Abstract
Tn501 random mutagenesis was applied to the Pseudomonas aeruginosa wild-type strain PAO1 to select for mutants hypersusceptible to aminoglycoside antimicrobial agents. One such mutant, called 19A, was found to be hypersusceptible to a wide range of antibiotics including aminoglycosides, β-lactams, fluoroquinolones, colistin, erythromycin, rifampin, and glycopeptides. Light microscopy of the mutant strain revealed abnormal morphology characterized by large, filamentous cells. The drug supersusceptibility of 19A was accompanied by loss of motility, reduced resistance to osmotic and heat shock stress, and impaired growth at low temperatures. The insertion site of the Tn501 transposon in mutant 19A has occurred in an open reading frame (PA5550 according to the PAO1 genome project), whose gene product shows amino acid sequence similarity to the DeoR family of transcriptional repressors. The gene, which we called glmR, is located between the glmS (PA5549) and glmU (PA5552) homologues of E. coli, responsible for the synthesis of UDP-N-acetylglucosamine-1-P, a precursor of both lipopolysaccharide (LPS) and peptidoglycan. We showed that GlmR represses the transcription of the adjacent glmS homologue (PA5549) in P. aeruginosa, possibly affecting the pool of precursors for peptidoglycan and LPS synthesis. To our knowledge GlmR is the first regulator in P. aeruginosa that affects susceptibility to a large variety of antibiotics and is therefore a potential target for novel anti-infective agents.
Pseudomonas aeruginosa is naturally more resistant to antibacterial agents than many other gram-negative species, including Escherichia coli. This poor susceptibility results from the complex interaction of several mechanisms, which tend to inactivate the antibiotics or prevent their intracellular accumulation to inhibitory levels (7). For instance, a significant protection against labile β-lactams and 3′hydroxy-aminoglycosides (kanamycin, neomycin) is provided by the chromosomally encoded enzymes AmpC and APH(3′), respectively (29). The natural resistance of P. aeruginosa also involves an active efflux process mediated by at least two multidrug transport systems. The first, named MexAB-OprM, is constitutively produced in wild-type bacteria and exhibits an extraordinarily broad substrate specificity including fluoroquinolones, β-lactams, macrolides, and tetracyclines (13, 14, 18). The second, MexXY, which requires the outer membrane protein OprM for its functioning in reference strain PAO1 (23, 38) and appears to extrude a smaller number of antibacterial agents (18), is produced only in response to some of its substrates such as aminoglycosides, tetracyclines, and macrolides (17).
All these mechanisms are potentiated by the relative impermeability of the outer membrane to both hydrophilic and hydrophobic drugs (26). The slow penetration of solutes into the cell interior is indeed expected to favor the detoxifying action of hydrolyzing enzymes or efflux pumps. This distinctive property of the outer membrane of P. aeruginosa has been attributed mostly to the structural heterogeneity of the major porin OprF, as only a small proportion of this protein forms open, functional diffusion channels through the lipid bilayer (4). In contrast to small hydrophilic solutes, hydrophobic or large amphiphilic antibiotics (for example, erythromycin, fusidic acid, and novobiocin) cannot efficiently cross the cell envelope via the porin uptake pathway. Furthermore, these agents, which would normally partition into classical phospholipid bilayers, cannot diffuse through the lipid domains of the outer membrane because of the very low fluidity of the lipopolysaccharide (LPS) outer leaflet (26). Adjacent LPS molecules are indeed maintained in a tightly packed arrangement by divalent cations (mainly Mg2+) that cross-bridge their multiple phosphate residues (26). Consequently, alterations of the LPS or removal of the divalent cations that stabilize the outer membrane may compromise the barrier function of the lipid bilayer (32, 44). For instance, polycationic antibiotics such as polymyxins, which displace the divalent cations from their LPS binding sites (8, 31), are able to promote their own passage through the outer membrane lipids and to render P. aeruginosa more susceptible to hydrophobic inhibitors (32).
Despite a growing bulk of information, the complex architecture and the physiological properties of the outer membrane of P. aeruginosa remain poorly understood. In particular, whether and how the bacterium can modulate its outer membrane permeability to environmental conditions remains to be clarified. In an attempt to better understand the mechanisms involved in the natural resistance of P. aeruginosa to antibiotics, a number of drug-supersusceptible mutants were isolated and characterized. These studies demonstrated the important contribution of the MexAB-OprM (35) and MexXY (17, 38) efflux systems to the defense of the bacterium against noxious products. The integrity of the outer membrane permeability barrier was also found to be essential, as quantitative and/or qualitative changes in the LPS may dramatically decrease levels of resistance to toxic agents (2, 12, 22). In the present work, we report on a novel gene, named glmR, probably involved in amino sugar metabolism, whose inactivation dramatically sensitizes P. aeruginosa to a large variety of antibiotics including macromolecules such as aminoglycosides, erythromycin, vancomycin, teicoplanin, and rifampin.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, media, and growth conditions
Wild-type, susceptible P. aeruginosa strain PAO1 (PT581) was obtained from B. W. Holloway. P. aeruginosa strain MT350 (43) was used as the donor strain for conjugational transfer of plasmid pMT1000 carrying transposon Tn501. P. aeruginosa PT581 was mutagenized by Tn501 insertion as described previously (38). E. coli DH10B [F− araD139 Δ(ara-leu)7696 galU galK Δ(lac)X74 mcrΔ(mrr-hsdRMS-mcrBC) deoR φ80d lacZΔM15 endA1 nupG recA1 rpsL] was used for plasmid propagation. The pilus-specific bacteriophages B9 and PO4 were kindly provided by R. T. Irvin (University of Alberta, Edmonton, Canada), and the LPS-specific phage E79tv2 (25) was a gift from D. Haas (University of Lausanne, Lausanne, Switzerland). LPS O-serotyping was performed by slide agglutination according to the Habs classification scheme, by using specific antisera obtained from Bio-Rad (Marnes La Coquette, France).
E. coli and P. aeruginosa strains were grown in Luria-Bertani (LB) broth or on LB agar (LBA) plates at 37°C unless specified otherwise in the text. When necessary, 15 μg of gentamicin/ml, 10 μg of tetracycline/ml, and 10 μg of HgCl2/ml for E. coli or 15 μg of gentamicin/ml, 30 μg of tetracycline/ml, and 12.5 μg of HgCl2/ml for P. aeruginosa were added to the growth media. For trans-complementation experiments, bacterial cultures of mutant 19A were supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce expression of the cloned glmR gene.
Antimicrobial susceptibility testing
Bacterial susceptibility to antibiotics was determined by the Kirby-Bauer diffusion method in Mueller-Hinton agar medium (Bio-Rad, Ivry-sur-Seine, France), according to the recommendations of the National Committee for Clinical Laboratory Standards (3). The inhibition zones around the disks of antibiotics (Bio-Rad) were measured precisely with a Sirscan automated image analyzer (I2A, Perols, France) from two different antibiograms for the wild-type strain PAO1 and mutant 19A, respectively. The disk loads were as follows: ticarcillin, 75 μg; aztreonam, 30 μg; imipenem, 10 μg; amikacin, 30 μg; gentamicin, 15 μg; ciprofloxacin, 5 μg; colistin, 50 μg; erythromycin, 15 IU; lincomycin, 15 μg; rifampin, 30 μg; vancomycin, 30 μg; teicoplanin, 30 μg; fusidic acid, 10 μg. Antibiotic gradient plates (5), prepared with Muller-Hinton agar medium, were used for comparing susceptibility levels between the wild type and the complemented strain 19A.
DNA techniques
Plasmid DNA was routinely purified by the alkaline lysis procedure (39) or by using a QIAGEN Plasmid Midi kit (Qiagen S.A.). Restriction endonucleases or T4 DNA ligase were from Stratagene Inc. Selected restriction fragments were isolated from agarose gels by using a MiniElute gel extraction kit (Qiagen S.A.) as recommended by the manufacturer. Transformation of E. coli and P. aeruginosa with plasmid DNA was performed either by conjugation or by heat shock treatment of CaCl2 competent cells. DNA sequencing was performed using universal primers or custom primers (MWG S.A.). Nucleotide sequences were determined by the dideoxy-chain termination method (40) with a 377A DNA sequencer (Applied Biosystems Division, Perkin-Elmer) at the sequencing facility of the Geneva Medical School. DNA sequences were edited with Navigator software (Perkin-Elmer) and compared with the BLASTN (version 2.02) program provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Plasmid constructions
Insertion of transposon Tn501 into the chromosome of mutant 19A was confirmed by PCR amplification of the merA gene of the Tn501 transposon. To determine the insertion site of Tn501 in the chromosome of 19A, the genomic DNA of the mutant was extracted and digested with the BamHI and HindIII endonucleases. Restriction fragments ranging from 8 to 15 kbp were separated on agarose gels, extracted, and purified by using the MiniElute gel extraction kit. These fragments were then ligated to phagemid vector pBlueScript II KS(+) (Stratagene Inc.) that had previously been cleaved with HindIII and BamHI. Hg-resistant clones were selected after transformation of E. coli strain DH10B on LB plates containing HgCl2. The presence of transposon Tn501 in one of the resistant clones, called pBSK19A, was confirmed by PCR amplification of the merA gene. The M13F universal primer and the custom primer Tn8105 (5′-TGGTGACGGCGGCTATCG-3′) were used to sequence the DNA region flanking Tn501 in plasmid pBSK19A. The glmR gene was amplified by PCR with primers gluc2R (5-AGCCGCTCACGGAGAGGTTCG-3′) and gluc2F (designed to create an EcoRI restriction site; 5′-GCGAATTCTCACCTTGCTGCCCTATCCC-3′). The glmR PCR product (1,292 bp) was digested with the EcoRI and PstI endonucleases to give a 1,082-bp fragment, which was subsequently cloned downstream of the inducible ptac promoter in plasmid vector pMMB207GM to yield plasmid pMBGL11. Plasmid pMMB207GM had previously been constructed (T. Köhler et al., unpublished data) by replacing a HindIII-DraI fragment carrying the chloramphenicol resistance marker in vector pMMB207 (24) with a HindIII-XmnI fragment of pJQ200 (36) carrying a gentamicin cassette. In parallel, we cloned a 482-bp BamHI-PstI DNA fragment from the glmR PCR product into the promotorless vector pMP220 (digested with BglII-PstI) to create a glmS::lacZ transcriptional fusion (recombinant plasmid pMPGL11BP). Both constructions, pMBGL11 and pMPGL11BP, were then conjugated into P. aeruginosa 19A and PT581 for trans-complementation and β-galactosidase assays.
β-Galactosidase assays
Wild-type strain PT581 and mutant 19A harboring plasmid pMPGL11BP were grown to the mid-exponential-growth phase in LB medium and assayed spectrophotometrically for β-galactosidase activity with ONPG (o-nitrophenyl-β-d-galactosidase) as a substrate, according to the method described by Miller (21).
Permeability assay with NPN
The permeability of the outer membrane to hydrophobic solutes was assessed in living cells of P. aeruginosa by using 1-N-phenylnaphthylamine (NPN) as described by Loh et al. (15). Briefly, cells were grown to an optical density at 600 nm (OD600) of 0.5 to 0.6, centrifuged at room temperature for 10 min at 2,000 × g, and resuspended in the same volume of 5 mM HEPES, pH 7.2. NPN was prepared freshly and added to the cell suspension at a final concentration of 5 μM. Fluorescence was monitored at room temperature in a Perkin-Elmer fluorospectrophotometer.
Stress assays
The susceptibilities of wild-type strain PT581 and mutant 19A to osmotic stress and heat shock were determined as described by Suh et al. (42). In heat shock experiments, cells were resuspended in M9 minimal medium. In osmotic stress experiments, the strains were grown in 35 ml of LB medium in 250-ml flasks to an OD600 of 0.3. At this point the cultures were divided into two parts; one was incubated with a 1.5 M final concentration of NaCl, and the other was incubated without NaCl.
Motility assays
Swarming was assayed on M9-based medium supplemented with 0.2% glucose, 2 mM MgSO4, trace elements, and 0.05% glutamate (instead of NH4Cl) as a nitrogen source and solidified with agar (0.5%) (11). For twitching motility assays, strains were inoculated from a fresh LBA plate with a toothpick to the bottom of an LBA plate containing LB medium solidified with 1% agar (41). After a 24-h incubation at 37°C, the agar was removed, and the zone of motility on the bottom of the petri dish was revealed by using a 0.25% Coomassie brilliant blue R250 staining solution (40% methanol, 10% acetic acid). Swimming motility was assayed on LB medium containing 0.3% agar. Strains were inoculated with a toothpick and incubated at room temperature for 24 h. Bacterial flagella were observed by light microscopy after staining with the SpotTest Flagella Stain reagent (Difco Laboratories, Detroit, Mich.) as recommended by the manufacturer.
Bacteriophage susceptibility assays
The pilus-specific bacteriophages B9 and PO4 and the LPS-specific phage E79tv2 were diluted from stock solutions into LB medium. Ten microliters of serial dilutions of each phage suspension were spotted separately onto a lawn of PT581 and 19A bacteria. Plates were incubated for 18 h at 37°C.
Outer membrane and LPS analysis
Outer membranes were extracted and purified as described by Michéa-Hamzehpour et al. (20). LPS extraction and analysis were carried out as described by Hitchcock and Brown (9). The LPS fractions were transferred to nitrocellulose membranes for Western blot analysis. Binding of the primary monoclonal antibodies MAb N1F10 (specific for A-band LPS) and MAb MF15-4 (specific for B-band LPS) (6), obtained from J. S. Lam (University of Guelph, Guelph, Canada), was detected with an alkaline phosphatase-conjugated second antibody and a Lumi-Light Western blotting substrate kit (Roche) for color development. Protein concentrations were determined by the bicinchoninic acid protein assay reagent (Pierce) according to the manufacturer's instructions.
RESULTS
Isolation of mutant 19A
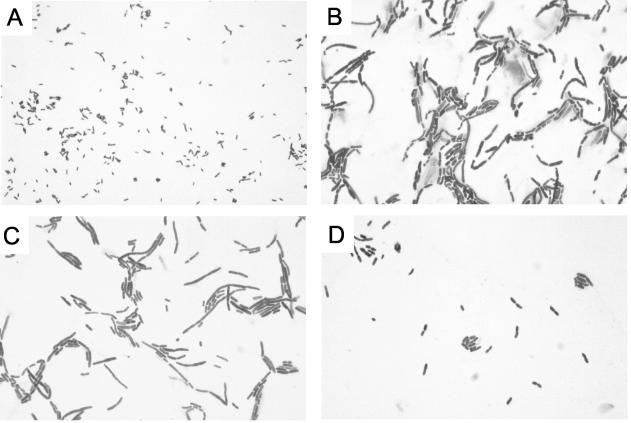
In a previous work, three Tn501 (HgCl2r) insertional mutants of wild-type strain PAO1 (PT581) had been selected for their hypersusceptibility to aminoglycosides (38). Determination of the levels of resistance of these mutants to various antimicrobial agents showed that one mutant, named 19A, was also more susceptible than PT581 to a wide range of antibiotics including β-lactams, fluoroquinolones, colistin, erythromycin, rifampin, and glycopeptides (Table 1). In contrast to wild-type PT581, 19A did not grow on culture media containing bile salts such as MacConkey or Drigalsky medium. Microscopic examination of 19A after staining with crystal violet revealed polymorphic bacterial cells with abnormal shapes ranging from small cocci to long, sinuous filaments of single or multiple elements, with central or terminal enlargements (Fig. 1B). Most of these bacteria were gigantic compared to the small rods of PT581 (Fig. 1A). These morphological patterns were similar to those observed when gram-negative bacteria are exposed to subinhibitory concentrations of cell wall-active antibiotics such as β-lactams (16, 49).
TABLE 1.
Antibiotic susceptibilities for the parent strain PT581 and mutant 19A
| Strain | Inhibition zone diam (mm)a
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RIF | VAN | TEI | ERY | AMK | GEN | CIP | COL | IMI | TIC | AZR | LIN | FUS | |
| PT581 | 17 | 6 | 6 | 6 | 20 | 15 | 34 | 18 | 25 | 27 | 31 | 6 | 6 |
| 19A | 38 | 15 | 14 | 21 | 35 | 30 | 41 | 26 | 42 | 38 | 40 | 6 | 6 |
RIF, rifampin; VAN, vancomycin; TEI, teicoplanin; ERY, erythromycin; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; COL, colistin; IMI, imipenem; TIC, ticarcillin; AZR, aztreonam; LIN, lincomycin; FUS, fusidic acid.
FIG. 1.
Light microscopic images of crystal violet-stained cells at a magnification of ×900. (A) PAO1 wild-type strain PT581; (B) hypersusceptible mutant 19A; (C) mutant 19A carrying the glmR plasmid pMBGL11; (D) mutant 19A carrying the glmR plasmid pMBGL11 and grown in the presence of 1 mM IPTG.
Mutant 19A is defective in a protein of the DeoR family
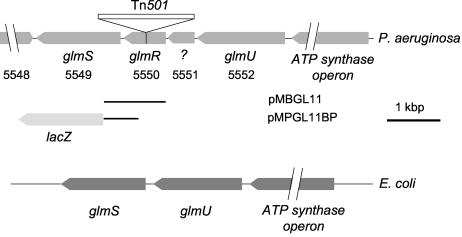
In order to identify the genetic alteration in 19A, we constructed a BamHI-HindIII genomic library of this mutant in phagemid pBSKSII+ and selected for Hgr transformants of E. coli containing DNA inserts with Tn501 (see Materials and Methods). Restriction mapping and PCR analysis of one of these clones revealed the presence of an insert carrying the mercury resistance marker flanked by a 1.5-kbp fragment from the 19A chromosome. Comparison of the nucleotide sequence of this 1.5-kbp DNA segment with the annotated genome of PAO1 (www.pseudomonas.com) allowed us to locate the Tn501 transposon in a 771-bp open reading frame (ORF) referenced as PA5550 (Fig. 2). This ORF is predicted to encode a 257-amino-acid polypeptide. Insertion of the transposon interrupts the coding sequence of PA5550 after position 146. A homology search using the BLAST algorithm (1) revealed significant sequence identity with members of the DeoR family of transcriptional regulators, most of which control sugar metabolism, such as the glucitol (SrlR, GutR) (50) and galactitol (GatR) (27) operon repressors of E. coli. High score matches were also found in the genomes of Pseudomonas putida (88% amino acid identity), Azotobacter vinelandii (86%), and Shewanella oneidensis (84%).
FIG. 2.
Schematic organization of the glm genes in the P. aeruginosa and E. coli chromosomes. ORFs are numbered according to the current annotation of the PAO1 genome (www.pseudomonas.com). DNA regions present on the glmR-expressing plasmid pMBGL11 and the glmS::lacZ fusion plasmid pMPGL11BP are indicated by solid lines.
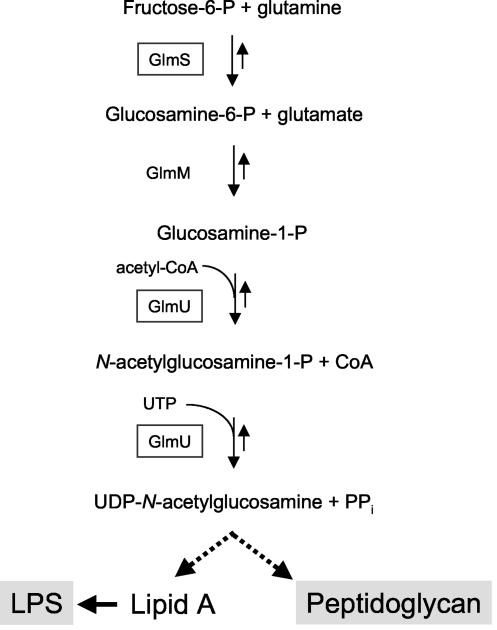
Immediately downstream of PA5550 in P. aeruginosa resides another ORF (PA5549), whose product is homologous to glucosamine-fructose-6-phosphate aminotransferase (or glucosamine-6-phosphate synthetase), encoded by the glmS gene in E. coli. Upstream of ORF PA5550 is an ORF (PA5551) coding for a putative 170-amino-acid protein that shows the M23/M37 endopeptidase signature (http://www.ebi.ac.uk/interpro/IEntry?ac=IPR002886#). Next to this gene, a third ORF, of 454 amino acids (PA5552), displays 56% amino acid sequence identity with the bifunctional GlmU enzyme, possessing glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities in E. coli. The GlmS and GlmU proteins are involved in the metabolism of amino sugars required for the synthesis of both LPS and peptidoglycan (Fig. 3). In contrast to their homologues in P. aeruginosa, the glmS and glmU genes are adjacent and belong to the same transcriptional unit in E. coli (Fig. 2). In reference to the nomenclature adopted for the glmS and glmU genes in E. coli, we propose to name the gene inactivated in mutant 19A (PA5550) glmR.
FIG. 3.
Biosynthetic pathway of UDP-N-acetylglucosamine in E. coli.
Complementation of mutant 19A
We attempted to complement mutant 19A by overexpressing the glmR gene under the control of the tac promoter from plasmid pMBGL11. Although the complemented strain grew faster in the presence of 1 mM IPTG on Mueller-Hinton agar medium at 37°C than mutant 19A harboring the vector alone, it still produced smaller colonies than the parent strain PT581 (data not shown). Under uninduced conditions, the complemented cells appeared as large bacilli upon microscopic examination (Fig. 1C) and were comparable to mutant 19A cells. However, when the complemented strain was grown in the presence of 1 mM IPTG, no filamentous structures were visible, although the individual cells were still larger than wild-type cells (Fig. 1D). In agreement with the findings of microscopic examinations, the antibiotic susceptibility of mutant 19A, could be only partially restored when glmR was expressed from plasmid pMBGL11 upon IPTG induction (Table 2). These data confirm that the observed phenotype of mutant 19A is linked to inactivation of the glmR gene and that partial complementation probably results from nonoptimal or untimely expression of glmR from the plasmid.
TABLE 2.
Antibiotic susceptibilities of strain PT581, mutant 19A, and the complemented mutant 19A
| Strain (plasmid) | Growth on antibiotic gradient plates (cm)a
|
||||
|---|---|---|---|---|---|
| RIF (4) | VAN (400) | ERY (400) | AMK (2) | NOV (400) | |
| PT581 | >8.0 | >8.0 | 4.8 | 6.8 | >8.0 |
| 19A(pMMB207GM) | 1.9 | 4.0 | 1.5 | 2.6 | 3.2 |
| 19A(pMBGL11) | 2.2 | >8.0 | 3.5 | 4.5 | >8.0 |
Susceptibility testing was performed using gradient plates. Growth across the gradient was recorded in centimeters; 8 cm was the maximum length of the plate. RIF, rifampin; VAN, vancomycin; ERY, erythromycin; AMK, amikacin; NOV, novobiocin. Numbers in parentheses following drug abbreviations are maximum concentrations of the antibiotics on the gradient plate in micrograms per milliliter.
GlmR acts as a transcriptional repressor
To determine whether glmR exerts direct control on the neighboring gene glmS, we measured the transcription of a glmS::lacZ fusion present on vector pMPGL11BP. β-Galactosidase activities were 55 times higher in mutant 19A (849.0 ± 30.0 Miller units) than in the parental strain PT581 (15.4 ± 0.5 Miller units). This strongly suggests that GlmR represses glmS expression at the level of transcription.
Surface structure and LPS analysis
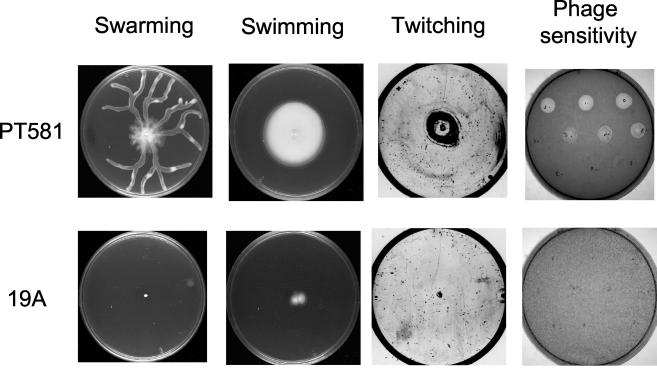
To further characterize mutant 19A, we examined surface structures known to depend in E. coli on LPS integrity, namely, flagella and pili (30). Plate assays showed that in contrast to the parental strain PT581, mutant 19A was unable to propagate by swimming, swarming, or twitching motility (Fig. 4). Swimming and twitching are known to rely on flagella and pili, respectively, in wild-type P. aeruginosa, while swarming requires both structures (11). Microscopic examination of stained cells confirmed the presence of flagellar structures in the parental strain PT581 but not in mutant 19A, which is in agreement with its swimming motility deficiency (data not shown). Furthermore, the lack of sensitivity to the pilus-specific bacteriophages B9 and PO4 suggested the absence of pili in the mutant (Fig. 4), explaining the inability of mutant 19A to exhibit swarming and twitching motility. On the other hand, mutant 19A showed normal sensitivity to the LPS-specific bacteriophage E79tv2 (data not shown), which recognizes the O-side chain of LPS. This observation is consistent with the fact that 19A, like its parent, PT581, could be easily typed by slide agglutination with an O5 antiserum, indicating an antigenically intact O-side chain of the LPS B-band.
FIG. 4.
Assays for motility and bacteriophage susceptibility of the P. aeruginosa parental strain PT581 and mutant 19A. Swimming plates were incubated for 24 h at room temperature. Swarming and twitching plates were incubated at 37°C for 24 and 48 h, respectively. Twitching motility was visualized after removal of the agar from the plate and staining of the spread zone with Coomassie blue. Sensitivities to the pilus-specific bacteriophage B9 or PO4 were tested by plating dilutions of phage stock onto an LBA plate seeded with 105 CFU of wild-type or mutant bacteria.
Outer membranes and LPS were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Silver-stained gels and immunoblots showed similar banding patterns of LPS in 19A and PT581. In addition, the two strains displayed similar outer membrane protein profiles. Western immunoblotting of total LPS, using A-band- and B-band-specific antibodies, revealed slightly smaller amounts of both A- and B-band LPS in mutant 19A than in the parental strain (data not shown).
Response of mutant 19A to environmental stress
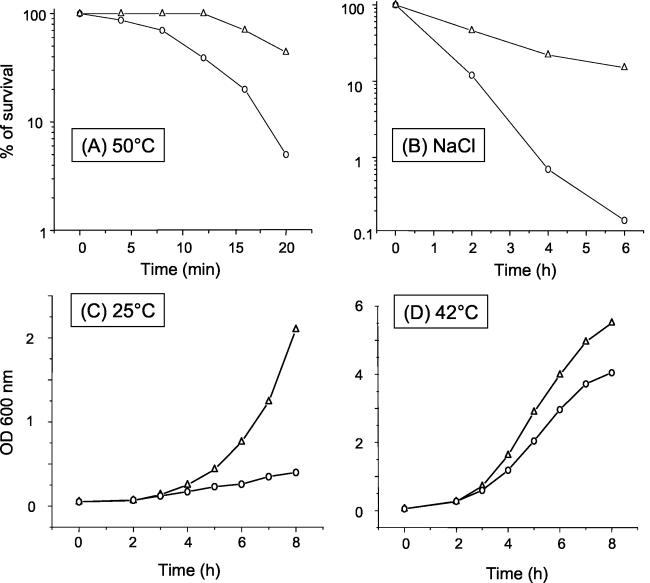
We investigated the abilities of 19A and PT581 to resist drastic changes in temperature and osmolarity. Stationary-phase cultures of both strains were exposed to a sudden shift in temperature, which was raised from 37 to 50°C. After 20 min at 50°C, lethality in the 19A culture was about 10 times higher than that observed in the parent culture (Fig. 5A). Exposure of mutant 19A to 1.5 M NaCl for 6 h resulted in a considerable decrease (almost 100-fold) in the number of viable cells relative to that in the wild type (Fig. 5B). Thus, inactivation of the glmR gene seriously impairs the capacity of mutant 19A to adapt rapidly to both temperature and osmotic variations.
FIG. 5.
Resistance to temperature or osmotic stress and growth at different temperatures of the P. aeruginosa parental strain PT581 and mutant 19A. (A) To assay cell survival after exposure to heat shock at 50°C, stationary-phase cultures of PT581 (○) and 19A (▵) were grown in LB medium, washed in M9 medium, and transferred to prewarmed tubes. The number of viable cells in each suspension was measured by plating aliquots on LB plates at each time point and determining the number of CFU after overnight incubation. Viability is expressed as a percentage of the number of CFU at time zero. (B) To assay cell survival after exposure to 1.5 M NaCl, stationary-phase cultures of PT581 (○) and 19A (▵) were diluted in LB broth to an OD600 of 0.3, divided in two, and incubated with agitation at 37°C in the presence or absence of 1.5 M NaCl. Cell viability was determined after 2, 4, and 6 h. One hundred percent viability corresponds to the number of CFU present immediately after resuspension of the cells in 1.5 M NaCl. Heat and osmotic shock experiments were performed at least three times with reproducible results. (C and D) Growth curves of the P. aeruginosa parental strain PT581 (○) and mutant 19A (▵) at 25°C (C) and at 42°C (D) were performed in LB medium, and absorbance was measured at 600 nm. Growth experiments were performed three times.
To determine the influence of temperature on bacterial growth, 19A and PT581 were cultured at 25, 37, and 42°C in LB broth. The two strains showed similar growth curves at 37°C (data not shown). However, at 25°C, the doubling time of mutant 19A was fivefold less than that of the parental strain (Fig. 5C). This difference was less pronounced but still significant (1.4-fold) when the bacteria were grown at 42°C (Fig. 5D). These results suggest that the adaptation of 19A to growth is affected primarily at low temperatures.
Permeability of the hydrophobic uptake pathway
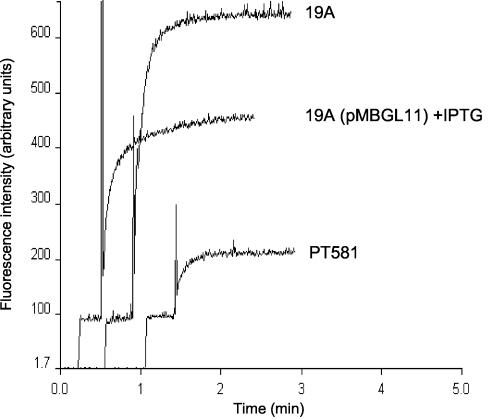
The lipid pathway of the outer membrane was investigated spectrophotometrically with NPN, a hydrophobic probe that fluoresces weakly in aqueous environments and strongly when dissolved in lipids (8, 47). As shown in Fig. 6, the accumulation levels of NPN at steady state were about threefold higher in the mutant than in the wild-type strain PT581. NPN uptake was reduced upon IPTG induction when glmR from plasmid pMBGL11 was expressed in mutant 19A. This clearly shows that inactivation of glmR leads to increased outer membrane permeability, a result in agreement with the increased susceptibility of mutant 19A to large antimicrobial molecules.
FIG. 6.
Outer membrane permeability of the P. aeruginosa parental strain PT581 and mutant 19A to NPN. Two milliliters of cell suspension was stirred in a cuvette before NPN was added. Fluorescence (excitation wavelength, 340 nm; emission wavelength, 420 nm; slit width, 5 nm) was recorded over time and is expressed in arbitrary units.
DISCUSSION
In this study, we report the isolation and characterization of a Tn501 transposon mutant of P. aeruginosa, called 19A, that is more susceptible than its parental strain, PAO1, to a large variety of antibacterial agents, including macromolecules such as glycopeptides, which usually permeate very poorly through the outer membranes of gram-negative bacilli (46). In support of a rupture of the outer membrane permeability barrier in 19A, NPN uptake turned out to be higher in the mutant than in the wild type. The drug supersusceptibility of 19A was also accompanied by loss of motility, reduced resistance to osmotic and heat shock stress, and impaired growth at low temperatures.
All these phenotypic alterations in 19A were associated with the inactivation of a so far uncharacterized gene (PA5550) of P. aeruginosa, which we called glmR. This gene encodes a transcriptional regulator that, as shown here, represses the expression of the enzyme glucosamine-fructose-6-phosphate aminotransferase (GlmS), involved in the early steps of the cell wall and LPS biosynthesis pathways (37). On the basis of protein sequence homology, the glmR gene was found to belong to the DeoR family of transcriptional regulators (51). Members of this family display a typical N-terminal domain containing a helix-turn-helix DNA binding motif and often interact with sugars as effector molecules, including, among others, fucose and glucitol (SrlR) (50). In E. coli, glmS and glmU are cotranscribed and are repressed by NagR when cells are grown in the presence of the amino sugars glucosamine and GlcNAc (33). On the other hand, glmS expression has been proposed to be induced in the presence of the amino sugar glucosamine-1-phosphate or UDP-N-acetyl-d-glucosamine (UDP-GlcNAc) (34). Interestingly, a BLAST search failed to identify a GlmR homologue in E. coli, since the protein most similar to GlmR was SrlR (41% amino acid identity), the repressor of the glucitol utilization operon (50). Conversely, no NagR homologue was found in the Pseudomonas PAO1 genome (Köhler et al., unpublished), suggesting differences in the regulation of amino sugar metabolism in these two species. Indeed, the amino sugars GlcNAc and glucosamine-6-phosphate fulfill several distinct functions in bacteria. They play a role as structural components of peptidoglycan and LPS in gram-negative species, and they are also valuable energy sources. A control mechanism for the transcription of genes involved in the anabolism and catabolism of amino sugars is therefore necessary to maintain a correct balance between both pathways. The repressor gene glmR could be part of such a regulatory circuit. Further investigations, including analyses of glmU and ORF PA5551 expression, are required to define more precisely the role of GlmR in LPS and cell wall synthesis in P. aeruginosa.
Inactivation of glmR in P. aeruginosa resulted in giant, polymorphic bacteria sensitive to osmotic stress. We noticed no drastic alterations in the outer membrane protein profile or LPS banding pattern of the mutant on regular sodium dodecyl sulfate gels. However, we cannot exclude subtle changes in the LPS core region. In E. coli, alteration of this core region resulted in hypersusceptibility to novobiocin and loss of flagella and pili (30), as observed in mutant 19A. omsA, envA1, and lpxA2 mutants of E. coli, defective in the first steps of the lipid A biosynthetic pathway (with mutations in the firA, lpxC, and lpxA genes, respectively), have been reported to contain reduced amounts of LPS in their outer membranes and to be hypersusceptible to many drugs, including hydrophobic and large peptide antibiotics (reviewed in reference 45). It has been assumed that phospholipid bilayer domains, which are more permeable to hydrophobic solutes, would compensate for the lack of LPS molecules in the outer membrane (45). If mutant 19A had an increased phospholipid content, this could explain its altered response to temperature shock and its reduced growth at 25°C (Fig. 5). In contrast to the omsA and lpxA2 alleles, the envA1 mutation results in bacteria exhibiting abnormal shapes, such as long chains of bacilli, because of a defect in septum formation (28).
Our results indicate that glmS is strongly overexpressed in the hypersusceptible mutant 19A as a result of glmR inactivation. Since the product of glmS (glucosamine-6-phosphate synthetase) is involved in the formation of glucosamine-6-phosphate, a precursor in the peptidoglycan and LPS biosynthesis pathways (Fig. 2), it is reasonable to assume that glmS overexpression somehow affects the levels of murein and LPS precursor pools. Feedback inhibition of enzymes of the peptidoglycan biosynthesis pathway such as UDP-GlcNAc enolpyruvoyltransferase or the MurD and MurE synthetases has been suggested for E. coli (48). An increase in the rate of input of glucosamine-6-phosphate and its metabolite UDP-GlcNAc might therefore be detrimental to murein synthesis and might lead to the formation of abnormal cells more sensitive to osmotic pressure variations. It is interesting that, in E. coli, increases in the pools of UDP-GlcNAc and its derivative UDP-N-acetylmuramyl-pentapeptide were associated with a reduction in the level of peptidoglycan cross-linking (19). On the other hand, competitive inhibition of glucosamine-6-phosphate synthetase (GlmS) by glutamine analogues, including bacilysin, was found to provoke morphological changes on E. coli similar to those induced by β-lactams (10). In P. aeruginosa, repression of glmS by cloned glmR did not completely restore wild-type levels of antibiotic resistance in mutant 19A, probably because glmS expression needs to be finely tuned in order to precisely adapt the pool of nucleotide precursors to murein synthesis.
In summary, we show that alteration of glmR in P. aeruginosa results in a complex, pleiotropic phenotype, including hypersusceptibility to a large number of antimicrobial agents. This probably results from perturbation of the peptidoglycan structure in the glmR mutant, as evidenced by its abnormal cell shape and response to osmotic stress. Furthermore, subtle modifications in the core LPS region and in the amounts of LPS and phospholipids might contribute to the antibiotic hypersusceptibility of mutant 19A and explain its loss of motility and its impaired adaptation to temperature changes. The study of such antibiotic-hypersusceptible mutants, especially in drug-resistant species such as P. aeruginosa or Acinetobacter spp, may be useful in the effort to find new drug targets and tailor innovative therapeutical agents.
Acknowledgments
We thank Ekatherina Charvalos for help with the cloning of glmR.
This work was supported by a grant to T.K. from the Swiss National Science Foundation.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angus, B. L., A. M. Carey, D. A. Caron, A. M. Kropinski, and R. E. Hancock. 1982. Outer membrane permeability in Pseudomonas aeruginosa: comparison of a wild type with an antibiotic-supersusceptible mutant. Antimicrob. Agents Chemother. 21:299-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balows, A., W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy. 1991. Manual of clinical microbiology, 5th ed. ASM Press, Washington, D.C.
- 4.Benz, R., and R. E. Hancock. 1981. Properties of the large ion-permeable pores formed from protein F of Pseudomonas aeruginosa in lipid bilayer membranes. Biochim. Biophys. Acta 646:298-308. [DOI] [PubMed] [Google Scholar]
- 5.Bryson, L., and W. Szybalski. 1952. Microbial selection. Science 116:45-51. [PubMed] [Google Scholar]
- 6.Burrows, L. L., D. F. Charter, and J. S. Lam. 1996. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol. Microbiol. 22:481-495. [DOI] [PubMed] [Google Scholar]
- 7.Hancock, R. E., and D. P. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updat. 3:247-255. [DOI] [PubMed] [Google Scholar]
- 8.Hancock, R. E., and P. G. Wong. 1984. Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 26:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenig, M., and E. P. Abraham. 1976. Antimicrobial activities and antagonists of bacilysin and anticapsin. J. Gen. Microbiol. 94:37-45. [DOI] [PubMed] [Google Scholar]
- 11.Köhler, T., L. Kocjancic-Curty, F. Barja, C. Van Delden, and J. C. Pechère. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 182:5990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kropinski, A. M., J. Kuzio, B. L. Angus, and R. E. Hancock. 1982. Chemical and chromatographic analysis of lipopolysaccharide from an antibiotic-supersusceptible mutant of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 21:310-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, X.-Z., D. M. Livermore, and H. Nikaido. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloramphenicol and norfloxacin. Antimicrob. Agents Chemother. 38:1732-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, X.-Z., D. Ma, D. M. Livermore, and H. Nikaido. 1994. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as contributing factor to β-lactam resistance. Antimicrob. Agents Chemother. 38:1742-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loh, B., C. Grant, and R. E. Hancock. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorian, V., and C. G. Gemmell. 1991. Effect of low antibiotic concentrations on bacteria: effects on ultrastructure, virulence, and susceptibility to immunodefenses, p. 493-555. In V. Lorian. (ed.), Antibiotics in laboratory medicine, 3rd ed. Williams and Wilkins, Baltimore, Md.
- 17.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Contribution of the MexX-MexY-oprM efflux system to intrinsic resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda, N., E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto, and T. Nishino. 2000. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-OprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 44:3322-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mengin-Lecreulx, D., E. Siegel, and J. van Heijenoort. 1989. Variations in UDP-N-acetylglucosamine and UDP-N-acetylmuramyl-pentapeptide pools in Escherichia coli after inhibition of protein synthesis. J. Bacteriol. 171:3282-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michéa-Hamzehpour, M., J.-C. Pechère, P. Plésiat, and T. Köhler. 1995. OprK and OprM define two genetically distinct multidrug efflux systems in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:2392-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Mills, B. J., and B. W. Holloway. 1976. Mutants of Pseudomonas aeruginosa that show specific hypersensitivity to aminoglycosides. Antimicrob. Agents Chemother. 10:411-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morales, V. M., A. Bäckman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 25.Morgan, A. F. 1979. Transduction of Pseudomonas aeruginosa with a mutant of bacteriophage E79. J. Bacteriol. 139:137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikaido, H., and R. E. W. Hancock. 1986. Outer membrane permeability of Pseudomonas aeruginosa, p. 145-193. In J. R. Sokatch and N. L. Ornston (ed.), The Bacteria. Academic Press, New York, N.Y.
- 27.Nobelmann, B., and J. W. Lengeler. 1995. Sequence of the gat operon for galactitol utilization from a wild-type strain EC3132 of Escherichia coli. Biochim. Biophys. Acta 1262:69-72. [DOI] [PubMed] [Google Scholar]
- 28.Normark, S. 1970. Genetics of a chain-forming mutant of Escherichia coli. Transduction and dominance of the envA gene mediating increased penetration to some antibacterial agents. Genet. Res. 16:63-78. [DOI] [PubMed] [Google Scholar]
- 29.Okii, M., S. Iyobe, and S. Mitsuhashi. 1983. Mapping of the gene specifying aminoglycoside 3′-phosphotransferase II on the Pseudomonas aeruginosa chromosome. J. Bacteriol. 155:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson, A. A., R. E. Hancock, and E. J. McGroarty. 1985. Binding of polycationic antibiotics and polyamines to lipopolysaccharides of Pseudomonas aeruginosa. J. Bacteriol. 164:1256-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plesiat, P., J. R. Aires, C. Godard, and T. Köhler. 1997. Use of steroids to monitor alterations in the outer membrane of Pseudomonas aeruginosa. J. Bacteriol. 179:7004-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plumbridge, J. 1995. Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 14:3958-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plumbridge, J. A., O. Cochet, J. M. Souza, M. M. Altamirano, M. L. Calcagno, and B. Badet. 1993. Coordinated regulation of amino sugar-synthesizing and -degrading enzymes in Escherichia coli K-12. J. Bacteriol. 175:4951-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 37.Raetz, C. R. H. 1996. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles, p. 1035-1063. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 38.Ramos-Aires, J., T. Köhler, H. Nikaido, and P. Plésiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semmler, A. B., C. B. Whitchurch, and J. S. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145:2863-2873. [DOI] [PubMed] [Google Scholar]
- 42.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuda, M., S. Harayama, and T. Iino. 1984. Tn501 insertion mutagenesis in Pseudomonas aeruginosa PAO. Mol. Gen. Genet. 196:494-500. [DOI] [PubMed] [Google Scholar]
- 44.Vaara, M. 1992. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56:395-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaara, M. 1993. Antibiotic-supersusceptible mutants of Escherichia coli and Salmonella typhimurium. Antimicrob. Agents Chemother. 37:2255-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaara, M. 1993. Outer membrane permeability barrier to azithromycin, clarithromycin, and roxithromycin in gram-negative enteric bacteria. Antimicrob. Agents Chemother. 37:354-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaara, M., W. Z. Plachy, and H. Nikaido. 1990. Partitioning of hydrophobic probes into lipopolysaccharide bilayers. Biochim. Biophys. Acta 1024:152-158. [DOI] [PubMed] [Google Scholar]
- 48.van Heijenoort, J. 1996. Murein synthesis, p. 1025-1034. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 49.Waisbren, S. J., D. J. Hurley, and B. A. Waisbren. 1980. Morphological expressions of antibiotic synergism against Pseudomonas aeruginosa as observed by scanning electron microscopy. Antimicrob. Agents Chemother. 18:969-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamada, M., and M. H. J. Saier. 1988. Positive and negative regulators for glucitol (gut) operon expression in Escherichia coli. J. Mol. Biol. 203:569-583. [DOI] [PubMed] [Google Scholar]
- 51.Zeng, G., S. Ye, and T. J. Larson. 1996. Repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J. Bacteriol. 178:7080-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]