Abstract
Codon usage bias (CUB) is an omnipresent phenomenon, which occurs in nearly all organisms. Previous studies of codon bias in Plasmodium species were based on a limited dataset. This study uses whole genome datasets for comparative genome analysis of six Plasmodium species using CUB and other related methods for the first time. Codon usage bias, compositional variation in translated amino acid frequency, effective number of codons and optimal codons are analyzed for P.falciparum, P.vivax, P.knowlesi, P.berghei, P.chabaudii and P.yoelli. A plot of effective number of codons versus GC3 shows their differential codon usage pattern arises due to a combination of mutational and translational selection pressure. The increased relative usage of adenine and thymine ending optimal codons in highly expressed genes of P.falciparum is the result of higher composition biased pressure, and usage of guanine and cytosine bases at third codon position can be explained by translational selection pressure acting on them. While higher usage of adenine and thymine bases at third codon position in optimal codons of P.vivax highlights the role of translational selection pressure apart from composition biased mutation pressure in shaping their codon usage pattern. The frequency of those amino acids that are encoded by AT ending codons are significantly high in P.falciparum due to action of high composition biased mutational pressure compared with other Plasmodium species. The CUB variation in the three rodent parasites, P.berghei, P.chabaudii and P.yoelli is strikingly similar to that of P.falciparum. The simian and human malarial parasite, P.knowlesi shows a variation in codon usage bias similar to P.vivax but on closer study there are differences confirmed by the method of Principal Component Analysis (PCA).
Abbreviations
CDS - Coding sequences, GC1 - GC composition at first site of codon, GC2 - GC composition at second site of codon, GC3 - GC composition at third site of codon, Ala - Alanine, Arg - Arginine, Asn - Asparagine, Asp - Aspartic acid, Cys - Cysteine, Gln - Glutamine Glu - Glutamic acid Gly - Glycine His - Histidine Ile - Isoleucine Leu - Leucine Lys - Lysine Met - Methionine Phe - Phenylalanine Pro - Proline Ser - Serine Thr - Threonine Trp - Tryptophan Tyr - Tyrosine Val - Valine.
Keywords: Codon usage bias (CUB), RSCU, tRNA, Optimal codon, Principal Component Analysis (PCA), Effective number of codon (ENC)
Background
The advent of next generation sequencing techniques has made a large amount of nucleotide sequence data available for analysis. Whole genome sequences of six malarial parasites: Plasmodium falciparum, Plasmodium chabaudii, Plasmodium yoelii, Plasmodium berghei, Plasmodium knowlesi and Plasmodium vivax are now available [1–5], and are extensively used in studying application of codon usage bias phenomenon for comparative genome analysis.
Apart from human host, Plasmodium species infect a wide range of vertebrate hosts including birds, rodents and primates. According to specific ecological and physiological requirements, Plasmodium species are non-randomly distributed across different ecological habitats. Codon usage analysis provides insight into the environmental adaptation and evolution of organisms [6]. It is well-known that sixty four codons coding for twenty-odd amino acids results in synonymous codons (different codons coding for the same amino acid). Eighteen out of twenty amino acids are encoded by multiple synonymous codons (exceptions being methionine and tryptophan) and their probability of occurrence is not equally distributed. This phenomenon was first explained by the “genome hypothesis” which suggested that this observed bias is species specific [7]. In most genomes, it is observed that synonymous codons are not used with equal frequency. Various genes show differential patterns of codon usage and these results in enhancing their efficiency and accuracy of translation into proteins [8–10]. The gene length, compositional constraints, expression level and RNA stability are the main factors responsible for influencing codon usage. In general, pattern of codon usage is determined by mutational pressure acting specifically over entire genome and selective forces acting on coding regions [11]. In some prokaryotes like E. coli and single cell eukaryotes like C.elegans, codon usage is determined by both mutational and translational selection pressure [12–14]. The intra-genomic and inter-genomic codon usage variability is mainly governed by various biological factors simultaneously but directional mutational pressure on DNA sequences and translational selection forces are the key role players [15].
Previous investigations on these parasites and their vectors [16] showed differences in their genomic architecture as well as ecological habitat. Codon distribution and factors shaping codon usage bias can be explained by understanding their usage patterns in Plasmodium species. The aim of this study is to understand a few questions. First - whether the codon usage pattern in Plasmodium genus is conserved or not; second - is the comparative analysis of six different Plasmodium species to understand their phylogenetic relationships possible on the basis of codon usage bias and third-to identify the role of mutational and translational selection forces using their genome sequence data for shaping codon usage pattern.
Codon usage pattern of Plasmodium species appears to be conserved at the genus level but on further analysis, this conservation level is less and changes gradually from species to species. The atypical A+T richness of the genome in P.falciparum leads to codon bias of these bases at the wobble position in all the coding sequences [17]. The genome of P.vivax has GC content close to fifty percent, hence the variation in codon usage bias is expected to be different and it is found to be so. The other four malarial parasite genomes were analyzed for the first time, and the codon usage bias of these rodent parasites, P.berghei, P.chabaudi and P.yoelii are very similar to that of P.falciparum. The simian and human parasite, P.knowlesi, however, shows behavior similar to P.vivax. This grouping of Plasmodium species were also confirmed by principal component analysis (PCA) using sequence data. PCA categorizes the studied species into different clusters, which are similar to their phylogenetic status [18]. Equilibrium among various forces like mutation pressure, translational selection and genetic drift are some of the factors responsible for explaining complex codon usage patterns in any species [19]. The highly expressed and less expressed genes of studied malarial species show significant differences in their codon usage bias. Our analysis based on CDS (complete coding sequences) and highly expressed genes of Plasmodium species find a more pronounced role of compositional mutational pressure instead of selection at translational level for shaping different codon usage pattern.
Methodology
Plasmodium Genome Sequences:
Complete coding sequences (CDSs) of six Plasmodium species (P.falciparum, P.vivax, P.knowlesi, P.yoelii, P.chabaudi and P.berghei) were taken from PlasmoDB database [20] and their redundancies were removed. Total number of CDSs analyzed: 5523 (P.falciparum), 5435 (P.vivax), 5197 (P.knowelsi), 7724 (P.yoelii), 251 (P.chabaudi) and 4904 CDSs (P.berghei). The whole genome sequence of P.falciparum and P.vivax were obtained from NCBI site [www.ncbi.nlm.nih.gov].
Codon usage analysis:
The frequency of codons (excluding stop codons) corresponding to each amino acid in the CDSs is used for codon usage analysis. Codon frequency, codon bias, amino acid frequency per thousand, GC, GC1, GC2 and GC3 percentage were calculated by using in-house code using Matlab 7.12.0 [www.mathworks.com].
Relative synonymous codon usage (RSCU):
RSCU value of each codon is the observed frequency of that codon divided by the expected frequency for synonymous codons of an amino acid using equal usage as a conjecture (see supplementary material).
Effective Number of Codons (ENC):
ENC is the most widely used estimator of codon usage bias [21] and provides the range of codon preferences in a gene. Its value lies among 20 to 61. ENC value of 61indicates equal codon usage for coding an amino acid and, its value is 20 when only single codon codes it (see supplementary material).
Detection of tRNA:
The tRNA genes of all Plasmodium species except P.vivax were taken from GeneDB database [http://www.genedb.org]. The tRNA genes of P.vivax were searched using tRNAscanSE [22] using default parameters. Their respective anti-codons were predicted using WebMGA server [http://www.weizhonglab. ucsd.edu]. The correlation between amino acid frequency of highly expressed genes and their respective tRNA copy number in Plasmodium species were analyzed using linear regression analysis model with significance level of P<0.05.
Highly and lowly expressed genes, and frequency of optimal codons were identified in six Plasmodium species using CodonW 1.3 (written by John Peden and taken from fttp://molbiol.ox.ac.uk/cu/codonW. tar.Z/).
Principal Component Analysis (PCA):
PCA is one of the most commonly used multivariate statistical techniques. Here PCA is used to analyze major trends in codon usage pattern of different Plasmodium species. It involves a mathematical procedure that reduces the original variables to a lower number of orthogonal transformed variables (principal components), without losing much of its information. Each species was represented as a 59 dimensional vector (the number of possible codons minus the two unique ones for methionine and tryptophan and the three stop codons) where each vector contains RSCU value of codons. Finally top 2 uncorrelated PCs having greatest variance, was taken in to consideration for analyzing the variation of codon usage bias within each species. Here statistical analysis was carried out using Microsoft® Excel 2007/ XLSTAT©-Pro (Version 2012, Addinsoft, Inc., Brooklyn, NY, USA).
Result and Discussion
Codon usage blueprint in Plasmodium species:
Codon usage summary of six Plasmodium species is given in Table 1 (see supplementary material). Though all the species have the same number of chromosomes, they differ in their genome size. GC percent of coding sequences (CDSs) is greater than their genomic GC percent in all six species, which is a usual trend. GC percent of coding sequences is one of the important factor affecting the codon usage in Plasmodium species and it varies from 23.73 to 46.21 percent. The GC1 and GC2 content of Plasmodium species coding sequences are generally higher than their GC3 content except for P.vivax and P. knowlesi. Genomic GC and GC at third synonymous position (GC3) are closely associated. Usage of G/C ending codons will increase the overall GC bias and decreases with increase in usage of A/T ending codons. This variation is observed because GC content is an important factor explaining the variation of genome wide pattern of codon usage in different species.
Among six Plasmodium species analyzed, P.falciparum and P.vivax are the most studied malaria causing species as they are the main pathogens affecting humans. A separate study is done here in context of GC3. GC3 content in P.falciprum is lowest and it is highest in P.vivax CDSs. Nonetheless apparently there is some heterogeneity in the dataset, given that the GC3 value ranges from 10 % to 32.5 % (Figure 1A) in case of P.falciparum and 10 % to 82.5 % in P.vivax (Figure 1B). This indicates that high variation in codon usage occurs between genes present in both species. This large range of variation in codon usage is probably due to differential mutational pressure acting on different coding regions of a genome during the course of evolution.
Figure 1.
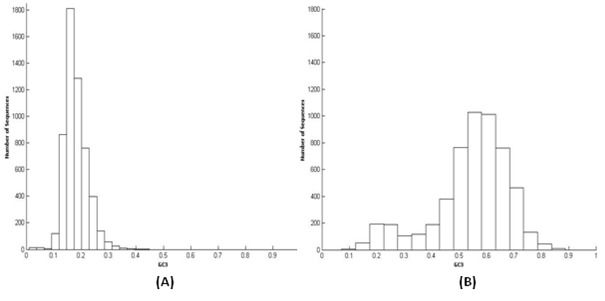
GC3 distribution in P.falciparum (A) and P.vivax (B), coding sequences
Codon usage data shown in Table 2 (see supplementary material) provides ample evidence of codon usage bias in six Plasmodium species. Here the codon count, RSCU values and tRNA copy numbers are calculated. It is expected that G/C ending codon usage should increase with increasing genomic GC bias. This trend is followed in CDSs of P.vivax, where most of the codons are G/C ending and other five Plasmodium species show preference towards A/T ending codons at the wobble position.
Transfer RNA is an adaptor molecule that decodes protein information residing in mRNA codons. Anti-codon of tRNA is responsible for encoding multiple codons differing only at wobble position. The total copy number of tRNA in Plasmodium species shows greater variation and its copy number is higher in case of P.falciparum (tRNA copy number: 72) and P.vivax (tRNA copy number: 72) when compared with the other species like P.knowlesi (tRNA copy number: 41), P.yoelii (tRNA copy number: 50), P.chabaudi (tRNA copy number: 53).
Arginine, an amino acid having six fold degenerate codons, uses more copy number of tRNA for preferred codons(AGA, AGG, CGT) and tRNA for less preferred codons (CGC, CGG) is absent in all chosen Plasmodium species. C and G ending codons are encoded by wobbling with T ending codons. Equal copy number of tRNA genes are available for all codons in leucine except CTT in P.vivax, CTA in P.knowlesi and CTC in rest of the species and wobbling takes place between A-T or T-A, and between C and G-ending codons. In serine, tRNA genes for AGC (less preferred) are more expressed rather than AGT (preferred codon). The tRNA for TCC and AGT codons are absent in all Plasmodium species and here wobbling takes place between T and C, and between C and G-ending codons. Wobbling phenomenon seems very common in all six fold degenerate codons of studied Plasmodium species. The four-fold degenerate codons encoding amino acid uses high copy number of tRNA genes in highly preferred codons among all Plasmodium species except P.vivax. Wobbling is observed between G/C and between T and A-ending codons in all Plasmodium species. Isoleucine has three fold degenerated codons where tRNA genes are expressed for highly preferred codons and C ending codons wobble with A or T bases. The amino acids: asparagine, aspartate, cysteine, histidine, phenylalanine and tyrosine, are coded by pyrimidine ending two fold codons. The tRNA copy number is higher in less preferred codons (exception is P.vivax) of Plasmodium species. Here wobbling plays a prominent role for coding XYT codons. The purine ending codons of amino acids: glutamine, lysine and glutamate show high copy number of tRNA for their highly preferred codons except glutamate in case of P.yoelii and wobbling does not occur for these amino acids.
Strength of codon bias:
Codon usage bias is the parameter that delineates the differences in the occurrence of synonymous codons in genomic coding sequences [23]. This codon bias is calculated for all coding sequences of six Plasmodium species. On analyzing codon usage bias of six different Plasmodium species, we see two distinct groups (Figure 2), four (P.falciparum, P.yoelii, P.chabaudi and P.berghei) following similar pattern in codon usage bias and the other two, P.vivax and P.knowlesi, varying together in a noticeably different manner. The pattern of codon usage bias within each group is remarkably similar. Although the species of the second group show a similarity in the overall codon bias pattern, but some prominent differences are also seen when detailed analysis is done. These differences in the codon bias pattern of all the six Plasmodium species are due to mutation and genetic drift as well as translation selection acting on coding sequences [17, 24]. Selection favors the preferred codons over the non-preferred ones. Nevertheless the existences of nonpreferred or non-optimal codons are due to the action of mutational and genetic drift forces [15].
Figure 2.
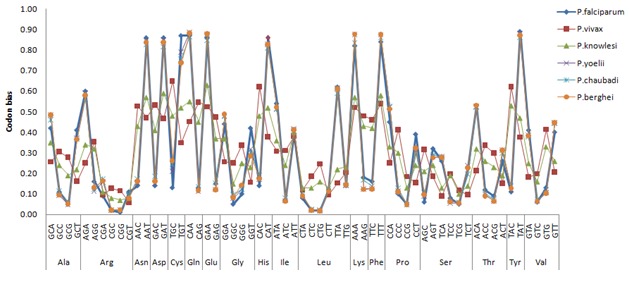
Comparative codon bias analysis in Plasmodium species.
Variation in translated amino acid frequency in Plasmodium species:
Proteins comprise of amino acid residues encoded by degenerate codons. Amino acid usage is calculated here for understanding the codon bias pattern in all six Plasmodium species. Amino acid frequency variations are clearly visible (Figure 3). Amino acid usage varies among different Plasmodium species but the overall picture represents higher usage of Asn, Asp, Glu, Ile, Leu, Lys and Ser residues considering whole proteome. This represents high adaptivity of these amino acid residues over evolutionary time. P.vivax and P.knowlesi genomes are found to be closely related [4], having nearly similar amino acid frequencies. These species shows higher usage of Ala, Val, Arg, Gly and Pro amino acids when. Compared to other Plasmodia species
Figure 3.
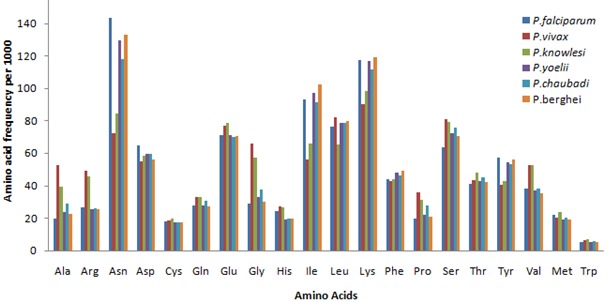
Amino acid frequency in different Plasmodium species
The occurrence of low-complexity regions in variable surface proteins of P.falciparum is well known [25]. Amino acid repeats require one particular preferred codon being chosen repeatedly to code for that particular amino acid. Therefore frequency of that particular codon becomes high. Amino acid content of proteome depends upon the symmetric GC pressure and AT pressure [26–28]. The P.falciparum proteome have high Asn- and Lys- rich low-complexity repeats and this is basically due to high AT bias compositional pressure which leads in increase of their frequencies. The higher usage of Asn and Lys amino acids in P.falciparum is clearly visible (Figure 4). While in case of P.vivax, amino acid residues (Ala, Arg, Gly and Pro) encoded by GC rich codons are higher in number and can be explained by GC pressure. A wide range of variation occurs in amino acid composition of only two species namely P.falciparum and P.vivax, compared with the others due to disparity in their genome composition.
Figure 4.
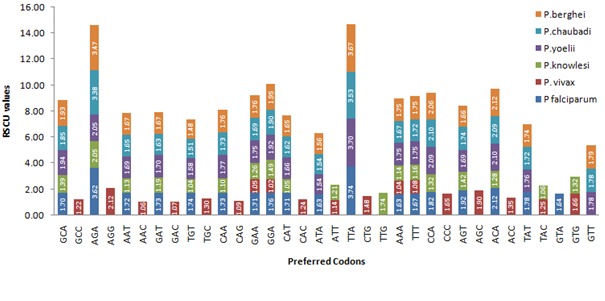
Graph showing preferred codons for each amino acid in six Plasmodium species. Numerical values inside blocks show RSCU values of preferred codons.
Determination of preferred codons for each amino acid in Plasmodium species:
Preferred codons are used at higher frequencies for encoding a particular amino acid. Generally, preferred codons encoding the same amino acid are similar in five Plasmodium species with the exception of P.vivax (Figure 5). Glu, Lys, and Phe shows identical preferred codons throughout the species and represented by A/T at the third codon position. For Ala and Val, P.falciparum has two alternative codons as optimal, but they differ by having A/T at the third place, providing a very good example of A/T wobble. Although P.vivax and P.knowlesi have high GC content, but preferred codon encoding Ile is ATT, not ATC and this might be due to C -> T mutations resulting from deamination of cytosine.
Figure 5.
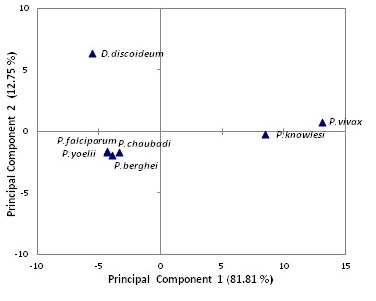
Principal component analyses of relative synonymous codon usage (RSCU) indices of seven species
Identification of codon usage preferences by PCA:
PCA was carried out to look at the similarities and differences of codon usage patterns in different Plasmodium species. Here Dictyostelium discoideum, having similar GC content to P.falciparum is also taken in this statistical analysis to understand the role of factors influencing codon usage bias. The 7 x 59 RSCU matrixes was created and used as initial data for PCA. The PC1 has the Eigen value of 48.27, so it shows higher variability of 81.81 % in codon usage. The first two PC vectors accounted for 94.56 % of cumulative variance in codon usage pattern. PC1 value was much bigger than the other component values, so it has a higher interpretation degree for explaining the total variability in codon usage.
The principal component analysis grouped the given species into three clusters according to their positions along first two principal components (Figure 5). Here PCA is done using RSCU values and variations in these values depend on their codon usage. These species occupy different ecological niche according to their ecological and physiological needs, resulting in wide separation of these species. Although P.falciparum and D.discoideum, are similar in their GC composition but they lie in different clusters when PC2 is taken in to consideration. This shows that apart from compositional constraint, few other factors like translational selection and tRNA availability are also responsible for shaping codon usage bias.
Relationship between ENC and GC3:
Based on the codon homozygosity, ENC is the most useful concept reflecting codon usage bias pattern in different organisms [29]. ENC values are calculated for coding sequences of six Plasmodium species (Table 1). Here the degree of codon usage bias in two species namely P.vivax and P.knowlesi is lowest altogether as measured by ENC. This implies almost random usage of synonymous codons, while the other four species have lower values of ENC; this implies almost random usage of synonymous codons, while the other four species have lower values of ENC, showing more biased usage of synonymous codons (Figure 6).
Figure 6.
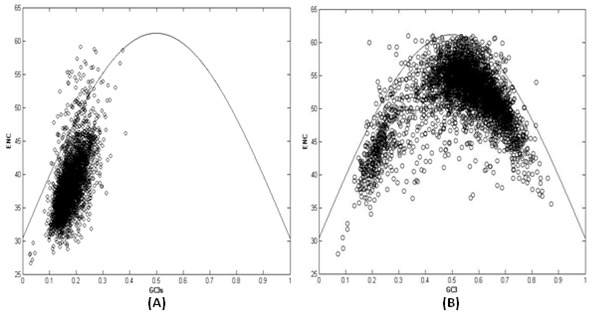
Graph showing the relationship between the effective number of codons (ENC) and the GC content of the third codon position (GC3) in P.falciparum (A) and P.vivax (B)
ENC and their corresponding GC3 values are used to demonstrate the role of dominant factors in shaping codon usage bias in Plasmodium species. The normal curve represents the relationship between ENC and GC3, in absence of selection pressure (Figure 6). In other words when GC-composition bias is not responsible for any codon usage bias, all genes must be lie on normal curve. Here P. falciparum shows extreme GC3 content from 3.02 % to 39 % with a mean of 17.4 % and a wide range of ENC variation from 26.62 to 59.15 with a mean value 38.22 among different coding genes (Figure 6A). Most of the genes of P.falciparum plotted on ENC-GC3 plot, lie near the extreme end of normal curve and group of genes with low ENC values comprises of putatively highly expressed genes and as a consequence, selection for codon usage can be easily seen. While in P.vivax, GC3 values vary from 7 % to 87 % with a mean value 54 % and their corresponding ENC values varies from 28 to 61, having 52.75 mean values (Figure 6B). ENC-GC3 plot clearly shows that variation of GC3 is greater in P.vivax and its correlation with ENC is less compared with that of P.falciparum; resulting in variation of codon usage among them. The average ENC value for all genes is less in P.falciparum and this shows overall codon usage bias is more in P.falciparum compared to P.vivax. Correlation analysis between ENC and GC3 shows higher correlation coefficient for P.falciparum (0.63) compared with that of P.vivax (0.21). This shows expression of genes in P.falciparum is more dependent on composition biased mutational pressure than P.vivax.
To highlight the role of various factors in shaping codon usage bias, we diversified our codon data between highly expressed and lowly expressed genes using their respective ENC values. Here 5 % of genes are taken from extreme ends. Significance of chi- square test is performed to visualize the variation of codon usage between two groups of gene. Optimal codons occur more frequently in highly expressed genes and their frequency is low in less expressed genes [13]. In case of P.falciparum, putative highly expressed genes show higher usage of 25 optimal codons. Adenine (36 %) and Thymine (36 %) are the dominant bases at third position in optimal codons of highly expressed genes and their occurrence is due to compositional-biased mutational pressure acting on P.falciparum genome. While presence of G3 (20 %) and C3 (12 %) in optimal codons of highly expressed genes predicts that G3 base is under higher translational selection pressure than C3.
P.vivax chromosomes have scattered regions of constant AT and GC-content, also called ‘isochore’ regions. Genes lying near the centromere are more GC rich and their richness subsequently decreases with increase in distance from centromere [4]. Genes residing near telomeric and sub-telomeric (high AT rich) shows codon usage equivalent to P.falciparum. Highly expressed genes of P.vivax contain 30 optimal codons, and are mostly A3 and T3 rich. Higher usage of A/T at the third position of optimal codons in highly expressed genes of relatively GC-rich P.vivax genome shows higher translational selection pressure acting at AT3 codons. The correlation between amino acid frequency of highly expressed genes and their respective tRNA copy number is weak, and this hints at limitation of the role of translational selection in shaping codon usage bias in Plasmodium species. The correlation coefficient between amino acid frequency of highly expressed genes and their respective tRNA is slightly higher in case of P.vivax than P.falciparum. This represents the role of translational selection in shaping codon usage bias is also weak but more in P.vivax compared with P.falciparum.
Conclusion
Various factors affecting codon usage bias in Plasmodium species is studied here considering larger data-sets. A strong correlation is found among exonic GC content and their codon usage bias in six Plasmodium species. The GC content of a genome also explains much of the observed variation in amino acid frequencies, effective number of codons and other factors related to codon usage. The separation of each aspect of codon usage behavior of studied species classifies them into two separate lineages. This diversification is clearly seen via PCA. Our PCA result shows a large evolutionary distance between P.falciparum and P. vivax and these results are also confirmed by previous phylogenetic studies [30]. The three rodent parasites, P.berghei, P.chabaudi and P.yoelii cluster together as expected and are close to P.falciparum; the genome-scale synteny among them, confirmed that these species evolved from a common ancestor [31]. Higher average ENC values, lower dependence on GC3 content and location of all genes on ENC-GC3 plot in P.vivax, shows more random usage of synonymous codons compared with that of P.falciparum.
Correlation study of ENC-GC3 graph among these species explains that P.falciparum gene expression level is highly dependent on its biased genome composition. The optimal codons of highly expressed genes in both, P.falciparum and P.vivax, show higher usage of A/T ending codons. This stresses higher role of compositional biased mutational pressure rather than translational selection in shaping codon usage bias of these species. Amino acid usage of highly expressed genes and their tRNA copy numbers in studied Plasmodium species displays a weak correlation, hence shows a limiting role of selection at translational level for shaping codon usage. Our findings confirm that codon usage bias phenomenon differs for genes having differential expression level and factors responsible for it are also different in studied Plasmodium species. This study involves comparison of six Plasmodium species using codon usage bias and our main focus is on P.falciparum and P.vivax, as they are human malarial parasites. Although both of these species use composition biased mutational and translational selection pressure for shaping their codon usage, but their relative strength is different. The expression of genes in P.falciparum shows higher role of compositional mutation pressure than P.vivax, and though the role of translational selection for shaping codon usage bias is weak in both species, but its effect is higher in P.vivax as compared to P.falciparum. The CUB variation in the three rodent parasites, P.berghei, P.chabaudii and P.yoelli is strikingly similar to P.falciparum. The simian and human malarial parasite, P.knowlesi shows a variation in codon usage bias similar to P.vivax but on closer study there are differences confirmed by the PCA analysis.
Supplementary material
Footnotes
Citation:Yadav & Swati, Bioinformation 8(24): 1230-1239 (2012)
References
- 1.JM Carlton, et al. Nature. 2002;419:512. [Google Scholar]
- 2.MJ Gardner, et al. Nature. 2002;419:498. [Google Scholar]
- 3.JM Carlton, et al. Curr Issues Mol Biol. 2005;1:23. [PubMed] [Google Scholar]
- 4.JM Carlton, et al. Nature. 2008;455:757. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A Pain, et al. Nature. 2008;455:799. [Google Scholar]
- 6.MC Angellotti, et al. Nucleic Acids Res. 2007;35:132. doi: 10.1093/nar/gkl965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R Grantham, et al. Nucleic Acids Res. 1981;9:r43. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.EP Rocha. Genome Res. 2004;14:2279. doi: 10.1101/gr.2896904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R Hershberg, DA Petrov. Annu Rev Genet. 2008;42:287. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- 10.PM Sharp, et al. Phil Trans R Soc Lond B Biol Sci. 2010;365:1203. doi: 10.1098/rstb.2009.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.SL Chen, et al. Proc Natl Acad Sci. 2004;101:3480. [Google Scholar]
- 12.M Gouy, C Gautier. Nucleic Acids Res. 1982;10:7055. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.M Stenico, et al. Nucleic Acids Res. 1994;22:2437. doi: 10.1093/nar/22.13.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.PM Sharp, et al. Nucleic Acids Res. 2005;33:1141. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.JB Plotkin, G Kudla. Nature Rev Geneti. 2011;12:32. [Google Scholar]
- 16.AA Hanafi-Bojd, et al. Asian Pac J Trop Med. 2011;4:498. doi: 10.1016/S1995-7645(11)60134-X. [DOI] [PubMed] [Google Scholar]
- 17.H Musto, et al. J Mol Evol. 1999;49:27. [Google Scholar]
- 18.MK Yadav, et al. IJBST. 2010;3:46. [Google Scholar]
- 19.P Shah, MA Gilchrist. Proc Natl Acad Sci U S A. 2011;108:10231. [Google Scholar]
- 20.C Aurrecoechea, et al. Nucleic Acids Res. 2009;37:D539. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.F Wright, et al. Gene. 1990;87:23. [Google Scholar]
- 22.TM Lowe, SR Eddy. Nucleic Acids Res. 1997;25:955. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.SG Andersson, CG Kurland. Microbiol Rev. 1990;54:198. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.L Duret. Curr Opin Genet Dev. 2002;12:640. doi: 10.1016/s0959-437x(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 25.E Pizzi, C Frontali. Genome Res. 2001;11:218. doi: 10.1101/gr.152201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.N Sueoka. Proc Natl Acad Sci U S A. 1988;85:2653. [Google Scholar]
- 27.H Romero, et al. Gene. 2003;317:141. [Google Scholar]
- 28.Makedonka Mitreva, et al. Genome Biology. 2006;7:R75. doi: 10.1186/gb-2006-7-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders Fuglsang, et al. Biochem and Biophys Res Commun. 2004;317:957. doi: 10.1016/j.bbrc.2004.03.138. [DOI] [PubMed] [Google Scholar]
- 30.A Escalante, et al. Proc Natl Acad Sci U S A. 1998;95:8124. [Google Scholar]
- 31.Carlton, et al. Mol and Biochem Parasitol. 1998;93:285. doi: 10.1016/s0166-6851(98)00043-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


