Abstract
Diabetes is a metabolic disorder associated with either improper functioning of the beta-cells or wherein cells fail to use insulin properly. Insulin, the principal hormone regulates uptake of glucose from the blood into most of the cells except central nervous system. Therefore, deficiency of insulin or the insensitivity of its receptors plays a key role in all forms of diabetes. In the present work, attempt has been made to find out plant sources which show anti hyperglycaemic activity (AhG) (i.e. compounds that bring down the blood glucose level in the body). Ayurvedic plants showing AhG activity formed the basis of our study by using the platform of Computer Aided Drug Designing (CADD). Among 600 plants showing AhG activity, 500 compounds were selected and screened, out of which 243 compounds showed drug likeness property that can be used as therapeutic ligand/drug. Initial screening of such compounds was done based on their drug likeness or biochemical properties. Dynamic interaction of these molecules was captured through Protein-Ligand study. It also gave an insight of the binding pockets involved. Bench marking of all the parameters were done using the diabetic inhibitor drug, Glipizide. Pharmacokinetic studies of the compounds such as Aloins, Capparisine, Funiculosin and Rhein exhibited less toxicity on various levels of the body. As a conclusion these ligands can lay a foundation for a better anti-diabetic therapy.
Abbreviations
AhG - Anti hyperglycaemic, CADD - Computer Aided Drug Designing.
Keywords: Anti hyperglycaemic property, Protein-ligand interactions, Computer aided drug designing, Diabetic inhibitor, Pharmacokinetic parameters
Background
Ayurveda- “The Science of Life” goes way back to ancient days which aimed to integrate herbs and other plants - including oils and common spices for curing diseases. As a medical system, it includes herbal medicines, dietary therapies, physical therapies as well as psychological and spiritual healing [1]. Concept of Ayurvedic medicine is followed as it relates to the body's constitution (prakriti) and life forces (doshas) i.e. its Universal interconnectedness [2]. Ayurvedic medicine being a holistic approach to health and life makes a comparative study with the commercial drug and has added conventional therapies to their practices. Availability of various new drugs in the market showing side effects to our body emphasizes the need to explore natural plant sources that would play a remarkable and substitute role to address the problem of less toxic and costaccounting drug. Currently, more than 600 herbal formulae and 250 single plant drugs have been included in the pharmacy of Ayurvedic treatments [2]. Ethno-botanical studies of traditional herbal remedies used for diabetes around the world have identified more than 1,200 species of plants with antihyperglycaemic activity [1]. The pharmacopoeia of India forms the base for such kind of structure based study since it is rich in herbal treatments for diabetes [3]. Diabetes, an issue of international concern can lead to fatigue, disorders of organ systems / organs. It is generally accompanied with high level of blood sugar leading to polyuria, polydipsia and polyphagia and is classified into 3 types – Type 1, Type 2 and Gestational diabetes. Aiming to approach for a novel ayurvedic compound, we used target based drug discovery which follows on rationally identifying chemical compounds that should bind to a therapeutic target molecule – protein [4].
Concept of Computer Aided Drug Designing (CADD) was applied to understand atomic details of drug binding strength and specificity in order to identify or create a novel molecule that binds to a selected target and also optimizes the therapeutic index of an already available drug or lead compound [5]. The dynamic of these binding interactions was achieved through protein-ligand study. In these kinds of interactions, an atom/ion binds to the specific binding site of the protein. An important condition for binding is its active site, exhibiting the correct size and shape in order to optimally fit into a cavity exposed to the surface of the protein, i.e. the “binding pocket”. In our present bench marking protein-ligand interaction study, the protein Insulin is a transmembrane receptor which belongs to the large class of tyrosine kinase (Figure 1 a). It is made up of two alpha and beta subunits each. The beta subunits pass through the cellular membrane and are linked through disulfide bonds. The alpha and beta subunits are encoded by a single gene (INSR). The receptor initiates insulin action through the binding and activation of its cell-surface receptor [6].
Figure 1.
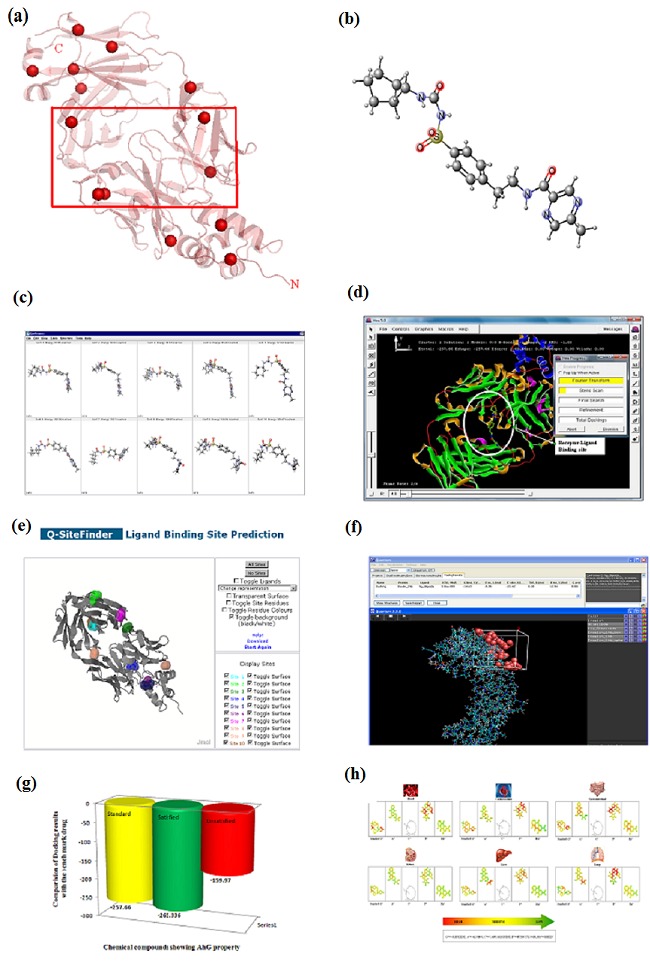
(a) Molecular secondary structure representation of Insulin receptor with various active sites highlighted with red spheres and important region of the protein captured with the red rectangular box; (b) three dimensional structural representation of bench mark drug – Glipizide; (c) Snap Shot of 10 energy minimized conformational structure for the bench mark drug – Glipizide, the first lowest value is of our interest; (d) schematic representation of Protein-Ligand Interaction (Phase I) study for Insulin receptor (protein) versus bench mark drug – Glipizide (ligand) using Hex docking software (version 5.1), (e) ligand binding pocket / site analysis for the Insulin receptor using Q-site server prediction; (f) schematic representation of Protein-Ligand Interaction (Phase II) study for Insulin receptor (protein) versus bench mark drug – Glipizide (ligand) using Quantum docking software. Here a grid (represented in white) has been constructed around the active binding site where the molecular dynamic interaction takes place; (g) comparative analysis of the binding score for the AhG compounds with Glipizide (represented in yellow bar). The red color bar represents the average Protein-Ligand Interaction (Phase I) score of the compounds eliminated. The green color bar represents the Protein-Ligand Interaction (Phase I) of the compounds accepted; (h) propotional toxicity analysis of screened AhG natural compounds on various body organs.Here green color signifies less, yellow medium and red more toxicity effect.
The ligand, Glipizide is a second generation oral blood-glucoselowering drug of the sulfonylurea class available in a Gastrointestinal Therapeutic System (GITS) (Figure 1 b). The mechanism of action is produced by blocking potassium channels in the beta cells of the Islets of Langerhans which appears to lower blood glucose acutely by stimulating the release of insulin from the pancreas. In this study, an attempt has been made to explore molecules of Ayurvedic plant origin that would play a role in anti-hyperglycaemic activity addressing the problem of Diabetes and act well than the existing drugs. For identifying the chemical structures having good inhibiting effects on specific targets with low toxicity and also for evaluating the drug likeness property of these AhG compounds, we used quantitative structure-activity relationships (i.e. partition coefficient log p value) which are detailed in subsequent sections.
Methodology
Screening of AhG compounds (Based on drug likeness / biochemical property):
Compounds possessing AhG activity were identified from their plant sources and their drug likeness and biochemical properties were checked using Lipinski's Rule of Five and Molinspiration respectively. Glipizide was choosen to be the most suitable benchmark/reference drug for this study [7].
a) Drug-likeness property (Lipinski's Rule of Five):
Christopher Lipinski's Rule of Five analysis helped to raise awareness about properties and structural features that makes molecules more or less drug-like. The guidelines predicted that poor absorption or permeation of an orally administered compound will meet the following criteria such as molecular mass > 500 Da, high lipophilicity (expressed as cLogP > 5), > 5 hydrogen bond donors and > 10 hydrogen bond acceptors [8]. The compounds not meeting the above mentioned criteria were considered for docking studies as shown in Table 1 (see supplementary material).
b) Biochemical property (Molinspiration):
Molinspiration – a cheminformatics tool for calculation of important molecular properties (for example logP, polar surface area, number of hydrogen bond donors and acceptors), prediction of bioactivity score for important drug targets i.e. GPCR ligand-based signaling cascade was used for the development of a new functional drug with increased binding selectivity profile and less undesirable effects [9], kinase inhibitors for development of selective inhibitors that can block or modulate diseased signaling pathways were considered a promising approach for drug development [10], ion channel modulators allowed the movement of charged particles across cell membranes and are important therapeutic targets which are modulated by a range of theraupetic drugs [11] and nuclear receptors for ligand activation of target genes for therapeutic drug development [12] along with possible molecular toxicity [13]. Analysis of Chemical and Bioactivity performed through cheminformatics tool is represented in (Table 1).
Energy Minimization of AhG compounds:
Marvin Sketch – an editor for drawing chemical structures, queries and reactions was used for energy minimization, which is important in molecular conformations/ligand (AhG compounds) in order to release any constraints such as torsional or steric, for proper binding to the receptor. Energy minimization generated 10 conformers, of which the one with least energy was considered for our study. As represented graphically in (Figure 1c) for the compound Glipizide, the conformer with least energy 110.8 kcal/mol was considered out of other 9 conformers.
Selection of Receptor:
In order to identify a molecular activator with potent antidiabetic effect, Insulin receptor (PDB ID: 3KR3) was selected based on factors like resolution > 1 and ≤ 3Å to avoid structure with basic contours of the protein chain, r-factor < 0.25 to provide a nearly perfect fit atomic model, length ≥ 100 to avoid peptides and X-ray crystallized structure to check the reliability of the protein receptor.
Protein-Ligand Interaction study (Phase I):
Protein-Ligand Interaction Phase I study involves the dynamic of ligand binded induced conformational change to the Insulin receptor by compairing the docking score of the benchmarked drug Glipizide (Figure 1d) with other AhG compounds (i.e. filteration based on better interaction score for further cheminformatics study (Table 1) using Hex software (Version 5.0) [14].
Binding site analysis:
a) Swiss PDB Viewer:
The receptor-ligand complex obtained through Phase I of Protein-Ligand Interaction study provided an insight of their structural alignments and a comparision of their active sites [15] through statistically significant amino acids contributing for the binding pocket (Table 1).
b) Q-Site Finder:
An energy based matrix captured the location of the ligandbinding sites on protein - insulin giving a view of its structural identification and comparision of its functional sites (Figure 1e) and thus a combined statistical analysis of the binding pockets for macro vs. micro molecules brought out the significant active site residue [16].
Protein-Ligand interaction study (Phase II):
The statistically potent amino-acid (combination of binding site analysis vs. H-bond distance i.e. Leucine – 32) served as an active site for the grid formation i.e. environment for the study of Protein-Ligand Interaction (Phase II) (Figure 1f), which enhances further stages of drug discovery, such as target identification, drug / lead hit identification and lead optimization using Quantum 3.0 [17]. A breakthrough into thermodynamic analysis of the interaction study between macro vs. micro molecules was achieved through the Gibbs free energy values whereas best structural fit and their chemical activity were studied through Root Mean Square Deviation [18]. Best fitted structure (lower Gibbs score /Root Mean Square Deviation) paved the platform of selection of the AhG compounds for pharmacokinetic study with reference to our benchmark drug – Glipizide (Table 1).
Pharmacokinetic study:
a) ADME Analysis:
The AhG molecules screened from Protein – Ligand Interaction study (Phase II) were subjected to pharmokinectic and pharmodynamic studies (Table 2) to capture the dosage level as well as the process of active drug components governing our body thus giving an insight into target site concentration and their theraupetic responses [19].
b) Toxicity analysis:
Mutagenesis plays an important role to decide the potential of the compounds for drug designing. AMES test (biological assay) aims to assay this potential only, thus briefing us whether these AhG compounds can be possible carcinogens or not [20]. Virtual lab provided us with the AMES value as predicted on the rat liver Table 2 (see supplementary material). Further, it also comprehended us with their bio-chemical effect on various body organells (Figure 1g).
Discussion
Prevention of diabetes is quite a difficult task considering our present way of living. To treat the imbalance of glucose level in the body one has to take complex decisions about treatment. Hence, considering prevention of diabetes is better than controlling it. The best known way to control diabetes is by medication through commercially available drug such as, Glipizide. In this study, we found four AhG compounds namely Aloins, Capparisine, Funiculosin and Rhein, which can be extracted from naturally available plant sources and can control diabetes in a manner similar to Glipizide or better than it. Also, these compounds found by us can serve as therapeutic drugs as well as dietary supplements.
AhG compounds targeting diabetes were taken for our study. Initial screening of these compounds was done based on Lipinski's rule of five, to check the drug likeness property. 500 compounds were selected, out of which 243 compounds showed drug likeness property that can be used as therapeutic ligand/drug. These molecules were subjected to Phase I Protein-Ligand interaction study followed by Chemical/Bioactivity analysis which suggested that out of 243 natural compounds, 6 compounds i.e. Aloins, Capparisinine, Daturilinol, Funiculosin, Rhein, and Tamarixetin possessed lower docking score than the reference drug (Glipizide) (Table 1). Lower the docking score of a compound; the more effective it is as an inhibitor (Figure 1h). For the grid construction in the Phase II study of Protein-Ligand Interaction, contribution of each amino acid was investigated (Table 1). Comparative analysis of ligand vs. receptor revealed Leucine (LEU) to be the most active contributor for the ligand binding site. Phase II of Protein-Ligand interaction study filtered out Daturilinol and Tamarixetin (Table 1) since they had high docking score compared to the bench mark drug. Further, LD50 values were also predicted for selecting reliable AhG molecule for ADME analysis. The reference investigational drug Glipizide is good in solubility, stability and absorption but showed no significant first pass metabolism in liver and intestine and no active transport. Plasma binding protein and volume of distribution prediction of Glipizide was unreliable in distribution analysis. The natural molecules Aloins, Capparisine, Funiculosin and Rhein were considered as final reliable molecules based on their ADME and Toxicity features (Table 2).
These molecules were considered as better ligands for insulin receptor based on their interaction, pharmacokinetics and pharmacodynamics features. Aloins are naturally occurring compound in Aloe barbadensis (Aloe vera). Aloe vera tends to show very positive results for skin treatments. Though it has anti-diabetic properties, but consuming aloe vera orally or taken internally causes active secretion of fluids and electrolytes, delays wound healing and is also responsible for spontaneous abortion in pregnant women if high dosages are taken frequently or could cause premature birth. It is recommended to take them in a minimal quantity and consuming a good amount of water will help in the metabolic activities to function smoothly. Capparisinine is a naturally occurring compound in Capparis decidua (Kerda) that has an efficient anti-diabetic property, consumption of which causes hypercholesterolemia, a low fat diet and poor bile flow, which can keep a check on the cholesterol levels as bile acts as an emulsifier in the presence of fats. Also, thinning the bile can help in raising the cholesterol levels to normal. It can be taken in small quantities as a form of spice in our daily diet. Funiculosin and Rhein are naturally occurring compounds in Senna occidentalis (Coffee senna/ Mogdad coffee). Funiculosin is the new antibiotic in market. Senna occidentalis can be used as a substitute for coffee as it doesn't contain caffeine. It is suggested not to take this during pregnancy as Funiculosin and Rhein are anthraquinones which have the ability to cause electrolyte abnormalities and uterine contractions during pregnancy and anthraquinones also have the ability to cross into breast milk during lactation. It also acts as an efficient antiviral, antitumor and antifungal in nature.
Hence, in the present study we can conclude saying that Funiculosin and Rhein can be considered as a better antidiabetic compound for their better results and lesser toxicity levels, followed by Capparsinine and Aloins. It is also essential to maintain a well balanced diet along with consumption of a good quantity of water and regular exercise is recommended (Table 1).
Conclusion
Through CADD, the AhG compounds were made to undergo screening process to find out efficient anti-diabetic drugs which were extracted from natural plant sources. The compounds which showed better drug like properties against diabetes when compared to our bench mark drug (Glipizide) were Aloins, Capparisine, Funiculosin and Rhein naturally found in Aloe barbadensis (Aloe vera), Capparis deciduas (Kerda) and Senna occidentalis (Coffee senna) respectively and can be consumed orally in limited quantities but are advised not to be consumed during pregnancy, heart problems and/on steroids or while undergoing diuretics treatment. These compounds can be taken as a daily diet prior to the onset of diabetes in a limited quantity.
Future prospective:
As the work is purely on computational analysis, we wish to connect a bridge between dry lab to wet lab in our further work by implementing our present analysis on model organisms like mice and monitor them to undergo a balanced diet along with the plant extract which is rich in the above mentioned compounds and test them for the blood glucose level in their body before and after physical activity like swimming. Further, we would like to concentrate on combining the compounds to understand the physical dosage of each on human beings to overcome diabetes using Support Vector Machine. Further Molecular Dynamic (MD) studies on these natural compounds will help us to analyse how well these ligand/micromolecules are complexed with the protein insulin.
Supplementary material
Acknowledgments
This research has been carried out in Maharani lakshmi Ammanni College For Women, DBT- BIF facility under BTIS (Biotechnology information system), DBT (Department of biotechnology), Ministry of Science and Technology, Government of India, India. We would also like to thank Dr. M B Nagaveni, dean life science and Mr. Sushil Kumar Middha, head bioinformatics for their moral support all through the work and allowing us to work with the commercial software Quantum (version 3.0).
Footnotes
Citation:Chatterjee et al, Bioinformation 8(24): 1195-1201 (2012)
References
- 1.A Chopra, et al. Medical Clin of North Am. 2002;86:88. [Google Scholar]
- 2.PM Barnes, et al. Natl Health Stat Report. 2008;12:1. [Google Scholar]
- 3. http://www.ayurveda.hu/api.html.
- 4.F Sams-Dodd, et al. Drug Discov Today. 2005;10:139. doi: 10.1016/S1359-6446(04)03316-1. [DOI] [PubMed] [Google Scholar]
- 5.AV Veselovsky, AS Ivanov. Curr drug targets Infect Disord. 2003;3:33. doi: 10.2174/1568005033342145. [DOI] [PubMed] [Google Scholar]
- 6.M Jin, et al. Future Med Chem. 2012;4:315. doi: 10.4155/fmc.11.180. [DOI] [PubMed] [Google Scholar]
- 7.HM Berman, et al. Nucleic acids res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CM Tice. Pest Manag Sci. 2001;57:3. doi: 10.1002/ps.263. [DOI] [PubMed] [Google Scholar]
- 9.E Stefan, et al. Nature Commun. 2011;2:598. [Google Scholar]
- 10.JJ Crawford, et al. Expert Opin Ther Pat. 2012;22:293. doi: 10.1517/13543776.2012.668758. [DOI] [PubMed] [Google Scholar]
- 11.JJ Clare, et al. Discov Med. 2010;9:253. [PubMed] [Google Scholar]
- 12.T Chen, et al. Curr opin chem boil. 2008;12:418. [Google Scholar]
- 13.MA Bakht, et al. Eur J Med Chem. 2010;45:5862. doi: 10.1016/j.ejmech.2010.07.069. [DOI] [PubMed] [Google Scholar]
- 14.G Macindoe, et al. Nucleic Acids Res. 2010;38:445. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.N Guex, MC Peitsch. Electrophoresis. 1997;18:2714. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 16.AT Laurie, RM Jackson. Bioinformatics. 2005;21:1908. [Google Scholar]
- 17.PV Bharatam, et al. Curr Pharm Des. 2007;13:3518. doi: 10.2174/138161207782794239. [DOI] [PubMed] [Google Scholar]
- 18.AM Rossi, CW Taylor. Nat Protoc. 2011;6:365. doi: 10.1038/nprot.2011.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SM He, et al. Current pharm des. 2011;17:357. doi: 10.2174/138161211795164194. [DOI] [PubMed] [Google Scholar]
- 20.GW Peng, WL Chiou. J Chromatography. 1990;531:3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


