Abstract
Pancreatic cancer is a highly lethal malignancy with few effective therapies. We performed exome sequencing and copy number analysis to define genomic aberrations in a prospectively accrued clinical cohort (n = 142) of early (stage I and II) sporadic pancreatic ductal adenocarcinoma. Detailed analysis of 99 informative tumours identified substantial heterogeneity with 2,016 non-silent mutations and 1,628 copy-number variations. We define 16 significantly mutated genes, reaffirming known mutations (KRAS, TP53, CDKN2A, SMAD4, MLL3, TGFBR2, ARID1A and SF3B1), and uncover novel mutated genes including additional genes involved in chromatin modification (EPC1 and ARID2), DNA damage repair (ATM) and other mechanisms (ZIM2, MAP2K4, NALCN, SLC16A4 and MAGEA6). Integrative analysis with in vitro functional data and animal models provided supportive evidence for potential roles for these genetic aberrations in carcinogenesis. Pathway-based analysis of recurrently mutated genes recapitulated clustering in core signalling pathways in pancreatic ductal adenocarcinoma, and identified new mutated genes in each pathway. We also identified frequent and diverse somatic aberrations in genes described traditionally as embryonic regulators of axon guidance, particularly SLIT/ROBO signalling, which was also evident in murine Sleeping Beauty transposon-mediated somatic mutagenesis models of pancreatic cancer, providing further supportive evidence for the potential involvement of axon guidance genes in pancreatic carcinogenesis.
Pancreatic cancer is the fourth leading cause of cancer death, with an overall 5-year survival rate of <5%, statistics that have not changed in almost 50 years1. Advances in neoadjuvant and adjuvant chemotherapeutic regimens have resulted in some improvement in outcome, but pancreatectomy remains the single most effective treatment modality for pancreatic cancer, and offers the only potential for cure. Only 20% of patients present with localized, non-metastatic disease which is suitable for resection2. Those who undergo resection and receive adjuvant therapy have a median survival of 12–22 months and a 5-year survival of 20–25%3. Existing systemic therapies are only modestly effective and the median survival for patients with metastatic disease remains 6 months. Genomic characterization of pancreatic ductal adenocarcinoma (PDAC), which accounts for over 90% of pancreatic cancer, has so far focused on targeted polymerase chain reaction (PCR)-based exome sequencing of primary and metastatic lesions propagated as xenografts or cell lines4. A deeper understanding of the underlying molecular pathophysiology of the clinical disease is needed to advance the development of effective therapeutic and early detection strategies.
Clinical cohort
A cohort of 142 consecutive patients with primary operable, untreated PDAC who underwent pancreatectomy with curative intent (pre-operative clinical stages I and II) were recruited, and consent was obtained for genomic sequencing through the Australian Pancreatic Cancer Genome Initiative (APGI), the Baylor College of Medicine Pancreatic Cancer Genome Project and the Ontario Institute for Cancer Research Pancreatic Cancer Genome Study (ABO collaboration) between June 2005 and June 2011 as part of the International Cancer Genome Consortium (ICGC)5. Detailed clinico-pathological characteristics of the cohort demonstrated features typical of resected PDAC with regard to tumour size, grade, lymph node metastasis and survival when compared to multiple retrospectively acquired cohorts6–8, defining the accrued population as representative of the clinical disease in the community (Supplementary Table 1 and Supplementary Fig. 1).
Cellularity and mutation detection
A major challenge in genomic sequencing is the low malignant epithelial cell content of many cancers, which can adversely impact on the sensitivity of mutation detection. Most sequencing studies so far have used samples with >70% tumour cellularity, or cell lines/xenografts4,9. To implement genomic sequencing approaches in clinical practice, it is imperative to efficiently and accurately detect actionable mutations in diagnostic clinical samples. We devised methodologies to overcome the challenges associated with extensive desmoplastic stroma that is characteristic of the majority of PDAC, and these strategies facilitated the discovery of novel molecular mechanisms in the pathophysiology of this disease. The cellularity of each primary sample was estimated through pathological review, deep amplicon-based sequencing of exons 2 and 3 of KRAS (average depth of 1,000×), and single nucleotide polymorphism (SNP) array-based cellularity estimates using a novel algorithm (qpure)10. KRAS mutations were identified in 93% of 142 cases and tumour cellularity ranged from 5% to 85% with a mean of 38% (Supplementary Table 2, Supplementary Figs 2 and 3, and Supplementary Methods).
To inform cellularity thresholds for subsequent analyses, we defined the impact of stromal DNA content on mutation detection by exome capturing and sequencing different mixtures of cancer cell line and matched germline DNA (100%, 80%, 60%, 40%, 20% and 10% cell line DNA) when sequenced to a depth of 70× coverage. Using these data as a standard, the median sensitivity to detect true positives across all samples in the cohort with greater than 20% epithelial cellularity was estimated at 45% (Supplementary Table 3). An informative cohort of 99 patients who had greater than 20% cellularity and/or ≥10 validated somatic mutations was taken forward for further analysis.
Mutation detection and CNV analysis
We performed hybrid-selection-based capture and sequencing of the entire exomes of tumour and matched normal DNA derived from all 142 patients using a combination of capture systems and next-generation sequencing platforms (see Supplementary Methods). The sequence depths at each site (APGI 65×, BCM 104× and OICR 205×) were adopted to ensure suitable sensitivity across their respective cohorts (Supplementary Table 3). In the informative 99 samples, we detected 2,627 high-confidence mutations, 2,016 of which were non-silent (Table 1). A total of 1,502 of these events (1,350 non-silent) were independently validated via an orthogonal sequencing method (see Supplementary Methods). The average number of mutations detected per patient was 26 (range 1–116), consistent with the expected sensitivity based on cellularity estimates and previous studies4,11 (Supplementary Table 2). We confirmed the high prevalence of genetic aberrations known to be important in PDAC and observed mutations in 38 of the 79 genes (48% overlap) that occurred more than once previously reported by ref. 4, and 186 of all 998 mutated genes (19% overlap) in that study. We also defined a large number of novel mutations (1,456 genes), most of which occurred at low frequency (see Supplementary Tables 4– 6 and Supplementary Fig. 4 for detailed comparisons). The observed transversion/transition rates in the cohort correlated closely with those previously reported in PDAC cell lines and xenografts (Supplementary Table 7).
Table 1.
Mutations in pancreatic ductal adenocarcinoma (n = 99)
| Mutation class | Total |
|---|---|
| Missense | 1,684 |
| Nonsense | 99 |
| Splice site | 89 |
| Insertion/deletion | 144 |
| Non-silent | 2,016 |
| Silent | 611 |
Significant mutated gene analysis12 of genes with non-silent mutations that occurred in 2 or more individual cancers identified 16 genes in the top 20 mutated genes in 2 of 3 stringent analytical approaches (Table 2, Supplementary Table 8 and Supplementary Methods) and reaffirmed the importance of mutations known to occur in PDAC: KRAS, TP53, CDKN2A, SMAD4, MLL3, TGFBR2, ARID1A and SF3B1. Novel significantly mutated genes included additional genes involved in chromatin modification (EPC1 and ARID2) and ATM, recently implicated as a PDAC susceptibility gene through bi-allelic inactivation in a case of familial PDAC (germline mutation and loss of heterozygosity (LOH) in the tumour)13. Aberrations of ATM occurred in 8% of our cohort (mutated in 5%, LOH or loss in 5%, with two patients exhibiting both mutation and LOH or loss) and mutations detected in other genes not previously reported: ZIM2, MAP2K4, NALCN, SLC16A4 and MAGEA6 (Table 2). GISTIC2.014 identified 30 genes affected by copy-number alterations (Q value <0.0001) and included losses of CDKN2A and SMAD4 (Supplementary Table 4).
Table 2.
Significantly mutated genes in pancreatic ductal adenocarcinoma
| Gene symbol | Gene name and protein function | SB mutagenesis* | shRNA† |
|---|---|---|---|
| KRAS | Oncogene; GTPase; activation of MAPK activity | Yes | Yes |
| TP53 | Tumour suppressor p53; DNA damage response | – | Yes |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A; G1/S transition of mitotic cell cycle; tumour suppressor | Yes | – |
| SMAD4 | Mothers against decapentaplegic homologue 4; BMP signalling pathway | Yes | Yes |
| MLL3 | Myeloid/lymphoid or mixed-lineage leukaemia protein 3; DNA binding; regulation of transcription | Yes | Yes |
| TGFBR2 | Transforming growth factor-β receptor type II; regulation of growth | Yes | – |
| ARID1A | AT-rich interactive domain-containing protein 1A; SWI/SNF complex; chromatin modification | Yes | Yes |
| ARID2 | AT-rich interactive domain-containing protein 2; chromatin modification | Yes | – |
| EPC1 | Enhancer of polycomb homologue 1; histone acetylation | Yes | – |
| ATM | Ataxia telangiectasia mutated; DNA damage response | – | Yes |
| SF3B1 | Splicing factor 3B subunit 1; nuclear mRNA splicing | – | Yes |
| ZIM2 | Zinc finger imprinted 2; regulation of transcription | – | Yes |
| MAP2K4 | Dual specificity mitogen-activated protein kinase kinase 4; Toll-like receptor signalling pathway | Yes | Yes |
| NALCN | Sodium leak channel non-selective protein; sodium channel activity | – | Yes |
| SLC16A4 | Solute carrier family 16 member 4; monocarboxylate transporter | – | Yes |
| MAGEA6 | Melanoma-associated antigen 6; protein binding | – | ND |
Pathways in pancreatic cancer
To better understand potential underlying mechanisms of importance in PDAC, we performed a series of pathway analyses using genes that were recurrently mutated in two or more individuals using GeneGO15, and identified mechanisms known to be importantin cancer: G1/S checkpoint machinery (P = 1.49 × 10–3), apoptosis (P = 1.32 × 10–4), regulation of angiogenesis (P = 7.72 × 10–4) and TGF-β signalling (P = 9.50 × 10–4). Interestingly, novel gene signatures were enriched in our cohort, including axon guidance (P = 5.30 × 10–5) (Supplementary Table 9). The inclusion of mutation data for 24 cases from ref. 4 strengthened the association of axon guidance (P = 3.3 × 10–7), and was more evident still when all mutated genes in our data set were used as input (P = 4.67 × 10–8).
Functional relevance of genomic events
Differentiating somatic driving events of carcinogenesis from passenger mutations is a major challenge in cancer genomics16. Despite significant advances in computational algorithms, experimental evidence of functional relevance is paramount. We used data from three published experimental biological screens to infer functional consequences for the individual genomic events and the pathways we identified. These included data from two independent Sleeping Beauty transposon (SB) mutagenesis screens in Kras transgenic mouse models of PDAC17,18 and an in vitro short hairpin RNA (shRNA) screen which examined the consequences of downregulating 11,194 putative cancer genes on survival in a panel of 102 cell lines (13 pancreatic)19 (Supplementary Methods and Supplementary Figs 5 and 6). Data from these screens confirmed the functional importance of KRAS, TP53, CDKN2A and SMAD4 mutations and attributed potential functional relevance to most significantly mutated genes—MLL3, TGFBR2, SF3B1, EPC1, ARID1A, ARID2, MAP2K4, ATM, NALCN, ZIM2, SLC16A4 (Table 2)—and many genes mutated at low frequency (Supplementary Table 4).
Pathway analysis of high confidence insertions in SB transposon mutagenesis screens demonstrated enrichment for axon guidance genes (P = 1.6 × 10–3), providing independent supportive evidence for a potential role in the pathogenesis of PDAC. In these screens, 14 genes involved in axon guidance pathways were detected (5 genes common to both). In addition, a further 32 genes were mutated in at least one SB pancreatic tumour (out of 21) but did not meet the significance threshold with the stringent analyses that were applied17 (Supplementary Tables 10 and 11).
Axon guidance pathway genes
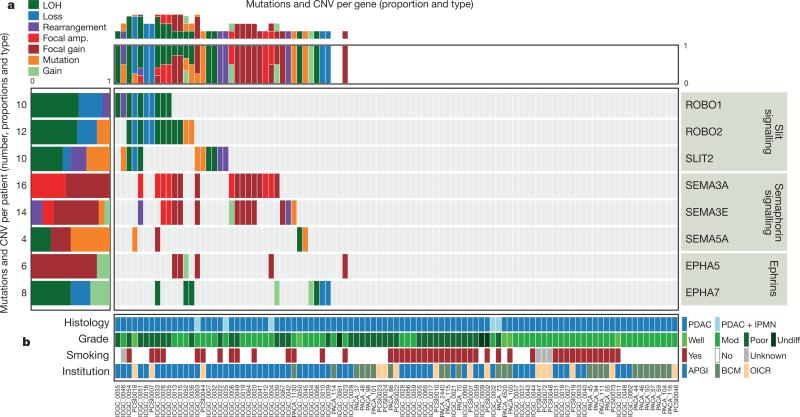
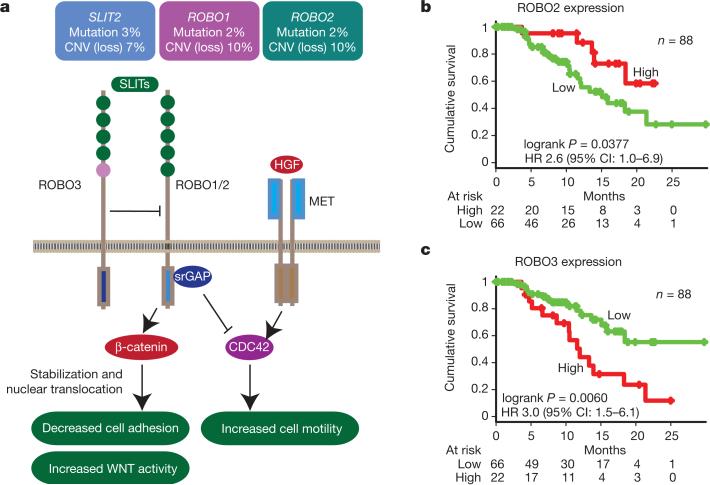
The class of genes traditionally described for their roles in axon guidance (semaphorins, slits, netrins and ephrins) are important regulators of normal neuronal migration and positioning during embryonic development. More recently, they have been implicated in cancer cell growth, survival, invasion and angiogenesis20; however, the incidence of aberrations in these genes in cancer is largely unknown. We identified recurrent mutations and copy-number variations (CNVs) of axon guidance pathway genes in this cohort (Fig. 1 and Supplementary Table 4): SLIT2 and ROBO2 mutations were present in 5% of patients, with focal copy-number losses of ROBO1, and SLIT2 detected by GISTIC2.0 analysis and confirmed by manual review, potentially having an impact on a further 15% of the cohort, suggesting that aberrant SLIT/ROBO signalling is potentially a common feature of PDAC (Figs 1 and 2). In addition, we used targeted PCR-based sequencing of an additional 30 cases of PDAC for axon guidance genes and identified mutations in ROBO1 in two patients and additional mutations in SLIT2 and ROBO2 (one patient each). Low mRNA expression of the ROBO2 receptor was associated with poor patient survival (P = 0.04). Furthermore, high mRNA expression of ROBO3, a known inhibitor of ROBO2 signalling21, demonstrated an appropriate reciprocal inverse association with poor survival (P < 0.006) (Fig. 2).
Figure 1. Mutations and copy number variation in axon guidance genes.
Axon guidance pathway genes with recurrent mutations and/or copy-number changes defined by GISTIC2.0 analysis (Q < 0.2), and manually reviewed for focal alterations. a, SNV and CNV frequency per patient with gene-centric summary (left) and patient-centric summary (top); numbers of patients with mutations and proportion of each event are presented. Please see Supplementary Table 4 for further details. b, Clinico-pathological variables for individual patients. APGI, Australian Pancreatic Cancer Genome Initiative; BCM, Baylor College of Medicine; IPMN, intraductal papillary mucinous neoplasm; Mod, moderately differentiated; OICR, Ontario Institute for Cancer Research; PDAC, pancreatic ductal adenocarcinoma; Undiff, undifferentiated.
Figure 2. SLIT/ROBO signalling in pancreatic ductal adenocarcinoma.
a, SLIT/ROBO signalling normally enhances β-catenin complex formation with E-cadherin and suppresses WNT signalling activity. Loss of ROBO1/2 signalling promotes stabilization of β-catenin, which decreases E-cadherin complex formation and cell adhesion and augments WNT signalling activity through increased nuclear translocation of β-catenin. In addition, SLIT/ROBO signalling can downregulate MET signalling activity; loss of ROBO signalling activity promotes MET signalling downstream and may have an impact on therapeutic strategies aimed at inhibiting MET activity at the receptor level. (Adapted from ref. 20.) Aberrations in SLIT2 and/or ROBO1/2 affected 23% of patients (6% mutated with 1 patient showing mutations in both SLIT2 and ROBO2), with 18% demonstrating CNV corresponding to loss of the gene. b, c, High expression of SLIT receptor ROBO2 was associated with a better prognosis (b), and high expression of ROBO3, an inhibitor of ROBO2, showed an inverse relationship, with high levels associated with poor survival (c). HR, hazard ratio.
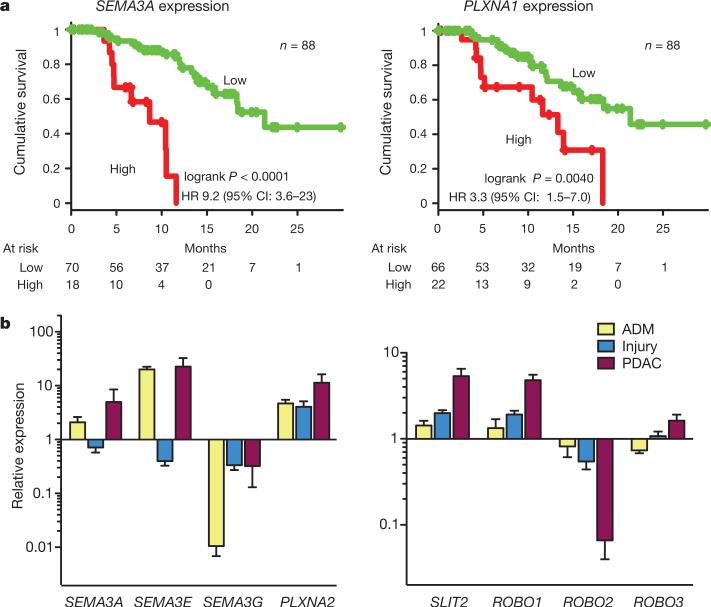
Class 3 semaphorins (SEMA3A and SEMA3E) exhibited significant amplification in 18% of patients and an additional 3% harboured mutations (Fig. 1). Semaphorins signal through neuropilin and plexin receptors to elicit their effects22. SEMA3A amplification correlated with high mRNA expression on microarray (P = 0.03), and high mRNA expression of SEMA3A and PLXNA1, another molecule central to semaphorin signalling, were both associated with poor patient survival on univariate analysis (Fig. 3a), and were independently prognostic on multivariate analyses with clinico-pathological variables (Supplementary Table 12).
Figure 3. Axon guidance genes in human and murine pancreatic ductal adenocarcinoma.
a, Kaplan–Meier survival curves showing co-segregation of aberrant expression of components of semaphorin signalling with outcome. Amplification at SEMA3A and PLXNA1 loci was associated with high mRNA expression and both are independent poor prognostic factors. b, Quantitative RT–PCR for components of semaphorin and SLIT/ROBO signalling in murine models of early (acinar-to-ductal metaplasia (ADM) and pancreatic injury) and established PDAC in genetically engineered mice with a Pdx1-promoter-driven activating mutation of Kras and mutant Tp53 allele (Pdx1-Cre; LSL-KrasG12D; LSL-Trp53R172H). Error bars represent standard error of the mean (see Supplementary Table 15 for details).
To elucidate further the significance of the observed CNV events, we reviewed copy number, CNV segment size and changes in heterozygosity of axon guidance genes in a recent independent CNV analysis of 39 fine-needle aspiration biopsies23 and the 16 PDAC cell lines in the CONAN database (http://www.sanger.ac.uk/cosmic)24. Overall, the predominant changes recapitulated our studies, showing frequent focal losses within genes involved in SLIT/ROBO signalling, and gains in genes involved in canonical semaphorin signalling (Supplementary Tables 4, 13 and 14).
To assess whether dysregulation of axon guidance genes is associated with early neoplastic transformation, as are many developmental signalling pathways, we examined mRNA expression in murine models of early pancreatic carcinogenesis (in vitro acinar-to-ductal metaplasia and in vivo pancreatic injury). Expression levels of components of SLIT/ROBO and semaphorin signalling changed progressively from normal pancreas, through acinar-to-ductal metaplasia and pancreatic injury to genetically engineered murine PDAC, indicating a role for the dysregulation of these axon guidance genes in tumour initiation and progression (Fig. 3b and Supplementary Table 15).
Discussion
We devised methodologies to optimize mutation detection for clinical samples in a large cohort of patients and reaffirm known mutations in PDAC, better define their prevalence in a large cohort of early PDAC, and identify potential novel drivers in this disease. Somatic mutations in ATM were identified in a significant proportion of patients (8%), highlighting the importance of BRCA-mediated DNA damage repair mechanisms in sporadic PDAC as well as familial disease13. Previously, mutations in individual genes involved in chromatin remodelling such as ARID1A25 have been described and additional genes identified here (EPC1 and ARID2) infer that chromatin remodelling may have an important role in PDAC, along with other cancer types26.
Novel mutations in genes traditionally described for their roles in axon guidance were also observed by a combination of genomic data and supportive experimental evidence from independent murine SB mutagenesis screens. Axon guidance is integral to organogenesis, regeneration, wound healing and other basic cellular processes22,27. The widespread genomic aberrations observed here in axon guidance genes suggests that they may have a role in PDAC, joining mounting evidence in other cancers20,28, including a recent report demonstrating ROBO2 mutations in liver-fluke-associated cholangiocarcinoma29. In addition, evidence from cancers of the lung, breast, kidney and cervix implicate aberrant SLIT/ROBO signalling in carcinogenesis20; Robo1 knockout mice develop bronchial hyperplasia and focal dysplasia, and inactivation of Slit2 and Slit3 leads to the development of hyperplastic disorganized lesions in the breast20. Upregulation of MET and WNT signalling have important roles in PDAC, and recent data indicate that SLIT/ROBO signalling modulates MET and WNT signalling activity through CDC42 and β-catenin, respectively20. Loss of SLIT/ROBO signalling can potentially be an alternative mechanism for deregulating these pathways downstream of their receptors, and in addition could influence the activity of inhibitors that target these upstream components, for example, MET inhibitors (Fig. 2).
Class 3 semaphorins are the only secreted semaphorins in vertebrates. They regulate cell growth, invasiveness and angiogenesis, and are highly expressed in metastatic cells in many cancer types30,31. Although aberrant semaphorin signalling in cancer seems to be organ specific32, our finding that high expression of SEMA3A and its receptor PLXNA1 co-segregates with poor patient survival is supported by a previous study that reported this association and also demonstrated promotion of invasiveness of PDAC cell lines by SEMA3A31. Therapeutics targeting molecules involved in axon guidance have been developed as potential strategies to facilitate neuronal regeneration after injury33, but are yet to be assessed for their role in cancer treatment.
As illustrated here, global genomic analysis of large, well-annotated and clinically homogeneous cohorts of patients can identify mechanisms that are common among genomically diverse cancers, and will be pivotal in the development of novel therapeutic strategies that are guided by the determination of the molecular phenotype of individual patients34. Future work will be required to determine which key components, when damaged, drive the disease, and these mechanisms will need to be assessed in molecularly well-characterized preclinical models35. The potential therapeutic strategies identified will then require testing in appropriate clinical trials that are specifically designed to target subsets of patients stratified according to well-defined molecular markers36,37.
METHODS SUMMARY
Sample acquisition and processing
Samples used were prospectively acquired and restricted to primary operable, non-pretreated pancreatic ductal adenocarcinoma. Representative sections were reviewed independently by at least one additional pathologist with specific expertise in pancreatic diseases. Samples either had full face frozen sectioning performed in optimal cutting temperature (OCT) medium, or the ends excised and processed in formalin to verify the presence of carcinoma in the sample to be sequenced and to estimate the percentage of malignant epithelial nuclei in the sample relative to stromal nuclei. Macrodissection was performed if required to excise areas that did not contain malignant epithelium.
Sequencing
Cellularity of each tumour sample was estimated with pathology review, deep sequencing of KRAS and a method developed using genome-wide SNP array data (qpure10). Exon capture was performed using the SureSelect II or Nimblegen capture methods and paired-end sequenced on the SOLiD (v4) or GAII/HiSeq platforms. Somatic mutations were called and then verified on the Ion Torrent Personal Genome Machine (Life Technologies Corporation) and 454 (Hoffman–La Roche Limited).
Analysis
Significantly mutated genes were identified using the Genome MuSiC package12. DNA copy number analyses were performed using the Illumina HumanOmni1 Quad genotyping arrays and GenoCN software. Recurrent and significant copy number changes were identified using GISTIC2.014. Functional enrichment of gene categories was assessed using the Metacore package (Thomson-Reuters Corporation) and the MSigDB v3.0 database38. All sample information and data for mutation, copy number and expression analyses were submitted to the ICGC DCC at http://dcc.icgc.org/. A complete description of the materials and methods including approvals for human research and animal experimentation is provided in Supplementary Information.
Supplementary Material
Acknowledgements
This paper is dedicated to Robert L. Sutherland who died on 10 October 2012 of pancreatic cancer. We would like to thank C. Axford, D. Gwynne, M.-A. Brancato, S. Rowe, M. Thomas, S. Simpson and G. Hammond for central coordination of the Australian Pancreatic Cancer Genome Initiative, data management and quality control; M. Martyn-Smith, L. Braatvedt, H. Tang, V. Papangelis and M. Beilin for biospecimen acquisition; and W. Waterson, J. Shepperd, E. Campbell and E. Glasov for their efforts at the Queensland Centre for Medical Genomics. We also thank M. B. Hodgin, M. Debeljak and D. Trusty for technical assistance at Johns Hopkins University, and J. Lau, M. Karaus, K. Rabe, L. Zhang and T. Smyrk at the Mayo Clinic. We acknowledge the following funding support: National Health and Medical Research Council of Australia (NHMRC; 631701, 535903, 427601, 535914); Australian Government: Department of Innovation, Industry, Science, Research and Tertiary Education (DIISRTE); Australian Cancer Research Foundation (ACRF); Queensland Government (NIRAP); University of Queensland; Cancer Council NSW (SRP06-01; ICGC09-01; SRP11-01); Cancer Institute NSW (06/ECF/1-24, 09/CDF/2-40, 07/CDF/1-03, 10/CRF/1-01, 08/RSA/1-15, 07/CDF/1-28, 10/CDF/2-26,10/FRL/2-03, 06/RSA/1-05, 09/RIG/1-02, 10/TPG/1-04, 11/REG/1-10, 11/CDF/3-26); Garvan Institute of Medical Research; Avner Nahmani Pancreatic Cancer Research Foundation; R.T. Hall Trust; Petre Foundation; Jane Hemstritch in memory of Philip Hemstritch; Gastroenterological Society of Australia (GESA); American Association for Cancer Research (AACR) Landon Foundation – INNOVATOR Award; Royal Australasian College of Surgeons (RACS); Royal Australasian College of Physicians (RACP); Royal College of Pathologists of Australasia (RCPA); HGSC-BCM: NHGRI U54 HG003273; CPRIT grant RP101353-P7 (Tumor Banking for Genomic Research and Clinical Translation Site 1); The Ontario Institute for Cancer Research; The Ontario Ministry of Economic Development andInnovation; Canada Foundationfor Innovation; Pancreatic Cancer Genetic Epidemiology Consortium, NIH grant R01 CA97075; The Agency for Science, Technology, and Research (Singapore); University of Verona and Italian Ministry of University (FIRB RBAP10AHJB), Rome, Italy; Cancer Research UK; Wellcome Trust; CPRIT (Cancer Prevention Research Institute of Texas); NIH P50CA062924 (SPORE) and P01CA134292 (PPG); The Sol Goldman Pancreatic Cancer Research Center; NCI grant P50 CA102701 (Mayo Clinic SPORE in Pancreatic Cancer) and NCI grant R01 CA97075 (Pancreatic Cancer Genetic Epidemiology Consortium); NIH SPORE grant 2P50CA101955 (UMN/UAB), and AIRC (Associazione Italiana Ricerca sul Cancro) 5xmille grant 12182, Italy.
Australian Pancreatic Cancer Genome Initiative
The Kinghorn Cancer Centre, Garvan Institute of Medical Research Andrew V. Biankin1, Amber L. Johns1, Amanda Mawson1, David K. Chang1, Christopher J. Scarlett1, Mary-Anne L. Brancato1, Sarah J. Rowe1, Skye L. Simpson1, Mona Martyn-Smith1, Michelle T. Thomas1, Lorraine A. Chantrill1, Venessa T. Chin1, Angela Chou1, Mark J. Cowley1, Jeremy L. Humphris1, Marc D. Jones1, R. Scott Mead1, Adnan M. Nagrial1, Marina Pajic1, Jessica Pettit1, Mark Pinese1, Ilse Rooman1, Jianmin Wu1, Jiang Tao1, Renee DiPietro1, Clare Watson1, Rachel Wong1, Andreia V. Pinho1, Marc Giry-Laterriere1, Roger J. Daly1, Elizabeth A. Musgrove1, Robert L. Sutherland1; Queensland Center for Medical Genomics, Institute for Molecular Bioscience Sean M. Grimmond2, Nicola Waddell2, Karin S. Kassahn2, David K. Miller2, Peter J. Wilson2, Ann-Marie Patch2, Sarah Song2, Ivon Harliwong2, Senel Idrisoglu2, Craig Nourse2, Ehsan Nourbakhsh2, Suzanne Manning2, Shivangi Wani2, Milena Gongora2, Matthew Anderson2, Oliver Holmes2, Conrad Leonard2, Darrin Taylor2, Scott Wood2, Qinying Xu2, Katia Nones2, J. Lynn Fink2, Angelika Christ2, Tim Bruxner2, Nicole Cloonan2, Felicity Newell2, John V. Pearson2; Royal North Shore Hospital Jaswinder S. Samra3, Anthony J. Gill3, Nick Pavlakis3, Alex Guminski3, Christopher Toon3; Bankstown Hospital Andrew V. Biankin4, Ray Asghari4, Neil D. Merrett4, David K. Chang4, Darren A. Pavey4, Amitabha Das4; Liverpool Hospital Peter H. Cosman5, Kasim Ismail5, Chelsie O'Connor5; Westmead Hospital Vincent W. Lam6, Duncan McLeod6, Henry C. Pleass6, Arthur Richardson6, Virginia James6; Royal Prince Alfred Hospital James G. Kench7, Caroline L. Cooper7, David Joseph7, Charbel Sandroussi7, Michael Crawford7, James Gallagher7; Fremantle Hospital Michael Texler8, Cindy Forrest8, Andrew Laycock8, Krishna P. Epari8, Mo Ballal8, David R. Fletcher8, Sanjay Mukhedkar8; Sir Charles Gairdner Hospital Nigel A. Spry9, Bastiaan DeBoer9, Ming Chai9; St John of God Healthcare Nikolajs Zeps10, Maria Beilin10, Kynan Feeney10; Royal Adelaide Hospital Nam Q. Nguyen11, Andrew R. Ruszkiewicz11, Chris Worthley11, Chuan P. Tan11, Tamara Debrencini11; Flinders Medical Centre John Chen12, Mark E. Brooke-Smith12, Virginia Papangelis12; Greenslopes Private Hospital Henry Tang13, Andrew P. Barbour13; Envoi Pathology Andrew D. Clouston14, Patrick Martin14; Princess Alexandria Hospital Thomas J. O'Rourke15, Amy Chiang15, Jonathan W. Fawcett15, Kellee Slater15, Shinn Yeung15, Michael Hatzifotis15, Peter Hodgkinson15; Austin Hospital Christopher Christophi16, Mehrdad Nikfarjam16, Angela Mountain16; Victorian Cancer Biobank17; Johns Hopkins Medical Institutes James R. Eshleman18, Ralph H. Hruban18, Anirban Maitra18, Christine A. Iacobuzio-Donahue18, Richard D. Schulick18, Christopher L. Wolfgang18, Richard A. Morgan18, Mary B. Hodgin18; ARC-NET Center for Applied Research on Cancer Aldo Scarpa19, Rita T. Lawlor19, Paola Capelli19, Stefania Beghelli19, Vincenzo Corbo19, Maria Scardoni19, Paolo Pederzoli19, Giampaolo Tortora19, Claudio Bassi19; University of California, San Francisco Margaret A. Tempero20
1The Kinghorn Cancer Centre, Cancer Research Program, Garvan Institute of Medical Research, 370 Victoria Street, Darlinghurst, Sydney, New South Wales 2010, Australia. 2Queensland Center for Medical Genomics, Institute for Molecular Bioscience, University of Queensland, St Lucia, Queensland 4072, Australia. 3Royal North Shore Hospital, Westbourne Street, St Leonards, New South Wales 2065, Australia. 4Bankstown Hospital, Eldridge Road, Bankstown, New South Wales 2200, Australia. 5Liverpool Hospital, Elizabeth Street, Liverpool, New South Wales 2170, Australia. 6Westmead Hospital, Hawkesbury and Darcy Roads, Westmead, New South Wales 2145, Australia. 7Royal Prince Alfred Hospital, Missenden Road, Camperdown, New South Wales 2050, Australia. 8Fremantle Hospital, Alma Street, Fremantle, Western Australia 6959, Australia. 9Sir Charles Gairdner Hospital, Hospital Avenue, Nedlands, Western Australia 6009, Australia. 10St John of God Healthcare, 12 Salvado Road, Subiaco, Western Australia 6008, Australia. 11Royal Adelaide Hospital, North Terrace, Adelaide, South Australia 5000, Australia. 12Flinders Medical Centre, Flinders Drive, Bedford Park, South Australia 5042, Australia. 13Greenslopes Private Hospital, Newdegate Street, Greenslopes, Queensland 4120, Australia. 14Envoi Pathology, 1/49 Butterfield Street, Herston, Queensland 4006, Australia. 15Princess Alexandria Hospital, Cornwall Street & Ipswich Road, Woolloongabba, Queensland 4102, Australia. 16Austin Hospital, 145 Studley Road, Heidelberg, Victoria 3084, Australia. 17Victorian Cancer Biobank, 1 Rathdowne Street, Carlton, Victoria 3053, Australia. 18Johns Hopkins Medical Institutes, 600 North Wolfe Street, Baltimore, Maryland 21287, USA. 19ARC-NET Center for Applied Research on Cancer, University of Verona, Via dell'Artigliere, 19 37129 Verona 37134, Italy. 20University of California, San Francisco, 1600 Divisadero Street, San Francisco, California 94115, USA.
Footnotes
Author Contributions The research network comprising the Australian Pancreatic Cancer Genome Initiative, the Baylor College of Medicine Cancer Genome Project and the Ontario Institute for Cancer Research Pancreatic Cancer Genome Study (ABO collaboration) contributed collectively to this study as part of the International Cancer Genome Consortium. Biospecimens werecollected at affiliated hospitals and processed at each biospecimen core resource centre. Data generation and analyses were performed bythe genomesequencing centres, cancer genomecharacterizationcentres and genome data analysis centres. Investigator contributions are as follows: S.M.G., A.V.B., J.V.P., R.L.S., R.A.G., D.A.W., M.-C.G., J.D.M., L.D.S and T.J.H. (project leaders); A.V.B., S.M.G. and R.L.S. (writing team); A.L.J., J.V.P., P.J.W., J.L.F., C.L., M.A., O.H., J.G.R., D.T., C.X., S.Wo., F.N., S.So., G.K. and W.K. (bioinformatics/databases); D.K.M., I.H., S.I., C.N., S.M., A.Chr., T.Br., S.Wa., E.N., B.B.G., D.M.M., Y.Q.W., Y.H., L.R.L., H.D., R. E. D., R.S.M. and M.W. (sequencing); N.W., K.S.K., J.V.P., A.-M.P., K.N., N.C., M.G., P.J.W., M.J.C., M.P., J.W., N.K., F.Z., J.D., K.C., C.J.B., L.B.M., D.P., R.E.D., R.D.B., T.Be. and C.K.Y. (mutation, copy number and gene expression analysis); A.L.J., D.K.C., M.D.J., M.P., C.J.S., E.K.C., C.T., A.M.N., E.S.H., V.T.C., L.A.C., E.N., J.S.S., J.L.H., C.T., N.B. and M.Sc. (sample processing and quality control); A.J.G., J.G.K., R.H.H., C.A.I.-D., A.Cho., A.Mai., J.R.E., P.C. and A.S. (pathology assessment); J.W., M.J.C., M.P., C.K.Y. and mutation analysis team (network/pathway analysis and functional data integration); K.M.M., N.A.J., N.G.C., P.A.P.-M., D.J.A., D.A.L., L.F.A.W., A.G.R., D.A.T., R.J.D., I.R., A.V.P., E.A.M., R.L.S., R.H.H. and A.Maw. (functional screens); E.N.,A.L.J., J.S.S., A.J.G., J.G.K., N.D.M., A.B., K.E.,N.Q.N., N.Z., W.E.F.,F.C.B., S.E.H., G.E.A., L.M., L.T., M.Sam., K.B., A.B., D.P., A.P., N.B., R.D.B., R.E.D., C.Y., S.Se., N.O., D.M., M-S.T., P.A.S., G.M.P., S.G., L.D.S., C.A.I.-D., R.D.S., C.L.W., R.A.M., R.T.L., S.B., V.C., M.Sca., C.B., M.A.T., G.T., A.S. and J.R.E. (sample collection and clinical annotation); D.K.C., M.P., C.J.S., E.S.H., J.A.L., R.J.D., A.V.P. and I.R. (preclinical models).
Author Information BAM files and associated metadata in XML format have been uploaded to the European Genome-phenome Archive (EGA; http://www.ebi.ac.uk/ega) under accession numbers EGAS00001000154 and EGAS00001000343. Additional sequence data is located at dbGAP accession number phs000516.v1.p1. Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper.
The authors declare no competing financial interests.
Supplementary Information is available in the online version of the paper.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Butturini G, et al. Influence of resection margins and treatment on survival in patients with pancreatic cancer: meta-analysis of randomized controlled trials. Arch. Surg. 2008;143:75–83. doi: 10.1001/archsurg.2007.17. [DOI] [PubMed] [Google Scholar]
- 3.Neoptolemos JP, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. J. Am. Med. Assoc. 2010;304:1073–1081. doi: 10.1001/jama.2010.1275. [DOI] [PubMed] [Google Scholar]
- 4.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Cancer Genome Consortium International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biankin AV, et al. Expression of S100A2 calcium-binding protein predicts response to pancreatectomy for pancreatic cancer. Gastroenterology. 2009;137:558–568. doi: 10.1053/j.gastro.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Chang DK, et al. Margin clearance and outcome in resected pancreatic cancer. J. Clin. Oncol. 2009;27:2855–2862. doi: 10.1200/JCO.2008.20.5104. [DOI] [PubMed] [Google Scholar]
- 8.Jamieson NB, et al. A prospective comparison of the prognostic value of tumor- and patient-related factors in patients undergoing potentially curative surgery for pancreatic ductal adenocarcinoma. Ann. Surg. Oncol. 2011;18:2318–2328. doi: 10.1245/s10434-011-1560-3. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, et al. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res. 2012;22:208–219. doi: 10.1101/gr.123109.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song S, et al. qpure: A tool to estimate tumor cellularity from genome-wide single-nucleotide polymorphism profiles. PLoS ONE. 2012;7:e45835. doi: 10.1371/journal.pone.0045835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuel N, Hudson TJ. The molecular and cellular heterogeneity of pancreatic ductal adenocarcinoma. Nature Rev. Gastroenterol. Hepatol. 2012;9:77–87. doi: 10.1038/nrgastro.2011.215. [DOI] [PubMed] [Google Scholar]
- 12.Dees ND, et al. MuSiC: Identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts NJ, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2011;2:41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun W, et al. Integrated study of copy number states and genotype calls using high-density SNP arrays. Nucleic Acids Res. 2009;37:5365–5377. doi: 10.1093/nar/gkp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann KM, et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc. Natl Acad. Sci. USA. 2012;109:5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez-Mancera PA, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines revealslineage-specific dependencies inovariancancer. Proc. Natl Acad. Sci. USA. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehlen P, Delloye-Bourgeois C, Chedotal A. Novel roles for Slits and netrins: axon guidance cues as anticancer targets? Nature Rev. Cancer. 2011;11:188–197. doi: 10.1038/nrc3005. [DOI] [PubMed] [Google Scholar]
- 21.Sabatier C, et al. The divergent Robo family protein rig-1/Robo3 is a negative regulator of slit responsiveness required for midline crossing by commissural axons. Cell. 2004;117:157–169. doi: 10.1016/s0092-8674(04)00303-4. [DOI] [PubMed] [Google Scholar]
- 22.Trusolino L, Comoglio PM. Scatter-factor and semaphorin receptors: cell signalling for invasive growth. Nature Rev. Cancer. 2002;2:289–300. doi: 10.1038/nrc779. [DOI] [PubMed] [Google Scholar]
- 23.Birnbaum DJ, et al. Genome profiling of pancreatic adenocarcinoma. Genes Chromosom. Cancer. 2011;50:456–465. doi: 10.1002/gcc.20870. [DOI] [PubMed] [Google Scholar]
- 24.Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones S, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum. Mutat. 2012;33:100–103. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comoglio PM, Trusolino L. Invasive growth: from development to metastasis. J. Clin. Invest. 2002;109:857–862. doi: 10.1172/JCI15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chédotal A, Kerjan G, Moreau-Fauvarque C. The brain within the tumor: new roles for axon guidance molecules in cancers. Cell Death Differ. 2005;12:1044–1056. doi: 10.1038/sj.cdd.4401707. [DOI] [PubMed] [Google Scholar]
- 29.Ong CK, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nature Genet. 2012;44:690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 30.Capparuccia L, Tamagnone L. Semaphorin signaling in cancer cells and in cells of the tumor microenvironment–two sides of a coin. J. Cell Sci. 2009;122:1723–1736. doi: 10.1242/jcs.030197. [DOI] [PubMed] [Google Scholar]
- 31.Müller MW, et al. Association of axon guidance factor semaphorin 3A with poor outcome in pancreatic cancer. Int. J. Cancer. 2007;121:2421–2433. doi: 10.1002/ijc.22949. [DOI] [PubMed] [Google Scholar]
- 32.Ellis LM. The role of neuropilins in cancer. Mol. Cancer Ther. 2006;5:1099–1107. doi: 10.1158/1535-7163.MCT-05-0538. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi K, et al. In vitro and in vivo characterization of a novel semaphorin 3A inhibitor, SM-216289 or xanthofulvin. J. Biol. Chem. 2003;278:42985–42991. doi: 10.1074/jbc.M302395200. [DOI] [PubMed] [Google Scholar]
- 34.Cao Y, DePinho RA, Ernst M, Vousden K. Cancer research: past, present and future. Nature Rev. Cancer. 2011;11:749–754. doi: 10.1038/nrc3138. [DOI] [PubMed] [Google Scholar]
- 35.Pajic M, Scarlett CJ, Chang DK, Sutherland RL, Biankin AV. Preclinical strategies to define predictive biomarkers for therapeutically relevant cancer subtypes. Hum. Genet. 2011;130:93–101. doi: 10.1007/s00439-011-0990-0. [DOI] [PubMed] [Google Scholar]
- 36.Biankin AV, Hudson TJ. Somatic variation and cancer: therapies lost in the mix. Hum. Genet. 2011;130:79–91. doi: 10.1007/s00439-011-1010-0. [DOI] [PubMed] [Google Scholar]
- 37.Kris MG, Meropol NJ, Winer EP, editors. Accelerating Progress Against Cancer: ASCO's Blueprint for Transforming Clinical and Translational Cancer Research. Am. Soc. Clin. Oncol.; 2011. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.