Abstract
Disease and Gene Annotations database (DGA, http://dga.nubic.northwestern.edu) is a collaborative effort aiming to provide a comprehensive and integrative annotation of the human genes in disease network context by integrating computable controlled vocabulary of the Disease Ontology (DO version 3 revision 2510, which has 8043 inherited, developmental and acquired human diseases), NCBI Gene Reference Into Function (GeneRIF) and molecular interaction network (MIN). DGA integrates these resources together using semantic mappings to build an integrative set of disease-to-gene and gene-to-gene relationships with excellent coverage based on current knowledge. DGA is kept current by periodically reparsing DO, GeneRIF, and MINs. DGA provides a user-friendly and interactive web interface system enabling users to efficiently query, download and visualize the DO tree structure and annotations as a tree, a network graph or a tabular list. To facilitate integrative analysis, DGA provides a web service Application Programming Interface for integration with external analytic tools.
INTRODUCTION
Understanding underlying mechanisms of human disease is a fundamental driver for biomedical research. Simple genetic diseases fit well in the ‘one gene-one disease’ rubric, and many of these diseases have been successfully addressed with molecular therapeutics. However, complex diseases (those with multiple genetic etiologies, highly variable penetrance and significant diet or environmental components) have been less tractable using molecular reductionism. Complex diseases require network/system-centric and integrative investigation and modeling (1,2). Our ability to build multi-layer multi-component networks is largely due to the development of high-throughput/omics technologies that can broadly probe a biological system to delineate the molecular underpinnings of disease. These networks will in turn help identify new disease–gene relations and reveal novel molecular targets for potential therapeutic intervention.
However, there are hurdles to overcome in achieving the integrated systems approach. Some of these include the management, integration and synchronization of an ever-expanding set of experimental data generated by these high-throughput techniques. More specifically, these data are heterogeneous, produced by multiple technical platforms (each with unique analytical characteristics), stored in diverse formats and arising from a variety of biological models and experimental designs. Each of these layers of differences makes fundamental integration and knowledge generation difficult. One way to address these difficulties is to integrate data that are directly comparable and extract the knowledge from those comparisons, and then enable the integration of those facts at a more general and disease-related level.
To highlight the data integration problem a bit further, current databases of gene–disease associations, such as Online Mendelian Inheritance in Man (OMIM) (3), Genetic Association Database (GAD) (4), Human Gene Mutation Database (HGMD) (5), and database of Genotypes and Phenotypes (dbGaP) (6), have the following limitations. First, these existing databases focus on one layer of the network, typically focused on annotating a gene with aberrant phenotype information based on single mutations. This approach is extremely useful but does not enable assessing disease–gene associations in the context of biological networks. For complex diseases, there may be mis-regulation of one or more gene expression regulatory network, disruption of normal protein–protein interactions, novel genetic interactions or signaling network changes. Understanding the impact of a given change from a systems biology standpoint is not currently enabled by these databases. Second, disease terms are interrelated in a conceptual hierarchy that is reflected in the Disease Ontology (DO) structure. None of the existing disease–gene association databases use a formal ontology or attempt to provide an integrated molecular interaction network (MIN) to describe contributions of a given aberrant association with one or more diseases. This limits the potential for comprehensive computational analysis from any one of these source databases. In addition, most of the databases are based on manual or semi-computerized curation and do not provide a mechanism for automated updates (4,7,8). Third, textual descriptions in OMIM make further inferences by computational tools difficult, although the human expert review is of tremendous value for genetic counselors and physicians. Fourth, GAD and dbGaP are limited to results from genome-wide association studies, and HGMD is limited to mutations only.
To overcome these obstacles, DGA provides an integrated environment to facilitate the analysis of disease–gene associations and explore potential gene interactions shared among multiple diseases. To enable the exploration of these data, there are three key interwoven modules: DO (9), the Electronic Annotator (EA) and the Molecular Interaction Network Integrator (MINI). DO is a community-driven open-source ontology to represent human disease and was used as backbone for annotating the human genes from a disease perspective. In the EA module, we provide the results of semantically annotating human genes with disease descriptors by using National Center for Biomedical Ontology (NCBO) Annotator service (10) and NCBI Gene Reference Into Function (GeneRIF). The MINI module integrates disease–gene annotations with additional biological network information including 8566549 human gene/protein interactions by using PSICQUIC web service (11). Overall, DGA provides an integrated resource for interrogating the results of ontology-based text mining and network analysis methods from a gene, protein or disease perspective. Behind the scenes, DGA is updated through a fully automated annotation process and therefore maintains a current integrated view of these resources.
OVERVIEW OF THE DGA SYSTEM ARCHITECTURE
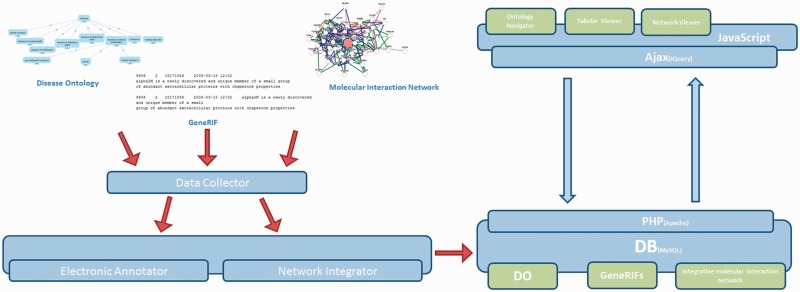
The DGA system is implemented using PHP, MySQL, JavaScript and Cytoscape Web (12). DGA consists of five system components (Figure 1): (i) The Data Collector responsible for gathering GeneRIF, DO and molecular interaction data. This module periodically probes for the latest update of data and can perform either incremental or full imports from each of the configured sources. (ii) The Electronic Annotator is responsible for orchestrating the submission of GeneRIF and DO information to NCBO Annotator and subsequent integration of these results to build the relationships between diseases and genes. (iii) The Network Integrator is responsible for integrating biological network data with disease–gene annotation and stores this information in a graph-based data structure, enabling fast and efficient user examination of these data. (iv) A relational database for maintaining these data and operational data such as the last time a given association was updated, processing information and general state information. (v) The Web Interface for querying and visualizing DGA data.
Figure 1.
DGA system architecture.
DGA DISEASE ONTOLOGY
The DGA uses DO as foundation to organize the disease-related annotations using the conceptual framework of the ontology. DO is an Open Biomedical Ontology and is available at the OBO Foundry, the DO source forge site and from the NCBO Bioportal. DO provides a unifying disease-focused structure, which can be used to map human disease knowledge between datasets such as patient records and large-scale genome, sequencing and microbiome projects. DO delineates a semantically computable structure of inherited, environmental and infectious human disease that is based on a manually curated subset of the Unified Medical Language System (UMLS) and includes terms from other sources as well. Since 2003, DO has undergone three major version updates and currently contains 8043 unique disease terms.
Similar to the graph structure of Gene Ontology, the DO is also organized as a directed acyclic graph where nodes are disease terms and edges denote the relationships between the disease terms. Every term/node is unique, assigned an identifier prefixed with ‘DOID:’ and contains textual description and external references to well-established well-adopted terminologies that contain disease and disease-related concepts such as UMLS (13), Medical Subject Headings (MeSH), Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT) (3), OMIM (14), the NCI Thesaurus (NCIt) (15) and International Classification of Diseases (ICD). The relationship/edge between terms/nodes is represented as standard defined formulation: ‘is_a’ based on the OBO format. For instance, the term ‘myelophthisic anemia’ assigned as DOID: 2354 has definition ‘A myeloma and anemia that is located in some people with diseases that affect the bone marrow.’ and has external references to OMIM2009_05_01:MTHU012207 and ‘is_a’ term ‘aplastic anemia’ with ‘DOID: 12449’.
DGA ELECTRONIC ANNOTATOR
The DGA EA orchestrates the mining of disease information from the NCBI GeneRIF database using DO terms and the NCBO Annotator and the re-association of the mined information with disease terms and genes. A GeneRIF statement consists of concise textual descriptions (up to 250 characters) of the function of a gene. The GeneRIF database is available from the NCBI Gene database (ftp://ftp.ncbi.nih.gov/gene/GeneRIF/). Every GeneRIF statement includes an NCBI Gene ID and a PubMed ID, creating a short biological evidence annotation coupling a gene with a publication. NCBI provides frequent updates to GeneRIFs based on manual and automated processes and provides open access to GeneRIFs for the community. We previously demonstrated that GeneRIFs were a good source of disease annotations (16). However, the method used previously, a standalone-java application, has proven difficult to maintain, update and integrate with other resources.
To overcome these hurdles, the DGA uses the NCBO annotator, which is an online tool providing text mining services using biomedical ontologies. The electronic annotation process is composed of three main steps: (i) Collecting GeneRIF statements and submitting them to NCBO annotator for annotation with DO. (ii) NCBO Annotator annotation, wherein the NCBO annotator creates annotation(s) in the raw GeneRIF text based on syntactic word recognition using a dictionary complied based on DO terms. (iii) Semantic expansion where additional annotation information is produced by taking advantage of the semantic relationships in DO such as the ‘is_a’ relation. The DGA EA automates the process of obtaining the latest release of the GeneRIF database from the NCBI, submitting each GeneRIF statement to the NCBO annotator, retrieving and post-analyzing mapping results and automates the removal of known mapping artifacts (quality control) to eliminate non-informative and incorrect mapping results (16).
DGA MINI
The DGA MINI is responsible for integrating multi-level biological networks with gene–disease annotation by collecting biological network information from the PSICQUIC web service. PSICQUIC not only provides a standard interface to query major molecular interaction resources but also provides a confidence score to help assess validity of the information. Through PSICQUIC, DGA MINI currently targets five different types of networks: physical and genetic interaction networks, co-expression, co-localization and protein-shared domain networks. PSIQUIC provides access to 8566549 human gene/protein interactions that have been integrated from six major molecular interaction databases, including GeneMania, BioGRID, IntAct, I2D, InnateDB and MINT (17–21). For example, the BioGRID database is a comprehensive and freely accessible online resource of physical and genetic interaction from 38 organisms. The GeneMania is a fast-heuristic-based-algorithm to integrate multiple functional association networks from five organisms, with 120 644 180 interactions in total. Furthermore, the disease–gene and gene–gene association confidence and strength are important for using the resource. PSIQUIC score and GeneMania interaction weight provide preliminary information for this. DGA also integrates this information into the result sets to provide a more quantitative score for assessing the confidence of a given molecular interaction. For disease–gene associations, DGA provides a metric for strength by ranking annotation associations by the count of the number of GeneRIF statements that supports a given association. This score indicates how well each annotation is documented and supported by independent publications, although this simple score will be biased toward well-funded research areas. In the DGA network view, the width of edge connecting a disease and gene is proportional to this count of GeneRIF statements. The DGA MINI orchestrates an automated pipeline to periodically collect the latest information from these online repositories through PSICQUIC and provides a flexible data storage that can be easily extended to new interaction networks in the future such as disease–drug interaction network as those become available in a consistent and structured resource.
DGA WEB SYSTEM AND APPLICATION
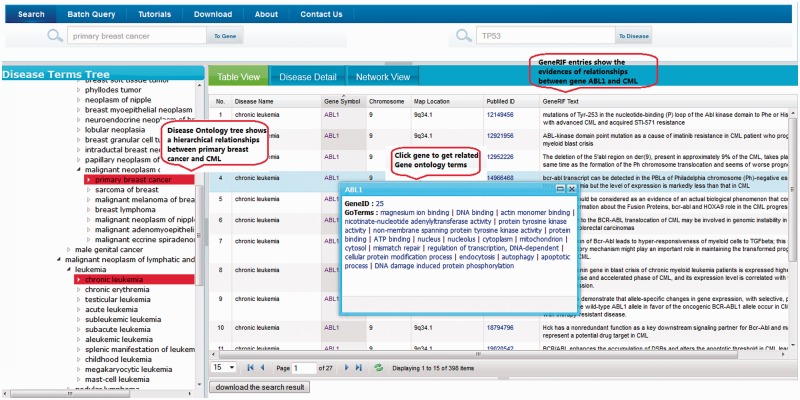
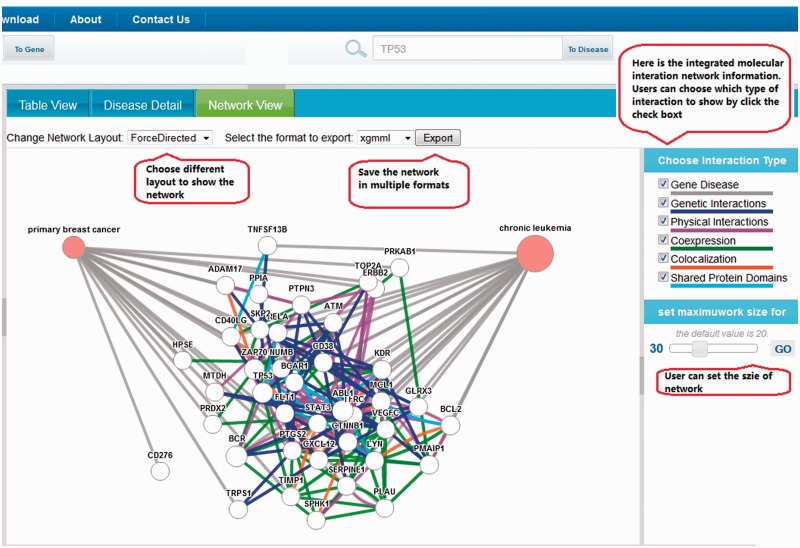
The DGA provides web-based interface that is intuitive and flexible for different types of users. The DGA web interface provides not only traditional text search functionality but also an ontology navigator (Figure 2), and a network graph-based navigator (Figure 3). User can search DGA for disease–gene relationships by either inputting single or multiple disease terms or gene name or synonyms. The results of the query can be visualized in several ways: (i) Through the hierarchical ontology navigator (left panel of Figure 2). (ii) Through the table view (Figure 2), which can be sorted by column name. These results can be downloaded as a CSV file by clicking ‘download’ button. (iii) The network graph navigator (Figure 3), which lays out the results as a network diagram, where nodes are disease terms or gene names and edges show connections based on disease–gene evidence from GeneRIFs or PSICQUIC biological interaction network data. In the graph, edges connecting two disease nodes through a common gene infer potential disease–disease associations. Both the network graph navigator and ontology navigator are interactive and enable the exploration of the relationships. For instance, clicking on a disease term in the ontology navigator or in the network navigator graph, the corresponding disease node and its associated gene nodes will be highlighted. The network graph can be exported in multiple formats, including PDF, PNG and xgmml files [xgmml is supported by Cytoscape (22)]. The DGA database also provides a consistent web service Application Programming Interface (API) that is accessible through RESTful calls. Any programming language supporting RESTful calls can be used to access the DGA API.
Figure 2.
DGA web interface (A) Disease Ontology Tree and disease detail information and Search results shown in tabular view and functionality of tabular view.
Figure 3.
Search result shown in network view.
USE CASE
Querying genes associated with a disease
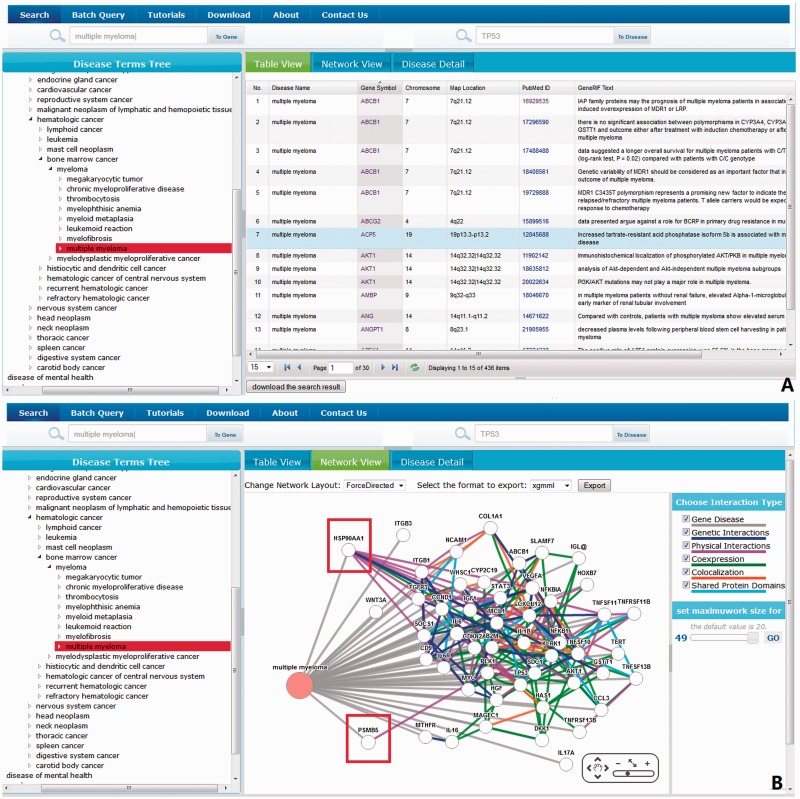
We are often asked to identify genes associated with a given disease, or diseases associated with a set of genes, often from a gene expression experiment. We present an example of querying multiple myeloma (MM)-associated genes based on searching for MM. MM is a cancer of plasma cells and occurs primarily in people older than 50, and has an incidence of 1–4 per 100 000 people per year (23). Figure 4A shows the results of searching for genes associated with multiple myeloma. This was done by typing ‘multiple myeloma’ (but without the quotes) in the web interface. DGA shows the MM-associated genes documented by GeneRIF entries (436 in total shown in tabular view Figure 4A). Clicking on the network view tab, we can switch to a network visualization of MM–gene relationships (Figure 4B). In the network canvas, PSMB 5 is connected to MM. PSMB 5 is the target of bortezomib, which is a therapeutic proteasome inhibitor for treating MM. Further, we can see that heat shock protein 90 (HSP90/HSP90AA1) is also connected with MM nodes. Interestingly, a class of drugs known as HSP90 inhibitors has shown some promising effects for treating MM as a single agent or potential combined therapeutic method with bortezomib (24,25). Furthermore, DGA shows not only the key genes related to MM but also interaction type information (Figure 4B) between them, which will be useful for further exploring the molecular mechanisms underlying the gene–disease associations.
Figure 4.
Searching for multiple myeloma (MM)-related genes. (A) An overview of MM-related genes shown in tabular view. (B) An overview of MM-related genes shown in network view. Visualization of an integrated network of all types of interactions between these genes, including genetic, physical, co-expression, co-localization and shared protein domain (PSMB5 and HSP90 are highlighted in red frames).
Exploring disease–disease association
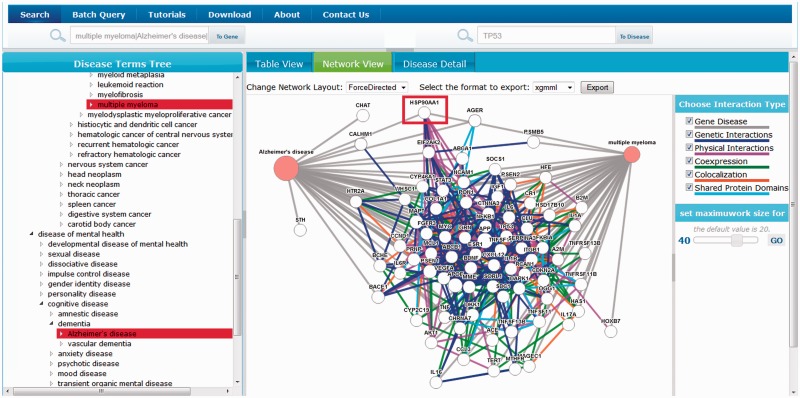
Based on the previous example, we further examine the genes involved in MM and how they overlap with genes involved in Alzheimer’s disease. After including Alzheimer’s disease in our query, we can examine the genes shared by these two diseases in the network view (Figure 5). In particular, the HSP90 (HSPAA1 protein) is connected to both diseases. This finding confirms that recent studies show that HSP90 may play a role in neurodegeneration and suggest that HSP90 inhibitors may be potentially beneficial in both neurodegenerative diseases and MM (26). These findings indicate that DGA will be useful for target discovery and for drug repositioning.
Figure 5.
Searching for genes associated with both MM and Alzheimer’s disease.
DISCUSSION AND FUTURE WORK
DGA is an integrative resource that provides human gene annotations incorporating DO terms and MIN results. DGA complements current disease–gene annotation databases by implementing a computable automated network-oriented system. The ontology structure of DGA allows the direct exploration of the integrated annotation knowledgebase. The fully automated DGA annotation pipeline will make it easy to maintain and update this resource, even in the face of ever-increasing data. The DO-based disease relationships allow the exploration of disease–gene, gene–gene and disease–disease associations in a systems biology framework. The current DGA framework and visualization tools can expose existing relationships between diseases by showing the disease shared genes. DGA will provide valuable resource for exploring drug repositioning opportunities. Through the flexible PSICQUIC web service, the DGA can easily integrate new MINs, for instance, transcription factor information that can be used to target gene regulation, microRNA regulation and pathway-level networks from KEGG (27).
FUNDING
Seed grant from Huazhong University of Science and Technology, National Natural Science Foundation China (NSFC) [31171275 and 61272410]; the National Institutes of Health—National Center for Research Resources (NCRR) [R01RR025342, in part] under the ARRA mechanism and in part by Award Number UL1RR025741 and UL1RR025011 from the Clinical and Translational Science Award (CTSA) program of the National Center for Research Resources (NCRR), National Institutes of Health, the SPORE in Prostate Cancer award [3P50CA090386]; the Cancer Center Support Grant award [3P30CA060553] from the National Cancer Institute of the National Institutes of Health. Funding for open access charge: Seed grant from Huazhong University of Science and Technology, National Natural Science Foundation China (NSFC) [31171275, 61272410 and 30771839].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
This research was supported by Chinese National Science Foundation C060704, in part by the National Institutes of Health—National Center for Research Resources (NCRR) (R01RR025342) under the ARRA mechanism and in part by Award Number UL1RR025741 and UL1RR025011 from the Clinical and Translational Science Award (CTSA) program of the National Center for Research Resources (NCRR), National Institutes of Health, the SPORE in Prostate Cancer award 3P50CA090386 and the Cancer Center Support Grant award 3P30CA060553 from the National Cancer Institute of the National Institutes of Health.
REFERENCES
- 1.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 2.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spackman KA, Campbell KE, Cote RA. SNOMED RT: a reference terminology for health care. Proc. AMIA Annu. Fall Symp. 1997:640–644. [PMC free article] [PubMed] [Google Scholar]
- 4.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat. Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 5.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The human gene mutation database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stenson PD, Ball EV, Mort M, Phillips AD, Shaw K, Cooper DN. The Human gene mutation database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr. Protoc. Bioinformatics. 2012 doi: 10.1002/0471250953.bi0113s39. Chapter 1, Unit 1.13. [DOI] [PubMed] [Google Scholar]
- 8.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schriml LM, Arze C, Nadendla S, Chang YW, Mazaitis M, Felix V, Feng G, Kibbe WA. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40:D940–D946. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonquet C, Shah NH, Musen MA. The open biomedical annotator. Summit Translat. Bioinforma. 2009;2009:56–60. [PMC free article] [PubMed] [Google Scholar]
- 11.Aranda B, Blankenburg H, Kerrien S, Brinkman FS, Ceol A, Chautard E, Dana JM, De Las Rivas J, Dumousseau M, Galeota E, et al. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat. Methods. 2011;8:528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphreys BL, Lindberg DA, Schoolman HM, Barnett GO. The Unified Medical Language System: an informatics research collaboration. J. Am. Med. Inform. Assoc. 1998;5:1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sioutos N, de Coronado S, Haber MW, Hartel FW, Shaiu WL, Wright LW. NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information. J. Biomed. Inform. 2007;40:30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Osborne JD, Flatow J, Holko M, Lin SM, Kibbe WA, Zhu LJ, Danila MI, Feng G, Chisholm RL. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10(Suppl.1):S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–D846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8:R95. doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl.1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raab MS, Podar K, Breitkreutz I, Richardson PG, Anderson KC. Multiple myeloma. Lancet. 2009;374:324–339. doi: 10.1016/S0140-6736(09)60221-X. [DOI] [PubMed] [Google Scholar]
- 24.Richardson PG, Mitsiades CS, Laubach JP, Lonial S, Chanan-Khan AA, Anderson KC. Inhibition of heat shock protein 90 (HSP90) as a therapeutic strategy for the treatment of myeloma and other cancers. Br. J. Haematol. 2011;152:367–379. doi: 10.1111/j.1365-2141.2010.08360.x. [DOI] [PubMed] [Google Scholar]
- 25.Ishii T, Seike T, Nakashima T, Juliger S, Maharaj L, Soga S, Akinaga S, Cavenagh J, Joel S, Shiotsu Y. Anti-tumor activity against multiple myeloma by combination of KW-2478, an Hsp90 inhibitor, with bortezomib. Blood Cancer J. 2012;2:e68. doi: 10.1038/bcj.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo W, Sun W, Taldone T, Rodina A, Chiosis G. Heat shock protein 90 in neurodegenerative diseases. Mol. Neurodegener. 2010;5:24. doi: 10.1186/1750-1326-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]