M., A. Tamakoshi, and K. Yamane. 1997. A 32 kb nucleotide sequence from the region of the lincomycin-resistance gene (22 degrees-25 degrees) of the Bacillus subtilis chromosome and identification of the site of the lin-. mutation. Microbiology 143:2775-2782. [DOI] [PubMed] [Google Scholar]
- 17.Kumano, M., M. Fujita, K. Nakamura, M. Murata, R. Ohki, and K. Yamane. 2003. Lincomycin resistance mutations in two regions immediately downstream of the −10 region of lmr promoter cause overexpression of a putative multidrug efflux pump in Bacillus subtilis mutants. Antimicrob. Agents Chemother. 47:432-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, et al. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 19.Lee, E. H., and W. M. Shafer. 1999. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33:839-845. [DOI] [PubMed] [Google Scholar]
- 20.Lomovskaya, O., and K. Lewis. 1992. Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89:8938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 22.Markham, P. N., and A. A. Neyfakh. 2001. Efflux-mediated drug resistance in gram-positive bacteria. Curr. Opin. Microbiol. 4:509-514. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell, B. A., M. H. Brown, and R. A. Skurray. 1998. QacA multidrug efflux pump from Staphylococcus aureus: comparative analysis of resistance to diamidines, biguanidines, and guanylhydrazones. Antimicrob. Agents Chemother. 42:475-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moellering, R. C., Jr. 1998. Antibiotic resistance: lessons for the future. Clin. Infect. Dis. 27(Suppl. 1):S135-S140. [DOI] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 1997. Approved standard M7-A4. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Neyfakh, A. A., C. M. Borsch, and G. W. Kaatz. 1993. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob. Agents Chemother. 37:128-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Schuler, G. D., S. F. Altschul, and D. J. Lipman. 1991. A workbench for multiple alignment construction and analysis. Proteins 9:180-190. [DOI] [PubMed] [Google Scholar]
- 30.Shivakumar, A. G., T. J. Gryczan, Y. I. Kozlov, and D. Dubnau. 1980. Organization of the pE194 genome. Mol. Gen. Genet. 179:241-252. [DOI] [PubMed] [Google Scholar]
- 31.Smith, T. L., M. L. Pearson, K. R Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, W. R. Jarvis, et al. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 32.Tercero, J. A., R. A. Lacalle, and A. Jimenez. 1993. The pur8 gene from the pur cluster of Streptomyces alboniger encodes a highly hydrophobic polypeptide which confers resistance to puromycin. Eur. J. Biochem. 218:963-971. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trucksis, M., J. S. Wolfson, and D. C. Hooper. 1991. A novel locus conferring fluoroquinolone resistance in Staphylococcus aureus. J. Bacteriol. 173:5854-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Veen, H. W., and W. N. Konings. 1998. The ABC family of multidrug transporters in microorganisms. Biochim. Biophys. Acta 1365:31-36. [DOI] [PubMed] [Google Scholar]
- 36.Yamada, H., S. Kurose-Hamada, Y. Fukuda, J. Mitsuyama, M. Takahata, S. Minami, Y. Watanabe, and H. Narita. 1997. Quinolone susceptibility of norA-disrupted Staphylococcus aureus. Antimicrob. Agents Chemother. 41:2308-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida, H., M. Bogaki, S. Nakamura, K. Ubukata, and M. Konno. 1990. Nucleotide sequence and characterization of the Staphylococcus aureus norA gene, which confers resistance to quinolones. J. Bacteriol. 172:6942-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, J. L., L. Grinius, and D. C. Hooper. 2002. NorA functions as a multidrug efflux protein in both cytoplasmic membrane vesicles and reconstituted proteoliposomes. J. Bacteriol. 184:1370-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, L., F. Fan, L. M. Palmer, M. A. Lonetto, C. Petit, L. L. Voelker, A. St. John, B. Bankosky, M. Rosenberg, and D. McDevitt. 2000. Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255:297-305. [DOI] [PubMed] [Google Scholar]
- 40.Zheleznova, E. E., P. Markham, R. Edgar, E. Bibi, A. A. Neyfakh, and G. Brennan. 2000. A structure-based mechanism for drug binding by multidrug transporters. Trends Biochem. Sci. 25:39-43. [DOI] [PubMed] [Google Scholar]