Abstract
The NaV1.7 voltage-gated sodium channel is critical for pain signaling in humans. Gain-of-function mutations are associated with several pain syndromes including inherited erythromelalgia (IEM). Most IEM patients with NaV1.7 mutations are resistant to pharmacotherapy, but carbamazepine (CBZ) normalizes activation of NaV1.7-V400M mutant channels from a family with CBZ-responsive IEM. Here we show that structural modeling and mutant cycle analysis predict pharmacoresponsiveness to CBZ of a NaV1.7 mutant channel that substitutes a residue 159 amino acids distant from V400M in the channel peptide. Structural modeling reveals that this IEM mutation (S241T) is only 2.4-angstrom (Å) apart from V400M in the folded NaV1.7 channel and mutant cycle analysis demonstrates that V400M is energetically coupled to S241T during channel activation. We further show that the atomic proximity and energetic coupling of V400M and S241T are paralleled by pharmacological coupling, as CBZ at therapeutic concentration (30 μM) causes a depolarizing shift of S214T mutant channel activation curve, similar to that previously reported for V400M mutant channel. This pharmacoresponsiveness of S241T to CBZ was further evident at a cellular level, where CBZ normalized the hyperexcitability of dorsal root ganglion (DRG) neurons expressing S241T mutant channel. We suggest that a similar approach might facilitate screening for amino acid variants of a variety of channels that confer enhanced pharmacoresponsiveness on the channel.
Introduction
Chronic pain affects more than one-quarter of Americans and pain in nearly 40% of these patients is not relieved by currently available drugs1. Recent studies have demonstrated that NaV1.7, a voltage-gated sodium channel preferentially expressed in dorsal root ganglia (DRG) and sympathetic ganglia neurons, is a central player in pain transduction2–5. Loss-of-function NaV1.7 mutations cause congenital indifference to pain (CIP)6, while gain-of-function NaV1.7 mutations produce several painful syndromes including inherited erythromelalgia (IEM), a disorder in which, as a result of hyperpolarized activation of NaV1.7, threshold is reduced and evoked firing frequency increased in pain-signaling DRG neurons, leading to intense burning pain4, 7, 8. These human studies have validated NaV1.7 as a key molecular target for the development of new therapeutics for the treatment of pain syndromes.
IEM is characterically unresponsive to existing pain therapies7. Against this background of pharmaco-unresponsiveness, multiple affected members of one family, carrying the NaV1.7 -V400M mutation, have responded favorably to carbamazepine (CBZ)9, a state-dependent sodium channel blocker used mainly for the treatment of epilepsy and trigeminal neuralgia10. Our previous study demonstrates that therapeutic concentrations of CBZ normalize the hyperpolarizing shift of the Na 1.7-V400M mutant channel activation curve9, while not affecting activation curve of wild-type channels. As the shift of voltage-dependence of activation is the strongest contributor to the hyperexcitability of DRG neurons expressing IEM mutations11, it is believed that the normalization of the activation curve in the V400M mutant channel by CBZ underlies the beneficial effect of CBZ in this family9. These findings suggest the possibility of personalized pharmacotherapy based upon a pharmacogenomics approach. However, to achieve this goal, a better understanding of the structure-function relationship of the human NaV1.7 channel mutations that underlies drug responsiveness and development of pre-clinical assays of pharmacoresponsiveness are needed.
Recently, the crystal structure of a bacterial voltage-gated sodium channel has been solved, shedding new light on structural elements for channel gating and drug accessibility12. Starting from modeling the human NaV1.7 channel to understand the interaction between different mutations, we demonstrated in this proof-of-principle study that a combined approach of structural modeling, thermodynamic mutant cycle analysis, and voltage- and current-clamp recording of NaV1.7 mutant channels can predict pharmacoresponsiveness of a NaV1.7 mutant channel to CBZ, suggesting a potential novel approach to screen for amino acid variants of a variety of channels that confer enhanced pharmacoresponsiveness on the channel.
Results
V400 and S241 show atomic proximity in a human NaV 1.7 channel structural model
Previous observations from linear protein sequence suggest that IEM mutations of Na 1.7 channel tend to cluster4 but their proximity to each other within the folded structure is not clear. To better understand the structure-function relationship among IEM mutations, we constructed a structural model of transmembrane helices of human NaV1.7 channel based on the recently solved crystal structure of a bacterial voltage-gated sodium channel12.
The human NaV1.7 structural model was constructed using the GPCR-ITASSER program13, an extension of the well-established I-TASSER algorithm14, 15, incorporating a composite set of transmembrane specific force fields and constraints16. The accuracy of the individual domain models can be reliably estimated by the confidence score (C score) that has a high correlation to the actual TM-score17. A model with TM-score >0.5 indicates a high similarity between the predicted model and the native structure18, 19. The estimated TM-scores based on C-score for our models of domains I–IV are 0.575, 0.805, 0.681 and 0.722, respectively.
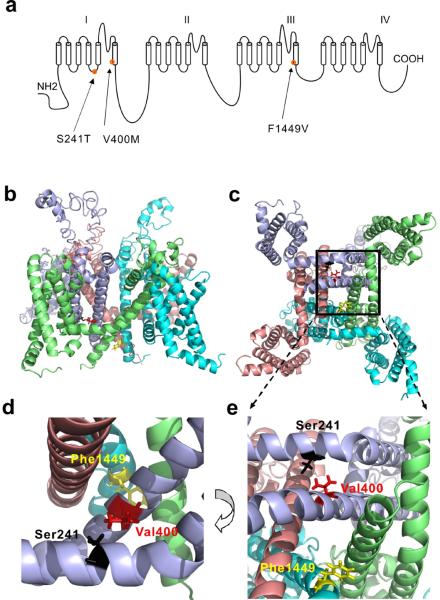
The linear schematic of NaV1.7 channel is shown in Fig.1a and the assembled structural models are shown in Fig.1b (intra-membrane view) and Fig.1c (cytosolic view). We are interested in V400M mutation of S6 helix because it is known to be pharmacoresponsive to CBZ9. Reasoning that the location of V400M in the folded channel might be a critical factor for its pharmacoresponsiveness, we thus look for IEM mutations that are structurally close to V400M. Our structural model revealed that the location of another IEM mutation (S241T) of S4-5 linker is close to that of V400M. The distance between S241 and V400 is only 2.4 –angstrom (Å) apart measured from the closest hydrogen in the folded channel. The side chain of V400 points toward the S4-5 linker, where S241 is located (Fig.1d,e). Zoom-in views of the DI/S6 show the location of the V400 and S241 (Fig.1d,e). Although the effect of the S241T mutation on channel activation has been linked to the increase in the size of the side chain at position 24120, the pharmacoresponsiveness of S241T mutant channel has not been previously assessed.
Figure 1. Structural modeling of transmembrane domains of human NaV1.7 channel.
(a) Linear schematic of the human NaV1.7 channel topology showing the mutations S241T, V400M and F1449V. (b) Intra-membrane view of structural model of NaV1.7 channel transmembrane domains. Domain I, lightblue; Domain II, salmon; Domain III, cyan; Domain IV, lime. (c) Cyotsolic view of the structural model of NaV1.7 channel transmembrane domains. Boxed area containing S241, V400 and F1449 residues was enlarged in (e). (d) Close-up intra-membrane view of the area containing V400, S241 and F1449 residues. (e) Close-up cytosolic view of the boxed area of (c). V400, S241 and F1449 were shown as stick and colored red, black, and yellow, respectively.
Waxman
Another IEM mutation (F1449V)21 was selected for analysis because it is located at the cytoplasmic end of S6 helix, analogous to the location of V400M mutation (Fig.1a). Based on homology modeling to potassium channel structure, F1449 has been suggested to contribute to a hydrophobic ring at the cytoplasmic pore opening, which acts as the channel's activation gate22. Modeling of F1449 using the bacterial sodium channel structure recapitulated the hydrophobic ring. However, like the S241T mutant, the pharmacoresponsiveness of F1449V mutant channel is also unknown.
V400M and S241T, but not V400M and F1449V, are energetically coupled during channel activation
Previous studies show that all of these IEM mutations hyperpolarize channel activation4, 9, 20, 22. However, it is not clear whether these mutations work independently or in concert to affect the same or different aspect of channel activation. To further understand the structure-function relationships among these IEM mutations, we asked whether the location information of NaV1.7 channel mutations found in our structural modeling might indicate their mechanistic coupling during channel activation.
Thermodynamic mutant cycle analysis is a well-established approach to measure the independence or coupling of two mutations of a given protein23, 24. We studied the coupling of V400M, a mutation located in the S6 helix, with either a structurally close mutation in the S4-5 linker (S241T) or with a mutation in a different S6 helix (F1449V). Constructs carrying double mutations, e.g. V400M and S241T or V400M and F1449V, were created (named as VM/ST and VM/FV, respectively) to perform mutant cycle analysis. Voltage-dependence of activation was determined for NaV1.7 WT channel, single mutations (V400M, F1449V, and S241T) and double mutations (VM/ST and VM/FV) in transiently transfected HEK293 cells. If two mutations are energetically coupled or affect channel activation through the same or shared mechanism, then the double mutation would not have an additive effect on channel activation compared to the single mutations. However, if two mutations are not energetically coupled or affect the channel independently through distinct mechanisms, then the double mutation would have an additive effect on channel activation compared to the two single mutations.
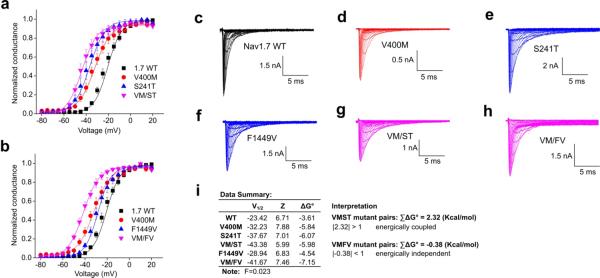
The voltage-dependence of activation protocol was applied and data from WT, the mutant pairs to be evaluated, and the corresponding double mutant were collected and fitted with the Boltzmann equation to determine the V1/2 (the voltage at half-activation) and Z (proportional to the slope at half-activation)(Fig.2a, b). Representative raw traces are shown in Fig.2c–h. As shown in Fig.2a, the activation curve of double mutation VM/ST was very close to that of the single mutations, indicating a non-additive effect and suggesting that V400M and S241T mutations alter activation via a common, shared mechanism. In contrast, the activation curve of double mutation VM/FV was more hyperpolarized relative to the single mutations (Fig.2b), indicating an additive effect of the two mutations and suggesting that the V400M and F1449V mutations alter activation via different, energetically-independent mechanisms.
Figure 2. Mutant cycle analysis of voltage-dependence of activation of NaV1.7 mutations.
(a) Voltage-dependence of activation curves of NaV1.7 WT, V400M, S241T, and VM/ST double mutant channels. Curves were Boltzmann fits of the data. (b) Voltage-dependence of activation curves of NaV1.7 WT, V400M, F1449V, and VM/FV double mutant channels. (c–h) Representative traces of current families recorded from HEK293 cells expressing WT (c), V400M (d), S241T (e), F1449V (f), VM/ST double mutant (g), and VM/FV double mutant (h) channels. (i) Data summary for the mutant cycle analysis and interpretation (n=4–5). ΣΔG° was used to determine the level of additivity and was calculated based on the equation described in methods.
Waxman
To quantitatively assess these effects, the energy change on channel activation caused by a single or double mutation was calculated (Fig.2i). For each single or double mutation, V1/2 and Z were used to determine the free energy of activation (ΔG°). The change in free energy of mutants relative to the WT channel was designated as ΔΔG° and coupling free energy (magnitude of non-additivity) was ΣΔG°. Based on several published studies 23–28, we set the following standard: if |ΣΔG°| > 1 kcal/mol, we accepted the two mutations as coupled (non-additive) whereas if |ΣΔG°| < 1 kcal/mol, we accepted the two mutations as independent (additive). We calculated the ΣΔG° for all the mutation pairs and found that for the V400M: S241T mutation pair, ΣΔG°=2.32 kcal/mol, and for V400M: F1449V mutation pair, the ΣΔG°=−0.38 kcal/mol (Fig.2i). These data are in good agreement with the fitted curves, and strongly suggest that V400M and S241T are energetically coupled, while V400M and F1449V are energetically independent during channel activation.
CBZ depolarizes S241T but not F1449V mutant channels activation curve
Structural modeling showed atomic proximity between V400 and S241 and mutant cycle analysis demonstrated that S241T is energetically coupled with V400M during channel activation. Pretreatment of cells with a therapeutic concentration of CBZ (30 μM) is known to cause a depolarizing shift of activation of V400M but not WT channels 9. While residues within the S4-S5 linkers of sodium channels have not been implicated in forming the local anesthetic binding site 29, we hypothesized that the atomic proximity and energetic coupling of V400M and S241T might be paralleled by pharmacological coupling so that they might share pharmacoresponsiveness to CBZ.
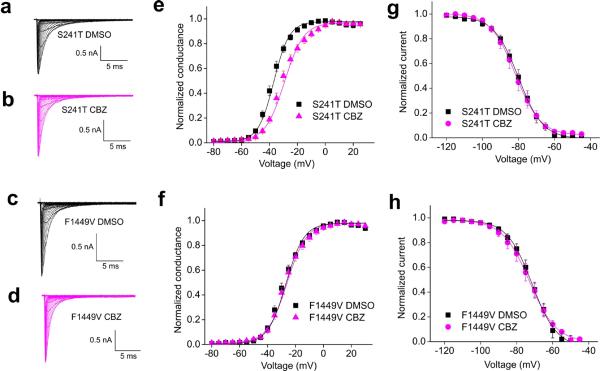
To test this hypothesis, the effect of CBZ (30 μM) pretreatment, initiated 30 min prior to recording 9, on each mutants activation curve was measured in HEK293 cells expressing S241T or F1449V mutant channels. Representative recordings are shown in Fig.3a–d. Notably, CBZ treatment caused a significant depolarizing shift in the V1/2 of voltage-dependence of activation of S241T mutant channels by 7.0 mV compared to DMSO treated cells (DMSO: −37.6±1.0 mV, CBZ: −30.5±1.3 mV, P<0.01, Fig.3e, Table 1). In contrast, a similar CBZ treatment to HEK293 cells expressing F1449V mutant channel did not show a detectable effect on channel activation compared with DMSO treatment (activation V1/2 for CBZ treatment: −27.1 ± 1.5 mV; activation V1/2 for DMSO treatment: −28.8 ± 1.1 mV; P>0.05, Fig.3f, Table 1).
Figure 3. Comparison of voltage-dependence of activation and fast inactivation of S241T and F1449V mutant channels with or without CBZ exposure.
(a–d) Representative traces of current families recorded using the activation protocol from HEK293 cells expressing S241T mutant channel treated with DMSO (a), or with CBZ (b); F1449V mutant channel treated with DMSO (c), or with CBZ (d). (e) The averaged voltage-dependence of activation of S241T mutant channel treated with DMSO or CBZ (30 μM) was plotted and fitted with Boltzmann equation. A depolarizing shift of activation of 7.0 mV was observed when S241T mutant channel was treated with CBZ. (f) The averaged voltage-dependence of activation of F1449V mutant channel treated with DMSO or CBZ was plotted and fitted with Boltzmann equation. No notable shift in activation curve of F1449V mutant channel was observed. (g–h) The voltage dependence of fast inactivation of S241T (g) or F1449V (h) mutant channel treated with DMSO or CBZ was plotted and fitted with Bolzmann equation. No notable shift was observed.
Waxman
Table 1.
Effects of CBZ on channel properties
| activation V1/2 (mV) | activation slope | fast inactivation V1/2 (mV) | fast inactivation slope | |
|---|---|---|---|---|
| S241T DMSO | −37.6±1.0 (n=10) | 6.1±0.2 | −79.9±1.7 (n=6) | 6.2±0.2 |
| S241T CBZ | −30.5±1.3 (n=13)** | 6.7±0.5 | −81.5±2.1 (n=6) | 6.1±0.1 |
| F1449V DMSO | −28.8±1.1 (n=9) | 6.9±0.5 | −71.6±1.5 (n=6) | 6.5±0.2 |
| F1449V CBZ | −27.1±1.5 (n=8) | 7.1±0.4 | −72.7±2.2 (n=8) | 6.9±0.2 |
significant difference was found between S241T DMSO and S241T CBZ activation V1/2, P<0.01
Enhancement of voltage-dependence of fast-inactivation (a hyperpolarizing shift of the fast-inactivation V1/2) is the classic mechanism of action for CBZ 10. To determine whether CBZ might have a similar effect on the fast-inactivation properties of S241T or F1449V mutant channels, HEK293 cells expressing S241T or F1449V mutant channels were treated with DMSO or CBZ (30 μM) as mentioned earlier. Fast-inactivation was assessed and this protocol demonstrated a fast-inactivation V1/2 for S241T of −79.9±1.7 mV, and a fast inactivation V1/2 for F1449 of −71.6±1.5 mV with DMSO treatment (Table 1), very close to those of our previous studies without any treatments 20, 22. CBZ (30 μM) did not significantly alter the V1/2 for either S241T or F1449V mutant channels (−81.5±2.1 mV, −72.7±2.2 mV, respectively; P>0.05 compared with DMSO treatment, Fig.3g,h).
Expression of S241T mutant channel leads to DRG neuron hyperexcitability
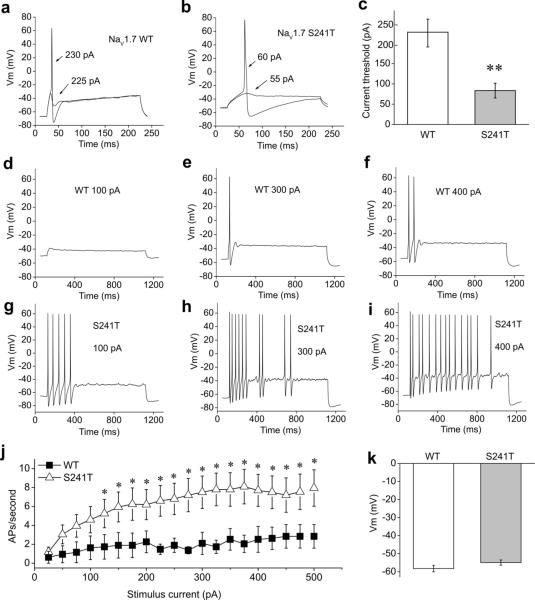
To study the effects of S241T mutant channel on nociceptor excitability, small (20–28 μM diameter) DRG neurons from postnatal day 1–5 rats were isolated and transfected with either human NaV1.7 WT or S241T mutant channels. DRG neuron excitability was assessed by current-clamp recording two days after transfection. Current threshold, the injection current required to produce a single all-or-none action potential, was determined by applying depolarizing currents of increasing magnitude (5 pA increments) for 200 ms. For the neuron transfected with WT channels shown in Fig. 4a, current injection smaller than 225 pA generated small, graded membrane potential depolarization, with the first all-or-none action potential elicited by a current injection of 230 pA. In contrast, a neuron expressing the S241T mutant channel fired the first action potential in response to a current injection of 60 pA (Fig.4b). On average, the expression of S241T mutant caused a significant ~2.7-fold reduction of action potential threshold (83.5±18.2 pA, n=20) compared to DRG neurons expressing WT channel (227.6±36.7 pA, n= 19, P<0.01, Fig.4c).
Figure 4. Current-clamp analysis of DRG neurons expressing WT or S241T mutant channel.
(a) Representative DRG neuron expressing NaV1.7 WT channel showed sub-threshold response to 225 pA current injection and subsequent action potential evoked by injection of 230 pA, which was the current threshold for this neuron. (b) Representative DRG neuron expressing NaV1.7-S241T mutant channel showed sub-threshold response to 55 pA current injection and subsequent action potential evoked by injection of 60 pA. (c) Comparison of current threshold for DRG neurons expressing WT and S241T mutant channels. Expression of S241T channel reduced current threshold significantly (P<0.01). Current threshold for WT: (227.6±36.7 pA, n= 19); for S241T: (83.5±18.2 pA, n=20). (d–f) responses of a representative DRG neuron expressing WT channel to 1s long depolarization current steps at 100 (d), 300 (e) and 400 (f) pA current injection. (g–i) Responses of a representative DRG neuron expressing S241T mutant channel to 1s long depolarization current steps at 100 (g), 300 (h) and 400 (i) pA current injection. The difference in responses is apparent across this range. (j) The averaged number of action potentials between DRG neurons expressing WT and S241T mutant channel was compared. The response of DRG neurons expressing WT channel to current injection was significantly different compared with DRG neurons expressing S241T mutant channel across a range (125–500 pA) of step current injections (P<0.05). (k) Averaged membrane potentials between DRG neurons expressing WT or S241T mutant channel were not statistically different.
Waxman
The firing frequency of DRG neurons was also assessed by a series of 1 sec long current injections from 25 to 500 pA in 25 pA increments. For DRG neurons expressing WT channel, no action potentials or only a single action potential was elicited in response to depolarizing current injection, with an occasional second spike in response to stronger stimuli (Fig.4d–e). In contrast, DRG neurons expressing S241T mutant channel fired repetitively in response to low and high current injections (Fig.4g–i). To compare the response of DRG neurons expressing WT or S241T channels, the number of spikes elicited by 1 sec long current injection at various stimulus intensities was averaged and plotted (Fig.4j). DRG neurons expressing S241T mutant channel fired significantly more spikes than DRG neurons expressing WT channel at all stimulus intensities ≥125 pA (Fig.4j). Since resting membrane potential (RMP) is known to affect DRG neurons excitability 30, we also assessed the RMP of DRG neurons expressing WT or S241T mutant channels and no difference regarding RMPs was found (Fig.4k). Taken together, these data show that the expression of S241T mutant channels induces DRG neuron hyperexcitability.
CBZ normalizes the hyperexcitability of DRG neurons expressing S241T but not F1449V mutant channels
To test the prediction from our structural modeling and mutant cycle analysis, we assessed the effect of CBZ on the hyperexcitability of DRG neurons expressing the S241T or F1449V mutant channels using current-clamp recording. DRG neurons were treated with either DMSO or CBZ (30 μM) initiated 30 min prior to recording as mentioned above.
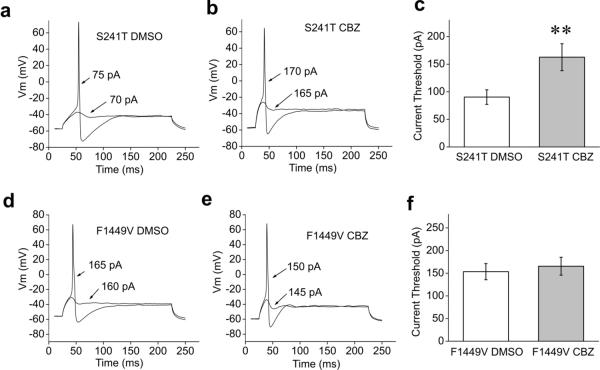
Current threshold was assessed first and the response of a representative neuron expressing S241T mutant channel treated with DMSO is shown in Fig. 5a. The current threshold for this neuron was 75 pA (Fig.5a). In contrast, the current threshold of a representative neuron expressing S241T mutant channels treated with CBZ is shown in Fig.5b; the current threshold for this neuron is 170 pA. On average, CBZ treatment resulted in a significant ~two-fold increase in current threshold for DRG neurons expressing S241T mutant channel (CBZ treatment: 162.7±24.4 pA, n=28; DMSO treatment: 90.4±13.2 pA, n=27, P<0.01; Fig. 5c), which indicates that CBZ treatment is likely to attenuate the excitability of DRG neuron expressing S241T mutant channel.
Figure 5. Comparison of current threshold of CBZ or DMSO treated DRG neurons expressing S241T or F1449V mutant channels.
(a–b) The sub- and supra-threshold responses of representative DRG neurons expressing S241T mutant channel treated with DMSO (a) or 30 μM CBZ (b) are shown. (c) Comparison of current threshold for DRG neurons expressing S241T mutant channel treated with DMSO or 30 μM CBZ. The CBZ treatment increased the current threshold significantly (P<0.01). Current threshold for DMSO treated DRG neurons: 90.4±13.2 pA (n=27); for CBZ treated DRG neurons: 162.7±24.4 pA (n=28). (d–e) The sub- and supra-threshold of DRG neurons expressing F1449V mutant channel treated with DMSO (d) or 30 μM CBZ (e) are shown. (f) Comparison of current threshold for DRG neurons expressing F1449V mutant channel with the treatment of DMSO (153.5±17.9 pA, n=29) or 30 μM CBZ (165.5±19.7 pA, n=28). No significant difference was found (P>0.05).
Waxman
Our previous studies have demonstrated that the expression of F1449V mutant channel in DRG neurons reduces the current threshold and increases the firing frequency of action potentials in response to sustained depolarizing stimuli 21. As we found that CBZ did not produce a depolarizing shift in F1449V mutant channel activation curve (Fig. 3f), we hypothesized that CBZ would not have a beneficial effect on firing properties of DRG neurons expressing F1449V mutant channel. Again we assess the current threshold first and as illustrated by the recording in Fig.5d,e, the current threshold for action potential firing in DRG neurons expressing F1449V mutant channels was similar between CBZ and DMSO treatments. For the population of neurons in these experiments, no significant difference was found for the current threshold (DMSO, 153.5±17.9 pA, n=29; CBZ, 165.5±19.7 pA, n=28, P>0.05, Fig.5f).
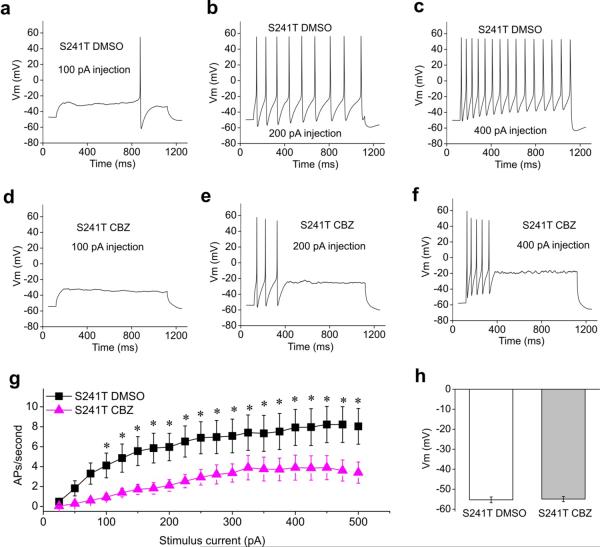
We further tested the effect of CBZ treatment on the firing frequency of DRG neurons expressing either the S241T or F1449V mutant channels. Robust repetitive firing could be seen for neurons expressing S241T mutant channel treated with DMSO (Fig. 6a–c). In contrast, pretreatment with CBZ dramatically reduced the number of action potentials in DRG neurons expressing S241T mutant channel (Fig. 6 d–f). There was a statistically significant reduction of firing frequency at all stimulus intensities ≥ 100 pA (P<0.05, Fig. 6g). Note that this effect was not confounded by changes in RMPs since CBZ treatments did not affect RMPs of DRG neurons expressing mutant channels tested in this study (Fig. 6h, and 7h). Taken together, our data demonstrate that CBZ attenuates the hyperexcitability of DRG neurons expressing S241T mutant channel.
Figure 6. Comparison of firing frequency and membrane potential of CBZ or DMSO treated DRG neurons expressing S241T mutant channel.
(a–c) Responses of a representative DRG neuron expressing S241T mutant channel treated with DMSO to 1sec long depolarization current steps at 100 (a), 200 (b) and 400 (c) pA current injection. (d–f) Similar recording from a representative DRG neuron expressing S241T mutant channel treated with 30 μM CBZ at 100 (d), 300 (e), 400 (f) pA current injection. (g) The averaged response for DRG neurons expressing S241T mutant channel treated with DMSO or CBZ were summarized. CBZ statistically reduced firing frequency starting from 100 pA current injection (P<0.05). (h) Averaged resting membrane potential between DRG neurons expressing S241T mutant channel treated with DMSO or CBZ were not statistically different (DMSO, −55.3±1.4 mV, n=27; CBZ, −54.9±1.3 mV, n=28, P>0.05).
Waxman
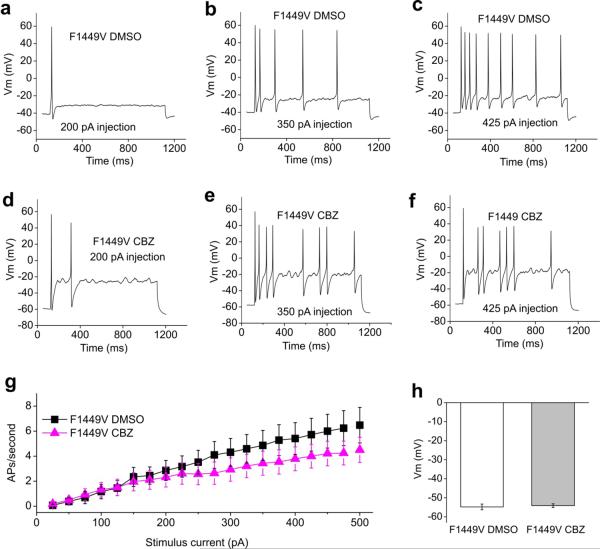
Figure 7. Comparison of firing frequency and membrane potential of CBZ or DMSO treated DRG Neurons expressing F1449V mutant channel.
(a–c) Responses of a representative DRG neurons expressing F1449V mutant channel treated with DMSO to 1s long depolarization current steps at 200 (a), 350 (b) and 425 (c) pA current injection. (d–f) Similar recording from a representative DRG neuron expressing F1449V mutant channel treated with CBZ at 200 (d), 350 (e), and 425 (f) pA current injection. (g) The averaged response of the firing frequency for DRG neurons expressing F1449V mutant channel treated with DMSO or CBZ were compared and no statistical difference was found across the entire range (P>0.05). (h) Averaged resting membrane potentials between DRG neurons expressing F1449V mutant channel treated with DMSO or CBZ were not statistically different (DMSO, −54.8±1.5 mV, n=29; CBZ, −54.1±1.0 mV, n=28, P>0.05).
Waxman
In contrast, CBZ did not attenuate the abnormal repetitive firing of DRG neurons expressing the F1449V mutant channel. Repetitive firing was observed for neurons expressing F1449V mutant channel treated with DMSO (Fig. 7a–c) as well as treated with CBZ (Fig.7d–f). Across a range of current injections, the firing frequency for CBZ and DMSO treated neurons was comparable in response to current injection ≤ 200 pA (Fig.7g). While the firing frequency started to diverge with stronger injection, the effect of CBZ did not reach statistical significance even at a current injection of 500 pA (Fig.7g). Together with our results on V400M and S241T mutant channels, these data suggest that the effect of CBZ is mutant-specific and agrees well with our predication from structural modeling and mutant cycle analysis.
Discussion
Using homology modeling, based upon the recently solved crystal structure of a bacterial voltage-gated sodium channel 12, we report that S241 within the DI/S4–5 linker, while 159 amino acids distant from V400 in the channel peptide, is located close to V400 in the folded structure of the human NaV1.7 voltage-gated sodium channel. Using mutant cycle analysis, we have demonstrated that the NaV1.7-V400M and -S241T mutant channels are energetically coupled during channel activation, suggesting that they contribute to the same feature of channel activation. As V400M mutation responds favorably to CBZ by shifting the activation curve of V400M 9, we reasoned that energetic coupling of V400M and S241T during channel activation might be paralleled by pharmacological coupling. Using voltage-clamp recording, we found that a therapeutic concentration of CBZ indeed normalizes the activation curve of S241T mutant channel. Moreover, we further show that this normalization of activation is accompanied by increased current threshold and reduced firing frequency of DRG neurons expressing S241T mutant channel. In contrast, the V400M and F1449V mutants were not energetically coupled, and CBZ had no detectable effect on activation of F1449V mutant channel or on the firing properties of DRG neurons expressing F1449V mutant channel.
Human and rodent studies have confirmed that the NaV1.7 channel is essential for pain signaling and have validated this channel as a drug target for the development of novel pain therapeutics 4. Structural and functional studies of mutant NaV1.7 channels may provide an insight into understanding mechanistic features of channel gating and its interaction with drugs. Previous studies of sodium channel structure and function were largely based on information obtained from the crystal structure of potassium channels 22, 25, 31, 32. The availability of the crystal structure of a bacterial voltage-gated sodium channel 12 permitted the construction of a more reliable mammalian voltage-gated sodium channel model. Additionally, recent enhancements to I-TASSER 17, a protein prediction package which has been successfully used to predict structure of many proteins including potassium channels for functional studies 33, 34, have provided improved predictions of membrane-bound proteins structure 15, 35. Taking advantage of these new developments, we constructed a homology model for the human NaV1.7 channel and used it in this study to investigate the participation of different residues in channel activation, and to make predictions about drug-channel interaction.
Our structural model revealed that the V400 side-chain points towards the DI/S4-5 linker and is atomically close to S241 residue located in this linker, suggesting that mutations of these two residues might affect the same step in channel activation. The interaction between S4–5 linker and S6 during channel gating has been suggested in a few studies including voltage-gated K+ (KV) channel 36–38 and bacterial voltage-gated sodium channel 12. Interestingly, the interaction site between S4–5 linker and S6 seems to be situated in the analogous location as V400M and S241T 36, 37. More recently, an all-atom molecular dynamics study of KV channel gating mechanism extends these findings by showing that channel activation imposes a pulling force on S4-5 linker, which perturbs the interaction between S4–5 linker and S6, leading to channel opening 39. On the other hand, F1449V may affect different steps in channel activation: the NaV1.7 structural model places F1449 at the cytoplasmic tip of the S6 helix, suggesting that F1449 contributes to a hydrophobic ring at the cytoplasmic vestibule of the pore, comprised of aromatic residues at equivalent positions in the S6 from four domains, acting as the activation gate of the channel. Taken together, we propose that V400M or S241T mutation may disrupt the tight packing of S4–5 linker with S6 pore forming helix, affecting channel activation, whereas the F1449V mutation may disrupt the hydrophobic ring and destabilize the pre-open state of the channel, causing a hyperpolarizing shift in channel activation via a different structural action 21, 22.
We used thermodynamic mutant cycle analysis, a well-established tool, to quantitatively assess the energetic independence or coupling of two mutations in affecting channel activation. Mutant cycle analysis has been used to understand transition of pore-elements during potassium channel gating 23, interactions between ion channels and toxins 24, 28, and identification of ion-pair-forming residues of the voltage sensor domain in a bacterial sodium channel (NaChBac) during voltage-gated activation 25–27. In this study, we show for the first time that mutant cycle analysis can be used to test the hypothesis that different pathogenic IEM mutations (S241T, V400M and F1449V), all of which cause a hyperpolarizing shift of channel activation, participate in different steps of channel gating and have distinct pharmacoresponsiveness. More importantly, this analysis allowed us to predict that S241T mutant channel can interact with CBZ in a similar fashion as V400M mutant channel due to their energetic coupling for channel activation.
Local anesthetic and antiepileptic drugs, including carbamazepine, are believed to have a common recognition site on sodium channels 40. Mutational analysis and functional studies have identified residues within S6 segments of domains III and IV that appear to be important in forming the drug binding site 29. In addition, study of NaV1.4 DI/S6 further shows that N434, located near the middle of the helix, is also important for drug binding 41. Accordingly, IEM mutation N395K of NaV1.7 channel, which corresponds to the N434 in NaV1.4 channel, attenuates the inhibitory effects of lidocaine on inactivated and resting NaV1.7 channel11. Results from above literature and others 29, 42 indicate that our effect of CBZ on NaV1.7-S241T and -V400M mutant channels is not likely to be attributed to direct effects of these mutations on the drug binding site. Alternatively, our data thus may be interpreted as suggesting a role for CBZ as a chemical chaperone for S241T and V400M mutant channels, rather than as a channel blocker. According to this hypothesis, CBZ-channel interaction may lead to stabilization of the S241T and V400M mutant channel in a WT-like conformation, and cause the depolarizing shift in activation, a biophysical change that contributes to attenuation of hyperexcitability of DRG neuron expressing mutant channels.
In summary, our results demonstrate that structural modeling and mutant cycle analysis can reveal the effect of sodium channel mutations on channel gating, and can provide useful information about the responses of mutant channels to pharmacotherapeutic agents. We suggest that a similar approach, initially screening channels of interest for amino acid variants that confer enhanced pharmacoresponsiveness to existing agents and then using these variants as “seeds” for further identification of other variants that display enhanced pharmacosensitivity, may permit identification of genomically-defined subgroups of the population that are pharmacoresponsive.
Methods
Structural modeling
Mammalian NaV1.7 sodium channel is made up of four transmembrane domains, named I, II, III, IV linked by cytoplasmic loops. The transmembrane helix structural model of human NaV1.7 sodium channel was constructed in two steps. First, each of the 4 transmembrane domains was modeled separately using an advanced membrane-bound protein predication algorithm GPCR-ITASSER 13, 16, 43, 44. Then, the whole channel model was built by structurally aligning four individual domain models to a global structural template of the recently solved bacterial sodium channel.
The GPCR-ITASSER algorithm is an advanced algorithm for transmembrane helical structure modeling. The program first uses a multiple threading procedure LOMETS 14 to identify the putative related template structures in the PDB. The structural fragments (mainly α-helices) are then excised from the threading templates and assembled into full-length models by replica exchange Monte Carlo simulations. In addition to the inherent I-TASSER force field, five transmembrane-protein specific energy terms were also used to describe the interactions between transmembrane domain and membrane 13. (1) Membrane repulsive energy is introduced for reducing the clash between intra- and extracellular domain and bilayer membrane. (2) Extracellular hydrophilic interactions for the hydrophilic interactions for residues inside and outside membrane. (3) Hydrophobic moment energy for accounting for the hydrophobic interaction between transmembrane helix and membrane. (4) Aromatic interactions for enhancing the specific interactions between aromatic-aromatic residues. (5) Cation-π interactions for specific non-covalent binding propensities between TM-helices. Finally, models of the lowest free-energy are identified by clustering the simulation trajectories using SPICER 45, which are refined at the atomic-level by a fragment-guided dynamic simulation program FG-MD 43.
We assembled the four trans-membrane domains models in a clock-wise order viewed from extra-cellular side as suggested by previous literature 46, 47. Every single domain model was aligned to the corresponding domain of the recently solved bacterial sodium channel by the structural alignment algorithm TM-align 48. The resultant four domain complete structural model was refined again by FG-MD 43 to accommodate inter-domain steric clashes and improve the model quality.
Plasmid preparation and HEK293 cell transfection
TTX resistant human NaV1.7 wild-type (WT) channel (hNaV1.7r) was used in this study and all other tested mutations were constructed on hNaV1.7r background using Quick Change XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). WT or the mutant channels were transfected into HEK293 cells together with human β-1 and β-2 subunit 49 using the Lipofectamine reagent (Invitrogen, Carlsbad, CA, USA). HEK293 cells were maintained in 1:1 Dulbecco's modified Eagle's media (DMEM)/F-12 supplemented with 10% fetal bovine serum (FBS, Hyclone) in a humidified 5% CO2 incubator at 37 °C. HEK293 cells were seeded onto poly-L-lysine coated glass coverslips (BD Biosciences, San Jose, CA, USA) in a 24 well plate 1 day before transfection. Recording was performed 1 day after transfection.
Voltage-clamp recording of sodium channel in HEK cells
Whole-cell voltage-clamp recordings were performed using the following solution. The extracellular solution contained the following (in mM): 140 NaCl, 3 KCl, 1 MgCl2, 1 CaCl2, 20 Dextrose and 10 HEPES, pH=7.3 with NaOH (320 mOsm adjusted with dextrose). The pipette solution contained the following (in mM): 140 Cs-Fluoride, 10 NaCl, 1.1 EGTA, 10 HEPES, 20 Dextrose, pH=7.3 with CsOH (310 mOsm adjusted with dextrose). Patch pipettes had a resistance of 1–2 mΩ when filled with pipette solution. After achieving the whole-cell recording configuration, the pipette and cell capacitance were manually minimized using the Axopatch 200B (Molecular Devices) compensation circuitry. Series resistance and prediction compensation (80–90%) was applied to reduce the voltage errors. The recorded currents were digitized using pClamp software (version 10) and a digidata 1440A interface (Molecular Devices) at a rate of 50 kHz after passing through a low-pass Bessel filter setting of 10 kHz. Linear leak and residual capacitance artifacts were subtracted out using a P/N method provided with the Clampex software. The recording was initiated after a 5 min equilibration period after breaking in whole-cell configuration. Carbamazepine (CBZ) was purchased from Sigma, dissolved in DMSO. For CBZ experiments, cells were treated with either CBZ (30 μM) or DMSO in incomplete medium without FBS for 30 min before recording and the CBZ or DMSO was maintained in bath solution during the recording as suggested previously 9.
Data analysis was performed using Clampfit (Molecular Devices) and Origin (Microcal Software). To generate activation curves, cells were held at −120 mV and stepped to potentials of −80 to +40 mV in 5 mV increments for 100 ms. Peak inward currents were automatically extracted by Origin and fitted with BoltzIV function to determine the half-activation (V1/2), activation curve slope at half-activation (Z) and reverse potential (ENa) for each recording. Conductance was calculated as G=I/(Vm-ENa) and were normalized by the maximum conductance value and fit with Boltzmann equation. To generate fast inactivation curves, cells were stepped to inactivating potentials from −120 to −40 mV for 500 ms followed by a 40 ms step to −10 mV. Peak inward currents obtained from fast inactivation were normalized by the maximum current amplitude and fitted with a Boltzmann equation.
Mutant cycle analysis
Double mutations (VM/ST and VM/FV) were created using site-directed mutagenesis kit. A total of six constructs (WT, V400M, S241T, F1449V, VM/ST, VM/FV) were expressed at HEK293 cells separately for patch-clamp analysis. The voltage-dependence of activation for these mutations were analyzed and a G–V curve was generated and fitted with Botzmann equation to obtain the voltage at half-activation (V1/2) and the activation curve slope at half-activation (Z). Using V1/2 and Z, the amount of free energy to switch the channel from closed to the open state was calculated as ΔG° (C→O) (kcal/mol) = –ZFV1/2. The additional free energy required for the mutant channel to open relative to the WT channel was calculated as ΔΔG° = Δ(ZFV1/2) = –F(ZmutV1/2mut –ZwtV1/2wt). The magnitude of nonadditivity (or the nonadditive coupling free energy) was calculated as ΣΔG° = ΔΔ(ZFV1/2) = –F[(ZwtV1/2wt–Zmut1V1/2mut1)–(Zmut2V1/2mut2–Zmut1,mut2V1/2 mut1, mut2)] 23–28.
Isolation and transfection of dorsal root ganglion (DRG) neurons
DRG of Sprague-Dawley rat pups (postnatal day 1–5) were isolated and then cultured as described previously 50, 51. Dissected ganglia were placed in ice-cold oxygenated complete saline solution (CSS), which contained the following (in mM): 137 NaCl, 5.3 KCl, 1 MgCl2, 25 sorbitol, 3 CaCl2 and 10 HEPES, pH 7.2. DRGs were then transferred to an oxygenated 37 °C CSS solution containing 1.5 mg/ml collagenase A (Roche Applied Science) and 0.6 mM EDTA and incubated with gentle agitation at 37 °C for 20 min. This solution was exchanged with an oxygenated, 37 °C CSS solution containing 1.5 mg/ml collagenase D (Roche Applied Science), 0.6 mM EDTA, and 30 U/ml papain (Worthington biochemicals) and was incubated with gentle agitation at 37 °C for 20 min. The solution was then aspirated and the ganglia were triturated in DRG media: DMEM/F12 (1:1) with 100 μ/ml penicillin, 0.1 mg/ml streptomycin (Invitrogen), and 10% FBS (Hyclone), which contained 1.5 mg/ml bovine serum albumin (Sigma-Aldrich) and 1.5 mg/ml trypsin inhibitor (Roche Applied Science).
WT, S241T or F1449V mutant channels were transiently transfected into the DRG neurons respectively, along with enhanced green fluorescent protein (GFP), by electroporation with a Nucleofector II (Amaxa) using basic Neuron SCN Nucleofector Kit and program “SCN basic program 6”. The ratio of sodium channel to GFP constructs was 10:1. The transfected neurons were allowed to recover for 5 min at 37 °C in 0.5 ml of Ca++-free DMEM. The cell suspension was then diluted with DRG media containing 1.5 mg/ml bovine serum albumin and 1.5 mg/ml trypsin inhibitor, 80 μl mixture was placed on 12 mm circular poly-d-lysine/laminin precoated coverslips (BD biosciences), and the cells were incubated at 37 °C in 5% CO2 for 30 min. DRG media (1 ml/well), supplemented with 50 ng/ml each of mouse NGF (Alomone Labs) and glial cell line-derived neurotrophic factor (GDNF, Peprotec), was then added to cells. The cells were thus maintained at 37 °C in a 5% CO2 incubator for further experiments.
Current-clamp recording in transfected DRG neurons
Whole-cell configuration was obtained in voltage-clamp mode before starting current-clamp recording. The pipette solution contained (in mM): 140 KCl, 0.5 EGTA, 3 Mg-ATP, 5 HEPES, 30 dextrose, pH7.3 with KOH (310 mOsmol/L). The extracellular bath solution contained (in mM): 140 NaCl, 3 KCl, 2 MgCl2, 2 CaCl2, 15 dextrose, 10 HEPES, pH 7.3 with NaOH (315 mOsmol/L). Recording was performed on transfected nociceptive neurons based on the morphology of small-diameter (20–28 μm) and round cell bodies that also exhibited GFP fluorescence. All recordings were performed 2 days after transfection. For CBZ experiments, neurons were treated with either CBZ (30 μM) or DMSO as mentioned above. Resting membrane potential and seal stability for each neuron was evaluated during a 30 sec long period. Current threshold for action potential generation was determined by a series of 200 ms depolarizing currents in 5 pA increments. Repetitive firing frequency was examined using a series of 1 sec long current steps from 25 to 500 pA in 25 pA increments. The interval between stimuli is 10 sec long. For firing frequency measurement, spikes with overshoot beyond 0 mV were counted as action potentials.
Data analysis
Data were analyzed with Clampfit 9.2 (Molecular Devices) and OriginPro 8.5 (Microcal Software). For statistical analysis, if samples obeyed a normal distribution, student's t-test was used. Non-parametric Mann-Whitney test was used when samples failed the normality test. Data was presented as means± SE. Statistical significance was accepted when P<0.05.
Acknowledgments
We thank Dr. Andrew Tan for his insights and comments on the manuscript. We thank Palak Shah and Bart Toftness for excellent technical assistance. This work was supported by the Medical Research Service and Rehabilitation Research Service, Dept. of Veterans Affairs and the Erythromelalgia Association (S.D.H. and S.G.W.); The Center for Neuroscience and Regeneration Research is a Collaboration of the Paralyzed Veterans of America with Yale University. J.Z and Y.Z are grateful to support from NSF Career Award (1027394) and NIGMS (GM083107 and GM084222).
Footnotes
Disclosure: The authors report no conflicts of interest.
Author contributions: Y.Y., S.D.H., M.E. and S.G.W designed the study. Y.Y. performed the experiments. Y. Y., S.D.H., and M.E. analyzed the data. J.Z. and Y.Z. provided new tools. L.T. provided new reagents. S.D.H. and S.G.W supervised the research. Y.Y., S.D.H., M.E. and S.G.W wrote the paper with inputs from all authors.
References
- 1.Relieving Pain in America: A Blueprint for Transforming Prevention, Care, Education, and Research. Washington (DC): 2011. [PubMed] [Google Scholar]
- 2.Waxman SG. Neurobiology: a channel sets the gain on pain. Nature. 2006;444:831–832. doi: 10.1038/444831a. [DOI] [PubMed] [Google Scholar]
- 3.Dib-Hajj SD, Cummins TR, Black JA, Waxman SG. From genes to pain: Na v 1.7 and human pain disorders. Trends Neurosci. 2007;30:555–563. doi: 10.1016/j.tins.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Dib-Hajj SD, Cummins TR, Black JA, Waxman SG. Sodium channels in normal and pathological pain. Annual review of neuroscience. 2010;33:325–347. doi: 10.1146/annurev-neuro-060909-153234. [DOI] [PubMed] [Google Scholar]
- 5.Minett MS, et al. Distinct Nav1.7-dependent pain sensations require different sets of sensory and sympathetic neurons. Nature communications. 2012;3:791. doi: 10.1038/ncomms1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox JJ, et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–898. doi: 10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drenth JP, Waxman SG. Mutations in sodium-channel gene SCN9A cause a spectrum of human genetic pain disorders. J Clin Invest. 2007;117:3603–3609. doi: 10.1172/JCI33297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han C, et al. Sporadic onset of erythermalgia: a gain-of-function mutation in Nav1.7. Ann Neurol. 2006;59:553–558. doi: 10.1002/ana.20776. [DOI] [PubMed] [Google Scholar]
- 9.Fischer TZ, et al. A novel Nav1.7 mutation producing carbamazepine-responsive erythromelalgia. Ann Neurol. 2009;65:733–741. doi: 10.1002/ana.21678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mantegazza M, Curia G, Biagini G, Ragsdale DS, Avoli M. Voltage-gated sodium channels as therapeutic targets in epilepsy and other neurological disorders. Lancet neurology. 2010;9:413–424. doi: 10.1016/S1474-4422(10)70059-4. [DOI] [PubMed] [Google Scholar]
- 11.Sheets PL, Jackson JO, 2nd, Waxman SG, Dib-Hajj SD, Cummins TR. A Nav1.7 channel mutation associated with hereditary erythromelalgia contributes to neuronal hyperexcitability and displays reduced lidocaine sensitivity. J Physiol. 2007;581:1019–1031. doi: 10.1113/jphysiol.2006.127027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:353–358. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Zhang Y. GPCR-ITASSER: A new composite algorithm for G protein-coupled receptor structure prediction and the application on human genome. Submitted. 2012 [Google Scholar]
- 14.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Research. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Zhang Y. GPCRRD: G protein-coupled receptor spatial restraint database for 3D structure modeling and function annotation. Bioinformatics. 2010;26:3004–3005. doi: 10.1093/bioinformatics/btq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. Journal of molecular biology. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 20.Lampert A, Dib-Hajj SD, Tyrrell L, Waxman SG. Size matters: Erythromelalgia mutation S241T in Nav1.7 alters channel gating. J Biol Chem. 2006;281:36029–36035. doi: 10.1074/jbc.M607637200. [DOI] [PubMed] [Google Scholar]
- 21.Dib-Hajj SD, et al. Gain-of-function mutation in Nav1.7 in familial erythromelalgia induces bursting of sensory neurons. Brain. 2005;128:1847–1854. doi: 10.1093/brain/awh514. [DOI] [PubMed] [Google Scholar]
- 22.Lampert A, et al. A pore-blocking hydrophobic motif at the cytoplasmic aperture of the closed-state Nav1.7 channel is disrupted by the erythromelalgia-associated F1449V mutation. J Biol Chem. 2008;283:24118–24127. doi: 10.1074/jbc.M802900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yifrach O, MacKinnon R. Energetics of pore opening in a voltage-gated K(+) channel. Cell. 2002;111:231–239. doi: 10.1016/s0092-8674(02)01013-9. [DOI] [PubMed] [Google Scholar]
- 24.Ranganathan R, Lewis JH, MacKinnon R. Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron. 1996;16:131–139. doi: 10.1016/s0896-6273(00)80030-6. [DOI] [PubMed] [Google Scholar]
- 25.DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci U S A. 2008;105:15142–15147. doi: 10.1073/pnas.0806486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci U S A. 2009;106:22498–22503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarov-Yarovoy V, et al. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci U S A. 2012;109:E93–102. doi: 10.1073/pnas.1118434109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rauer H, et al. Structure-guided transformation of charybdotoxin yields an analog that selectively targets Ca(2+)-activated over voltage-gated K(+) channels. J Biol Chem. 2000;275:1201–1208. doi: 10.1074/jbc.275.2.1201. [DOI] [PubMed] [Google Scholar]
- 29.Lipkind GM, Fozzard HA. Molecular model of anticonvulsant drug binding to the voltage-gated sodium channel inner pore. Molecular pharmacology. 2010;78:631–638. doi: 10.1124/mol.110.064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harty TP, et al. Na(V)1.7 mutant A863P in erythromelalgia: effects of altered activation and steady-state inactivation on excitability of nociceptive dorsal root ganglion neurons. J Neurosci. 2006;26:12566–12575. doi: 10.1523/JNEUROSCI.3424-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cestele S, et al. Structure and function of the voltage sensor of sodium channels probed by a beta -scorpion toxin. J Biol Chem. 2006;281:21332–21344. doi: 10.1074/jbc.M603814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, et al. Molecular basis and structural insight of vascular K(ATP) channel gating by S-glutathionylation. J Biol Chem. 2011;286:9298–9307. doi: 10.1074/jbc.M110.195123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Shi W, Cui N, Wu Z, Jiang C. Oxidative stress inhibits vascular K(ATP) channels by S-glutathionylation. J Biol Chem. 2010;285:38641–38648. doi: 10.1074/jbc.M110.162578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy A, Xu D, Poisson J, Zhang Y. A protocol for computer-based protein structure and function prediction. Journal of visualized experiments : JoVE. 2011:e3259. doi: 10.3791/3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labro AJ, et al. The S4-S5 linker of KCNQ1 channels forms a structural scaffold with the S6 segment controlling gate closure. The Journal of biological chemistry. 2011;286:717–725. doi: 10.1074/jbc.M110.146977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labro AJ, et al. Kv channel gating requires a compatible S4–S5 linker and bottom part of S6, constrained by non-interacting residues. The Journal of general physiology. 2008;132:667–680. doi: 10.1085/jgp.200810048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choveau FS, et al. KCNQ1 channels voltage dependence through a voltage-dependent binding of the S4–S5 linker to the pore domain. The Journal of biological chemistry. 2011;286:707–716. doi: 10.1074/jbc.M110.146324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jensen MO, et al. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 40.Catterall WA. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 41.Nau C, Wang SY, Strichartz GR, Wang GK. Point mutations at N434 in D1–S6 of mu1 Na(+) channels modulate binding affinity and stereoselectivity of local anesthetic enantiomers. Molecular pharmacology. 1999;56:404–413. doi: 10.1124/mol.56.2.404. [DOI] [PubMed] [Google Scholar]
- 42.Nau C, Wang GK. Interactions of local anesthetics with voltage-gated Na+ channels. The Journal of membrane biology. 2004;201:1–8. doi: 10.1007/s00232-004-0702-y. [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Liang Y, Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19:1784–1795. doi: 10.1016/j.str.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, Zhang Y. A novel side-chain orientation dependent potential derived from random-walk reference state for protein fold selection and structure prediction. PloS one. 2010;5:e15386. doi: 10.1371/journal.pone.0015386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Skolnick J. SPICKER: a clustering approach to identify near-native protein folds. Journal of Computational Chemistry. 2004;25:865–871. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 46.Li RA, et al. Clockwise domain arrangement of the sodium channel revealed by (mu)-conotoxin (GIIIA) docking orientation. J Biol Chem. 2001;276:11072–11077. doi: 10.1074/jbc.M010862200. [DOI] [PubMed] [Google Scholar]
- 47.Dudley SC, Jr., et al. mu-conotoxin GIIIA interactions with the voltage-gated Na(+) channel predict a clockwise arrangement of the domains. The Journal of general physiology. 2000;116:679–690. doi: 10.1085/jgp.116.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Research. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lossin C, Wang DW, Rhodes TH, Vanoye CG, George AL., Jr Molecular basis of an inherited epilepsy. Neuron. 2002;34:877–884. doi: 10.1016/s0896-6273(02)00714-6. [DOI] [PubMed] [Google Scholar]
- 50.Estacion M, et al. NaV1.7 gain-of-function mutations as a continuum: A1632E displays physiological changes associated with erythromelalgia and paroxysmal extreme pain disorder mutations and produces symptoms of both disorders. J Neurosci. 2008;28:11079–11088. doi: 10.1523/JNEUROSCI.3443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han C, et al. Early- and late-onset inherited erythromelalgia: genotype-phenotype correlation. Brain. 2009;132:1711–1722. doi: 10.1093/brain/awp078. [DOI] [PubMed] [Google Scholar]