Abstract
Many RNA molecules undergo complex maturation, involving e.g. excision from primary transcripts, removal of introns, post-transcriptional modification and polyadenylation. The level of mature, functional RNAs in the cell is controlled not only by the synthesis and maturation but also by degradation, which proceeds via many different routes. The systematization of data about RNA metabolic pathways and enzymes taking part in RNA maturation and degradation is essential for the full understanding of these processes. RNApathwaysDB, available online at http://iimcb.genesilico.pl/rnapathwaysdb, is an online resource about maturation and decay pathways involving RNA as the substrate. The current release presents information about reactions and enzymes that take part in the maturation and degradation of tRNA, rRNA and mRNA, and describes pathways in three model organisms: Escherichia coli, Saccharomyces cerevisiae and Homo sapiens. RNApathwaysDB can be queried with keywords, and sequences of protein enzymes involved in RNA processing can be searched with BLAST. Options for data presentation include pathway graphs and tables with enzymes and literature data. Structures of macromolecular complexes involving RNA and proteins that act on it are presented as ‘potato models’ using DrawBioPath—a new javascript tool.
INTRODUCTION
RNA performs many essential roles in living cells, including the transmission of genetic information between DNA and proteins, regulation of many processes and catalysis. In each of these cases, the function of the RNA depends on its nucleotide sequence. The sequence of the mature functional RNA is quite often different from that of the primary transcript, due to a combination of various processing events such as excision of functional units from the precursor molecule, removal of introns (splicing), joining of different units (trans-splicing), addition of nucleoside residues absent from the template (by capping, editing or polyadenylation) and changing the chemical identity of individual residues (by editing or modification) [reviews: (1–5)]. The function of RNA depends also on its localization, and formation of higher order structures and complexes with other molecules, in particular proteins and other RNAs (6–9).
Functional RNA molecules that are no longer needed or exhibit flaws due to damage or improper processing, folding or assembly into functional complexes, are eliminated from the cell by degradation, which can also occur by many different pathways (10–12). RNA decay removes the by-products of gene expression, including excised introns and other RNA pieces released during RNA processing. Finally, RNA degradation pathways eliminate intergenic, intragenic, promoter-associated and antisense RNAs that arise either as regulatory RNAs or transcriptional noise. The efficiency and specificity of RNA degradation is ensured primarily by a broad spectrum of various endo- and exoribonucleases (1,13) but also is dependent on the formation of specific structures by the RNAs and RNA–protein complexes involving regulatory factors (14) and can be regulated, thus providing means to regulate gene expression and protein levels.
The biogenesis of functional RNAs as well as RNA decay in both eukaryotic and prokaryotic cells involves a series of chemical, structural and spatial alterations, in which RNA interacts with various cellular factors, in particular with enzymes that catalyse reactions with RNA molecules as substrates and products (15). It has to be emphasized that RNA processing pathways interact with each other and with other cellular processes, e.g. they can share some steps and/or proteins involved (1,16). One RNA molecule may be also subject to several enzymatic processes at the same time. Therefore, knowledge of both the entire network of RNA processing events and proteins responsible for individual transformations is critical for our understanding of RNA metabolism. Many of the proteins involved in RNA processing are very well described (with quantitative and qualitative information regarding substrate specificity, kinetics, mechanisms of action and their 3D structure). However, there are still processes, for which an enzymatic activity is known or suspected to exist, but the genes/proteins/enzymes have not yet been characterized. The reconstruction of RNA metabolic pathways and networks can therefore highlight the elements of the system that evidently require additional information. The comparison of metabolic pathways between different species can in turn suggest homologous proteins that may be involved in similar activities.
The information on RNA processing events is scattered in the literature and thus far there has been no database dedicated to the storage and presentation of these data. To this end, we have developed the RNApathwaysDB (a database of RNA maturation and decay pathways) as a ‘one stop shop’ for information about RNA processing pathways and proteins involved in RNA metabolism from model organisms. While the first release is limited to mRNA, tRNA and rRNA metabolism in just three model organisms, we intend to expand it to include other RNAs and species.
DATABASE CONTENT
Based on a comprehensive survey of literature and information available in general-purpose databases, we have compiled the following datasets:
mRNA, tRNA and rRNA maturation and degradation pathways comprising RNA states and intermediates of processes, connected by particular transformations, such as reactions catalysed by enzymes, binding or release of components, or well-defined, functionally important conformational changes.
Proteins, enzymatic complexes and catalytic RNA molecules together with known structures involved in the above-mentioned transformations.
All data items have been curated manually and whenever possible and reasonable we provided references to the published experimental reports and/or to other databases. In particular we linked the entries in RNApathwaysDB to such databases as KEGG (http://www.genome.jp/kegg/) (17) or REACTOME (http://www.reactome.org) (18) in case of pathways or UniProt (http://www.uniprot.org/) (19), NCBI databases (20), BRENDA (http://www.brenda-enzymes.info) (21), PFAM (http://pfam.sanger.ac.uk/) (22) and InterPro (http://www.ebi.ac.uk/interpro/) (23) in case of proteins. RNA molecules are linked to the RFAM database (http://rfam.sanger.ac.uk/) (24), and macromolecular structures are linked to the Protein Data Bank (http://www.pdb.org) (25).
DATABASE ORGANIZATION AND ACCESS
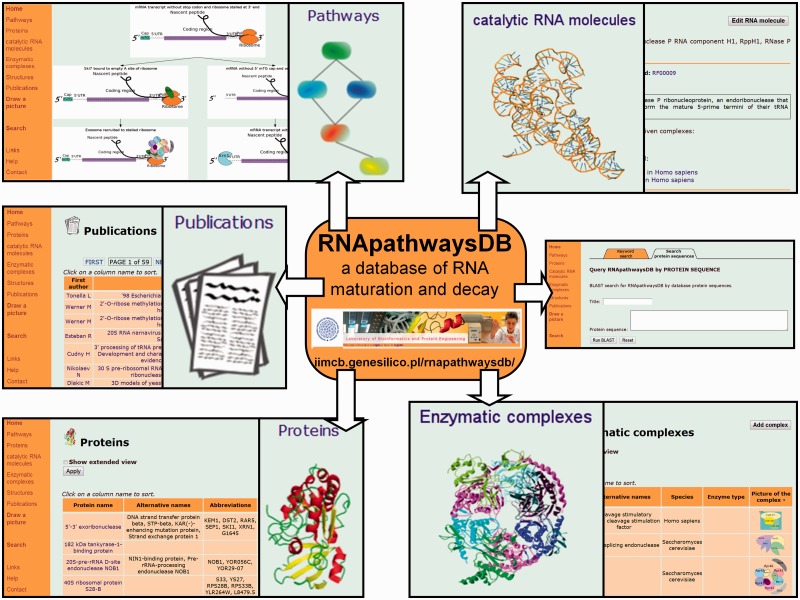
RNApathwaysDB is a relational database linking datasets mentioned above, and can be queried via six menus, ‘PATHWAYS’, ‘PROTEINS’, ‘CATALYTIC RNA MOLECULES’, ‘ENZYMATIC COMPLEXES’, ‘STRUCTURES’ and ‘PUBLICATIONS’ (Figure 1).
Figure 1.
Content of the RNApathwaysDB.
PATHWAYS: This menu provides access to data on different processes (pathways) involving mRNA, tRNA and rRNA, classified as maturation (from a primary transcript to a mature form) or degradation (decomposition of the mature form) for a set of model organisms. As of 25 September 2012 there are altogether 33 pathways for 3 model organisms: Escherichia coli, Saccharomyces cerevisiae and Homo sapiens. All pathways are visualized as graphs created with PyGraphviz (http://networkx.lanl.gov/pygraphviz). RNA states are represented as graph nodes whereas transitions between them (e.g. enzymatic reactions) are visualized as edges, shown as a directed arrow that links individual states. Each node and each edge are hyperlinked to static pages displaying basic information about stages of RNA processing and reactions leading from one stage to another. Pages comprise pictures and detailed information about proteins, enzymatic complexes and RNA molecules taking part in a given process. All types of data are linked to relevant publications, if available. As of 25 September 2012, there are 289 RNA states and 294 reactions in RNApathwaysDB.
PROTEINS: As of 25 September 2012, RNApathwaysDB stores information about 17, 85 and 128 proteins from E. coli, S. cerevisiae and H. sapiens, respectively. This dataset does not cover all proteins involved in RNA metabolism in these species, and is limited to proteins that are well-characterized and whose role in RNA metabolism or RNA degradation can be clearly defined and connected to one or more distinct steps in enzymatic transformation of RNAs that are covered by RNApathwaysDB. All proteins are linked to the genes that encode them. Data that have been collected is comprised of names of genes and proteins together with their sequences from the NCBI databases, 3D structures from the PDB and other details relevant for a particular entry, based on information from other databases (e.g. the type of enzymatic activity, the presence of isoforms, cellular/tissue and subcellular localization, together with links to the relevant database entries). For the eukaryotic mRNA maturation pathways involving splicing by the spliceosome (a large multi-component complex) we have not listed all individual parts of the system, but provided links to the Spliceosome Database (http://spliceosomedb.ucsc.edu) and to SpliProt3D (http://iimcb.genesilico.pl/SpliProt3D/) (26). Links to selected relevant publications are also provided.
CATALYTIC RNA MOLECULES: This section stores data about RNA molecules that are catalytically active and can act either separately or as elements of larger complexes. For each RNA molecule data such as molecule name, nucleotide sequence and a link to the homologous family in the RFAM database are provided, if available.
ENZYMATIC COMPLEXES: Currently, RNApathwaysDB stores information about 37 enzymatic complexes: 2 from E. coli, 25 from H. sapiens and 10 from S. cerevisiae. For each complex, RNApathwaysDB provides an overall description, a simplified 2D diagram (a ‘potato model’) and links to selected relevant publications, if available.
STRUCTURES: For those proteins present in RNApathwaysDB with NMR or X-ray structures available, atomic coordinates from the PDB are made available. Currently RNApathwaysDB includes 216 structural models.
PUBLICATIONS: Literature references to entries in PubMed (20) have been compiled into an additional dataset, currently comprising 1541 positions.
The full catalogue of data about RNA maturation and degradation pathways is a moving target. The current listings in RNApathwaysDB should be considered provisional and would not be fully complete for some time. We intend to update the information on RNA maturation and degradation pathways with information from new discoveries, e.g. new enzymatic steps that will be identified in the future. We also encourage the users to report any known reactions and enzymes that may be missing in RNApathwaysDB. In the future, we plan to expand the repertoire of pathways to include other types of RNAs, in particular small non-coding RNAs of various types. We also intend to extend the dataset to encompass additional model organisms (those with well-characterized RNA metabolism), e.g. representatives of Gram-positive bacteria and plants. Along with the expansion of the pathways we will also expand the associated datasets of proteins, RNAs and complexes. Therefore, we invite experts interested in inclusion or refinement of information for particular systems in RNApathwaysDB for collaboration on data gathering and curation.
IMPLEMENTATION
RNApathwaysDB has been implemented using the Django Web Framework (http://www.djangoproject.com/), and uses a MySQL relational database to store data. The features provided by the webpage are: user profile and a login, search tool with the possibility to search using a keyword or protein sequence, wiki-like pages, which enable editing of entries, and a link to a new javascript image drawing tool.
To view the content of the database there is no need to log in or register. However, editing of the content of the database requires registration. Logging in not only uncovers the wiki-like pages of all the entries but also gives access to the otherwise hidden administration site, which provides tools to add, edit or delete information. Users can also add comments and new references to existing records.
DrawBioPath is a new image drawing tool (accessible via the ‘draw a picture’ link in the main menu which redirects to http://drawbiopath.genesilico.pl/) that has been developed for creating graphical representations of metabolic pathways of DNA and RNA, and was used for creating all images in RNApathwaysDB. The tool is written in JavaScript and is based on the SVG-edit web editor (http://code.google.com/p/svg-edit/). The drawing engine uses the SVG format provided by the W3C consortium, which enables resizing the images without the loss of quality and makes it possible to modify the images with external tools for processing vector graphics, e.g. Inkscape or others. In contrast to the drawing tool available in REPAIRtoire (27), another database of nucleic acid metabolism, DrawBioPath is more comfortable and easier to use. DrawBioPath provides a library of shapes that can be used for drawing RNA and DNA molecules, proteins or irregular objects. Our tool provides a utility to upload a user’s graphics file in SVG format and to modify it using either a graphical interface or a text editor.
RNApathwaysDB can be queried in two ways: using a keyword or a protein sequence. Keyword search is a simple text search tool available in the main menu. It returns a structured list of the database entries that contain the query, with the part of the text corresponding to the query highlighted in red. Protein sequence search is available via the ‘SEARCH’ link in the main menu. A search using protein sequence is performed with BLAST program. There is also a utility that sends a protein sequence from a RNApathwaysDB protein entry to BLAST on the NCBI webserver.
DISCUSSION
The metabolism of nucleic acids is a very old and extensive field of research. Networks of relationships and interactions in RNA metabolic pathways are extremely complex and the amount of data is huge, however different pieces of information are scattered around different resources, in literature and in various databases. Information about RNA processing pathways and their components is available to some extent in general-purpose pathway databases such as KEGG, REACTOME, BioCyc (http://biocyc.org) (28) or WikiPathways (http://www.wikipathways.org/) (29). However, none of these databases contain a complete description of RNA maturation and degradation pathways. Thus, the vast quantity of data and the lack of a single bioinformatics resource dedicated to RNA processing and degradation prompted us to develop resource database with a WWW server that does not only gather the information dedicated to RNA processing pathways but also helps to systematize the knowledge and makes it easily accessible for lay users.
RNApathwaysDB contains more detailed information than the databases mentioned above including information on various aspects of RNA maturation, such as mRNA capping, splicing and polyadenylation, tRNA biosynthesis and rRNA maturation. The database also describes RNA degradation pathways, such as rapid tRNA decay, tRNA nuclear surveillance, mRNA decay pathways and rRNA quality control pathways. An important component of RNApathwaysDB is the dataset of enzymatic complexes together with their macromolecular structures visualized as ‘potato models’ that can be used for illustration e.g. in teaching. To our knowledge, there is no other resource available that provides such a range of information on RNA metabolism.
In the development of RNApathwaysDB we used our experience from the work on the MODOMICS database of RNA modification pathways (30) and from the more recent work on the REPAIRtoire database of DNA repair pathways (27). We hope that the RNApathways database will become as popular and useful for the community of researchers working on RNA processing pathways, as MODOMICS has become in the RNA modification field. In the future, we plan to integrate the databases on different aspects of RNA metabolism using a common data model and a joint interface. Another envisaged direction of development is to link generic RNA representations in RNApathwaysDB to particular mature RNA sequences that will be stored in the future RNAcentral database (31) and to prokaryotic and eukaryotic genome sequence databases, so e.g. cleavage sites in maturation reactions could be mapped onto the precursor RNA molecules.
AVAILABILITY
The content of the RNApathways database is freely available at the URL http://iimcb.genesilico.pl/rnapathwaysdb/. The software for generation of custom images for biological processes can be accessed at the URL http://drawbiopath.genesilico.pl/. Researchers interested in adding or curating data (proteins, features, complexes, pathways, etc.) or in implementing options that are not yet available are encouraged to contact J.M.B. (at iamb@genesilico.pl).
FUNDING
Foundation for Polish Science (FNP) [TEAM/2009-4/2 to J.M.B.]; Polish Ministry of Science and Higher Education (MNiSW) [POIG.02.03.00-00-003/09 to Bujnicki lab]; E.U. Framework Programme 7 [HEALTH-PROT, contract number 229676]; National Science Centre (NCN) [N N301 072 640 to K.Mil.]; German Academic Exchange Service (DAAD) [D/09/42768 to K.R.]; European Research Council (ERC) [StG grant RNA+P=123D to M.N. and J.M.B.]; statutory funds of the Adam Mickiewicz University (to K.Mil., K.Mik., A.L., M.S. and Z.B.). Funding for open access charge: E.U. Framework Programme 7 [HEALTH-PROT] and NCN [N N301 072 640].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Joanna Maria Kasprzak for stimulating discussions and for critical reading of this article. We are indebted to the authors of primary databases and services, whose content could be reused or linked to by RNApathwaysDB. We also thank all developers and curators of the MODOMICS database and REPAIRtoire database, whose feedback has been taken into account in the development of the RNApathwaysDB.
REFERENCES
- 1.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 2010;34:883–923. doi: 10.1111/j.1574-6976.2010.00242.x. [DOI] [PubMed] [Google Scholar]
- 2.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:22–31. doi: 10.1002/wrna.47. [DOI] [PubMed] [Google Scholar]
- 4.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA. 2011;2:611–631. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 5.Grosjean H. Fine-Tuning of RNA Functions by Modification and Editing. Berlin, Heidelberg: Springer-Verlag; 2005. [Google Scholar]
- 6.Fatica A, Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 7.Iglesias N, Stutz F. Regulation of mRNP dynamics along the export pathway. FEBS Lett. 2008;582:1987–1996. doi: 10.1016/j.febslet.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 8.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hocine S, Singer RH, Grunwald D. RNA processing and export. Cold Spring Harb. Perspect. Biol. 2010;2:a000752. doi: 10.1101/cshperspect.a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kushner SR. mRNA decay in prokaryotes and eukaryotes: different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- 11.Coller J, Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 12.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Tomecki R, Dziembowski A. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA. 2010;16:1692–1724. doi: 10.1261/rna.2237610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alonso CR. A complex ‘mRNA degradation code' controls gene expression during animal development. Trends Genet. 2012;28:78–88. doi: 10.1016/j.tig.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- 16.Beggs JD, Tollervey D. Crosstalk between RNA metabolic pathways: an RNOMICS approach. Nat. Rev. Mol. Cell Biol. 2005;6:423–429. doi: 10.1038/nrm1648. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.UniProt C. Reorganizing the protein space at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, Rother M, Sohngen C, Stelzer M, Thiele J, Schomburg D. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korneta I, Magnus M, Bujnicki JM. Structural bioinformatics of the human spliceosomal proteome. Nucleic Acids Res. 2012;40:7046–7065. doi: 10.1093/nar/gks347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milanowska K, Krwawicz J, Papaj G, Kosinski J, Poleszak K, Lesiak J, Osinska E, Rother K, Bujnicki JM. REPAIRtoire–a database of DNA repair pathways. Nucleic Acids Res. 2011;39:D788–D792. doi: 10.1093/nar/gkq1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelder T, van Iersel MP, Hanspers K, Kutmon M, Conklin BR, Evelo CT, Pico AR. WikiPathways: building research communities on biological pathways. Nucleic Acids Res. 2012;40:D1301–D1307. doi: 10.1093/nar/gkr1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 2009;37:D118–D121. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bateman A, Agrawal S, Birney E, Bruford EA, Bujnicki JM, Cochrane G, Cole JR, Dinger ME, Enright AJ, Gardner PP, et al. RNAcentral: a vision for an international database of RNA sequences. RNA. 2011;17:1941–1946. doi: 10.1261/rna.2750811. [DOI] [PMC free article] [PubMed] [Google Scholar]